Abstract
Neonatal mouse cochlear supporting cells have a limited ability to divide and trans-differentiate into hair cells, but this ability declines rapidly in the two weeks after birth. This decline is concomitant with the morphological and functional maturation of the organ of Corti prior to the onset of hearing. However, despite this association between maturation and loss of regenerative potential, little is known of the molecular changes that underlie these events. To identify these changes, we used RNA-seq to generate transcriptional profiles of purified cochlear supporting cells from 1- and 6-day-old mice. We found many significant changes in gene expression during this period, many of which were related to regulation of proliferation, differentiation of inner ear components and the maturation of the organ of Corti prior to the onset of hearing. One example of a change in regenerative potential of supporting cells is their robust production of hair cells in response to a blockade of the Notch signaling pathway at the time of birth, but a complete lack of response to such blockade just a few days later. By comparing our supporting cell transcriptomes to those of supporting cells cultured in the presence of Notch pathway inhibitors, we show that the transcriptional response to Notch blockade disappears almost completely in the first postnatal week. Our results offer some of the first molecular insights into the failure of hair cell regeneration in the mammalian cochlea.
Introduction
The death of auditory hair cells due to noise damage, ototoxins or aging is a principal cause of sensorineural hearing loss [1–3]. In contrast to other vertebrates, where supporting cells readily re-enter the cell cycle and generate hair cells after damage, the mature organ of Corti is unable to regenerate [1, 4–7]. However, recent studies suggest that neonatal mouse supporting cells retain a limited, transient capacity for regeneration. For example, neonatal mouse supporting cells are able to down-regulate cell cycle inhibitors, re-enter the cell cycle and generate hair cells in culture [8–10]. This cell cycle re-entry can be driven by activation of the Wnt signaling pathway [11–15] or by deletion of cell cycle regulators such as p27Kip1 [16]. Blockade of Notch signaling between hair cells and supporting cells can result in trans-differentiation of supporting cells into hair cells [13, 17–20]. Such trans-differentiation of supporting cells can also be observed at very low levels after hair cell killing [17, 21]. Finally, ectopic activation of the hair cell-specific transcription factor Atoh1 in supporting cells can drive their differentiation into hair cells [12, 22–24]. In all these cases, however, the capacity of mouse supporting cells to either divide or trans-differentiate into hair cells is lost between birth and the onset of hearing at two weeks of age [1, 9, 22, 23, 25].
All supporting cells in the mouse organ of Corti are generated prior to birth and undergo dramatic morphological changes, such as the elaboration of phalangeal processes, and formation of the reticular lamina and the tunnel of Corti [4, 26, 27]. This functional maturation of supporting cells, together with the decline in their regenerative ability over the first two weeks of postnatal life is likely to be reflected by transcriptional or epigenetic changes. To better understand the molecular basis for these changes, we performed an RNA-seq-based analysis of purified cochlear supporting cells from 1- and 6-day old mice. We find large scale gene expression changes consistent with morphological maturation including changes in the cytoskeleton and the extracellular matrix, together with changes in the gene regulatory network over this time period.
We and others have demonstrated that the ability of supporting cells to trans-differentiate into hair cells after Notch inhibition declines dramatically in the first postnatal week [19]. To understand this phenomenon, we performed RNA-seq analysis of purified supporting cells from new born (P0)-or 5-day old cochleas (P5) that had been cultured for 24 hours in the presence of the Notch inhibitor DAPT. Strikingly, we found that while over 2,000 transcripts were significantly altered as P0 supporting cells trans-differentiated into hair cells, only 20 transcripts changed significantly in P5 cochleas cultured in the same conditions. Our study has identified the transcriptional signature of supporting cell maturation and shows that the Notch pathway is greatly attenuated during the first postnatal week.
Materials and Methods
Experimental animals
LfngEGFP mice (Tg(Lfng-EGFP)HM340Gsat) were generated by the GENSAT project [28–30] and obtained from Dr. Nathaniel Heintz. Neonatal mouse pups were taken for organ cultures and cell sorting at postnatal day (P) 0, P1, P5 and P6. ICR or CF1 mice were used for in situ hybridization and for generating RNA from whole cochleas. The Baylor College of Medicine Institutional Animal Care and Use committee and the Faculty of Medicine of Universidad de Chile Bioethics committee approved all animal experiments. Genotyping for the LfngEGFP transgene was performed with primers to GFP (Forward primer: CGA AGG CTA CGT CCA GGA GCG CAC; Reverse primer GCA CGG GGC CGT CGC CGA TGG GGG TGT, yielding a 300bp band.
Cochlear isolation and culture
P0, P1, P5 and P6 cochlear explants were dissected and cultured as previously described [19]. Briefly, following euthanasia, mouse heads were bisected, the temporal bone was removed from the skull base and the otic capsule was removed with forceps to separate the intact membranous cochlea from the surrounding bony structures. For P0 and P1 animals, the cochlear duct was peeled away from the modiolus and the medial structures (Kölliker’s and Corti´s organs) were separated from the lateral wall, Reissner´s membrane and the stria vascularis. For P5 and P6 mice, the cochlear duct was gently separated from the modiolus by cutting between them with forceps to preserve the organ of Corti. The lateral wall, stria vascularis and Reissner´s membrane were then partially removed by cutting with a 27 gauge needle. P0 and P5 cochleas were cultured in DMEM/F12 (buffered with Hepes; Thermo Fisher Scientific) supplemented with B27 (Thermo Fisher Scientific), 1mM N-acetylcysteine (Sigma), 5 ng/ml EGF and 2.5 ng/ml FGF2 and 67 μg/ml penicillin. For Notch inhibition experiments, cultures were treated with 10μM DAPT (Gamma secretase inhibitor IX, Calbiochem EMD) or 0.04% v/v DMSO (vehicle control; Thermo Fisher Scientific) for 24 hours as previously described [19].
Immunostaining
Mid-modiolar 14μm frozen sections of paraformaldehyde-fixed temporal bones of LfngEGFP mice were washed, permeabilized and blocked in PBS containing 0.2% Triton X-100 and 10% donkey serum. They were then incubated with primary antibodies, washed in PBST (0.1% Triton X-100 in PBS) and incubated with secondary antibodies, followed by 3 washes in PBST. Primary antibodies used were rabbit polyclonal anti-MYOSIN VI (1:500; Proteus Biosciences), anti-GLAST (1:500; Abcam), anti-PROX1 (1:1000; EMD Millipore), anti-SOX2 (1:250; EMD Millipore) and anti-p75NTR (NGFR: 1:200; Advanced Targeting Systems AB- N01 AP). Secondary antibodies were Alexa Fluor 594 (Invitrogen). Images were obtained on an Axio Observer Zeiss microscope with an Apotome2 structured illumination attachment and analyzed in Axiovision 4.8 (Zeiss).
Cell Dissociation and FACS sorting
For supporting cell sorting, explants were washed in ice cold Ca2+, Mg2+-free -PBS and incubated at 37°C in EBSS solution containing papain (20U/ml), 1mM L-cysteine and 0.5mM EDTA (Worthington, Lakewood, NJ) for 8 minutes (P0 cultured explants) or 10 minutes (all other organs). The papain solution was removed and inactivated by gentle washes in ice cold PBS containing 2% FBS. The cells were dissociated by gently pipetting on ice and filtered with a cell strainer cap (BD Biosciences). The dissociated and filtered cells were sorted in a FACSAriaII cell sorter (BD Biosciences) at 4°C in PBS containing 2% FBS, using a 130μm nozzle. The cells were collected on the basis of their fluorescence gating in DMEM 5% FBS, spun down, lysed in RTL buffer (Qiagen) and stored at -80°C. For each RNAseq library, approximately 50,000 sorted cells were used from the freshly dissected cochleas, and 10,000 sorted cells were used from the cultured explants. The identity of sorted cells was confirmed using epifluorescence and qRT-PCR for hair cell and supporting cell markers. Duplicates samples were collected for each condition.
RNA Extraction
For RNASeq libraries, total RNA was extracted using Trizol (Thermo Fisher Scientific) and PelletPaint (Novagen EMD Millipore). Briefly, samples were homogenized in 300μl of Trizol and shaken with 60μl chloroform and 2μl PelletPaint and centrifuged for 15 minutes. The supernatant containing RNA was precipitated with 1 volume of isopropanol, centrifuged for 30 minutes, washed in 75% ethanol, resuspended in 100μl water and re-precipitated with 10μl 3M sodium acetate and 250μl ethanol. After a final wash in 75% ethanol, the pellet was allowed to dry, resuspended in 20μl RNAse-free water and stored at -80°C. For qRT-PCR, RNA was prepared using an RNeasy Micro kit (Qiagen).
RNA probe synthesis
Primer sets for each candidate gene were selected to target a 500–700 bp DNA fragment in a single exon of each gene for screen. A T7 RNA polymerase sequence (5’-GGATCCTAATACGACTCACTATAGGGAG-3’) was added to the 5’ end of each reverse primer. Primer sets used are listed in S1 Table. Mouse genomic DNA was used as the template for PCR. The PCR product of the correct size was purified with a PCR Purification Kit (Qiagen). Purified DNA was used as the template for RNA probe synthesis with T7 polymerase (Promega) using standard protocols [31].
In situ hybridization
The in situ hybridization procedure for frozen sections was carried out as recently described [19], and for whole mounts as described in [32]. A probe concentration of 1μg/ml was used for sections and 0.3–0.8μg/ml for whole mounts.
qRT-PCR
cDNA was prepared using approximately 100–500ng of total RNA, random primers and the SuperScript III First-Strand Synthesis System (Invitrogen). qPCR reactions were performed with SYBRGreen PCR Master Mix in a StepOne Plus Real Time PCR machine (Applied Biosystems), using in the reaction at 0.3 to 0.6 ng/μl cDNA and 50nM primers. Primers sequences used in this study were: Myo6: Forward primer—5’ -TGTTAAGGCAGGTTCCTTGAAG-3’, Reverse primer—5’ -ACACCAGCTACAACTCGAAAC-3’, Prox1: Forward primer—5’-CGTTACGGGAGTTTTTCAATG-3’, Reverse primer—5’-CCTTGTAAATGGCCTTCTTCCA-3’, Gapdh: Forward primer—5’-AGGTCGGTGTGAACGGATTTG-3’, Reverse primer—5’-TGTAGACCATGTAGTTGAGGTCA-3’. Gapdh was used as reference gene. Statistical analysis of qRT-PCR data was performed with a Mann-Whitney test using PAST statistical software.
RNA sequencing
Duplicate samples of GFP+ and GFP- sorted cells were prepared for each experimental condition. 10,000–60,000 sorted cells were used as starting material to generate approximately 100–600 ng RNA, as measured by a Nanodrop spectrophotometer. cDNA libraries for RNAseq were generated using RNA Seq Truseq RNA sample preparation kit v2 (Illumina) following the “low sample” protocol according to the manufacturer's instructions for mRNA extraction, cDNA synthesis, indexing and amplification. cDNA generated from less than 20,000 cells received an initial amplification using the NuGen Ovation Kit. The quality and integrity of RNA samples and the final quality of the sequencing libraries was checked by electrophenogram in an Agilent Bioanalyzer. Paired-end sequencing was performed in HiSeq2000 sequencing platform (Illumina). Fastq files of paired end reads have been deposited in the NCBI GEO database, Accession No. GSE83357.
Bioinformatic analysis
Analysis of the sequencing reads was performed by two different approaches. (1) Reads were mapped to the Mus musculus NCBI build37.2 iGenome (Ilumina) using TopHat 2.0 software [33, 34] and the mapped reads were quantitated and compared using Cufflinks 2.0 providing differential gene expression data and statistics. (2) Reads were aligned to the Mus musculus Ensembl mm9 iGenome (Ilumina) using TopHat 1.4.1 software and the number of reads per gene and per library was obtained using DESeq program. From the 24 cDNA libraries (12 duplicates) 50–200 million of paired end reads were obtained from each library. Of those reads 86 to 99% were correctly mapped and 73 to 93% were properly paired. After comparing the level of expression of each gene within each pair of related libraries, the most significant differentially expressed genes (DEG) were annotated and analyzed separately for both approaches. In order to find enriched genes in freshly isolated Lfng-EGFP+ cells, a significantly DEG was considered to have an RPKM higher than 3000, Fold Change (FC) higher than 4 and p value and FDR < 0.01. The level of DEG significance for comparing cultured explants (treated or not with DAPT) sample libraries was a FC higher than 2 and a p value and FDR < 0.01. To evaluate our Notch inhibition experiments where cultures were treated with DAPT or DMSO (in which supporting cells may trans-differentiate into hair cells), we used our consensus gene list of P1 Lfng-EGFP+ cells to represent supporting cells. This list was used to identify supporting cell genes down-regulated by DAPT treatment. All the complete transcriptomes obtained in this study have been included in S2 and S3 Tables.
