Significance
Tandem MS is a technique for compound identification in untargeted metabolomics experiments. Because of a lack of reference spectra, most molecules cannot be identified, and many spectra cannot be used. We present MS2LDA, an unsupervised method (inspired by text-mining) that extracts common patterns of mass fragments and neutral losses—Mass2Motifs—from collections of fragmentation spectra. Structurally characterized Mass2Motifs can be used to annotate molecules for which no reference spectra exist and expose biochemical relationships between molecules. For four beer extracts, without training data, we show that, with 30 structurally characterized Mass2Motifs, we can annotate approximately three times as many molecules as with library matching. These Mass2Motifs were validated in reference spectra from Global Natural Products Social Molecular Networking (GNPS) and MassBank.
Keywords: metabolomics, mass spectrometry, fragmentation, bioinformatics, topic modeling
Abstract
The potential of untargeted metabolomics to answer important questions across the life sciences is hindered because of a paucity of computational tools that enable extraction of key biochemically relevant information. Available tools focus on using mass spectrometry fragmentation spectra to identify molecules whose behavior suggests they are relevant to the system under study. Unfortunately, fragmentation spectra cannot identify molecules in isolation but require authentic standards or databases of known fragmented molecules. Fragmentation spectra are, however, replete with information pertaining to the biochemical processes present, much of which is currently neglected. Here, we present an analytical workflow that exploits all fragmentation data from a given experiment to extract biochemically relevant features in an unsupervised manner. We demonstrate that an algorithm originally used for text mining, latent Dirichlet allocation, can be adapted to handle metabolomics datasets. Our approach extracts biochemically relevant molecular substructures (“Mass2Motifs”) from spectra as sets of co-occurring molecular fragments and neutral losses. The analysis allows us to isolate molecular substructures, whose presence allows molecules to be grouped based on shared substructures regardless of classical spectral similarity. These substructures, in turn, support putative de novo structural annotation of molecules. Combining this spectral connectivity to orthogonal correlations (e.g., common abundance changes under system perturbation) significantly enhances our ability to provide mechanistic explanations for biological behavior.
Mass spectrometry (MS)-based metabolomics aims to capture the entire small-molecule composition of biological systems. Analysis of MS metabolomics data are challenging as many molecules cannot be identified from their mass (e.g., isobaric molecules, and isomers) (1–3). Separation by liquid chromatography before MS (LC-MS) can add discriminatory information but does not solve the problem as isomers can exhibit similar chromatographic behavior, and chromatographic retention time is currently unpredictable.
Fragmentation spectra have been used to partially overcome this problem (4–6). Most tools compare individual fragmentation spectra to reference spectra (5, 7) stored in public databases, for example, MassBank (8) or Human Metabolome Database (9), and are thus constrained by the limited number of reference spectra (10–12). Poor identification coverage can result in poor biochemical insight. We propose a method that analyzes all acquired fragmentation spectra to expose underlying biochemistry without relying on metabolite identification, inspired by machine-learning techniques developed initially for text processing (13).
The paucity of techniques that share information across fragmentation spectra can be explained by the complexity of fragmentation data (14). One example, “Molecular Networking,” clusters MS1 peaks by their MS2 spectral similarity such that one structurally annotated metabolite in a cluster facilitates structural annotation of its neighbors (15, 16). However, spectral features causing the clustering must be extracted manually, and only MS2 spectra with high overall spectral similarity are grouped. Another package, MS2Analyzer (17) mines MS2 spectra for specific features defined by the user (i.e., mass fragments and neutral losses). Some will be common to many experiments (e.g., CO or H2O losses), but sample-specific features are easily overlooked. Although Molecular Networking requires no user intervention, it may fail to group molecules that share small substructures, whereas MS2Analyzer can find all molecules that share a particular set of features provided they are user specified. Our approach, MS2LDA, which is based on latent Dirichlet allocation (LDA) (13), retains the benefits of both of these approaches while losing the shortfalls—it can find relevant substructures based on the co-occurrence of mass fragments and neutral losses, and group the molecules accordingly. Although adapted to other domains [e.g., genomics (18) and transcriptomics (19)], LDA has never been used to exploit the parallels between MS2 data and text.
