Abstract
Quorum sensing (QS) regulates many natural phenotypes (e.q. virulence, biofilm formation, antibiotic resistance), and its components, when incorporated into synthetic genetic circuits, enable user-directed phenotypes. We created a library of Escherichia coli lsr operon promoters using error-prone PCR (ePCR) and selected for promoters that provided E. coli with higher tetracycline resistance over the native promoter when placed upstream of the tet(C) gene. Among the fourteen clones identified, we found several mutations in the binding sites of QS repressor, LsrR. Using site-directed mutagenesis we restored all p-lsrR-box sites to the native sequence in order to maintain LsrR repression of the promoter, preserving the other mutations for analysis. Two promoter variants, EP01rec and EP14rec, were discovered exhibiting enhanced protein expression. In turn, these variants retained their ability to exhibit the LsrR-mediated QS switching activity. Their sequences suggest regulatory linkage between CytR (CRP repressor) and LsrR. These promoters improve upon the native system and exhibit advantages over synthetic QS promoters previously reported. Incorporation of these promoters will facilitate future applications of QS-regulation in synthetic biology and metabolic engineering.
INTRODUCTION
Quorum sensing is a process of cell-cell communication that allows bacteria to enumerate their cell density and modify their behaviors (such as virulence, biofilm formation and antibiotic resistance) in a collective manner. To do this, bacteria ‘talk’ to each other using small molecules called autoinducers (1). Autoinducer-2 (AI-2) has attracted significant attention because its terminal synthase (LuxS) and its signal transduction cascade (Lsr regulon) are widely conserved among Eubacteria (2–4). For example, the Enterobacteriaceae, Escherichia coli and Salmonella enterica serovar Typhimurium share similarities involving detection and production of AI-2, as they possess homologous machinery to control quorum sensing based on AI-2 levels. The bidirectional quorum-sensing E. coli lsr regulon, lsrRK-lsrACDBFG is responsible for controlling the expression of genes involved in the perception and degradation of AI-2 (5). The lsr operon (lsrACDBFG) coordinates the expression of genes involved in the transport and degradation of AI-2. The four genes lsrACDB produce the ABC transporter components, which are responsible for AI-2 uptake (5–7); the lsrFG genes are responsible for the degradation of its active form, phospho-AI-2 (8,9), and its subsequent assimilation into central carbon metabolism (8–10). In Salmonella Typhimurium, there is an additional lsrE gene, which encodes a putative sugar epimerase (11). In both E. coli and Salmonella, lsrK and lsrR serve to coordinate the induction and repression of the lsr operon (8,11). LsrR represses transcription of the operon and itself by directly binding to two LsrR binding ‘boxes’ within the lsr promoter region (4). LsrR is released in the presence of phospho-AI-2, which, in turn, is the phosphorylated product of the LsrK kinase (4,8) and AI-2.
Because they mediate communication among various bacteria and their genetic circuitry is relatively well understood, QS-based circuits have been engineered for use in widely varied application areas: biochemicals production, sensor development, infectious disease, tissue engineering, and mixed-species fermentations (12–21). Also, both AHL (N-acyl-homoserine lactones) and AI-2 based species communication systems have been developed as tools for exogenously controlling bacterial phenotype and protein expression (22,23). For example, the luxCDABE operon of the bioluminescent bacterium Photorhabdus luminescens, based on AHL, has proven to be an exceptional transcriptional reporter for expression in high-GC bacteria (12). AHL-actuated promoters were incorporated into many synthetic biology strategies for guiding cell behaviors (24–33), including an elaborate circuit that enables the sensing, tracking, and targeting of Pseudomonas aeruginosa, an important human pathogen (20,34).
The native lsr quorum sensing regulon, on the other hand, has received comparatively less attention, even though it is the native system of E. coli, which could to be viewed as one of the ‘workhorses’ of microbial synthetic biology. The lsr promoter operon was rewired by Tsao and colleagues (2010) (13) to act as an autonomous inducer for the expression of recombinant proteins. In their two-plasmid system, the E. coli lsrACDBFG promoter region served as a trigger of T7 RNA polymerase expression, which, in turn ‘amplified’ target protein expression from commercially available pET vectors (13). Due to the fact that the lsrACDBFG operon promoter in E. coli [-307 to +92 relative to the start codon of lsrA] is known to be very weak (13), we created a library of lsr operon promoters through directed evolution using the error-prone PCR (ePCR). Our objective was to discover promoter sequences that were superior to the native system that also required no signal amplification (via T7 polymerase). For this, we constructed a plasmid (pLSR) for the expression of two gene reporters, gfp and tet(C) under lsr regulon control (in the direction of the lsrACDBFG operon as opposed to lsrRK). Our mutagenic library yielded two mutant lsr promoters, EP01rec and EP14rec, that demonstrated greater strength than the wild type lsr promoter. Sequencing and subsequent expression analyses revealed mutations responsible for the increase in promoter strength. We also identified what is believed to be a CytR binding site within the lsr operon promoter sequence. Importantly, evolved lsr promoters (EP01rec and EP14rec) retain the same properties of the wild type lsr promoter: induction via AI-2 and repression by LsrR.
MATERIALS AND METHODS
Strains and media
NEB turbo electrocompetent E. coli, DH5α (NEB), LW6, and LW7 (6) strains (Supplementary Table S1) were grown in Luria-Bertani (LB) medium at 37°C for DNA manipulation or expression experiments. Media were supplemented with chloramphenicol (34 ug/ml) to maintain the pLSR plasmid and/or tetracycline (2.5, 5, 10 or 20 ug/ml) to select the lsr operon promoter mutant candidates from the library. Miller assay experiments were performed using 50 ug/ml ampicillin to maintain the LW7 (6) strain transformed with pLW11 (6), pPH01 or pPH14 (Supplementary Table S1).
Plasmid and library creation
pTS40 is a plasmid that carries the CloDF13 replication origin (20–40 copies/cell) and under the control of the ampC promoter expresses bicistronic, gfp_mut2 and tet (C). This plasmid confers chloramphenicol antibiotic resistance from pTS1 (35). In order to remove the ampC promoter and ampR gene from this plasmid and replace with the E. coli lsrACDBFG operon promoter region, pTS40 was digested using the restriction site PvuI present in the ampR sequence to linearize the plasmid. Primers pTS40delampR_F and pTS40delampR_R (Supplementary Table S2) were used in a PCR to exclude a sequence fragment containing both the ampR gene and ampC promoter and also to insert at the restriction sites PvuI and SpeI. The 399-bp E. coli lsrACDBFG operon promoter region [−307 to +92 relative to the start codon of lsrA] (6) was amplified with the primers lsrpF_PvuI and lsrpR_SpeI (Supplementary Table S2) and cloned into the pTS40 backbone containing the same restriction sites inserted through PCR. This final plasmid (pLSR) was used as a template to generate the lsr operon promoter mutant library (Supplementary Table S1).