Gene Ontology (GO) analysis
The lists of differentially expressed genes in each experiment were uploaded into the DAVID bioinformatics suite [35–38] for gene ontology analysis. We obtained GO terms related to biological processes, and the long list of terms was uploaded to REVIGO [39] using default parameters to summarize it in representative terms. The threshold to detect redundancies used was set arbitrarily to 0.1. The REVIGO output was complemented with DAVID output in order to create merged tables. In all tables only represented terms with p<0.01 were included.
Results
RNA-seq analysis identifies mouse supporting cell transcripts differentially expressed in the first postnatal week
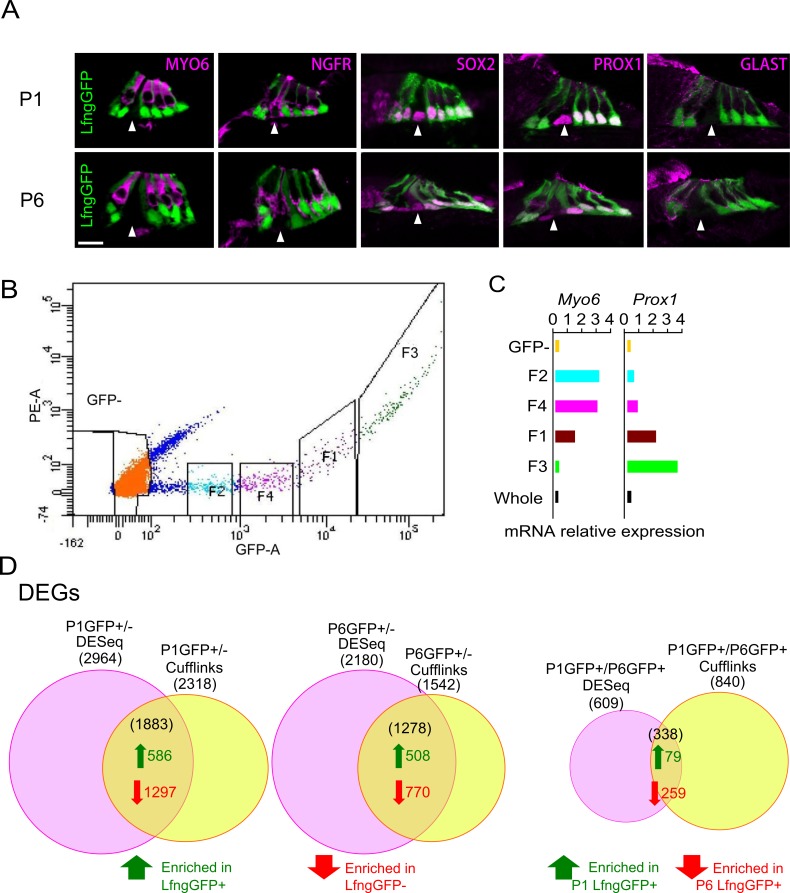
Despite widespread interest in stimulating supporting cells to trans-differentiate into hair cells, relatively little is known about the genes expressed in supporting cells as they differentiate and mature. To identify supporting cell-specific transcripts in the neonatal mouse cochlea, we used Lfng-GFP BAC transgenic mice (Fig 1A) from the GENSAT project [28–30] to purify supporting cells by fluorescence-activated cell sorting. These mice show specific and strong GFP expression in most of the supporting cell types in the P1 and P6 cochlea, labeling border cells, inner phalangeal cells, outer pillar cells and all three rows of Deiters’ cells (Fig 1A). FACS analysis of the dissociated cell population revealed different populations with varying degrees of GFP fluorescence (Fig 1B). We collected only the brightest GFP+ cells for further analysis (Fig 1B “F3” fraction). Some cells expressed detectable but low levels of GFP fluorescence (Fig 1B; F1, F2 and F4 fractions) but were not collected for analysis, as they are likely to be inner pillar cells and inner hair cells that have been previously shown to express very low levels of this transgene [40]. We performed a preliminary validation of our collected fractions by examining expression of the hair cell transcript Myo6 and the supporting cell transcript Prox1 by Q-PCR (Fig 1C). The F3 fraction that was used for all subsequent analyses had the highest relative levels of Prox1 expression and the lowest relative expression of Myo6. We obtained an average of 580±142 and 430±133 Lfng-EGFP+ cells per cochlea from P1 and P6 mice respectively. We prepared total RNA cDNA libraries from 50,000 cells per replicate from P1 and P6 mice and sequenced each library.
Fig 1. Isolation of neonatal supporting cells and analysis by RNA-seq.
(A) Sections through postnatal day 1 and 6 LFng-GFP transgenic mice. At both ages, the GFP transgene is expressed in border cells, inner phalangeal cells, outer pillar cells and all three rows of Deiters’ cells, but not inner pillar cells (arrowheads). Sections are counterstained with antibodies to MYOSIN VI to show hair cells and SOX2, PROX1, GLAST or NGFR to show distinct types of supporting cells (magenta). Scale bar 20 μm. (B) Representative FACS profile for sorting of P1 LFng-GFP transgenic cochleas. GFP intensity is shown on the x-axis, with four fractions (F1-F4) identified on the sorting profile. (C) QPCR analysis of the different fractions for expression of a hair cell marker (Myo6) and a supporting cell marker (Prox1). Cells falling in F3, which contained the highest Prox1 signal and lowest Myo6 signal were used in subsequent experiments. (D) Identification of transcripts enriched in the GFP+ fractions at P1 (left) and P6 (center) using the intersection of differential gene expression between GFP+ and GFP- cells analyzed with DESeq and Cufflinks. 586 consensus GFP+ transcripts were identified at P1 and 508 at P6. The right Venn diagram shows the intersection of enriched GFP+ transcripts differentially expressed between P1 and P6 analyzed with DESeq and Cufflinks. 79 transcripts were enriched in P1 GFP+ cells compared to P6 GFP+ cells, whereas 259 transcripts were enriched in P6 GFP+ cells compared to P1 GFP+ cells. p < 0.01, q < 0.01 and log2(fold change) > 2.
We compared the transcriptomes of GFP+ cells from P1 and P6 mice with those of the corresponding GFP- populations. It is well-established that different methods to analyze RNA-seq data can give significantly different results. We therefore used two different analysis programs, Cufflinks [34] and DESeq [41] to normalize and compare differences in the mapped reads from our populations. We identified transcripts that were differentially expressed in GFP+ supporting cells compared to their GFP- counterparts. Specifically, we found 2,964 significant differentially expressed genes (DEG) in the DESeq output and 2,318 DEG using Cufflinks for P1, and 2,180 DEG by DESeq and 1,542 DEG by Cufflinks for P6 (Fig 1D). We found a number of transcripts that were previously shown to be specifically expressed in supporting cells, such as Lfng itself, Hes5, Fgfr3, Prox1, Sox2, Lgr5, Cdkn1b/p27Kip1, and the Notch1 receptor (S4 Table). We then focused on genes that were shown to be differentially expressed by both methods of analysis using similar criteria (p<0.01; FDR<0.01 and a fold change of at least 4). We identified 586 transcripts that were enriched in P1 GFP+ supporting cells compared to their GFP- counterparts and 508 transcripts that were enriched in P6 GFP+ supporting cells compared to GFP- cells (Fig 1D). We filtered these gene lists to generate candidates for further analysis by setting an arbitrary expression value of > 3000 RPKM. This yielded 277 supporting cell-enriched genes in the P1 cochlea and 202 at P6 (Tables 1 and 2 and S5 Table).
Table 1. Top 20 genes enriched in Lfng-GFP+ cells at P1.
| Rank | Gene symbol | Mean GFP+ (RPKM) | Fold change (GFP+ vs GFP-) | p-adj |
|---|---|---|---|---|
| 1 | Cysltr2 | 4171.25 | 342.83 | 1.5E-135 |
| 2 | Hes5 | 6663.27 | 306.73 | 1.68E-16 |
| 3 | Umodl1 | 9261.97 | 227.61 | 2.68E-82 |
| 4 | Fgf3 | 3272.03 | 219.78 | 5.68E-146 |
| 5 | Trhr | 5138.74 | 171.67 | 1.89E-236 |
| 6 | Gm5887 | 7392.25 | 154.81 | 7.94E-254 |
| 7 | Pkd2l1 | 8263.65 | 138.71 | 9.75E-255 |
| 8 | Fndc7 | 5642.82 | 137.82 | 8.69E-171 |
| 9 | Pdzk1ip1 | 6160.10 | 125.68 | 1.33E-37 |
| 10 | Slitrk6 | 47604.15 | 120.46 | 3.23E-167 |
| 11 | Lfng | 109921.44 | 112.4 | 2.5E-252 |
| 12 | Agr3 | 8355.38 | 104.39 | 6.16E-237 |
| 13 | Inhba | 4837.93 | 100.2 | 9.14E-208 |
| 14 | Slc6a14 | 3419.69 | 95.49 | 6.99E-184 |
| 15 | Fgfr3 | 86721.79 | 92.49 | 1.44E-275 |
| 16 | Nr4a3 | 8069.48 | 91.92 | 1.34E-79 |
| 17 | Ntf3 | 6712.00 | 78.59 | 9.04E-57 |
| 18 | Syt6 | 16012.90 | 75.08 | 9.37E-239 |
| 19 | Slc27a2 | 5244.69 | 70.04 | 1.46E-193 |
| 20 | Rab3b | 8196.90 | 67.84 | 7.35E-163 |
List of the 20 genes showing the greatest fold-change enrichment in P1 Lfng-GFP+ cells compared their GFP- counterparts. The mean expression level (RPKM) is shown, together with the fold change versus GFP- cells. p-adj = adjusted p-value for the difference between GFP+ and GFP- populations.
Table 2. Top 20 genes enriched in Lfng-GFP+ cells at P6.
| Rank | Gene symbol | Mean GFP+ (RPKM) | Fold change (GFP+ vs GFP-) | p-adj |
|---|---|---|---|---|
| 1 | Umodl1 | 87697.47 | 1022.11 | 1.86E-149 |
| 2 | Gm5887 | 17389.04 | 850.84 | 1.06E-148 |
| 3 | Crhr1 | 10718.38 | 310.72 | 1.17E-120 |
| 4 | Agr3 | 4234.31 | 201.36 | 9.31E-24 |
| 5 | Tsga14 | 79752.75 | 139.51 | 1.52E-104 |
| 6 | Wnt7a | 7143.40 | 125.77 | 1.26E-35 |
| 7 | Rassf6 | 7298.55 | 77.97 | 2.34E-25 |
| 8 | Sapcd2 | 15279.43 | 50.42 | 1.4E-13 |
| 9 | Dgkb | 5928.65 | 50.39 | 5.61E-69 |
| 10 | Slc6a14 | 4914.60 | 45.72 | 1.62E-15 |
| 11 | Gdf6 | 31099.52 | 45.01 | 2.29E-15 |
| 12 | Emx2 | 3373.32 | 42.51 | 2.23E-19 |
| 13 | Tmprss3 | 15912.06 | 42.17 | 1.45E-66 |
| 14 | Tppp | 4647.09 | 41.33 | 1.33E-62 |
| 15 | Otog | 126262.45 | 37.63 | 5.36E-67 |
| 16 | Slitrk6 | 35908.27 | 36.59 | 1.69E-64 |
| 17 | Rnd1 | 5759.25 | 34.17 | 4.57E-06 |
| 18 | Fgfr3 | 49608.94 | 33.2 | 1.18E-18 |
| 19 | Prox1 | 4088.87 | 32.69 | 4.72E-09 |
| 20 | Rorb | 4155.72 | 31.4 | 3.5E-55 |
List of the 20 genes showing the greatest fold-change enrichment in P6 Lfng-GFP+ cells compared their GFP- counterparts. Ranking is based on the size of the fold change. The mean expression level (RPKM) is shown, together with the fold change versus GFP- cells. p-adj = adjusted p-value for the difference between GFP+ and GFP- populations
While 907 of the genes enriched in supporting cells were expressed at both P1 and P6, 976 were expressed only at P1 and 371 were expressed only at P6. Consistent with maturation of supporting cells prior to the onset of hearing, the expression of many supporting cell-specific genes changed during this developmental period. We identified differentially expressed supporting cell genes by comparing P1 and P6 Lfng-GFP+ samples using the same combined analysis with DESeq and Cufflinks described above. We identified 338 differentially expressed genes, of which 79 were enriched in P1 GFP+ supporting cells compared to their P6 counterparts and 259 genes that were enriched in P6 GFP+ supporting cells compared to P1 supporting cells (Fig 1D, Tables 3 and 4 and S6 Table).