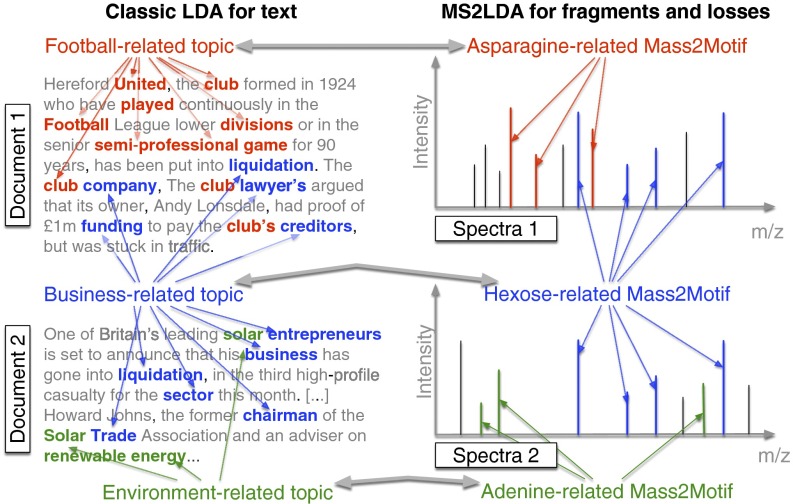
Fragmentation spectra contain recurring patterns of fragments and losses due to common biological substructures (e.g., a hexose unit, or a carboxyl group loss). We assume each observed spectrum is composed of one or more such substructures, an assumption successfully used in other workflows (6, 20); however, no unsupervised strategy exists that finds mass fragmental-based substructures without training data. Fig. 1 demonstrates the parallels between text and fragmentation data. LDA decomposes documents into topics based on co-occurring words, whereas MS2LDA decomposes fragmentation spectra into blocks of co-occurring fragments and losses, referred to as “Mass2Motifs.” Using all of the fragmentation spectra generated by data-dependent mass fragmentation analysis (DDA), MS2LDA learns the conserved substructures (the Mass2Motifs) and the decomposition of the fragmentation spectra into Mass2Motifs.
Fig. 1.
Analogy between LDA for text and MS2LDA. LDA finds topics interpreted as “football related,” “business-related,” and “environment related.” MS2LDA finds sets of concurring mass fragments or losses (Mass2Motifs) that can be interpreted as “Asparagine-related,” “Hexose-related,” and “Adenine-related.”
Our analysis pipeline (SI Appendix, section 1) performs data preprocessing, extracts Mass2Motifs, and allows interactive exploration of the results. Through the analyses of four beer extracts, we show that, without labeled training data or metabolite identification, MS2LDA extracts mass patterns indicative of biological substructures that can be structurally annotated, some of which are pathway related. These can aid in the putative de novo annotation or functional classification of otherwise-unidentifiable molecules. Many more molecules can be annotated in this way than through comparison with reference spectra. Grouping of molecules based on common substructures is particularly useful for hypothesis-generating research. For example, hypotheses as to the source of variation in metabolite abundances can be obtained by linking MS1 abundance changes to the presence of common substructures.
MS2LDA
Data, in the form of .mzXML (full scan) and .mzML (fragmentation) files, are preprocessed using XCMS (21) and MzMatch (22) for peak detection and RMassBank (23) for detecting MS1–MS2 pairs, before matrix formation by aligning MS2 features across different spectra. The resulting matrix has MS2 features (fragments and losses) as rows, and MS2 peaks as columns. The values in the matrix are the MS2 feature intensities, which are subsequently transformed into integer “counts” (SI Appendix, section S1).
For LDA inference, we have implemented both collapsed Gibbs sampling (24) and variational Bayes (13) in Python. The output is a set of Mass2Motifs and assignments of Mass2Motifs to each MS1 peak. In addition, we provide an optional elemental formula assignment step (25–27) to assign candidate elemental formulas to the MS2 features and MS1 peaks. On a laptop (Intel Core i7; 16-GB RAM), running the workflow for one beer sample takes around 20 min for the feature extractions, and between 1 h (Gibbs sampling) and 30 min (variational Bayes) for the inference. The LDA output can be explored in the MS2LDAvis module [customized from LDAvis (28)]. Full details are provided in SI Appendix, section S1. We used MS2LDAvis to inspect Mass2Motifs with degree 10 (i.e., that were present in 10 or more spectra) and structurally characterized them (assigned a substructural annotation) at varying levels of confidence (SI Appendix, section S2.1) through expert knowledge and matching of the Mass2Motif spectra to reference spectra in MzCloud (www.mzcloud.org).
Results
The MS2LDA workflow was independently applied to four beer extracts. After preprocessing, each sample consisted of around 1,000 MS peaks in both positive and negative ionization mode (SI Appendix, section S2.2). Three hundred Mass2Motifs were extracted for each data file and checked for biochemical relevance. Thirty to 40 Mass2Motifs in each of the positive ionization mode files were structurally characterized (SI Appendix, Table S-4) and diverse biochemically relevant substructures found included histidine, phenylalanine, adenine, hexose units, and structural features such as water or carboxyl group loss.