Error-Prone PCR (ePCR) was employed to obtain the mutant library containing a vast diversity of lsr promoter mutants. For this, the pLSR plasmid harboring the wild type lsr operon promoter was used as template, and also two oligonucleotides LsrEP_F and LsrEP_R (Supplementary Table S2) flanked by PvuI and SpeI restriction sites, respectively, were used as forward and reverse primers. The conditions to perform EP PCR were performed according to a previously published protocol with some modifications (36). In this study, three reactions of 50 μl reaction mixture contained 5 μl of 10X PCR buffer -Mg, 0.8 MnCl2, 5 mM MgCl2, 1mM dATP, 1 mM dTTP, 0.2 mM dCTP, 0.2 mM dGTP, 10 pmol of each primer, 1 ng of template plasmid and 2.5 U recombinant Taq DNA Polymerase (Life Technologies) were performed to create bias upon sequence amplification. The ePCR was conducted in a C-1000 Touch™ Thermal Cycler (Biorad) for 35 cycles consisting of denaturation at 94°C for 30 s, annealing at 58°C for 1 min, and extension at 72°C for 2 min. The ePCR products were digested with PvuI and SpeI and then ligated at 16°C overnight with pLSR previously digested with the same restriction enzymes and transformed into library efficiency NEB turbo electrocompetent E. coli (New England BioLabs).
Library characterization
For selection, lsr operon promoter library plasmid DNA was isolated from an aliquot of NEB turbo electrocompetent E. coli, and 50 ng of purified plasmid was used to transform strain LW6 (Supplementary Table S1). The transformed cells were plated on LB agar containing 34 μg/mL chloramphenicol and then recovered en masse using a sweep buffer (LB containing 2% glucose and 15% glycerol) and stored in aliquots at −80°C. These cells were first plated on a 24.5 cm × 24.5 cm LB agar plate containing 34 μg/ml chloramphenicol. Then, colonies on this plate were recovered into sweep buffer and plated at a density of 106 cells/ml on a 24.5 cm × 24.5 cm LB media plate containing 34 μg/mL chloramphenicol and 20 ug/ml of tetracycline. A total of 14 library clones formed from this selection plate were individually recovered into LB liquid media containing 34 μg/ml chloramphenicol and incubated in a shaker at 37°C. After 16 h of incubation, the plasmid DNA from each clone that survived the selection was isolated to be sequenced and analyzed through alignment with the wild type lsr operon promoter as a reference.
In silico analyses
All of the 14 clones that were able to grow in media containing 20 μg/ml of tetracycline were sequenced and aligned using Clustal Omega (37) with the wild-type lsr operon promoter sequence as a reference. To find potential CytR binding sites, we searched for the corresponding consensus sequence (consisting of a right motif and/or left motif and specified by Pedersen and Valentin-Hansen (38) in the Lsr intergenic region using the Biostrings package (39) of the open source software Bioconductor. Here, the left and right portions of the consensus CytR sequences were compiled into position probability matrices. A position weight matrix (PWM) was generated by the PWM function, which outputs a 4×8 matrix (4 possible nucleotides {A, C, G, T} × motif base length of 8) with each element in the matrix representing a score relative to the possibility that nucleotide i would be located at position j. The matrix was scaled so that the total maximum score is 1. The PWM was then applied to the matchPWM function, producing the top scoring sequences from among those in the intergenic region between lsrR and lsrA start codons. This procedure was applied to each, the right and left motifs.
Restoring p-lsrR-box sequence region and putative CytR-binding site
PCR-driven overlap extension protocol (40) was used to restore the original nucleotides into the p-lsrR-box (4) that was corrupted through ePCR (on all the fourteen clones selected through 20 μg/ml tetracycline). Mutagenic primers (Supplementary Table S3) and lsr operon promoter flanking primers (lsrpF_PvuI and lsrpR_SpeI) (Supplementary Table S2) were used to generate intermediate overlapping PCR products that were combined to produce a full-length product using flanking primers. These fragments were cloned into pLSR between PvuI and SpeI sites. We adopted the same protocol used above to restore a mutated nucleotide found in both clones 1 and 14 inside of the putative CytR-binding site (38). For this, we used a specific mutagenic primer pair, mutAG_F1 and mutAG_R1, and mutAG_F14 and mutAG_R14 to generate intermediate overlapping PCR for clones 1 and 14, respectively (Supplementary Table S3). To produce a full-length product we employed the same primers used to amplify lsr promoter, lsrpF_PvuI and lsrpR_SpeI (Supplementary Table S2). All the full-length products were cloned into pLSR and transformed into E. coli DH5α.
Promoter strength metric
All fourteen clones containing the LsrR binding-site corrupted by ePCR and their restored counterparts, and also the two putative CytR binding-site sequence restored clones (EP01recAG and EP14recAG) had their in vivo promoter activities determined through experiments to measure tetracycline resistance and GFP fluorescence.
Tetracycline resistance
An lsrR knockout strain (LW6) with the wild type lsr operon promoter, the fourteen lsr operon promoter mutants obtained through 20 ug/ml tetracycline from library selection, and those cells with the restored LsrR-binding site were grown in liquid LB media containing 34 μg/mL chloramphenicol overnight in a shaker incubator at 37°C. Each sample was serially diluted in liquid LB media from 10−1 to 10−10 and then 3 μl of each serial dilution was spotted on LB media plates (OmniTray, 86 mm × 128 mm, Nunc) containing 34 μg/ml cloramphenicol and 0, 2.5, 5.0, 10 or 20 μg/ml of tetracycline. These plates were incubated at 37°C for 16 h.
GFP fluorescence
Escherichia coli LW6 harboring pLSR wild type or mutants (EP01rec, EP14rec, EP01recAG and EP14recAG) were grown overnight at 37ºC in 2 ml of LB medium supplemented with 34 μg/ml of chloramphenicol. Bacterial suspensions were then re-inoculated into 10 ml of fresh LB medium with chloramphenicol in order to have initial optical densities (OD600 nm) of 0.05. Cells were allowed to grow at 37ºC with shaking at 250 rpm. Bacterial cell samples (200 μl, technical triplicate) were collected at optical density (OD600 nm) ∼0.5 and 1.0. Samples were collected by centrifugation (1000 g, 5 min), washed, resuspended in PBS and kept on ice until flow cytometry and microscopy analysis. Flow cytometry analyses were performed using a FACSCanto II flow cytometer equipped with 488 nm, 633 nm, and 405 nm lasers (BD Biosciences, San Jose, CA, USA) and all flow cytometry data were analyzed with FACSDiva software (BD Biosciences). Side and forward scatter of bacterial suspensions were determined using semi-log scale SSC/FSC plots with a threshold of 5000. Voltage settings for the SSC, FSC and FITC channels were kept constant for all flow cytometry experiments. Bacterial suspensions were analyzed at a medium flow rate with a maximum of 1000 events per second for 75 s and a minimum of 50 000 events. Image-based cytometry analysis were performed using fluorescent microscopy (Olympus U-HGLGPS) with 20× objective and 1800 ms exposure. Positive cells for GFP fluorescence were compared with negative control (LW6 and wild type strain).