Table 3. Top 20 differentially expressed genes enriched in P1 Lfng-GFP+ cells versus P6 Lfng-GFP+ cells.
| Rank | Gene symbol | P1 Mean GFP+ (RPKM) | P6 Mean GFP+ (RPKM) | Fold change enrichment in P1 versus P6 | p-adj |
|---|---|---|---|---|---|
| 1 | Syt6 | 10312.10 | 146.09 | 70.52 | 8.13E-87 |
| 2 | Cysltr2 | 2686.38 | 81.44 | 32.90 | 2.99E-56 |
| 3 | Tmc5 | 5343.56 | 223.78 | 23.92 | 3.58E-52 |
| 4 | Tal1 | 2795.18 | 71.13 | 39.40 | 1.52E-48 |
| 5 | Pkd2l1 | 5321.86 | 285.92 | 18.64 | 2.28E-47 |
| 6 | March4 | 1390.15 | 45.89 | 30.27 | 1.81E-45 |
| 7 | Trhr | 3309.26 | 271.09 | 12.21 | 6.58E-36 |
| 8 | Angpt1 | 4046.49 | 387.49 | 10.41 | 5.43E-32 |
| 9 | Chst15 | 4208.43 | 392.24 | 10.70 | 6.02E-32 |
| 10 | Olfml3 | 7644.27 | 532.96 | 14.32 | 2.36E-27 |
| 11 | Chrng | 1009.93 | 87.34 | 11.55 | 6.5E-27 |
| 12 | Raver2 | 23804.79 | 3433.25 | 6.92 | 1.38E-25 |
| 13 | Fat3 | 3525.47 | 418.60 | 8.40 | 2.26E-25 |
| 14 | Cdh4 | 1950.64 | 231.45 | 8.46 | 1.14E-23 |
| 15 | Cxcl12 | 6129.68 | 847.05 | 7.26 | 1.02E-22 |
| 16 | Sox11 | 5314.63 | 764.77 | 6.96 | 1.41E-22 |
| 17 | St8sia2 | 6179.44 | 890.31 | 6.96 | 1.87E-22 |
| 18 | Trh | 1840.37 | 261.91 | 7.01 | 1.02E-21 |
| 19 | Dpysl4 | 9539.39 | 1636.75 | 5.82 | 2.46E-21 |
| 20 | Cyp26b1 | 52399.13 | 9464.61 | 5.54 | 2.26E-20 |
List of the 20 genes showing the greatest fold-change enrichment in P1 Lfng-GFP+ cells compared P6 Lfng-GFP+ cells. Ranking is based on the p value. The mean expression level (RPKM) is shown, together with the fold change of enrichment in P1 versus P6 cells. p-adj = adjusted p-value for the difference between P1 and P6 populations.
Table 4. Top 20 differentially expressed genes enriched in P6 Lfng-GFP+ cells versus P1 Lfng-GFP+ cells.
| Rank | Gene symbol | P1 Mean GFP+ (RPKM) | P6 Mean GFP+ (RPKM) | Fold change enrichment in P6 versus P1 | p-adj |
|---|---|---|---|---|---|
| 1 | Kcnj16 | 639.37 | 14990.18 | 23.43 | 3.11E-56 |
| 2 | Dgkb | 188.78 | 4453.61 | 23.59 | 1.29E-52 |
| 3 | Pcsk2 | 65.82 | 2669.74 | 40.50 | 3.66E-51 |
| 4 | Grip2 | 47.68 | 1375.26 | 28.84 | 6.34E-48 |
| 5 | Scd1 | 655.77 | 22369.52 | 34.06 | 1.07E-42 |
| 6 | Crhr1 | 624.41 | 8053.18 | 12.91 | 4.54E-39 |
| 7 | Crb2 | 109.85 | 2324.76 | 21.11 | 1.85E-38 |
| 8 | Rasgrp1 | 288.59 | 3520.79 | 12.21 | 2.45E-34 |
| 9 | Mmp28 | 98.40 | 1238.65 | 12.55 | 2.63E-30 |
| 10 | Bhlhe40 | 171.20 | 2946.79 | 17.27 | 1.19E-29 |
| 11 | Umodl1 | 5964.36 | 65915.69 | 11.08 | 2.59E-28 |
| 12 | Pla2g4e | 96.94 | 1113.05 | 11.47 | 2.88E-28 |
| 13 | Stat5a | 146.64 | 1498.22 | 10.20 | 3.54E-27 |
| 14 | Abcc12 | 129.82 | 1283.89 | 9.92 | 3.23E-25 |
| 15 | Aim1 | 237.75 | 1998.98 | 8.40 | 9.76E-25 |
| 16 | Hr | 966.69 | 6669.82 | 6.92 | 1.47E-24 |
| 17 | Cntfr | 85.05 | 1371.73 | 16.11 | 1.66E-24 |
| 18 | Sorl1 | 2809.49 | 18130.06 | 6.45 | 3.25E-24 |
| 19 | Col13a1 | 101.25 | 3235.28 | 32.00 | 3.36E-23 |
| 20 | Klf9 | 287.49 | 2141.53 | 7.46 | 4.03E-23 |
List of the 20 genes showing the greatest fold-change enrichment in P6 Lfng-GFP+ cells compared P1 Lfng-GFP+ cells. Ranking is based on the p value. The mean expression level (RPKM) is shown, together with the fold change of enrichment in P6 versus P1 cells. p-adj = adjusted p-value for the difference between P6 and P1 populations.
We performed gene ontology (GO) and pathway analyses on our filtered list of P1 and P6 supporting cell genes using the DAVID bioinformatics suite [35, 37]. The biological processes terms most significantly represented in our lists of P1 genes were “neuron differentiation” (GO:0030182), “cell fate commitment” (GO:0045165) and “cell adhesion” (GO:0007155; Table 5). In P6 supporting cells, the most significant terms were “inner ear development” (GO:0048839), “regulation of cellular component biogenesis” (GO: 0044087) and “neuron differentiation” (GO:0030182; Table 6). To interpret the long list of biological process represented in our analysis, we summarized them using the REVIGO suite [39], showing only the most significantly represented GO terms (p<0.01; Tables 5 and 6). We found that GO terms including “Notch pathway” (GO:0007219) or its regulation (GO:0008593), “cell projection organization” (GO:00330030) and “cell component movement” (GO:0006928) were present in our P1 lists but were less represented in the P6 gene lists. In contrast, terms associated with cellular organization, such as “cellular component biogenesis” (GO:0044087), “regulation of protein polymerization” (GO:0032271), “negative regulation of proliferation” (GO:0008285) or “cytoskeleton organization” (GO:0007010) were more represented in P6 supporting cells, consistent with the morphological maturation of supporting cells at this time. Finally, DAVID analysis of biological processes terms represented in the list of genes that significantly changed in supporting cells between P1 and P6 showed that the most significantly represented terms were cell adhesion (GO:0007155), extracellular structure organization (GO:0043062) and hormone metabolic processes (GO:0042445). To summarize the list of biological process terms we used the REVIGO suite as described above (Table 7).
Table 5. Biological processes (BP) GO terms represented in the genes enriched in P1 Lfng-GFP+ cells.
| Representative GO Term description (REVIGO Output) | Term ID | Included GO Term descriptions (DAVID Output) | Genes represented | log10 p value |
|---|---|---|---|---|
| Neuron Differentiation | GO:0030182 | Neuron differentiation | Enah, Fgfr3, Sox2, Uchl1, Rora, Jag1, Ephb1, Tgfb2, Nrcam, Efhd1, Tctn1, Sema3a, Ntf3, Ptprz1, Tgfbr1, Emx2, Ntng2, Rgnef, Notch3, Sall3, Notch1, Hes5, Ush1c, Id4, Slitrk6, Wnt7a, Igsf9 | -9.8358 |
| GO:0048839 | Inner ear development | Spry2, Fgfr3, Cdkn1b, Hes5, Sox2, Ush1c, Fgf10, Nr4a3, Jag1, Celsr1, Fzd6, Ptprq | -7.2417 | |
| GO:0048729 | Tissue morphogenesis | Enah, Fgfr3, Fgf10, Jag1, Celsr1, Nr4a3, Prox1, Fzd6, Tgfb2, Notch1, Sfrp1, Frem2, Lama5, Tctn1, Sema3a, Wnt7a | -5.6775 | |
| GO:0051094 | Positive regulation of developmental process | Tal1, Notch1, Fgfr3, Socs2, Ntf3, Sox2, Hey2, Jag1, Prox1, Igfbp3, Wnt7a, Tgfb2 | -3.5207 | |
| GO:0001709 | Cell fate determination | Ntf3, Hes5, Gata3, Cyp26b1, Prox1 | -3.229 | |
| GO:0035239 | Tube morphogenesis | Spry2, Notch1, Enah, Lama5, Fgf10, Cdh1, Tctn1, Nr4a3, Celsr1, Fzd6 | -3.045 | |
| GO:0007605 | Sensory perception of sound | Spry2, Cdkn1b, Otof, Sox2, Otog, Ush1c, Tectb | -2.8523 | |
| GO:0022612 | Gland morphogenesis | Notch1, Sfrp1, Lama5, Fgf10, Cdh1, Sema3a | -2.1185 | |
| GO:0001763 | Morphogenesis of a branching structure | Spry2, Notch1, Sfrp1, Lama5, Fgf10, Spint1, Sema3a | -2.0023 | |
| Cell Adhesion | GO:0007155 | Cell adhesion | Cldn7, Cldn9, Cldn6, Ninj1, Lmo7, Nedd9, Bcam, Cdh1, Tgfb2, Nrcam, Cd9, Fat3, Pvrl3, Ttyh1, Thbs1, F11r, Egfl6, Fblim1, Celsr2, Ptpru, Celsr1, Pgm5, Itga6, Hes5, Frem2, Lama5, Lsamp, Pkp3, Otog, Cntn1, Lamc2 | -9.2086 |
| GO:0016337 | Cell-cell adhesion | Cldn7, Cldn9, Cldn6, Lmo7, Celsr2, Cdh1, Celsr1, Ptpru, Tgfb2, Nrcam, Itga6, Fat3, Frem2, Pvrl3, Ttyh1 | -5.0319 | |
| Cell Projection Organization | GO:0030030 | Cell projection organization | Enah, Ntf3, Ptprz1, Uchl1, Ntng2, Rgnef, Ephb1, Tgfb2, Nrcam, Efhd1, Notch1, Itga6, Lama5, Ttyh1, Tctn1, Sema3a, Slitrk6, Igsf9 | -5.3666 |
| GO:0044087 | Regulation of cellular component biogenesis | Fmn1, Cdkn1b, Tgfbr1, Spnb1, Prox1, Wnt7a, Vill | -2.723 | |
| Notch Signaling Pathway | GO:0007219 | Notch signaling pathway | Notch3, Notch1, Hey1, Heyl, Hey2, Cntn1, Jag1 | -3.8925 |
| GO:0008593 | Regulation of Notch signaling pathway | Sox2, Hey2, Fgf10, Jag1 | -3.7913 | |
| GO:0016055 | Wnt receptor signaling pathway | Fzd9, Sfrp1, Ccdc88c, Kremen1, Celsr2, Frzb, Fzd4, Wnt7a, Fzd6 | -3.2014 | |
| GO:0007167 | Enzyme linked receptor protein signaling pathway | Ephb6, Fgfr3, Erbb4, Id1, Myo1e, Tgfbr1, Fgf10, Angpt1, Ephb1, Fgf3, Tgfb2 | -2.1651 | |
| Cell Motion | GO:0006928 | Cell motion | Enah, Ntf3, Tgfbr1, Emx2, Fgf10, Ephb1, Tgfb2, Nrcam, Cd9, Syne2, Itga6, Lama5, St14, Sema3a | -2.5456 |
Summary of biological processes GO terms that were significantly represented (p<0.01) in 277 genes enriched in P1 Lfng-EGFP+ cells (filtered lists: present in DESeq and Cufflinks outputs, RPKM>3000 and FC>4) relative to their GFP- counterparts. The biological processes represented were obtained with the DAVID bioinformatics suite and then summarized with the REVIGO tool.