The degree of Mass2Motifs (the number of spectra in which they occurred) varied from 1 to over 200, demonstrating that MS2LDA can extract both generic and specific structural features. The number of Mass2Motifs within each spectrum also varied (around 600 spectra in each file consisted of one Mass2Motif, 300 of two, 50 of three, and 20 of four or more). Across the four files, an average of 70% of spectra (SI Appendix, section S2.3) include at least one characterized Mass2Motif, demonstrating the power of MS2LDA for data reduction—that is, structurally characterizing just 30–40 of the discovered Mass2Motifs provides biochemical insight into 70% of the spectra. For comparison, we matched spectra to the MassBank and National Institute of Standards and Technology (NIST) libraries (SI Appendix, section S2.4) at a threshold of 90% normalized score, obtaining hits for only 25 and 6% of the spectra, respectively, demonstrating the wide coverage possible with MS2LDA.
Automatic, Unsupervised, Chemical Substructure Discovery.
Mass2Motifs cover a diverse set of biochemical features, including amino acid related (i.e., histidine, leucine, tryptophan, and tyrosine), nucleotide related (i.e., adenine, cytosine, and xanthine), and other molecules such as cinnamic acid, ferulic acid, ribose, and N-acetylputrescine. Mass2Motifs related to the same substructure or structural feature were consistently found across multiple beers (e.g., hexose-related Mass2Motifs were present in all positive-ionization mode files). Differences in degree and absence of some Mass2Motifs across the extracts show that MS2LDA captures variability in metabolic composition.
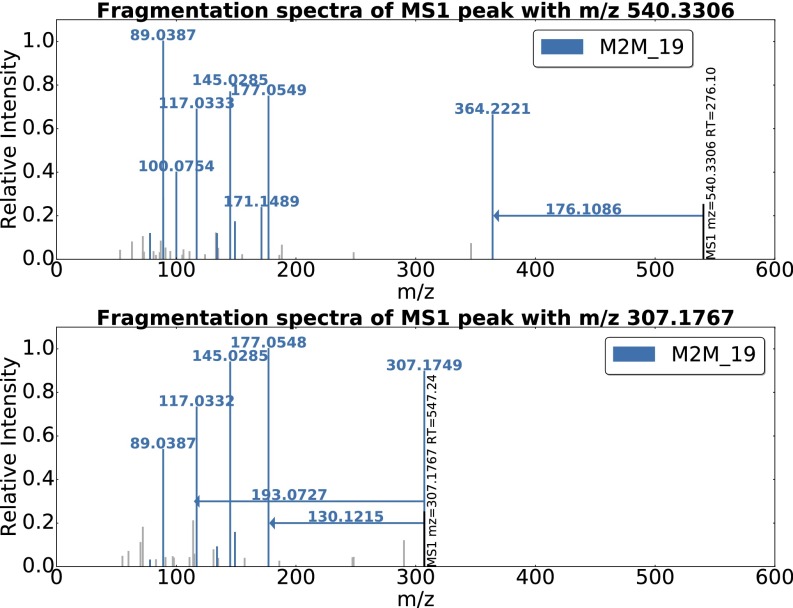
An example of ferulic acid (a compound present in cereals, an ingredient of beer) is given in Fig. 2. Two of the 11 spectra that include Mass2Motif 19 are shown. Conserved mass fragments are clearly visible across the two spectra. Unlike existing software, for example, MS2Analyzer (17), our method is unsupervised and has no need for prior knowledge about fragments of interest. It is of note that the neutral loss of the complete ferulic acid moiety was also included by MS2LDA, demonstrating that both fragments and losses can be present in a motif. MS2LDA is able to extract a relatively rare biochemically relevant pattern (present in 11 of the spectra), despite the individual spectra being quite different.
Fig. 2.
Two spectra, from the Beer3 positive-ionization mode file, each of which includes Mass2Motif 19, annotated as the plant-derived ferulic acid substructure. The mass fragments and neutral losses (arrows originating at the precursor ions) included in Mass2Motif 19 are highlighted in color. Fragments not explained by Mass2Motif 19 are light gray. The probabilistic nature of MS2LDA means that Mass2Motifs will not necessarily be identical in all spectra in which they appear.
Positive-ionization mode fragmentation spectra generally provide larger sets of conserved fragments but some Mass2Motifs, for example, those related to phosphate and sulfate groups [fragments at 78.9593 ([PO3]−) and 79.9575 ([SO3]−) m/z, respectively] were more easily identifiable in negative mode; an argument to use both ionization modes. Three of the characterized positive-mode Mass2Motifs pointed to the highly similar aromatic substructures of phenylethene, cinnamic acid (cinnamate), and phenylethyleneamine (i.e., [phenylalanine–CHOOH]), demonstrating discrimination of very similar yet functionally different substructures (SI Appendix, section S2.6).