Transcriptional analysis
Cultures inoculated as previously were grown for 3 h (OD600nm ∼ 0.5), and the total RNA was isolated using Trizol Max Bacterial RNA isolation kit (Ambion®, Life technologies) according to the manufacturer's instructions. In addition, RNA samples were treated with Amplification Grade DNAse I (Sigma-Aldrich) to eliminate possible DNA contamination. PCR primer sequences were designed using PrimerQuest® Design Tool (IDT) (Supplementary Table S2) and synthesized by IDT. The SensiFAST SYBR Hi-ROX One-Step kit (Bioline®) was used for first-strand cDNA synthesis and subsequent real-time PCR following to the manufacturer's instructions.
Real-time PCR conditions were carried out on an Applied Biosystems 7300 Real-Time PCR system using a two-step cycling protocol. Primers were used at a final concentration of 400 nM, and 25 ng of RNA was used as template in each 20-μl reaction. Each reaction was performed in triplicate, with outlying data removed for select samples. 16s rRNA was used as the endogenous housekeeping gene. To calculate the levels of gfp expression ΔCT values were calculated by the following equation: ΔCT = CT Target − CT Reference. The ΔΔCT value was calculated as ΔΔCT = ΔCT, sample − ΔCT, Wt where each DCT are represented by the difference between the Target and Reference (16srRNA) values, as above. Also, the relative quantification (RQ) is calculated as 2−ΔΔCT. Data plotted in figures also include the standard deviation (s) of each RQ, so that 2−ΔΔCT+s and 2−ΔΔCT-s represent the limits as indicated. Details of the 2−ΔΔCT method have been previously described (41,42). The relative quantification was based on the relative expression of gfp_mut2 versus 16S rRNA. Wild type lsr operon promoter CT's values for gfp were used as reference for all samples.
Cloning of the strongest lsr promoter variants, EP01rec and EP14rec into a low copy number plasmid
In order to clone the strongest lsr promoter mutants (EP01rec and EP14rec) into the low copy plasmid pFZY1 harboring galK’-lacZYA reporter segment (Supplementary Table S1), promoter sequences were amplified by PCR using the primers lsrpF_BamHI and lsrpR_HindIII (Supplementary Table S2) to create pPH01 and pPH14 (Supplementary Table S1). The plasmid pFZY1 and PCR fragments were digested with BamHI and HindIII, purified and then ligated using a T4 DNA ligase (New England Labs) at 16°C. The constructions pPH01 and pPH14, which are EP01rec and EP14rec cloned into pFZY1 were confirmed by sequencing.
Measurement of β-galactosidase activity to verify EP01rec and EP14rec promoters in presence of synthetic AI-2
Cultures of E. coli strain LW7 (6) harboring the plasmid pLW11 (6), pPH01 or pPH14 (Supplementary Table S1) containing an ampicillin resistance marker were grown overnight in LB media, and then diluted 100-fold (OD600 = 0.05) into fresh LB media supplemented with 50 μg/ml ampicillin. Cultures were incubated at 37°C with shaking at 250 rpm in flasks. When the OD600 reached approximately 0.2, the cultures were split into multiple 2 ml culture tubes and 40 μM of synthetic AI-2 was added (graciously provided by the H.O. Sintim Research Group, Purdue University). Cultures grew in the absence or presence of AI-2, and were removed at intervals of 2 and 4 h for determination of OD600 and β-galactosidase activity. Specific activity of β-galactosidase is expressed in Miller Units (43).
RESULTS AND DISCUSSION
Characterization of the promoter library variants
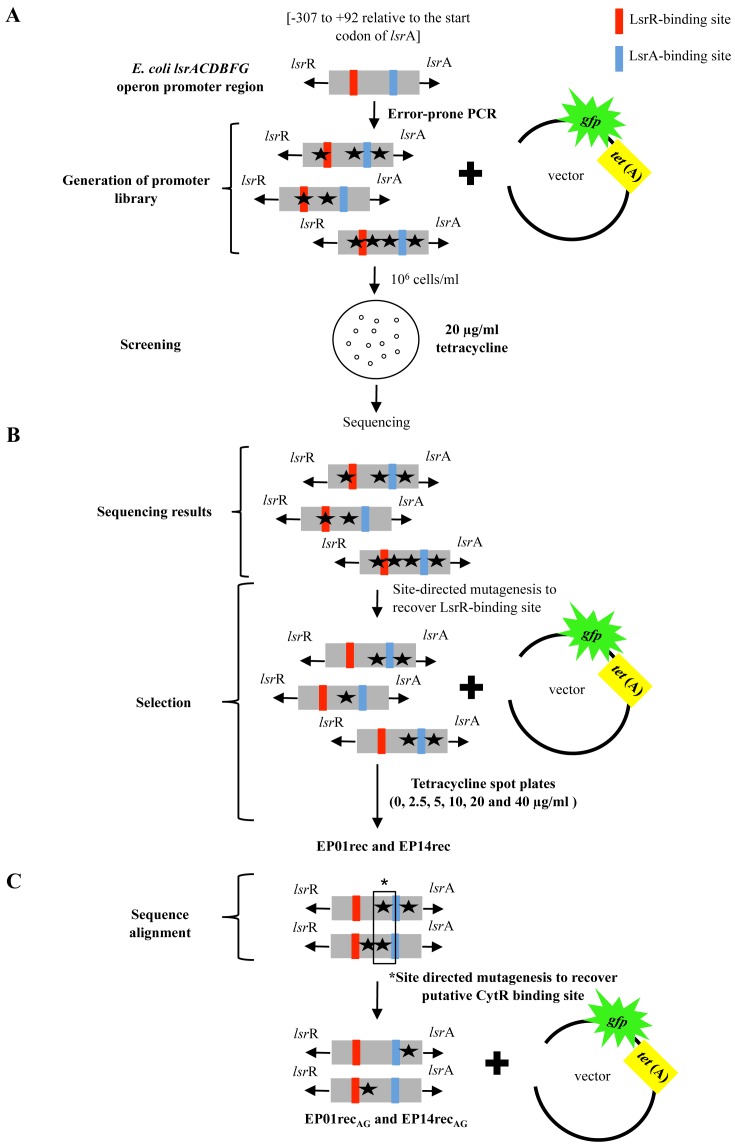
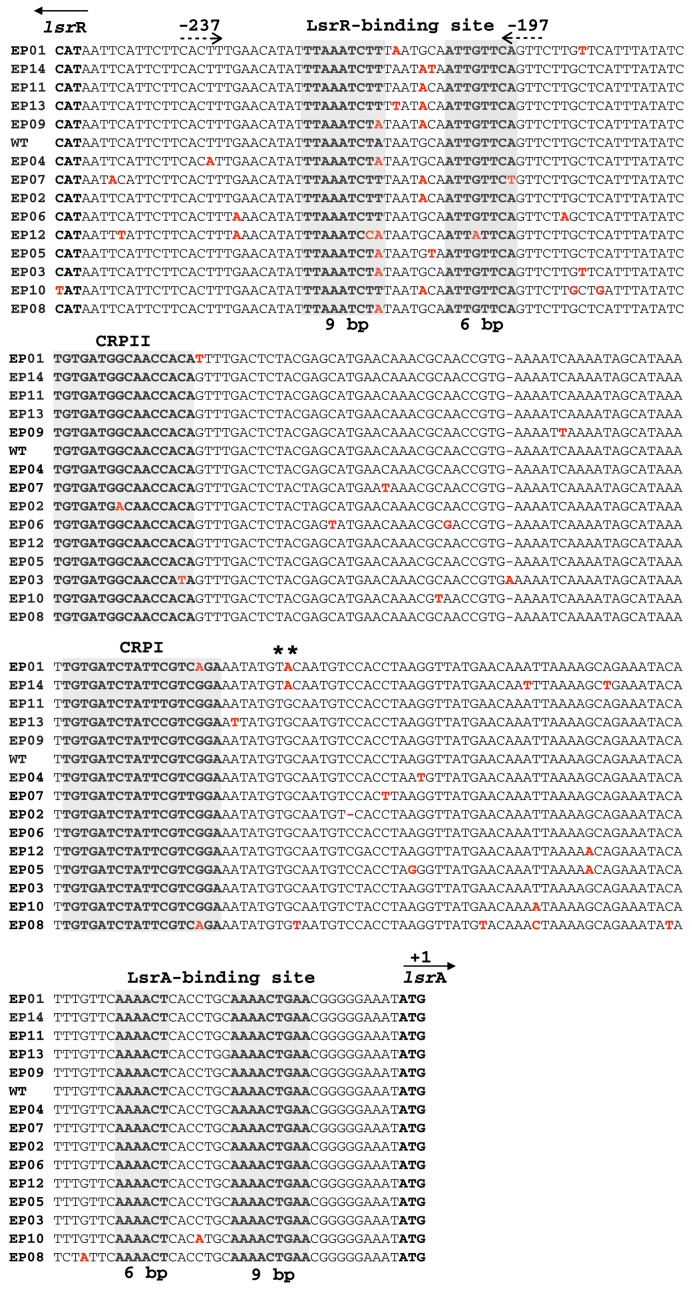
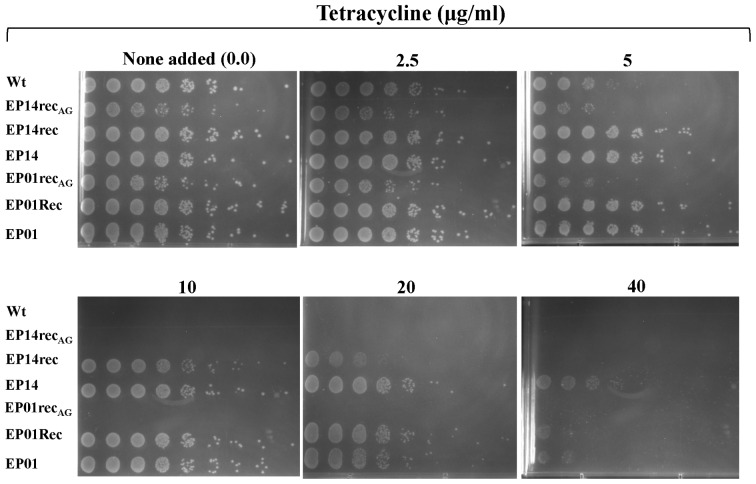