Table 6. Biological processes (BP) GO terms represented in the genes enriched in P6 Lfng-GFP+ cells.
| Representative GO Term description (REVIGO Output) | Term ID | Included GO Term descriptions (DAVID Output) | Genes represented | log10 p value |
|---|---|---|---|---|
| Inner Ear Development | GO:0048839 | Inner ear development | Spry2, Fgfr3, Cdkn1b, Sox2, Ush1c, Fgf10, Nr4a3, Jag1, Gjb6 | -5.363 |
| GO:0030182 | Neuron differentiation | Fgfr3, Ptprz1, Sox2, Uchl1, Emx2, Rorb, Jag1, Rora, Sall3, Nrcam, Efhd1, Ush1c, Id4, Stmn1, Slitrk6, Igsf9, Wnt7a | -5.2594 | |
| GO:0007605 | Sensory perception of sound | Spry2, Cdkn1b, Otof, Sox2, Otog, Ush1c, Gjb6, Tectb | -4.5074 | |
| GO:0051094 | Positive regulation of developmental process | Wnt7b, Fgfr3, Tnfrsf12a, Sox2, Hey2, Jag1, Prox1, Igfbp3, Vash2, Wnt7a | -3.3182 | |
| GO:0051130 | Positive regulation of cellular component organization | Cdkn1b, Tnfrsf12a, Tppp, Fgf10, Prox1, Wnt7a | -2.0054 | |
| Regulation of Cellular Component Biogenesis | GO:0044087 | Regulation of cellular component biogenesis | Cdkn1b, Gsn, Tppp, Capg, Stmn1, Prox1, Wnt7a, Epb4.9, Vill | -5.326 |
| GO:0032271 | Regulation of protein polymerization | Cdkn1b, Gsn, Tppp, Capg, Stmn1, Epb4.9, Vill | -4.5105 | |
| GO:0008593 | Regulation of Notch signaling pathway | Sox2, Hey2, Fgf10, Jag1 | -4.1932 | |
| GO:0051493 | Regulation of cytoskeleton organization | Cdkn1b, Gsn, Capg, Stmn1, Mid1ip1, Prox1, Epb4.9, Vill | -4.0483 | |
| GO:0051129 | Negative regulation of cellular component organization | Gsn, Snph, Capg, Stmn1, Mid1ip1, Epb4.9, Vill | -3.3247 | |
| GO:0043244 | Regulation of protein complex disassembly | Gsn, Capg, Mid1ip1, Epb4.9, Vill | -2.9542 | |
| GO:0033043 | Regulation of organelle organization | Cdkn1b, Gsn, Capg, Stmn1, Mid1ip1, Prox1, Epb4.9, Vill | -2.8818 | |
| GO:0008285 | Negative regulation of cell proliferation | Cd9, Spry2, Fgfr3, Cdkn1b, Gata3, Fgf10, Gjb6, Prox1, Igfbp3 | -2.5565 | |
| GO:0032970 | Regulation of actin filament-based process | Gsn, Capg, Prox1, Epb4.9, Vill | -2.3922 | |
| GO:0007010 | Cytoskeleton organization | Shroom1, Gsn, Tppp, Epb4.1, Ush1c, Stmn1, Prox1, Epb4.9, Vill, Tmod1 | -2.0821 | |
| Cell Adhesion | GO:0007155 | Cell adhesion | F11r, Cldn7, Cldn9, Col13a1, Tnfrsf12a, Cldn3, Nedd9, Fblim1, Bcam, Nrcam, Cd9, Wnt7b, Pvrl3, Pkp3, Otog, Ttyh1 | -3.0164 |
Summary of biological processes GO terms that were significantly represented (p<0.01) in 202 genes enriched in P6 LfngGFP+ cells (filtered lists: present in DESeq and Cufflinks outputs, RPKM>3000 and FC>4) relative to their GFP- counterparts. The biological processes represented were obtained with the DAVID bioinformatics suite and then summarized with the REVIGO tool.
Table 7. Biological processes (BP) GO terms significantly represented in the list of DEG between P1 and P6 Lfng-GFP+ cells.
| Representative GO Term description (REVIGO Output) | Term ID | Included GO Term descriptions (DAVID Output) | Genes represented | log10 p value |
|---|---|---|---|---|
| Cell adhesion | GO:0007155 | Cell adhesion | Ibsp, Col13a1, Tnc, Bcam, Vtn, Itgb3, Cdh3, Cdh4, Lama1, Wisp1, Fat3, Hes5, Itga8, Thbs1, Col8a1 | -4.1292 |
| GO:0030155 | Regulation of cell adhesion | Lama1, Stat5a, Vtn, Thbs1, Col8a1 | -2.264 | |
| Extracellular structure organization | GO:0043062 | Extracellular structure organization | Ibsp, Drp2, Itga8, Tnc, Eln, P2rx2, Vtn | -3.0499 |
| Hormone metabolic process | GO:0042445 | Hormone metabolic process | Aldh1a1, Pcsk2, Dio2, Chst8, Cyp26b1 | -2.3309 |
Summary of biological processes GO terms that were significantly represented (p<0.01) in 338 differentially expressed genes in P1 relative to P6 LfngGFP+ cells (present in DESeq and Cufflinks outputs, enriched in P1 or P6 and FC>4). The biological processes represented were obtained with the DAVID bioinformatics suite and then summarized with the REVIGO tool.
To further understand the genes responsible for supporting cell maturation, we compared our P1 and P6 transcriptomes, focusing on genes related to cell fate commitment, cellular component biogenesis and cytoskeletal organization. As a first step in assembling a supporting cell gene regulatory network, we found 36 transcription factors expressed significantly in P1 supporting cells, of which only nine (Sox2, Sox6, Prox1, Tal1, Isl2, Atoh1, Hes5, Sall1, and Gata3) were related to cell fate commitment on the basis of gene ontology. At P6 we found 32 enriched transcription factors, with only five related to cell fate commitment (Hes5, Sall1, Gata3, Sox2 and Prox1). Comparing both ages showed seven transcription factors (Tal1, Atoh1, Hes5, Egr4, Sox11, Gm98, and Hmga2) changing significantly from P1 to P6.
We also looked for known downstream genes that might be associated with supporting cell maturation. Potential candidates enriched in P6 supporting cells included microtubule associated genes (Tsga14, Stmn1, Shroom1 and Tppp) [42–45] actin microfilament associated genes (Shroom1, Capg and Gsn) [44, 46], and Tnfrsf12a, a gene implicated in extracellular matrix adhesion [47]. Of these genes, only Shroom1 and Tppp were also enriched in P1 supporting cells. Finally, we found 20 known deafness genes enriched in supporting cells at P1 (Tmprss3, Cabp2, Ush1c, Ildr1, Otog, Smp, Pou4f3, Gipc3, Grxcr2, Grhl2, Otof, Loxhd1, Strc, Grxcr1, Myo3a, Ptprq, Myh14, Marveld2, Pcdh15 and Fam65b), 18 enriched at P6 (Tmprss3, Smpx, Otog, P2rx2, Ildr1, Strc, Loxhd1, Ush1c, Pou4f3, Slc26a5, Grxcr2, Pcdh15, Grhl2, Otof, Ceacam16, Tmc1, Marveld2 and Gjb6) and just five changing significantly between P1 to P6 (Ceacam16, Tnc, Otoa, Slc26a5 and P2rx2).
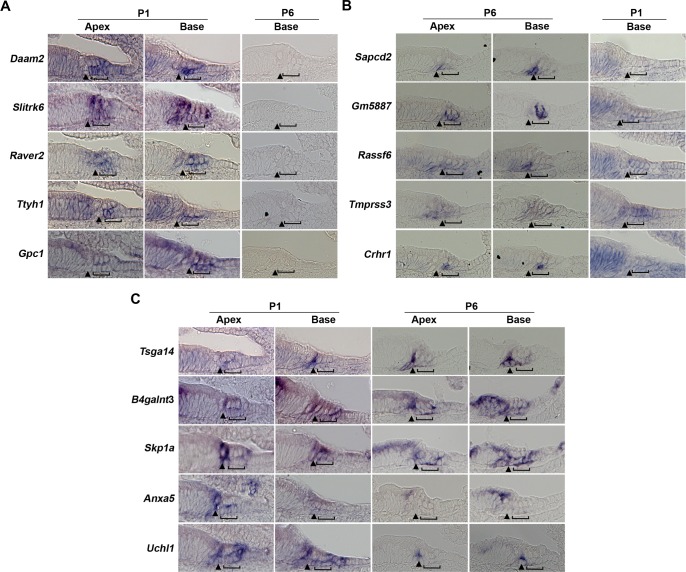
To further validate the expression of our supporting cell-specific genes, we examined the expression of selected genes on cochlear sections of P1 and P6 mice by in situ hybridization. We chose genes on the basis of their RPKM values and significant enrichment at either age (Fig 2; S7 Table). We previously used in situ hybridization to validate hair cell-specific transcripts, and found that only about 50% of hair cell-enriched transcripts gave detectable, hair cell-specific expression patterns [48]. Similarly, in our supporting cell analysis, we found many genes enriched in supporting cells on the basis of RNA-seq that gave broader patterns of expression in other cochlear cell types (S7 Table). We evaluated five genes that were specifically expressed at P1 (Daam2, Raver2, Slitrk6, Ttyh1 and Gpc1), five genes specifically expressed at P6 (Sapcd2, Gm5887, Rassf6, Tmprss3 and Crhr1) and five genes expressed at both ages (Tsga14, B4galnt3, Skp1a, Anxa5 and Uchl1). Of those only Slitrk6 expression has been previously characterized in the cochlea [49]. In situ analysis revealed a wide variety of expression patterns, with some genes expressed broadly in most supporting cell types (B4galnt3, Skp1a) at both ages, others restricted to one supporting cell subtype at both ages (e.g. Tsga14 in pillar cells), whereas other genes were broadly expressed at only one age (Slitrk6 at P1) or restricted to a particular sub-population at only one age (Gm5887 in Deiters’ cells at P6). We also observed apical-basal gradients of expression for some genes at both ages (Fig 2). In summary, in addition to validating our RNA-seq data, our in situ hybridization revealed the presence of spatially and temporally varying and sub-population-specific gene expression for many of our supporting cell transcripts.
Fig 2. In situ validation of supporting cell-specific transcripts.
Examples of supporting cell genes enriched in (A) P1 supporting cells with basal P6 sections to show negative expression (B) P6 supporting cells with basal P1 sections to show negative expression (C) Both P1 and P6 supporting cells, with sections to show both apical (less mature) and basal (more mature) regions at each age. Brackets: Deiters’ cells; Arrowhead: pillar cell region; horizontal line, greater epithelial ridge region.
Mouse cochlear supporting cells become almost completely unresponsive to blockade of Notch signaling by six days of age
The Notch signaling pathway plays a critical role in establishing hair cell and supporting cell identity during the differentiation of inner ear sensory tissue [50]. Specifically, hair cells expressing the DLL1, DLL3 and JAG2 ligands deliver a Notch signal to neighboring supporting cells that prevents their differentiation into hair cells. Accordingly, genetic inactivation of Notch receptors, their ligands or downstream transcriptional effectors in the inner ear causes an over-production of hair cells at the expense of supporting cells [51–56]. Moreover, blockade of the Notch signaling pathway through pharmacological inhibition of the gamma secretase complex required for Notch receptor cleavage or antibody blockade of Notch receptors themselves can also cause supporting cells to trans-differentiate into hair cells [13, 17–20]. However, the ability of Notch inhibition to cause supporting cell trans-differentiation decline rapidly with age, and can even be observed in supporting cells of different maturational states along the basal-apical axis of the neonatal cochlea [19]. We have previously shown that the expression of some components of the Notch pathway is reduced in mouse cochlear supporting cells in the first postnatal week [19]. However, it is formally possible that significant numbers of transcriptional changes still occur when the Notch pathway is blocked in P6 supporting cells, but these are nevertheless insufficient to drive trans-differentiation to hair cells.