Structurally Characterized Mass2Motifs Validated in Authentic Standards.
Reference molecules in the beer extracts were identified based on chromatographic coelution and corresponding exact mass. As their identity is known, we can validate our structurally characterized Mass2Motifs. Of the 45 reference molecules, we could identify, 38 included one or more characterized Mass2Motifs, 32 of which were validated (i.e., do indeed include the relevant substructure), despite the fact that the Mass2Motif was characterized without a reference molecule identification.
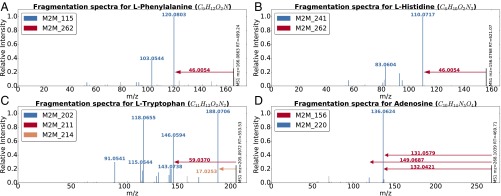
Some examples are provided in Fig. 3. The spectra for phenylalanine (Fig. 3A) and histidine (Fig. 3B) share Mass2Motif 262, indicating the presence of a free (underivatized) carboxylic acid group. The loss of CHOOH (Mass2Motif 262) is in fact a common characteristic for many other underivatized amino acids and free organic acids and was associated with 10 of the 18 identified amino acid structures [the remaining 8 prefer alternative fragmentation routes—e.g., see the amine loss (Mass2Motif 214) in tryptophan, Fig. 3C]. The other Mass2Motifs (115, 241) in Fig. 3 A and B are related to phenylalanine and histidine, respectively (more details in SI Appendix, section S2.7). Fig. 3D is the MS2 spectrum of adenosine, which consists of an adenine molecule conjugated to a ribose sugar molecule. The two associated Mass2Motifs (156, 220) represent these two biochemically relevant structural features (i.e., adenine substructure and a ribose sugar loss).
Fig. 3.
Mass2Motif spectra of identified metabolites: (A) l-histidine, (B) l-phenylalanine, (C) l-tryptophan, and (D) adenosine. Characterized motifs are indicated by color. Full details of the mentioned Mass2Motifs can be found in SI Appendix, section S2.7.
Spectra can include multiple Mass2Motifs. In each of Fig. 3 A–D, we observe two or more Mass2Motifs. We know of no other method that can do this without training spectra consisting of known structures, or prior knowledge of interesting feature combinations. Multiple Mass2Motifs can also explain the same feature in one spectrum, that is, the fragments 110.0717 (C5H8N3, [M+H]+) and 120.0803 (C8H10N, [M+H]+) in Fig. 3 A and B are explained by Mass2Motifs 241 and 115 and also by the 46.0054 loss (CHOOH) of Mass2Motif 262. This demonstrates the manner in which MS2LDA decomposes molecules into their constituent building blocks, allowing for de novo metabolite annotation.
Mass2Motifs Aid de Novo Metabolite Annotation.
On average, 70% of the fragmented MS1 features are explained by at least one structurally characterized Mass2Motif and can therefore be automatically classified. For comparison, we performed spectral matching using the NIST MS/MS database for small molecules (chemdata.nist.gov/mass-spc/msms-search/) and MassBank (8) on seven of the metabolites annotated via the ferulic acid Mass2Motif. Only one returned a ferulic acid-related hit, despite the clear presence of ferulic acid in all spectra (Fig. 2). The Mass2Motif itself can be represented as a spectrum and be subjected to spectral matching, resulting in transferulic acid as the best hit (hinting at the possibility of automatic Mass2Motif annotation). Spectra that are explained by the Mass2Motifs related to histidine, tyrosine, and tryptophan were also subjected to spectral matching. From 39 metabolites annotated with help of MS2LDA, 7 resulted in correct hits with another 8 producing structurally related hits (SI Appendix, section S2.4). These results clearly demonstrate the annotative power of MS2LDA, through which annotations can be made by matching only small portions of the spectra and therefore allowing annotation (classification) of molecules not present in databases. In summary, our experiments show that MS2LDA is able to annotate approximately three times as many metabolites as spectral matching. In addition, MS2LDA can annotate and group spectra based on neutral losses (e.g., the loss of CHOOH), which is not possible with spectral matching.