Figure 1 depicts the general scheme for design and construction of promoter libraries and the methodology for selecting promoters with superior expression characteristics (as noted in Materials and Methods). The original library contained approximately 106 members and was challenged to grow in the presence of 20 μg/ml tetracycline (Figure 1A). Fourteen clones, denoted EP (error-prone), survived tetracycline selection and were sequenced. The mutations were analyzed by sequence alignment (Figure 1B). In Figure 2, we found that most of the mutations were concentrated very close to or along the 6-bp (AACAAT) and 9-bp (AAGATTTAA) sequences of the p-lsrR-box, which are important for LsrR binding of the lsr promoter (4). In fact, most clones showed mutations in the sequence region (-214 to -208 relative to lsrA start codon) located between the two p-lsrR-box sequences, 6-bp (AACAAT) and 9-bp (AAGATTTAA). Aiming to isolate those mutations that contributed to increased lsr promoter strength but that also maintained LsrR repression, we performed site-directed mutagenesis in all the fourteen clones (Figure 1B). We replaced the mutations that were found along the region between the two 6-bp and 9-bp sequence in the p-lsrR-box (TAATGCA) (Figure 2) with the native sequence (lsr promoter wild type sequence). All fourteen clones with restored p-lsrR-box (named EPrec), were selected again against tetracycline again to verify the influence of these mutations on the promoter strength (Figure 1B). Interestingly, after this new selection only two clones were able to grow in high concentration of tetracycline (20 μg/ml), EP01rec and EP14rec (Figures 1C and 4), indicating that except for these two promoter mutants, the mutations within the p-lsrR-box were presumably responsible for the increase in promoter strength in the other 12 clones. This might, in part, be caused by the mutations located in the p-lsrR-box affecting the ability of LsrR repressor to bind, promoting leaky expression in the mutant lsr promoter.
Figure 1.
Schematic of the experimental design used to obtain and to characterize lsr operon promoter mutants from the E. coli lsrACDBFG operon promoter region. (A) Generation of promoter library by ePCR. The strongest promoters were selected based on their ability to confer survival in the presence of 20 μg/ml tetracycline and sequenced to identify the mutations. Surviving clones were denoted as EPxx, where xx refers to the colony number. (B) Identification of mutations on the 14 clones selected against 20 μg/ml tetracycline (EP01-EP14) and site-directed mutagenesis to revert mutations found in the LsrR-binding site region (these clones designated as EPxxrec). These clones were tested for growth at different concentrations of tetracycline (2.5, 5.0, 10, 20 and 40 μg/ml). (C) Sequencing analysis of the two lsr promoters (EP01rec and EP14rec) that maintained the ability to survive in 20 μg/ml tetracycline. Site-directed mutagenesis was used to restore the single mutual nucleotide found to be mutated in both clones, EP01rec and EP14rec, in the putative CytR-binding site sequence (G→A).
Figure 2.
Alignment of lsr promoter mutants versus wild type using Clustal Omega (37). Mutations are in red. Gray boxes highlight p-lsrR-boxes, CRPII, CRPI and p-lsrA-boxes. The methionine coded by the ATG start codon in the lsrR and lsrA gene, respectively are represented by solid arrows (←/→). (∗∗) indicates the location of the mutation found in both mutants, EP01rec and EP14rec sequences (G→A), and the mutation found in the EP08rec sequence (C→T) at the same location. Site-directed mutagenesis was performed on the region marked by the dashed arrows (<− −/− −>) in order to revert the mutations along the p-lsrR box (−237 to −197 upstream of the lsrA start codon).