We first assessed the expression of known Notch pathway genes in our P1 and P6 data sets. We found many Notch pathway genes enriched in supporting cells, and many of these were down-regulated between P1 and P6 as previously described [19] (Table 8). To test whether changes in Notch responsiveness of supporting cells between birth and P6 are associated with changes in the transcriptional response to Notch blockade, we cultured cochleas from newborn (P0) or P5 mice in the presence or absence of the gamma secretase inhibitor DAPT for 24 hours as previously described [19]. Our previous work showed that DAPT, which prevents gamma secretase-mediated cleavage of many proteins in addition to Notch receptors, has the same effect on neonatal cochlear cultures as Notch1 blocking antibodies [19]. At the end of the culture period, we dissociated the cultured cochleas and purified GFP+ and GFP- cells by fluorescence-activated cell sorting. We reasoned that culturing P0 and P5 cochleas for 24 hours was the most appropriate way to compare their transcriptomes to the acutely isolated P1 and P6 supporting cells described above. The GFP label was stable and retained by supporting cells during this 24 hour period of Notch blockade, allowing us to isolate supporting cells as they began to trans-differentiate into hair cells (S1A–S1C Fig). We isolated between 500–600 supporting cells per cochlea in each condition (DMSO or DAPT) at P1, and between 300–400 supporting cells per cochlea at P6. Between 10,000–20,000 purified supporting cells were used to create RNA-seq libraries at each age and condition, with duplicate samples being analyzed. GFP+ and GFP- cell populations from P0 and P5 supporting cells cultured in DAPT or DMSO vehicle were analyzed by RNA-seq. Mapped and normalized data sets were compared to identify supporting cell transcripts that were significantly up- or down-regulated in P0 supporting cells that were treated with DAPT compared to DMSO, and a similar analysis was performed for our P5 samples.
Table 8. Notch and Wnt pathway genes expressed in Lfng-GFP+ cells at P1 and P6.
| Postnatal Day 1 | Postnatal Day 6 | |||||
|---|---|---|---|---|---|---|
| Genes | Expression in Lfng-GFP+ (RPKM) | Fold change (GFP+vs GFP-) | p-adj | Expression in Lfng-GFP+ (RPKM) | Fold change (GFP+vs GFP-) | p-adj |
| Lfng | 109921.44 | 112.40 | 2.5E-252 | 117587 | 81.79 | 2.31E-10 |
| Hey1 | 22093.29 | 25.21 | 6.75E-160 | 9892 | 4.35 | 0.000014 |
| Jag1 | 81967.23 | 19.58 | 2.12E-105 | 47865 | 17.31 | 9.94E-43 |
| Hey2 | 7358.57 | 12.92 | 1.09E-102 | 6735 | 10.27 | 1.79E-24 |
| Mycl1 | 7233.13 | 10.13 | 1.11E-86 | 6790 | 7.76 | 4.09E-08 |
| Notch1 | 47252.67 | 4.36 | 1E-44 | 14809.73 | 1.650383 | 0.208748 |
| Notch3 | 21210.78 | 4.03 | 2.78E-38 | 8181.368 | 2.125937 | 0.000264 |
| Cntn1 | 9585.18 | 5.58 | 1.92E-34 | 4419 | 4.25 | 1.78E-12 |
| Mfng | 1254.47 | 6.30 | 9.05E-17 | 731 | 5.14 | 9.28E-08 |
| Hes5 | 6663.27 | 306.73 | 1.68E-16 | 737 | 174.97 | 4.39E-24 |
| Hr | 1500.99 | 4.81 | 1.34E-11 | 8878.668 | 3.452246 | 2.05E-09 |
| Lor | 364.97 | 8.87 | 9.93E-08 | 45.26282 | 1.175772 | 1 |
| Wnt7a | 10897.69 | 46.84 | 1.3E-190 | 7143.40 | 125.77 | 1.26E-35 |
| Fzd4 | 10204.53 | 35.42 | 7.98E-66 | 3761.40 | 5.64 | 2.77E-16 |
| Frzb | 13318.59 | 18.55 | 1.41E-134 | 2707.837 | 0.901449 | 1 |
| Kremen1 | 55934.39 | 16.31 | 1.17E-134 | 50543.90 | 5.94 | 2.14E-18 |
| Sfrp1 | 53947.70 | 9.40 | 1.6E-87 | 17549.5299 | 6.98 | 3.18E-22 |
| Fzd6 | 6100.32 | 4.52 | 2.92E-41 | 7130.342 | 3.684441 | 4.31E-10 |
| Wnt7b | 2444.61 | 4.26 | 1.43E-11 | 3047.78699 | 7.61 | 1.5E-13 |
Known members of the Notch and Wnt signaling pathways expressed in either P1 or P6 LFng-GFP+ cells. The mean expression level (RPKM) is shown, together with the fold change versus GFP- cells. p-adj = adjusted p-value for the difference between GFP+ and GFP- populations at each age.
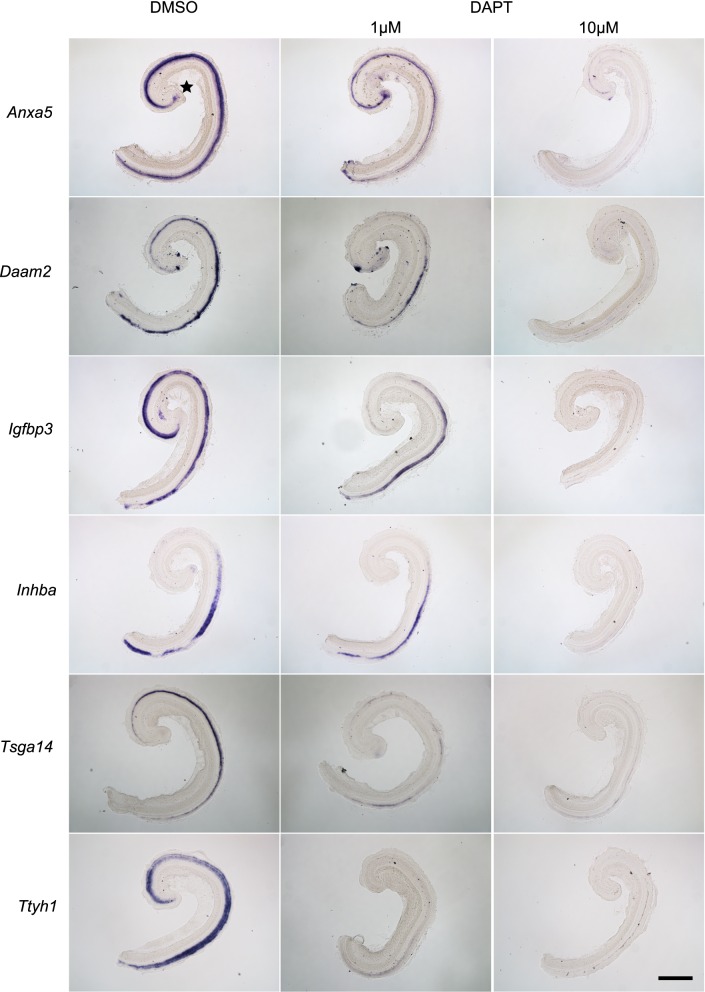
We used DESeq to identify 2,088 supporting cell-specific genes that were significantly changed after DAPT treatment of P0 cochleas (S1D Fig), with 1032 transcripts being down-regulated (Table 9) and 1056 transcripts up-regulated (Table 10 and S8 Table). In agreement with the observed trans-differentiation of neonatal supporting cells after Notch blockade, 237 of the transcripts up-regulated by DAPT were among the 304 genes we had previously shown to be strongly enriched in hair cells [48] and having RPKM higher than 3000. Moreover, consistent with our previous observation that DAPT treatment of cochlear cultures largely works through the Notch pathway [19], GO analysis of the down-regulated supporting cell-specific genes after DAPT treatment featured many Notch pathway members (Table 11). In stark contrast, when P5 cochleas were treated with DAPT, we only observed 20 supporting cell-specific genes that were changed significantly, with 2 transcripts up-regulated and 18 down-regulated (S1D Fig and Table 12). Moreover, many of these genes were expressed at extremely low levels compared to younger supporting cells–for example, although Hes5 still showed a significant down-regulation after DAPT treatment of P5 cochleas, its normalized expression (in RPKM) changed from 120 to 3 after DAPT treatment, whereas its levels in P1 supporting cells was over 6,600 (Table 1). To validate our data sets, we cultured P0 cochleas in the presence or absence of DAPT for 24 hours and then analyzed changes in some of the supporting cell-specific genes by whole mount in situ hybridization (Fig 3). Of the genes evaluated, Ttyh1 exhibited the strongest change in in situ signal, consistent with the RNA-seq data where its normalized level in RPKM dropped from 13941 to 1689. Similarly Anxa5, Daam2, Inhba, Igfbp3 and Tsga14 also decreased and resembled their decrease in RPKM seen after DAPT treatment. Consistent with our previous observations that the apex of the neonatal cochlea is more sensitive to Notch inhibition than the base [19], the genes evaluated showed more significant decreases in signal in the apex of the DAPT-treated cochleas than the base with the exception of Anxa5 that showed a more diffuse decrease in the signal from apex to base. Taken together, our data confirm that the Notch signaling pathway is functionally dismantled in supporting cells between P1 and P6 such that Notch inhibition leads to barely perceptible transcriptional changes in P6 mature supporting cells.
Table 9. Top 20 down-regulated genes after DAPT treatment in P0 Lfng-GFP+ cells.
| Rank | Gene symbol | DMSO Mean GFP+ (RPKM) | DAPT Mean GFP+ (RPKM) | Fold change DAPT vs DMSO | p-adj |
|---|---|---|---|---|---|
| 1 | Tal1 | 2983.37 | 2.56 | 0.0009 | 8.77E-181 |
| 2 | Cysltr2 | 3834.07 | 22.16 | 0.0058 | 9.07E-148 |
| 3 | Pdzk1ip1 | 2814.84 | 19.02 | 0.0068 | 5.70E-127 |
| 4 | Heyl | 4828.46 | 224.34 | 0.0464 | 1.55E-102 |
| 5 | Prss23 | 4249.27 | 215.01 | 0.0508 | 1.72E-95 |
| 6 | Gstm1 | 4572.23 | 331.06 | 0.0723 | 2.42E-80 |
| 7 | Gm10800 | 74333.61 | 8438.92 | 0.1134 | 2.45E-77 |
| 8 | 2310022B05Rik | 8186.49 | 813.73 | 0.0994 | 3.14E-74 |
| 9 | Dgat2 | 1061.61 | 21.63 | 0.0203 | 5.69E-70 |
| 10 | Syt6 | 6254.68 | 431.84 | 0.0689 | 6.15E-70 |
| 11 | Slitrk2 | 1253.99 | 38.20 | 0.0304 | 3.02E-69 |
| 12 | Ttyh1 | 13940.83 | 1688.84 | 0.1207 | 8.12E-68 |
| 13 | Pdgfrb | 2128.79 | 118.91 | 0.0559 | 2.74E-67 |
| 14 | Ccdc80 | 3066.68 | 252.98 | 0.0825 | 2.10E-66 |
| 15 | AC131780.2 | 6376.16 | 723.02 | 0.1134 | 2.31E-64 |
| 16 | Rab3b | 5451.58 | 600.43 | 0.1103 | 8.92E-64 |
| 17 | Nrarp | 1389.90 | 68.84 | 0.0494 | 2.34E-61 |
| 18 | Dkk3 | 3824.89 | 424.10 | 0.1111 | 2.32E-58 |
| 19 | Sema5a | 2652.79 | 252.04 | 0.0947 | 6.94E-56 |
| 20 | Eno1 | 3363.46 | 382.22 | 0.1134 | 3.34E-55 |
List of the 20 genes showing the greatest down-regulation in P0 Lfng-GFP+ cells after 24 hours DAPT treatment compared P0 Lfng-GFP+ cells treated with DMSO vehicle. Ranking is based on the p value. The mean expression level (RPKM) is shown, together with the fold change versus DMSO-treated cells. p-adj = adjusted p-value for the difference between DAPT and DMSO-treated populations.