To further assess the use of the structurally characterized Mass2Motifs in metabolite annotation, we used MS2LDA to decompose 1,953 and 5,670 spectra from MassBank and the Global Natural Products Social Molecular Networking (GNPS) (15), respectively, into 500 Mass2Motifs each. These datasets are those used for training in ref. 6. In contrast to the beer data, none of these spectra is derived from Orbitrap instruments. The structural identity of all metabolites is known, providing a ground truth. In both cases, the Mass2Motifs characterized from beer were included in the analysis and kept fixed, whereas all other Mass2Motifs are learned during LDA inference (details in SI Appendix, section S2.8). This therefore assesses the extent to which structurally characterized Mass2Motifs in one analysis can be used for metabolite annotations in another (from another instrument type). We manually verified all metabolites that include the formerly characterized Mass2Motifs and found that, at a probability threshold of 0.1, 81.5 and 63.3% of substructure annotations (for MassBank and GNPS, respectively) were validated (see SI Appendix, section S2.8, Fig. S-12, for detailed analysis of Mass2Motifs). In total, 694 (MassBank) and 613 (GNPS) spectra were found to have one or more validated substructure annotations (note that this is based solely on the Mass2Motifs annotated in the beer analysis, demonstrating a wide coverage from a small number of Mass2Motifs). MS2LDA also discovered MassBank- and GNPS-related substructures, complementary to those found in beer, showing its generic use. We repeated the analysis on a complex biological mixture (a human urine sample) and matched the Mass2Motifs discovered in beer to those found in urine. Matched standards in the urine are then used to validate the Mass2Motifs structural characterizations. At the 0.1 threshold, 74.3% of characterizations were validated. These results clearly demonstrate the potential of MS2LDA for substructure annotation.
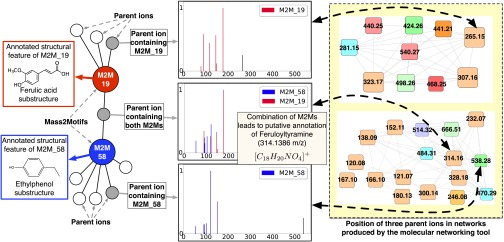
One illustrative example of annotation with MS2LDA is provided in Fig. 4. A subset of the network produced by MS2LDAvis (SI Appendix, section S1.4) is shown, consisting of molecules related to two Mass2Motifs (ferulic acid and ethylphenol). All but one molecule includes just one of the Mass2Motifs, but one belongs to both (the fragments belonging to each Mass2Motif are clearly visible). The presence of both Mass2Motifs allows us to putatively annotate it as feruloyltyramine (314.1386 m/z; [C18H20NO4]+) despite spectral matching producing no relevant hits (SI Appendix, Table S-9). The output of Molecular Networking (15, 29) is shown on the Right of Fig. 4 (described in SI Appendix, section S2.9). This produces clusters interpretable as ferulic acid and ethylphenol related, but as each molecule can belong to only one cluster, feruloyltyramine is assigned to the ethylphenol cluster and its relationship with ferulic acid is lost. Allowing each spectra to include multiple Mass2Motifs thus gives far greater potential in making de novo structural annotations of molecules. A lower perplexity of the LDA model compared with a standard multinomial model supports these results (SI Appendix, section S2.10). The phenomenon of individual spectra containing multiple correct substructure annotations is widespread. In the MassBank and GNPS datasets, we counted the number of spectra associated with one, two, three, and four different manually validated annotations from the beer-characterized Mass2Motifs. Of the 694 MassBank spectra (613 GNPS) that had one or more validated substructure annotations, 212 (GNPS 34) had two or more; 39 (GNPS 4), three or more; and 3, four (GNPS 0) (SI Appendix, Fig. S-14).
Fig. 4.
Mass2Motifs 19 and 58 were found to be representative of ferulic acid and ethylphenol, respectively. Eleven and 42 MS1 features in the Beer3 dataset were explained by those two Mass2Motifs. Of those, one was explained by both, aiding in its annotation as feruloyltyramine (314.1386 m/z; [C18H20NO4]+). On the Right of the plot, we show the clusters containing these MS1 features created using the molecular networking tool (15) [Top, ferulic acid; Bottom, tyramine (ethylphenol)]. Node coloring and size are irrelevant here. The compound containing both Mass2Motifs is forced into the ethylphenol cluster, losing its relationship with ferulic acid.
Differential Expression of Mass2Motifs Reveals Biochemical Changes Across Samples.