Figure 4.
Growth of LW6 (ΔlsrR) (6) transformed with pLSR harboring the wild-type lsr operon promoter (Wt) and its mutants: EP01, EP14, EP01rec, EP14rec, E01recAG and EP14recAG in the presence of different concentrations of tetracycline. Serial dilutions of saturated cultures from 10−9 (rightmost) to 10−1 (leftmost) were spotted on OmniTray LB agar plates supplemented with with 34 μg/ml chloramphenicol and the indicated concentration of tetracycline. Growth imaged after 16 h of incubation at 37°C.
Sequence alignment analysis of the fourteen clones revealed that EP01rec and EP14rec shared a mutation (TGTGCAAT→TGTACAAT) located very close to the CRPI sequence box (Figure 2), which led us to hypothesize that this mutation G→A might be important for enabling the observed increase in lsr promoter strength. Due to this mutation being located near the CRPI sequence box, we searched for promoter elements related to CRP, such as CytR-binding site sequences, which had not been previously identified in the lsr promoter sequence.
The CytR repressor and CRP bind cooperatively to several promoters in E. coli to repress transcription initiation (38). Interestingly, Pederson and Valentin-Hansen (1997) (38) have already demonstrated sequences showing homology to the octameric motifs 5′-AATGT/CAAC-3 and 5′-GTTGCATT-3′, respectively termed left (L) and right (R) half-sites on the deoP2 E. coli promoter sequence (in absence of cAMP-CRP). The right (R) half-site (5′-GTTGCATT-3′) described by Pederson and Valentin-Hansen (1997) (38) also has a consensus sequence (-TGCA-), which is present in the same location as the mutual mutation 5′- TG-(TACA)-AT-3 on both EP01rec and EP14rec found here.
Thus, we combined the in vitro assay findings of Pedersen and Valentin-Hansen (1997) (38) with the following in silico analysis in order to examine if a CytR-binding site exists in the E. coli lsrACDBFG operon promoter region [-307 to +92 relative to the start codon of lsrA].
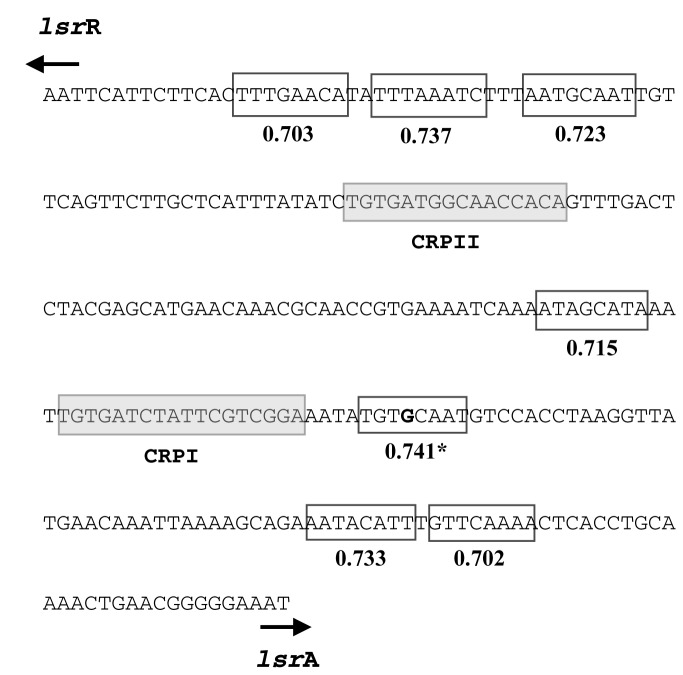
We searched for putative CytR binding sites corresponding to a consensus sequence (consisting of a right motif and/or left motif and specified by Pedersen and Valentin-Hansen (38) in the Lsr intergenic region using the Biostrings package (39) of the open source software Bioconductor. Pedersen and Valentin-Hansen (1997) (38) isolated CytR binding sites, without the presence of cAMP-CRP, in 46 fragments. Using the matchPWM function in the Biostrings package of Bioconductor, we found 7 matches for the right motif (Supplementary Table S4) in wild type lsr promoter sequence. The highest scoring sequence was the TGTGCAAT sequence (score = 0.741), which is the site of the (TGTGCAAT→TGTACAAT) mutation in EP01rec and EP14rec. We suggest that the higher expression levels afforded by our two mutated promoters is caused by the single (G→A) mutation, and that this is found in the putative CytR-binding site. Interestingly, this same sequence was also recognized as a match for the left motif (score = 0.765, the sixth highest score for the left motif) (Suplementary Table S4, Figure 3). Our in silico results showing all high scores for the left motif sequence, as well the top scores for both motifs in the lsrR (reverse-complement) direction, can be found in Figure 4.
Figure 3.
The top seven matches for the right motif (lsrA direction) based on in silico analysis performed in combination with in vitro results obtained using the deoP2 E. coli promoter reported by Pederson and Valentin-Hansen (1997) (38). The highest scoring sequence was TGTGCAAT (score = 0.741) which is the motif where the mutual mutation was found in EP01 and EP14 (TGTACAAT). This same sequence also scored high as a match for the left motif (score = 0.765—sixth highest score for the left motif), see Supplementary Table S4.
Correspondingly, we next performed site directed mutagenesis in the putative CytR-binding site in order to revert the (TGTACAAT) mutation back to wild type (TGTGCAAT) (Figures 1C; 2 and 3, Supplementary Table S4). The resultant mutants, EP01recAG and EP14recAG, were tested to ascertain the ability of these mutants and EP01rec and EP14rec to grow at different tetracycline concentrations (0, 2.5, 5, 10 and 20 and 40 μg/ml). We found EP01recAG and EP14recAG both had decreased ability to grow at high concentrations of tetracycline when compared with EP01rec and EP14rec, demonstrating similarity with the wild type lsrACDBFG operon promoter region (Figure 4). These results, therefore, confirmed that the G→A mutation present in the TGTGCAAT sequence (Figures 2 and 3 and Supplementary Table S4), located very close to the CRPI binding site and that might be associated with the CytR binding, is pivotal to increasing the strength of the evolved promoters obtained from the library created in this study. Interestingly, the EP08rec clone was found to possess a mutation at the directly adjacent nucleotide C→T (TGTGCAA→TGTGTAA) found mutated in EP01rec and EP14rec (Figure 2).
Pederson and Valentin-Hansen (38) have demonstrated through qualitative interactions of CytR with DNA, using DNaseI and dimethyl sulfate (DMS) footprinting at repressor concentrations that saturate the binding site, that interaction of CytR with the octameric motifs AATGTAAC and GTTGCATT invariably protects the central guanine from DMS methylation, consistent with the well conserved G at this position in the deoP2 E. coli promoter sequence.
Experimental validation of this hypothesis involving CytR contact point in Lsr promoter sequence in a more focused study is needed, however.