Table 10. Top 20 up-regulated genes after DAPT treatment in P0 Lfng-GFP+ cells.
| Rank | Gene symbol | DMSO Mean GFP+ (RPKM) | DAPT Mean GFP+ (RPKM) | Fold change DAPT vs DMSO | p-adj |
|---|---|---|---|---|---|
| 1 | AC123795.1 | 134.87 | 18009.79 | 133.44 | 3.21E-186 |
| 2 | Scn11a | 83.16 | 6687.08 | 80.45 | 8.07E-178 |
| 3 | Ush2a | 685.76 | 26222.34 | 38.32 | 1.77E-168 |
| 4 | Fam70b | 207.23 | 7185.86 | 34.78 | 9.23E-138 |
| 5 | Vwa5b2 | 101.49 | 4564.69 | 44.94 | 7.78E-136 |
| 6 | Srrm4 | 249.11 | 6795.38 | 27.28 | 9.01E-124 |
| 7 | P2rx3 | 49.28 | 2547.23 | 51.63 | 4.06E-115 |
| 8 | Gas6 | 251.21 | 5474.75 | 21.86 | 3.31E-97 |
| 9 | Myo3a | 365.31 | 6215.50 | 17.03 | 1.57E-96 |
| 10 | Thsd7b | 457.96 | 11936.04 | 25.99 | 5.86E-95 |
| 11 | Eya2 | 33.79 | 1702.55 | 50.56 | 5.03E-94 |
| 12 | 6330503K22Rik | 1044.82 | 19337.03 | 18.51 | 4.62E-88 |
| 13 | Gadd45g | 31.74 | 1444.82 | 45.57 | 6.97E-84 |
| 14 | Mobkl2b | 307.36 | 4600.97 | 14.93 | 8.34E-84 |
| 15 | Myo7a | 348.85 | 5023.23 | 14.42 | 2.21E-83 |
| 16 | Atoh1 | 133.28 | 5021.72 | 37.79 | 2.49E-79 |
| 17 | Atp8a1 | 2348.26 | 22393.66 | 9.51 | 7.43E-79 |
| 18 | Pole | 1944.59 | 18385.23 | 9.45 | 3.89E-77 |
| 19 | Jag2 | 98.59 | 3275.67 | 33.13 | 1.01E-75 |
| 20 | Slc16a5 | 58.11 | 1531.35 | 26.35 | 1.14E-71 |
List of the 20 genes showing the greatest up-regulation in P0 Lfng-GFP+ cells after 24 hours DAPT treatment compared P0 Lfng-GFP+ cells treated with DMSO vehicle. Ranking is based on the p value. The mean expression level (RPKM) is shown, together with the fold change versus DMSO-treated cells. p-adj = adjusted p-value for the difference between DAPT and DMSO-treated populations.
Table 11. Biological processes (BP) GO terms represented in the list of transcripts down-regulated after DAPT treatment of P0 Lfng-GFP+cells that were also enriched in P1 Lfng-GFP+cells.
| Representative GO Term description (REVIGO Output) | Term ID | Included GO Term descriptions (DAVID Output) | Genes represented | log10 p value |
|---|---|---|---|---|
| Notch signaling pathway | GO:0007219 | Notch signaling pathway | Notch3, Notch1, Hey1, Heyl, Hey2, Cntn1, Jag1 | -6.4222 |
| GO:0008593 | Regulation of Notch signaling pathway | Hey2, Fgf10, Jag1 | -3.1195 | |
| GO:0016055 | Wnt signaling pathway | Fzd9, Sfrp1, Frzb, Fzd4, Wnt7a | -2.3026 | |
| Cell fate commitment | GO:0045165 | Cell fate commitment | Notch3, Tal1, Notch1, Erbb4, Hes5, Fgf10, Sox6, Tgfb2 | -4.91 |
| GO:0048729 | Tissue morphogenesis | Notch1, Sfrp1, Fgf10, Sema3a, Nr4a3, Jag1, Wnt7a, Tgfb2 | -3.5865 | |
| GO:0051094 | Positive regulation of developmental process | Tal1, Notch1, Hey2, Jag1, Igfbp3, Wnt7a, Tgfb2 | -3.0288 | |
| Cell adhesion | GO:0007155 | Cell adhesion | Egfl6, Fblim1, Tgfb2, Cd9, Pgm5, Itga6, Hes5, Fat3, Lsamp, Ttyh1, Cntn1, Lamc2, Thbs1 | -4.4589 |
| Cell projection organization | GO:0030030 | Cell projection organization | Notch1, Itga6, Ptprz1, Ttyh1, Sema3a, Slitrk6, Tgfb2 | -2.1669 |
Summary of biological processes GO terms that were significantly represented (p<0.01) in 97 genes enriched in P1 LfngGFP+ cells (filtered list: present in both DESeq and Cufflinks outputs, RPKM>3000, FC>4) and down regulated in P0 LfngGFP+ DAPT 24 hours treated cells (present in DESeq or Cufflinks outputs and FC>2) relative to their DMSO treated counterparts. The biological processes represented were obtained with the DAVID bioinformatics suite and then summarized with the REVIGO tool.
Table 12. All significantly up- or down-regulated genes after DAPT treatment in P5 Lfng-GFP+ cells.
| Rank | Gene symbol | DMSO Mean GFP+ (RPKM) | DAPT Mean GFP+ (RPKM) | Fold change DAPT vs DMSO | p-adj |
|---|---|---|---|---|---|
| 1 | Fabp7 † | 2378.03 | 558.00 | 0.23 | 3.89E-10 |
| 2 | Ttyh1 | 5568.39 | 1810.79 | 0.33 | 1.06E-07 |
| 3 | 2310022B05Rik | 3060.16 | 1069.78 | 0.35 | 1.43E-06 |
| 4 | Hes5 † | 120.67 | 3.21 | 0.03 | 1.43E-06 |
| 5 | Fzd9 | 698.27 | 171.68 | 0.25 | 1.89E-06 |
| 6 | Ppp1r2 | 8051.64 | 3088.33 | 0.38 | 5.43E-06 |
| 7 | Slc6a14 | 1948.23 | 716.35 | 0.37 | 5.62E-05 |
| 8 | Cysltr2 | 157.51 | 11.46 | 0.07 | 0.00011759 |
| 9 | Efr3b | 866.96 | 302.29 | 0.35 | 0.0002018 |
| 10 | Sez6l | 523.69 | 148.11 | 0.28 | 0.0002018 |
| 11 | Ednrb | 1686.11 | 622.61 | 0.37 | 0.0002018 |
| 12 | Daam2 | 1579.09 | 376.46 | 0.24 | 0.00037462 |
| 13 | Eno1 | 2183.19 | 929.87 | 0.43 | 0.00057668 |
| 14 | Slc2a9 | 346.76 | 53.92 | 0.15 | 0.00122798 |
| 15 | Pak6 † | 141.12 | 16.16 | 0.11 | 0.00431559 |
| 16 | RP23-218C2.3 | 75.13 | 3.41 | 0.05 | 0.00637716 |
| 17 | Srebf1 † | 3809.42 | 1780.78 | 0.47 | 0.00637716 |
| 18 | 2610020H08Rik † | 3075.63 | 1446.24 | 0.47 | 0.0068551 |
| 19 | Dpysl2 | 1661.82 | 3693.20 | 2.22 | 0.00120881 |
| 20 | Irf4 † | 21.31 | 188.19 | 8.82 | 0.0050224 |
List of all genes showing the significant up- or down-regulation in P5 Lfng-GFP+ cells after 24 hours DAPT treatment compared P5 Lfng-GFP+ cells treated with DMSO vehicle. Ranking is based on the p value. The mean expression level (RPKM) is shown, together with the fold change versus DMSO-treated cells. p-adj = adjusted p-value for the difference between DAPT and DMSO-treated populations.
† = genes also showing a significant change in DAPT-treated P0 cells (p<0.01).
Fig 3. Gene expression changes in P0 supporting cells in response to Notch inhibition.
Demonstration of rapid down-regulation by in situ hybridization of six supporting cell-specific genes in P0 cochleas after 24 hours of culture in DAPT versus DMSO controls. Scale Bar: 500 μm. Star: apex tip.
Discussion
Mammalian supporting cells undergo a remarkable morphological transformation before the onset of hearing that includes the formation of phalangeal processes by Deiters’ cells and inner phalangeal cells, the integration of supporting cell apical processes into the reticular lamina and the formation of the tunnel of Corti by pillar cells [27, 57, 58]. Although these morphological changes have been well-characterized, the molecular basis of this maturation is far less clear. While much recent attention has been paid to understanding the transcriptome of cochlear and vestibular hair cells, far less is known about the genes expressed by supporting cells as they differentiate and mature. Since the loss of regenerative potential of mouse supporting cells occurs over a similar time period as their maturation [1, 58], it is likely that identifying maturational changes in the supporting cell transcriptome may provide insights into the failure of mammalian hair cell regeneration.
We have characterized the transcriptome of mouse cochlear supporting cells in the first postnatal week. Using LFng-GFP transgenic mice, we were able to isolate all major supporting cell populations from P1 and P6 mice with the exception of inner pillar cells (Fig 1A). Using stringent criteria, we identified approximately 500 genes that were significantly enriched in P1 and P6 supporting cells. Although the gene expression profiles differed significantly between P1 to P6, there were many genes expressed in supporting cells at both stages. Instead of a radically different gene expression profile that might be expected between different cell types, the changes observed from P1 to P6 with GO analysis instead showed maturational changes of genes involved in regulation of the cell cycle, differentiation, the cytoskeleton and extracellular matrix. We identified a number of factors associated with maturation that has not been previously identified in the ear, such as Tsga14, Stmn1, Tppp, Shroom1, Capg, Gsn and Tnfrsf12a. Of note, one of the transcripts most strongly enriched in supporting cells at both P1 and P6 was the Fgfr3 FGF receptor. FGFR3 signaling in pillar and Deiters’ cells is important for both the induction and maintenance of the identity of these supporting cells [59–62]. Strikingly, this signaling continues to be required for supporting cell identity in postnatal life, as mutations that change FGFR3 ligand specificity can cause Deiters’ cell–pillar cell transformations after the onset of hearing [63].
Our in situ analysis revealed a large amount of heterogeneity in expression patterns between different supporting cell genes. Although some genes were expressed in many supporting cell types, others were confined to just a few cell types such as pillar cells. In particular, many of the genes enriched in P6 supporting cells were localized to either pillar cells (Sapcd2, Crhr1, Rassf6) or Deiters’ cells (Gm5887). Recent advances in single cell transcriptome analysis have started to provide insights into supporting cell sub-populations [40, 64], and the availability of new markers for individual supporting cell types will allow for a more detailed characterization of their transcriptomes. Moreover, the known variations in hair cell gene expression along the tonotopic axis of the cochlea [65–68] are likely to have counterparts in the surrounding supporting cells, and it is likely that single cell analysis of supporting cell transcriptomes may reveal such gradients in the future.
Our study also confirms the importance of expression-based validation of RNA-seq data. In a previous study, we showed that only about 50% of genes shown to be enriched in neonatal hair cells gave specific or detectable signal in hair cells by in situ hybridization [48]. In the present study, we once again showed that only approximately 50% of genes identified as being enriched in supporting cells actually gave a specific in situ signal. While this may be due in part on technical artifacts arising from the choice of in situ probes or hybridization conditions, it is also a consequence of focusing on the degree of differential rather than absolute expression, whereby broadly expressed genes may still be expressed at higher levels in supporting cells.
Both the Notch and Wnt pathways have been suggested to regulate the differentiation of hair cells and supporting cells [50, 69] and manipulation of both pathways has been reported to increase both supporting cell proliferation and trans-differentiation into hair cells [13, 50, 52]. We previously showed that the mouse cochlear supporting cells become unresponsive to Notch inhibition in the first postnatal week, and that at least some components of the Notch pathway are down-regulated in supporting cells during this period [19]. Our current data confirms a clear down-regulation of signaling components in both the Notch and Wnt pathways between P1 and P6 (Table 8), which suggests that the number of transcriptional changes in supporting cells following Notch inhibition in P5 cochlear cultures was likely to be small. Our RNA-seq data of DAPT-treated cochleas now provide a striking confirmation of this hypothesis: although over 2,000 genes were either up- or down-regulated in supporting cells by DAPT treatment of P0 cochlear tissue, we only saw 20 genes change in treated P5 cochlear supporting cells. Moreover, of these 20 genes, only 13 had expression levels >500 RPKM. These results suggest that towards the end of the first postnatal week, blockade of the Notch pathway has essentially no effect on supporting cell transcription. However, we would caution that our experiments were performed on intact cochlear tissue in which hair cells remained alive, and it is possible that the loss of hair cells might trigger changes in supporting cells in which the Notch pathway was wholly or partly re-engaged. Indeed, a recent study suggest that treatment of the noise-damaged adult cochlea with gamma secretase inhibitors promoted some trans-differentiation of supporting cells into hair cells [70]. It remains to be determined conclusively whether this effect is specific to the Notch pathway, and whether hair cell loss is necessary for this response to occur. It is also notable that adult vestibular supporting cells are capable of expressing at least some markers of hair cells after gamma secretase treatment [71, 72]. It will therefore be profitable to compare the transcriptomes of supporting cells from different vestibular organs to their cochlear counterparts at different ages, and to compare the epigenetic state of hair cell loci in these different populations of supporting cells as a first step to understand the molecular basis for the different propensities of these cells for trans-differentiation into hair cells.