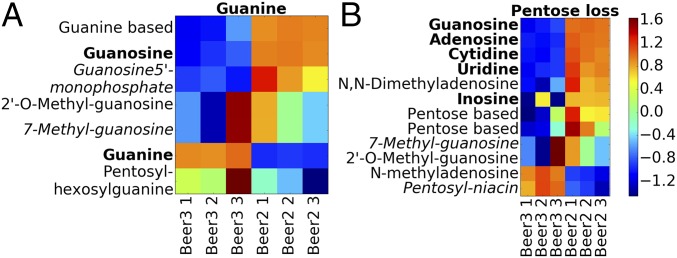
Annotating more metabolites is beneficial when investigating the changes in metabolite intensity across multiple samples. As MS2LDA groups metabolites in a biochemically relevant manner, we can go a step further and consider the differential expression (DE) of Mass2Motifs in a manner similar to approaches taken in transcriptomics where it is common to consider the shared DE of a group of related transcripts as indicative of their contribution to a common aspect of cellular biology (30). For example, consider a standard metabolomics experiment comparing MS1 intensities across multiple replicates of two conditions. After the MS1 peaks have been matched across samples, those that share a Mass2Motif (defined in a single MS2LDA analysis of one of the samples or an additional pooled sample) can be grouped, and the DE of the groups computed. To demonstrate, we compared three full-scan replicates of beers 2 and 3 using MS1 groupings defined by the Mass2Motifs from the MS2LDA analysis of Beer3. DE of groups was assessed using PLAGE (31). Fig. 5A shows MS1 peaks associated with a guanine-related Mass2Motif suggesting that, in Beer3, free guanine is more abundant, whereas in Beer2, guanine conjugates dominate. Similarly, molecules associated with the pentose Mass2Motif (Fig. 5B) show DE between beers 2 and 3. We investigated whether or not similar outcomes could be achieved with spectral similarity clustering. However, the 12 pentose-related metabolites were distributed across 10 clusters hiding the correlated intensity change (see SI Appendix, section S2.11, for more examples).
Fig. 5.
Log fold-change heat maps for the (A) guanine and (B) pentose loss Mass2Motifs. Each row is an MS1 peak, and columns represent samples. Bold names could be matched to a reference compound. Detailed annotations of metabolites can be found in SI Appendix, Table S-18.
Discussion
MS2LDA was inspired by the idea that conserved fragments and neutral losses can be indicative of metabolite substructures and the implied parallel with topic modeling of text. No alternative tools exist that allow for the unsupervised substructure mining from MS fragmentation data while also allowing for multiple such substructures to be present within one metabolite. MS2LDA can group molecules that share substructures without high similarity across their entire MS2 spectra. It reduces complex fragmentation datasets into metabolites explained by one or more patterns of concurring mass fragments or neutral losses—Mass2Motifs.
MS2LDA relies on reliable matching of MS1 peaks to MS2 spectra and works best for complex mixtures where a large number of metabolites are fragmented and information-rich MS2 spectra are available (e.g., generated by ramped or stepped collision energy). High-resolution MS fragmentation can differentiate mass fragments and neutral losses even at low mass range of 50–70 m/z (SI Appendix, section S2.12). Manual structural characterization of many Mass2Motifs is straightforward, and the structural features or substructures can be propagated to all connected MS2 spectra. Based on initial experiments, automated Mass2Motif annotation is promising (19 of the characterized positive-mode beer Mass2Motifs were correctly annotated, despite the fact that losses are not currently supported by spectral matching tools and had to be omitted; SI Appendix, section S2.13).
Metabolite annotation and identification is a bottleneck in high-throughput metabolomics. MS2LDA can assist by automatically assigning possible substructures to a fragmented LC-MS peak via the Mass2Motifs present in its MS2 spectrum. MS2LDA can thus quickly classify MS1 peaks into functional classes without knowing the complete structure of the metabolite. On average, over 70% of the fragmented metabolites were explained by one or more structurally annotated Mass2Motifs, a massive improvement on results reported in a recent study, again using beer as an exemplar, where only 2–3% of the high-abundance differentially expressed molecular features could be classified (11). Validation on data from the MassBank and GNPS databases also demonstrated the validity of our structurally characterized Mass2Motifs and also showed how fixed Mass2Motifs characterized in one analysis could be used in other datasets, even those produced from different laboratories on different instruments. In addition, the biochemically relevant metabolite grouping provided by MS2LDA allows us to identify Mass2Motifs that are enriched with metabolites with correlated intensity variation.
Computationally, MS2LDA is more costly than simpler tools, but not prohibitively so. For example, using variational Bayesian inference, the GNPS dataset (5,670 spectra) could be decomposed into 500 Mass2Motifs in approximately 4 h on a laptop. As LDA has been used on very large text corpora [e.g., 3.3 million documents from Wikipedia (32)], the technology exists to comfortably scale this type of analysis to larger metabolomic datasets. In addition, we envisage MS2LDA being used in conjunction with a standard MS1 analysis via fragmentation of a pooled sample from which Mass2Motifs can be linked to MS1 intensity variability as described in Differential Expression of Mass2Motifs Reveals Biochemical Changes Across Samples.