Strength of EP01rec and EP14rec promoter variants in comparison with wild type E. coli lsrACDBFG operon promoter region
In order to measure and compare the expression levels among the lsr promoter mutants, we evaluated both tet (C) and gfp_mut2 expression to determine the promoters’ strength. The pLSR plasmid was constructed using a pTS40 backbone, which contains two reporter genes, gfp_mut2 and tet (C), resulting in bicistronic expression under the lsr promoter, either wild type or mutant.
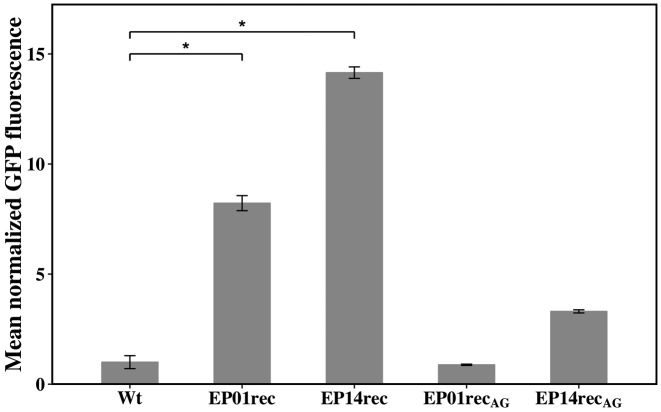
Our earlier library screenings and selections were performed using the tet (C) gene because it facilitated rapid searching for the strongest promoter candidates from a library containing around 106 members (based on the ability of these clones to survive in high concentrations of tetracycline). In order to confirm lsr promoter strength, we performed assays to detect and measure GFP expression (Figure 5), which is encoded by the first gene after the promoter sequence. Samples taken at early and late exponential phases (OD600nm0.5 and 1.0) were evaluated for GFP expression (represented by mean fluorescence using FACS). In all cases, the p-lsrR-box sites restored promoters (EP01rec and EP14rec) resulted in higher GFP fluorescence than their respective controls. There was no difference between the OD600nm 0.5 and 1.0 (data are not shown). Statistical significance was determined with ANOVA and the Tukey-Kramer method, adopting a significance level of α = 0.001. EP01rec and EP14rec showed 8-fold and 14-fold more GFP than the wild type lsr promoter, respectively. As expected, EP01recAG and EP14recAG achieved GFP expression levels comparable with the wild type lsr promoter.
Figure 5.
Representative FACS histograms show the mean normalized GFP fluorescence from LW6 cells transformed with expression vectors containing the wild type E. coli lsrACDBFG operon promoter region (Wt) and the mutants EP01rec, EP14rec, EP01recAG and EP14recAG. GFP expression was measured at two different growth points, OD600nm of 0.5 and 1.0 (only 0.5 shown here). These independent experiments, following β-gal experiments, were performed in technical triplicate and the error bars represent the population's standard deviation. The LW6 strain fluorescence was used as a negative control to set FACS’ voltage and gating, and the Wt sample (E. coli W3110) was used as a normalization factor for data analysis. The statistical significance level was determined to be α = 0.001 (indicated by *) using ANOVA and the Tukey-Kramer method. Fluorescence microscopy images of the FACS samples are also provided (see Supplementary Figure S1).
These same samples, as analyzed via FACS, were also visualized using a 20x microscope fluorescence field. EP01rec and EP14rec were brighter than other lsr promoter mutants (Supplementary Figure S1), confirming the data obtained through FACS. Results obtained investigating GFP levels corroborated the data obtained through the tetracycline resistance assays (Figure 4). As noted earlier, and reconfirmed here with analysis of GFP expression, the single nucleotide found mutated in the putative CytR-binding site sequence (TGTGCAAT→ TGTACAAT) in the strongest promoter mutants, EP01rec and EP14rec was important for increasing the strength of these promoters.
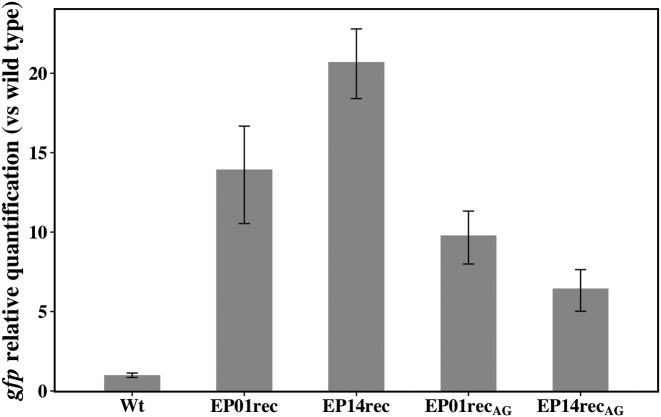
Finally, quantitative PCR (qPCR) was also performed to measure gfp_mut2 transcriptional levels obtained from each of the lsr promoter mutants (EP01rec, EP01recAG, EP14rec, and EP14recAG) versus the wild-type lsr promoter (Figure 6). The results corroborate the phenotypic patterns (Figure 6). We detected higher levels of gfp mRNA in EP01rec and EP14rec when compared to the wild type lsr promoter, and also to EP01recAG and EP14recAG. The gfp transcriptional levels found with the wild type lsr promoter were lower than the levels found with EP01recAG and EP14recAG and are consistent with the GFP expression data. Recall, however, that other mutations are present in the EP01recAG and EP14recAG clones that may have influenced their levels relative to the wild type lsr promoter. Interestingly, the gfp transcriptional pattern for EP01recAG and EP14recAG (Figure 6) does not precisely replicate the GFP expression pattern (Figure 5) and that the difference in transcriptional levels is less than expected when one compares EP01rec and EP14rec to EP01recAG and EP14recAG.
Figure 6.
Representative qPCR shows the gfp relative quantification from LW6 transformed with wild type E. coli lsrACDBFG operon promoter region (Wt) its mutants EP01rec, EP14rec, EP01recAG and EP14recAG. Total RNA was extracted at OD600nm ∼ 0.5 (∼3 h). This representative independent experiment was performed in technical triplicate and the error bars represent the standard deviation.
Inducible expression based in AI-2 quorum sensing components
Initial switching experiments using the lsr operon promoter mutants, as well as the wild type promoter cloned into the original pLSR plasmid were carried out in E. coli luxS and lsrR mutants as hosts. In the case of the luxS mutant, we added exogenous AI-2 to stimulate expression and in the case of the lsrR mutant, we looked for a step increase in expression per cell as cultures ensued. Interestingly, in these cases, there was minimal, if any, apparent repression, even after adding 0.2% glucose in LB agar media (data not shown). Perhaps this was an artifact of the pLSR plasmid carrying the CloDF13 replication origin (relatively high 20–40 copies/cell), which is originally from the pTS40 plasmid backbone (35). Thus, LsrR repressor levels, present in low or no copy number per cell might have been insufficient to repress the multi-copy lsr promoter.