Supporting Information
(A): Surface preps of LfngGFP+ cochlear explants. Sensory epithelium retains its GFP expression after culture for 24 hours in either the gamma secretase inhibitor DAPT or DMSO vehicle control. (B): Diagram of the supporting cells recognized from the top of the epithelium. (C): Diagram of the experimental design. P0 or P5 cochleas were dissected and cultured. Explants were then dissociated and sorted for GFP fluorescence and the GFP+ and GFP- fractions used to make RNA-seq libraries. (D): Summary of differentially expressed transcript comparisons in each experimental condition.
(TIF)
For each gene, forward and reverse PCR primers are given, together with the predicted size of the band generated from PCR with mouse genomic DNA. All reverse primers contain a T7 polymerase sequence at their 5’ end (GGATCCTAATACGACTCACTATAGGGAG).
(XLSX)
Sheet 1 shows the analysis performed on the data with DESeq for both P1 and P6 cells. Sheet 2 shows the analysis performed with Cufflinks on P1 cells and Sheet 3 shows the same Cufflinks analysis on P6 cells.
(XLSX)
Sheet 1 shows the analysis performed on the data with DESeq for both P0 and P5 cells. Sheet 2 shows the analysis performed with Cufflinks on P0 cells and Sheet 3 shows the same Cufflinks analysis on P5 cells.
(XLSX)
The gene name is indicated, together with the expression level (reads per kilobase of transcript per million mapped reads; RPKM; DESeq output only) and its fold change compared to GFP- cells. p-adj = adjusted p-value for the difference between GFP+ and GFP- populations.
(DOCX)
Consensus lists of genes enriched in FACS sorted Lfng-GFP+ cells from postnatal day 1 (P1; 1884 genes) and postnatal day 6 (P6; 1278 genes) mouse cochlea compared to Lfng-GFP-negative cells. Analysis of the sequencing reads was performed by two different approaches. (1) Reads were mapped to the Mus musculus NCBI build37.2 iGenome (Ilumina) using TopHat 2.0 software (Trapnell et al., 2009; Trapnell et al., 2012) and the mapped reads were quantitated and compared using Cufflinks 2.0 providing differential gene expression data and statistics. (2) Reads were aligned to the Mus musculus Ensembl mm9 iGenome (Ilumina) using TopHat 1.4.1 software and the number of reads per gene and per library was obtained using DESeq program. After comparing the level of expression of each gene within each pair of related libraries (GFP+ versus GFP- for P1 and P6 cells), the most significant differentially expressed genes (DEG) were annotated and analyzed separately for both approaches. A consensus list of DEGs common to both methods of analysis was then generated. A significantly DEG was considered to have an RPKM higher than 3000, Fold Change (FC) higher than 4 and p value and FDR < 0.01. Duplicate samples of Lfng-GFP+ and GFP- sorted cells were prepared for P1 and P6. Approximately 60,000 sorted cells were as starting material to generate approximately 100–600 ng RNA (measured by Nanodrop spectrophotometer). cDNA libraries for RNAseq were generated using RNA Seq Truseq RNA sample preparation kit v2 (Illumina) following the low sample protocol for RNA extraction, cDNA synthesis, indexing and amplification. The quality and integrity of RNA samples and the final quality of the sequencing libraries was checked by electrophenogram in an Agilent Bioanalyzer. Paired-end sequencing was performed in HiSeq2000 sequencing platform (Illumina). Fastq files of paired end reads have been deposited in the NCBI GEO database, Accession No. GSE83357.
(XLS)
Consensus list of genes enriched in Lfng-GFP+ supporting cells that were differentially expressed between P1 and P6. Data was obtained from the analysis described in S5 Table caption above, but now genes enriched in supporting cells were compared for changes between P1 and P6.
(XLS)
For each gene, its expression at P1 and P6 (RPKM) together with the fold enrichment between GFP+ and GFP- cell populations is shown, together with expression pattern in the cochlea (SC, supporting cell; HC, hair cell; GER, greater epithelial ridge; SV, stria vascularis; Ubi, ubiquitous expression; No, no detectable signal; ND = not determined). NS: p>0.01.
(DOCX)
P0 Lfng-GFP cochlear cultures were maintained for 24 hours in DAPT or DMSO, followed by FACS sorting for GFP fluorescence. DESeq was used to identify transcripts enriched in cultures treated with either DAPT or DMSO. The table shows a list of 2088 genes enriched in Lfng-GFP+ cells that were significantly altered in DAPT-treated cultures compared to DMSO controls. Up-regulated transcripts after DAPT treatment are highlighted in red; down-regulated transcripts are highlighted in green. Duplicate samples of LfngGFP+ and GFP- sorted cells were prepared for postnatal day 0 (P0) and postnatal day 5 (P5) cultured for 24 hours in DMSO or DAPT (10uM). 10,000–20,000 sorted cells were used as starting material to generate approximately 100–600 ng RNA (measured by Nanodrop spectrophotometer). cDNA received an initial amplification using the NuGen Ovation Kit. cDNA libraries for RNAseq were generated using RNA Seq Truseq RNA sample preparation kit v2 (Illumina) following the low sample protocol for RNA extraction, cDNA synthesis, indexing and amplification. The quality and integrity of RNA samples and the final quality of the sequencing libraries was checked by electrophenogram in an Agilent Bioanalyzer. Paired-end sequencing was performed in HiSeq2000 sequencing platform (Illumina). Fastq files of paired end reads have been deposited in the NCBI GEO database, Accession No. GSE83357. Reads were aligned to the Mus musculus Ensembl mm9 iGenome (Ilumina) using TopHat 1.4.1 software and the number of reads per gene and per library was obtained using DESeq program. After comparing the level of expression of each gene within each pair of related libraries, the most significant differentially expressed genes (DEG) were annotated and analyzed separately for both approaches. In order to find genes significantly changing in isolated Lfng-EGFP+ cells obtained from cultured explants (treated or not with DAPT), the level of DEG significance was a FC higher than 2 and a p value and FDR < 0.01.
(XLS)
Acknowledgments
We thank Alyssa Crowder and Huiling Li for excellent technical assistance and members of the Groves lab for their advice and comments. We also thank Nat Heintz for providing the Lfng-GFP line.
Data Availability
Fastq files of paired end reads for all RNA-seq data have been deposited in the NCBI GEO database, Accession No. GSE83357.
Funding Statement
This project was supported in part by (1) The Genomic and RNA Profiling Core and the RNA In Situ Core at Baylor College of Medicine with the expert assistance of Dr. Lisa D. White, Ph.D., Cecilia Ljunberg, Ph.D., and funding from the NIH grant P30HD024064 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development; (2) The Cytometry and Cell Sorting Core at Baylor College of Medicine with the expert assistance of Joel M. Sederstrom and funding from the NIH grants P30 AI036211, P30 CA125123, and S10 RR024574; (3) NIH Grants DC006185 and DC011657 to A.K.G.; (4) Department of Defense Grant DOD W81XWH-11-2-004 to A.K.G.; (5) A Hearing Restoration Project consortium grant from the Hearing Health Foundation to A.K.G.; and (6) An Initiation into Research grant FONDECYT 11130247 from CONICYT (Chile) to J.C.M.
References
- 1.Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood). 2010;235(4):434–46. Epub 2010/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong AC, Ryan AF. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front Aging Neurosci. 2015;7:58 PubMed Central PMCID: PMC4404918. 10.3389/fnagi.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang CH, Schrepfer T, Schacht J. Age-related hearing impairment and the triad of acquired hearing loss. Front Cell Neurosci. 2015;9:276 PubMed Central PMCID: PMC4515558. 10.3389/fncel.2015.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng AG. Sensory hair cell development and regeneration: similarities and differences. Development. 2015;142(9):1561–71. PubMed Central PMCID: PMC4419275. 10.1242/dev.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monroe JD, Rajadinakaran G, Smith ME. Sensory hair cell death and regeneration in fishes. Front Cell Neurosci. 2015;9:131 PubMed Central PMCID: PMC4404912. 10.3389/fncel.2015.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51(6–7):633–47. Epub 2007/09/25. 10.1387/ijdb.072408js [DOI] [PubMed] [Google Scholar]
- 7.Thomas ED, Cruz IA, Hailey DW, Raible DW. There and back again: development and regeneration of the zebrafish lateral line system. Wiley Interdiscip Rev Dev Biol. 2015;4(1):1–16. PubMed Central PMCID: PMC4268111. 10.1002/wdev.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, Geleoc GS, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8(1):18–31. Epub 2006/12/16. 10.1007/s10162-006-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441(7096):984–7. Epub 2006/06/23. 10.1038/nature04849 [DOI] [PubMed] [Google Scholar]
- 10.White PM, Stone JS, Groves AK, Segil N. EGFR signaling is required for regenerative proliferation in the cochlea: conservation in birds and mammals. Dev Biol. 2012;363(1):191–200. Epub 2012/01/11. 10.1016/j.ydbio.2011.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai R, Kuo B, Wang T, Liaw EJ, Xia A, Jan TA, et al. Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc Natl Acad Sci U S A. 2012;109(21):8167–72. PubMed Central PMCID: PMC3361451. 10.1073/pnas.1202774109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo BR, Baldwin EM, Layman WS, Taketo MM, Zuo J. In Vivo Cochlear Hair Cell Generation and Survival by Coactivation of beta-Catenin and Atoh1. J Neurosci. 2015;35(30):10786–98. PubMed Central PMCID: PMC4518053. 10.1523/JNEUROSCI.0967-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Wu J, Yang J, Sun S, Chai R, Chen ZY, et al. Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc Natl Acad Sci U S A. 2015;112(1):166–71. PubMed Central PMCID: PMC4291673. 10.1073/pnas.1415901112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roccio M, Hahnewald S, Perny M, Senn P. Cell cycle reactivation of cochlear progenitor cells in neonatal FUCCI mice by a GSK3 small molecule inhibitor. Sci Rep. 2015;5:17886 PubMed Central PMCID: PMC4672274. 10.1038/srep17886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi F, Hu L, Edge AS. Generation of hair cells in neonatal mice by beta-catenin overexpression in Lgr5-positive cochlear progenitors. Proc Natl Acad Sci U S A. 2013;110(34):13851–6. PubMed Central PMCID: PMC3752268. 10.1073/pnas.1219952110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters BJ, Liu Z, Crabtree M, Coak E, Cox BC, Zuo J. Auditory hair cell-specific deletion of p27Kip1 in postnatal mice promotes cell-autonomous generation of new hair cells and normal hearing. J Neurosci. 2014;34(47):15751–63. PubMed Central PMCID: PMC4236404. 10.1523/JNEUROSCI.3200-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Reports. 2014;2(3):311–22. PubMed Central PMCID: PMC3964281. 10.1016/j.stemcr.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16(1):58–69. Epub 2009/01/22. 10.1016/j.devcel.2008.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maass JC, Gu R, Basch ML, Waldhaus J, Lopez EM, Xia A, et al. Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front Cell Neurosci. 2015;9:110 PubMed Central PMCID: PMC4379755. 10.3389/fncel.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takebayashi S, Yamamoto N, Yabe D, Fukuda H, Kojima K, Ito J, et al. Multiple roles of Notch signaling in cochlear development. Dev Biol. 2007;307(1):165–78. Epub 2007/05/29. 10.1016/j.ydbio.2007.04.035 [DOI] [PubMed] [Google Scholar]
- 21.Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141(4):816–29. PubMed Central PMCID: PMC3912828. 10.1242/dev.103036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32(19):6699–710. 10.1523/JNEUROSCI.5420-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Dearman JA, Cox BC, Walters BJ, Zhang L, Ayrault O, et al. Age-dependent in vivo conversion of mouse cochlear pillar and deiters' cells to immature hair cells by atoh1 ectopic expression. J Neurosci. 2012;32(19):6600–10. Epub 2012/05/11. 10.1523/JNEUROSCI.0818-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Fang J, Dearman J, Zhang L, Zuo J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic Atoh1 expression. PLoS One. 2014;9(2):e89377 PubMed Central PMCID: PMC3930725. 10.1371/journal.pone.0089377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters BJ, Zuo J. Postnatal development, maturation and aging in the mouse cochlea and their effects on hair cell regeneration. Hear Res. 2013;297:68–83. PubMed Central PMCID: PMC3594364. 10.1016/j.heares.2012.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim DJ, Anniko M. Developmental morphology of the mouse inner ear. A scanning electron microscopic observation. Acta Otolaryngol Suppl. 1985;422:1–69. [PubMed] [Google Scholar]