The MS2LDA approach is markedly different from other analysis tools as multiple Mass2Motifs can be associated with one metabolite, and determination of the fragments/neutral losses that are part of a conserved structural motif is unsupervised. Our proposed focus on mining the MS2 fragmentation data alone to aid in identification of functional classes of metabolites is unique and complementary to existing use of fragmentation data. We anticipate MS2LDA to be particularly useful in research areas such as clinical/pharmaco and nutritional metabolomics, environmental analysis, and natural products research, as it can quickly recognize substructure patterns related to drugs and food-derived metabolites in an unsupervised way. Although we have demonstrated MS2LDA on DDA data, we see no reason why it would not work on data-independent acquisition data in which fragments have been matched to MS1 ions using, for example, MS-DIAL (33).
Materials and Methods
All data and code are available from dx.doi.org/10.5525/gla.researchdata.313.
Materials.
Four beer samples were used as representative of diverse complex mixtures (SI Appendix, section S3). Ten milliliters of beer were sampled directly after opening and stored at C before extraction. After thawing, (i) 200L of beer was mixed with 600L of methanol/chloroform, (ii) sonicated for 5 min at room temperature; (iii) and centrifuged for 5 min (12,000 g) at room temperature. The supernatants were stored at C. Urine fragmentation data from an earlier approved and published study on metabolite annotation of urinary metabolites were used for validation purposes (16). HPLC-grade methanol, acetonitrile, and analytical reagent-grade chloroform were acquired from Fisher Scientific. HPLC-grade H2O was purchased from VWR Chemicals. Formic acid (for MS) and ammonium carbonate were acquired from Fluka Analytical (Sigma-Aldrich).
Methods.
A Thermo Scientific Ultimate 3000 RSLCnano liquid chromatography system (Thermo Scientific) was coupled to a Thermo Scientific Q-Exactive Orbitrap mass spectrometer equipped with a HESI II interface (Thermo Scientific). Thermo Xcalibur Tune software (version 2.5) was used for instrument control and data acquisition. Column temperature was maintained at 25 °C. The hydrophilic interaction liquid chromatography (HILIC) separation was performed with a SeQuant ZIC-pHILIC column (150 × 4.6 mm, 5 m) equipped with the corresponding precolumn (Merck Sequant). A linear LC gradient was conducted from 80% B to 20% B over 15 min, followed by a 2-min wash with 5% B, and 7-min reequilibration with 80% B, where solvent B is acetonitrile and solvent A is 20 mM ammonium carbonate in water. The flow rate was 300 L/min, column temperature held at 25 °C, injection volume was 10 L, and samples were maintained at 4 °C in the autosampler (1). Samples were measured in randomized order (34) (SI Appendix, section S4). MS and MS/MS settings can be found in SI Appendix, section S5. For positive and negative-ionization combined fragmentation mode, the duty cycles consisted of a full scan in positive-ionization mode, followed by a TopN data-dependent MS/MS (MS2) fragmentation event taking the 10 most abundant ion species not on the dynamic exclusion list, followed by the same two scan events in negative mode. MS/MS fragmentation spectra were acquired using stepped higher collision dissociation combining 25.2, 60.0, and 94.8 normalized collision energies in one MS2 scan. In full-scan mode, the duty cycle consisted of two full-scan events. The duty cycles for positive- and negative-ionization separate fragmentation modes, respectively, consisted of one full-scan (MS1) event and one Top10 MS/MS (MS2) fragmentation event.
Supplementary Material
Acknowledgments
We thank Dr. Emma Schymanski (RMassBank), Dr. Tony Larson (xcmsFragments), and Dr. Samuel Bertrandt (the seven golden rules) for assistance with implementation of the mentioned R scripts; Kai Duhrkop for providing us the GNPS and MassBank spectra used in the CSI:FingerID paper; and Dr. Niels van den Broek for helpful discussions on acquisition of fragmentation spectra. This study was supported by Wellcome Trust Grant 105614/Z/14/Z (to J.J.J.v.d.H.), funded by Wellcome Trust Center for Molecular Parasitology Grant 104111/Z/14/Z (to M.P.B.), supported by a Scottish Information and Computer Science Alliance PhD studentship (J.W.), and supported by Biotechnology and Biological Sciences Research Council Grant BB/L018616/1 (to S.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608041113/-/DCSupplemental.