Accordingly, in order to more carefully evaluate whether both lsr promoter mutants, EP01rec and EP14rec, had similar switching characteristics as the native promoter, we recloned the entire promoter regions into a lower copy number vector, the single copy plasmid pFZY1. pFZY1 is a galK-lacZYA transcriptional fusional vector that expresses LacZ under an inserted promoter (44). We transformed pPH01, pPH14 (Supplementary Table S1) or pLW11 (6), which contains the wild type lsr promoter cloned into pFZY1, into the LW7 strain, which is a luxS knockout (Supplementary Table S1).
Miller assays were then performed to measure LacZ expression levels under the three promoters: wild type, EP01rec and EP14rec, in both the presence and absence of AI-2. That is, we measured LacZ expression levels after we allowed the cells to reach OD600nm ∼ 0.2 and added 40 μM AI-2, continued growth and retested. This mid-exponential phase OD scenario is aligned with native QS signaling. Thus, most of lsr promoter elements such as AI-2 uptake (lsrACBD), modification (phosphorylation by LsrK) and degradation (lsrFG) should have been at appropriate levels for subsequent analysis of engineered lsr promoter activity.
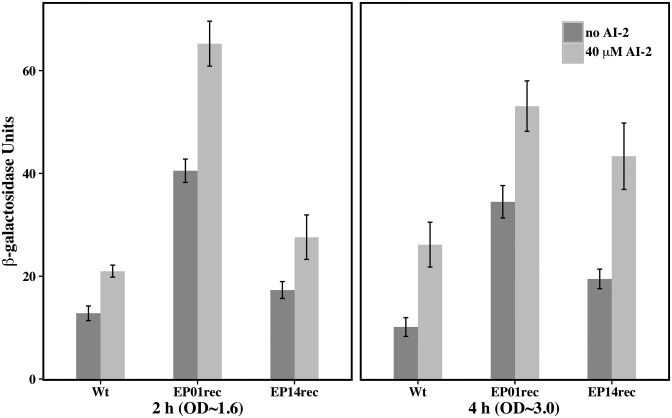
As expected, after 2 and 4 h of AI-2 induction all three of the promoters showed higher LacZ expression levels when compared with the same experiments performed in absence of inducer (Figure 7). After 2 h of AI-2 induction, EP01rec showed a nearly 3-fold increase in LacZ expression (Miller units) when compared with wild type. EP14rec exhibited only a 1.5-fold increase (Figure 7). Then, at later times, the EP14rec promoter exhibited over a 2-fold increase. In both cases, the influence of LsrR repression (indicated by amplification upon addition of AI-2), was exhibited. Experimental results obtained with E. coli cells carrying both multi and single copy vectors for lsr promoter driven gene expression were successful in that engineered promoter sequences exhibited greater expression while still retaining the LsrR-repression capability. Subsequent use of these evolved promoters EP01rec and EP14rec as simple expression vectors or as elements of additional genetic circuits will naturally require consideration of the availability of LsrR, LsrK and other AI-2 mediating components (13).
Figure 7.
Transcriptional regulation of the wild type and mutant E. coli lsr operon promoters EP01rec and EP14rec. LW7 cells (ΔluxS) carries the single copy plasmids pLW11 (wild type), pPH01 (EP01rec) or pPH14 (EP14rec) expressing β-galactosidase. Panels (A) and (B) represent different time points during cell growth (2 and 4 h); aliquots were collected for measurement of OD600 and β-galactosidase activity. The data displayed represent one of two comparable experiments performed independently. Data are plotted as means ± standard deviations of technical triplicates.
CONCLUSIONS
By creating a library of lsr operon promoter mutants through directed evolution we were able to obtain mutant promoters with activity stronger than the wild type promoter. Most of the mutants (12 out of the 14) showed increased strength that was related to mutations found near or within the LsrR-binding site; not just in the 6-bp (AACAAT) and 9-bp (AAGATTTAA) sequences of the p-lsrR-box region. Even though the p-lsrR-box sequence region has been shown to be essential to the LsrR-binding site (4), our results indicate that more nucleotides present in the vicinity of the proposed LsrR-binding site are also involved in controlling expression levels under this E. coli promoter.
In addition, we have isolated two lsr promoter mutants, EP01rec and EP14rec that behaved differently than the wild type. We identified a defining mutation in EP01rec and EP14rec responsible for the increase in lsr promoter strength. This point mutation is located very close to the CRPI sequence and was examined using in silico methods, where we found its association with the CytR binding site. The combination of experimental data from a previous study performed with the E. coli deoP2 promoter (38) and our in silico analysis revealed a possible association between increased promoter strength and a CytR-DNA-binding domain.
Finally, tests on inducible expression based on AI-2 QS were performed with EP01rec and EP14rec promoters. Data demonstrated that both promoters preserved their switching character based on the presence of AI-2. We note, however, that in order to demonstrate the full switching character of these promoters which are significantly stronger than the wild type, a single copy number plasmid was needed. This may be due to the need for enmeshing components of these vectors with other endogenous components (such as CytR or CRP) that are present at native levels. Thus, should maximum overexpression be an objective for application of these promoters, additional investigations are envisioned that more optimally accommodate native regulatory components and those of the synthetic construct. In general, our work represents progress toward engineering the Lsr promoter system into a tool for applied biotechnology. We can envision future applications such as expression of therapeutic proteins (45) through engineered probiotic bacteria (46), controlled by gut flora or even by pathogenic bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Defense Threat Reduction Agency (DTRA) [HDTRA1-13-1-00037]; US National Science Foundation [CBET #1160005]. Funding for open access charge: DTRA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Schauder S., Shokat K., Surette M.G., Bassler B.L. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 3.Quan D.N., Bentley W.E. Gene network homology in prokaryotes using a similarity search approach: queries of quorum sensing signal transduction. PLoS Comput. Biol. 2012;8:e1002637. doi: 10.1371/journal.pcbi.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue T., Zhao L., Sun H., Zhou X., Sun B. LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Res. 2009;19:1258–1268. doi: 10.1038/cr.2009.91. [DOI] [PubMed] [Google Scholar]
- 5.Xavier K.B., Bassler B.L. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Hashimoto Y., Tsao C.-Y., Valdes J.J., Bentley W.E. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 2005;187:2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Li J., March J.C., Valdes J.J., Bentley W.E. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J. Bacteriol. 2005;187:8350–8360. doi: 10.1128/JB.187.24.8350-8360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xavier K.B., Miller S.T., Lu W., Kim J.H., Rabinowitz J., Pelczer I., Semmelhack M.F., Bassler B.L. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2007;2:128–136. doi: 10.1021/cb600444h. [DOI] [PubMed] [Google Scholar]