References
- 1.van der Hooft JJJ, de Vos RCH, Ridder L, Vervoort J, Bino RJ. Structural elucidation of low abundant metabolites in complex sample matrices. Metabolomics. 2013;9(5):1009–1018. [Google Scholar]
- 2.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7(1):1–10. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn WB, et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2013;9(1):44–66. [Google Scholar]
- 4.Misra BB, der Hooft JJJ. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis. 2016;37(1):86–110. doi: 10.1002/elps.201500417. [DOI] [PubMed] [Google Scholar]
- 5.Hufsky F, Scheubert K, Böcker S. Computational mass spectrometry for small-molecule fragmentation. Trends Analyt Chem. 2014;53:41–48. doi: 10.1186/1758-2946-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dührkop K, Shen H, Meusel M, Rousu J, Böcker S. Searching molecular structure databases with tandem mass spectra using CSI: FingerID. Proc Natl Acad Sci USA. 2015;112(41):12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridder L, et al. Automatic chemical structure annotation of an LC–MSn based metabolic profile from green tea. Anal Chem. 2013;85(12):6033–6040. doi: 10.1021/ac400861a. [DOI] [PubMed] [Google Scholar]
- 8.Horai H, et al. MassBank: A public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 9.Wishart DS, et al. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2012;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridder L, et al. In silico prediction and automatic LC–MS(n) annotation of green tea metabolites in urine. Anal Chem. 2014;86(10):4767–4774. doi: 10.1021/ac403875b. [DOI] [PubMed] [Google Scholar]
- 11.Allen F, Greiner R, Wishart D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics. 2015;11(1):98–110. [Google Scholar]
- 12.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci USA. 2015;112(41):12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 14.Garg N, et al. Mass spectral similarity for untargeted metabolomics data analysis of complex mixtures. Int J Mass Spectrom. 2015;377:719–727. doi: 10.1016/j.ijms.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JY, et al. Molecular networking as a dereplication strategy. J Nat Prod. 2013;76(9):1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Hooft JJJ, Padmanabhan S, Burgess KEV, Barrett MP. Urinary antihypertensive drug metabolite screening using molecular networking coupled to high-resolution mass spectrometry fragmentation. Metabolomics. 2016;12(7):1–15. doi: 10.1007/s11306-016-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, Kind T, Yang D, Leon C, Fiehn O. Ms2analyzer: A software for small molecule substructure annotations from accurate tandem mass spectra. Anal Chem. 2014;86(21):10724–10731. doi: 10.1021/ac502818e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Hu X, Shen X, Rosen G. IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; Piscataway, NJ: 2010. Probabilistic topic modeling for genomic data interpretation; pp. 149–152. [Google Scholar]
- 19.Rogers S, Girolami M, Campbell C, Breitling R. The latent process decomposition of cDNA microarray data sets. IEEE ACM Trans Comput Biol Bioinformatics. 2005;2(2):143–156. doi: 10.1109/TCBB.2005.29. [DOI] [PubMed] [Google Scholar]
- 20.Sweeney DL. A data structure for rapid mass spectral searching. Mass Spectrom. 2014;3(Spec Iss 2):S0035–S0035. doi: 10.5702/massspectrometry.S0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 22.Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. PeakML/mzMatch: A file format, java library, R library, and tool-chain for mass spectrometry data analysis. Anal Chem. 2011;83(7):2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 23.Stravs MA, Schymanski EL, Singer HP, Hollender J. Automatic recalibration and processing of tandem mass spectra using formula annotation. J Mass Spectrom. 2013;48(1):89–99. doi: 10.1002/jms.3131. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths TL, Steyvers M. Finding scientific topics. Proc Natl Acad Sci USA. 2004;101(Suppl 1):5228–5235. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kind T, Fiehn O. Seven golden rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8(1):105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böcker S, Lipták Z. A fast and simple algorithm for the money changing problem. Algorithmica. 2007;48(4):413–432. [Google Scholar]
- 27.Böcker S, Letzel MC, Lipták Z, Pervukhin A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics. 2009;25(2):218–224. doi: 10.1093/bioinformatics/btn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievert C, Shirley KE. Proceedings of the Workshop on Interactive Language Learning, Visualization, and Interfaces. Association for Computational Linguistics; Stroudsburg, PA: 2014. LDAvis: A method for visualizing and interpreting topics; pp. 63–70. [Google Scholar]
- 29.Nguyen DD, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci USA. 2013;110(28):E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarca AL, Bhatti G, Romero R. A comparison of gene set analysis methods in terms of sensitivity, prioritization and specificity. PLoS One. 2013;8(11):e79217. doi: 10.1371/journal.pone.0079217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomfohr J, Lu J, Kepler TB. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6(1):225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman M, Blei D, Bach F. Online Learning for Latent Dirichlet Allocation. In: Lafferty JD, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, editors. Advances in Neural Information Processing Systems 23. NIPS Foundation; La Jolla, CA: 2010. pp. 1–9. [Google Scholar]
- 33.Tsugawa H, et al. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creek DJ, et al. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography–mass spectrometry: Improved metabolite identification by retention time prediction. Anal Chem. 2011;83(22):8703–8710. doi: 10.1021/ac2021823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.