- 9.Marques J.C., Lamosa P., Russell C., Ventura R., Maycock C., Semmelhack M.F., Miller S.T., Xavier K.B. Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): characterization of phospho-(S)-4, 5-dihydroxy-2, 3-pentanedione isomerization by LsrG protein. J. Biol. Chem. 2011;286:18331–18343. doi: 10.1074/jbc.M111.230227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marques J.C., Oh I.K., Ly D.C., Lamosa P., Ventura M.R., Miller S.T., Xavier K.B. LsrF, a coenzyme A-dependent thiolase, catalyzes the terminal step in processing the quorum sensing signal autoinducer-2. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14235–14240. doi: 10.1073/pnas.1408691111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taga M.E., Miller S.T., Bassler B.L. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 2003;50:1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- 12.Craney A., Hohenauer T., Xu Y., Navani N.K., Li Y., Nodwell J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 2007;35:e46. doi: 10.1093/nar/gkm086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsao C.-Y., Hooshangi S., Wu H.-C., Valdes J.J., Bentley W.E. Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab. Eng. 2010;12:291–297. doi: 10.1016/j.ymben.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Close D., Xu T., Smartt A., Rogers A., Crossley R., Price S., Ripp S., Sayler G. The evolution of the bacterial luciferase gene cassette (lux) as a real-time bioreporter. Sensors. 2012;12:732–752. doi: 10.3390/s120100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams B.L., Carter K.K., Guo M., Wu H.-C., Tsao C.-Y., Sintim H.O., Valdes J.J., Bentley W.E. Evolved quorum sensing regulator, LsrR, for altered switching functions. ACS Synth. Biol. 2013;3:210–219. doi: 10.1021/sb400068z. [DOI] [PubMed] [Google Scholar]
- 16.Wu H.-C., Tsao C.-Y., Quan D.N., Cheng Y., Servinsky M.D., Carter K.K., Jee K.J., Terrell J.L., Zargar A., Rubloff G.W., et al. Autonomous bacterial localization and gene expression based on nearby cell receptor density. Mol. Syst. Biol. 2013;9:636–636. doi: 10.1038/msb.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terrell J.L., Wu H.-C., Tsao C.-Y., Barber N.B., Servinsky M.D., Payne G.F., Bentley W.E. Nano-guided cell networks as conveyors of molecular communication. Nat. Commun. 2015;6:8500. doi: 10.1038/ncomms9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordonov T., Kim E., Cheng Y., Ben-Yoav H., Ghodssi R., Rubloff G., Yin J.-J., Payne G.F., Bentley W.E. Electronic modulation of biochemical signal generation. Nat. Nanotech. 2014;9:605–610. doi: 10.1038/nnano.2014.151. [DOI] [PubMed] [Google Scholar]
- 19.Tschirhart T., Zhou X.Y., Ueda H., Tsao C.-Y., Kim E., Payne G.F., Bentley W.E. Electrochemical Measurement of the β-Galactosidase Reporter from Live Cells: A Comparison to the Miller Assay. ACS Synth. Biol. 2015;5:28–35. doi: 10.1021/acssynbio.5b00073. [DOI] [PubMed] [Google Scholar]
- 20.Hwang I.Y., Tan M.H., Koh E., Ho C.L., Poh C.L., Chang M.W. Reprogramming Microbes to Be Pathogen-Seeking Killers. ACS Synth. Biol. 2013;3:228–237. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- 21.Hong S.H., Hegde M., Kim J., Wang X., Jayaraman A., Wood T.K. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat. Commun. 2012;3:613. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choudhary S., Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 2010;86:1267–1279. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 23.Tsao C.-Y., Wang L., Hashimoto Y., Yi H., March J.C., DeLisa M.P., Wood T.K., Valdes J.J., Bentley W.E. LuxS coexpression enhances yields of recombinant proteins in Escherichia coli in part through posttranscriptional control of GroEL. Appl. Environ. Microbiol. 2011;77:2141–2152. doi: 10.1128/AEM.02347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haseltine E.L., Arnold F.H. Implications of rewiring bacterial quorum sensing. Appl. Environ. Microbiol. 2008;74:437–445. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber W., Fussenegger M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 2012;13:21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu F., Menn D.J., Wang X. Quorum-sensing crosstalk-driven synthetic circuits: from unimodality to trimodality. Chem. Biol. 2014;21:1629–1638. doi: 10.1016/j.chembiol.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis R.M., Muller R.Y., Haynes K.A. Can the natural diversity of quorum-sensing advance synthetic biology. Front. Bioeng. Biotechnol. 2015;3:30. doi: 10.3389/fbioe.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soma Y., Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab. Eng. 2015;30:7–15. doi: 10.1016/j.ymben.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Tien S.-M., Hsu C.-Y., Chen B.-S. Engineering bacteria to search for specific concentrations of molecules by a systematic synthetic biology design method. PLoS ONE. 2016;11:e0152146. doi: 10.1371/journal.pone.0152146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott S.R., Hasty J. Quorum sensing communication modules for microbial consortia. ACS Synth. Biol. 2016 doi: 10.1021/acssynbio.5b00286. doi:10.1021/acssynbio.5b00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant P.K., Dalchau N., Brown J.R., Federici F., Rudge T.J., Yordanov B., Patange O., Phillips A., Haseloff J. Orthogonal intercellular signaling for programmed spatial behavior. Mol. Syst. Biol. 2016;12:849–849. doi: 10.15252/msb.20156590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chappell J., Jensen K., Freemont P.S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013;41:gkt052–3481. doi: 10.1093/nar/gkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saeidi N., Wong C.K., Lo T.-M., Nguyen H.X., Ling H., Leong S.S.J., Poh C.L., Chang M.W. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 2011;7:521–521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sohka T., Heins R.A., Phelan R.M., Greisler J.M., Townsend C.A., Ostermeier M. An externally tunable bacterial band-pass filter. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10135–10140. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S.K., Chou H.H., Pfleger B.F., Newman J.D., Yoshikuni Y., Keasling J.D. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl. Environ. Microbiol. 2007;73:5711–5715. doi: 10.1128/AEM.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen H., Valentin-Hansen P. Protein-induced fit: the CRP activator protein changes sequence-specific DNA recognition by the CytR repressor, a highly flexible LacI member. EMBO J. 1997;16:2108–2118. doi: 10.1093/emboj/16.8.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pages H., Gentleman R., Aboyoun P., DebRoy S. Biostrings: string objects representing biological sequences, and matching algorithms. 2014. R package version 2.36.4.
- 40.Heckman K.L., Pease L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 41.Livak K.J. ABI prism 7700 Sequence Detection System. 1997. http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf UserBulletin no.2. PE Applied Biosystems, AB website, bulletin reference: 4303859B 777802-002.
- 42.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Miller J.H. NY: Cold Spring Harbor Laboratory; 1972. Experiments in molecular genetics. [Google Scholar]
- 44.Koop A.H., Hartley M.E., Bourgeois S. A low-copy-number vector utilizing beta-galactosidase for the analysis of gene control elements. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J.A., Oliveira R.A., Djukovic A., Ubeda C., Xavier K.B. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 46.Duan F., March J.C. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc.. Natl. Acad. Sci. U.S.A. 2010;107:11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.