- 27.Raphael Y, Altschuler RA. Structure and innervation of the cochlea. Brain Res Bull. 2003;60(5–6):397–422. Epub 2003/06/06. [DOI] [PubMed] [Google Scholar]
- 28.Geschwind D. GENSAT: a genomic resource for neuroscience research. Lancet Neurol. 2004;3(2):82 [DOI] [PubMed] [Google Scholar]
- 29.Heintz N. Gene expression nervous system atlas (GENSAT). Nat Neurosci. 2004;7(5):483 10.1038/nn0504-483 [DOI] [PubMed] [Google Scholar]
- 30.Schmidt EF, Kus L, Gong S, Heintz N. BAC transgenic mice and the GENSAT database of engineered mouse strains. Cold Spring Harb Protoc. 2013;2013(3). [DOI] [PubMed] [Google Scholar]
- 31.Stern CD. Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr Top Dev Biol. 1998;36:223–43. [DOI] [PubMed] [Google Scholar]
- 32.Khatri SB, Groves AK. Expression of the Foxi2 and Foxi3 transcription factors during development of chicken sensory placodes and pharyngeal arches. Gene Expr Patterns. 2013;13(1–2):38–42. PubMed Central PMCID: PMC3562376. 10.1016/j.gep.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. PubMed Central PMCID: PMC2672628. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. PubMed Central PMCID: PMC3334321. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 36.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. PubMed Central PMCID: PMC2615629. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183 Epub 2007/09/06. 10.1186/gb-2007-8-9-r183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–75. PubMed Central PMCID: PMC1933169. 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800 PubMed Central PMCID: PMC3138752. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burns JC, Kelly MC, Hoa M, Morell RJ, Kelley MW. Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun. 2015;6:8557 PubMed Central PMCID: PMC4634134. 10.1038/ncomms9557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106 PubMed Central PMCID: PMC3218662. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–4. 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- 43.Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84(4):623–31. [DOI] [PubMed] [Google Scholar]
- 44.Lee C, Scherr HM, Wallingford JB. Shroom family proteins regulate gamma-tubulin distribution and microtubule architecture during epithelial cell shape change. Development. 2007;134(7):1431–41. 10.1242/dev.02828 [DOI] [PubMed] [Google Scholar]
- 45.Orosz F, Ovadi J. TPPP orthologs are ciliary proteins. FEBS Lett. 2008;582(27):3757–64. 10.1016/j.febslet.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 46.Barkalow K, Witke W, Kwiatkowski DJ, Hartwig JH. Coordinated regulation of platelet actin filament barbed ends by gelsolin and capping protein. J Cell Biol. 1996;134(2):389–99. PubMed Central PMCID: PMC2120875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang W, Xiao DZ, Wang Y, Shan ZX, Liu XY, Lin QX, et al. Fn14 promotes differentiation of human mesenchymal stem cells into heart valvular interstitial cells by phenotypic characterization. J Cell Physiol. 2014;229(5):580–7. 10.1002/jcp.24480 [DOI] [PubMed] [Google Scholar]
- 48.Cai T, Jen HI, Kang H, Klisch TJ, Zoghbi HY, Groves AK. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. J Neurosci. 2015;35(14):5870–83. PubMed Central PMCID: PMC4388939. 10.1523/JNEUROSCI.5083-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katayama K, A Z, Ota M, Y M, Inoue T, Fritzsch B, et al. Disorganized innervation and neuronal loss in the inner ear of Slitrk6-deficient mice. PLoS One. 2009;4(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiernan AE. Notch signaling during cell fate determination in the inner ear. Semin Cell Dev Biol. 2013;24(5):470–9. PubMed Central PMCID: PMC3725958. 10.1016/j.semcdb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham-McDonogh O. Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev Biol. 2008;316(1):87–99. Epub 2008/02/23. 10.1016/j.ydbio.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005;132(19):4353–62. Epub 2005/09/06. 10.1242/dev.02002 [DOI] [PubMed] [Google Scholar]
- 53.Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21(3):289–92. 10.1038/6804 [DOI] [PubMed] [Google Scholar]
- 54.Tateya T, Imayoshi I, Tateya I, Ito J, Kageyama R. Cooperative functions of Hes/Hey genes in auditory hair cell and supporting cell development. Dev Biol. 2011;352(2):329–40. 10.1016/j.ydbio.2011.01.038 [DOI] [PubMed] [Google Scholar]
- 55.Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127(21):4551–60. Epub 2000/10/12. [DOI] [PubMed] [Google Scholar]
- 56.Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, et al. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci. 2001;21(13):4712–20. Epub 2001/06/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leonova EV, Raphael Y. Organization of cell junctions and cytoskeleton in the reticular lamina in normal and ototoxically damaged organ of Corti. Hear Res. 1997;113(1–2):14–28. Epub 1997/12/05. [DOI] [PubMed] [Google Scholar]
- 58.Slepecky NB. Structure of the mammalian cochlea In: Dallos P, Popper AN, Fay RR, editors. The Cochlea. Springer Handbook of Auditory Research. New York: Springer-Verlag; 1996. p. 44–129. [Google Scholar]
- 59.Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134(16):3021–9. Epub 2007/07/20. 10.1242/dev.02874 [DOI] [PubMed] [Google Scholar]
- 60.Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of corti. J Neurosci. 2002;22(21):9368–77. Epub 2002/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, et al. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236(7):1905–17. Epub 2007/06/09. 10.1002/dvdy.21192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8(4):553–64. Epub 2005/04/06. 10.1016/j.devcel.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 63.Mansour SL, Li C, Urness LD. Genetic rescue of Muenke syndrome model hearing loss reveals prolonged FGF-dependent plasticity in cochlear supporting cell fates. Genes Dev. 2013;27(21):2320–31. PubMed Central PMCID: PMC3828518. 10.1101/gad.228957.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waldhaus J, Durruthy-Durruthy R, Heller S. Quantitative High-Resolution Cellular Map of the Organ of Corti. Cell Rep. 2015;11(9):1385–99. PubMed Central PMCID: PMC4465070. 10.1016/j.celrep.2015.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engel J, Braig C, Ruttiger L, Kuhn S, Zimmermann U, Blin N, et al. Two classes of outer hair cells along the tonotopic axis of the cochlea. Neuroscience. 2006;143(3):837–49. 10.1016/j.neuroscience.2006.08.060 [DOI] [PubMed] [Google Scholar]
- 66.Lelli A, Asai Y, Forge A, Holt JR, Geleoc GS. Tonotopic gradient in the developmental acquisition of sensory transduction in outer hair cells of the mouse cochlea. J Neurophysiol. 2009;101(6):2961–73. PubMed Central PMCID: PMC2694104. 10.1152/jn.00136.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mann ZF, Kelley MW. Development of tonotopy in the auditory periphery. Hear Res. 2011;276(1–2):2–15. Epub 2011/02/01. 10.1016/j.heares.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 68.Son EJ, Wu L, Yoon H, Kim S, Choi JY, Bok J. Developmental gene expression profiling along the tonotopic axis of the mouse cochlea. PLoS One. 2012;7(7):e40735 PubMed Central PMCID: PMC3395647. 10.1371/journal.pone.0040735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jansson L, Kim GS, Cheng AG. Making sense of Wnt signaling-linking hair cell regeneration to development. Front Cell Neurosci. 2015;9:66. PubMed Central PMCID: PMC4356074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77(1):58–69. 10.1016/j.neuron.2012.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin V, Golub JS, Nguyen TB, Hume CR, Oesterle EC, Stone JS. Inhibition of Notch activity promotes nonmitotic regeneration of hair cells in the adult mouse utricles. J Neurosci. 2011;31(43):15329–39. Epub 2011/10/28. 10.1523/JNEUROSCI.2057-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slowik AD, Bermingham-McDonogh O. Hair cell generation by notch inhibition in the adult mammalian cristae. J Assoc Res Otolaryngol. 2013;14(6):813–28. PubMed Central PMCID: PMC3825023. 10.1007/s10162-013-0414-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A): Surface preps of LfngGFP+ cochlear explants. Sensory epithelium retains its GFP expression after culture for 24 hours in either the gamma secretase inhibitor DAPT or DMSO vehicle control. (B): Diagram of the supporting cells recognized from the top of the epithelium. (C): Diagram of the experimental design. P0 or P5 cochleas were dissected and cultured. Explants were then dissociated and sorted for GFP fluorescence and the GFP+ and GFP- fractions used to make RNA-seq libraries. (D): Summary of differentially expressed transcript comparisons in each experimental condition.
(TIF)
For each gene, forward and reverse PCR primers are given, together with the predicted size of the band generated from PCR with mouse genomic DNA. All reverse primers contain a T7 polymerase sequence at their 5’ end (GGATCCTAATACGACTCACTATAGGGAG).
(XLSX)
Sheet 1 shows the analysis performed on the data with DESeq for both P1 and P6 cells. Sheet 2 shows the analysis performed with Cufflinks on P1 cells and Sheet 3 shows the same Cufflinks analysis on P6 cells.
(XLSX)
Sheet 1 shows the analysis performed on the data with DESeq for both P0 and P5 cells. Sheet 2 shows the analysis performed with Cufflinks on P0 cells and Sheet 3 shows the same Cufflinks analysis on P5 cells.
(XLSX)
The gene name is indicated, together with the expression level (reads per kilobase of transcript per million mapped reads; RPKM; DESeq output only) and its fold change compared to GFP- cells. p-adj = adjusted p-value for the difference between GFP+ and GFP- populations.
(DOCX)
Consensus lists of genes enriched in FACS sorted Lfng-GFP+ cells from postnatal day 1 (P1; 1884 genes) and postnatal day 6 (P6; 1278 genes) mouse cochlea compared to Lfng-GFP-negative cells. Analysis of the sequencing reads was performed by two different approaches. (1) Reads were mapped to the Mus musculus NCBI build37.2 iGenome (Ilumina) using TopHat 2.0 software (Trapnell et al., 2009; Trapnell et al., 2012) and the mapped reads were quantitated and compared using Cufflinks 2.0 providing differential gene expression data and statistics. (2) Reads were aligned to the Mus musculus Ensembl mm9 iGenome (Ilumina) using TopHat 1.4.1 software and the number of reads per gene and per library was obtained using DESeq program. After comparing the level of expression of each gene within each pair of related libraries (GFP+ versus GFP- for P1 and P6 cells), the most significant differentially expressed genes (DEG) were annotated and analyzed separately for both approaches. A consensus list of DEGs common to both methods of analysis was then generated. A significantly DEG was considered to have an RPKM higher than 3000, Fold Change (FC) higher than 4 and p value and FDR < 0.01. Duplicate samples of Lfng-GFP+ and GFP- sorted cells were prepared for P1 and P6. Approximately 60,000 sorted cells were as starting material to generate approximately 100–600 ng RNA (measured by Nanodrop spectrophotometer). cDNA libraries for RNAseq were generated using RNA Seq Truseq RNA sample preparation kit v2 (Illumina) following the low sample protocol for RNA extraction, cDNA synthesis, indexing and amplification. The quality and integrity of RNA samples and the final quality of the sequencing libraries was checked by electrophenogram in an Agilent Bioanalyzer. Paired-end sequencing was performed in HiSeq2000 sequencing platform (Illumina). Fastq files of paired end reads have been deposited in the NCBI GEO database, Accession No. GSE83357.
(XLS)
Consensus list of genes enriched in Lfng-GFP+ supporting cells that were differentially expressed between P1 and P6. Data was obtained from the analysis described in S5 Table caption above, but now genes enriched in supporting cells were compared for changes between P1 and P6.
(XLS)
For each gene, its expression at P1 and P6 (RPKM) together with the fold enrichment between GFP+ and GFP- cell populations is shown, together with expression pattern in the cochlea (SC, supporting cell; HC, hair cell; GER, greater epithelial ridge; SV, stria vascularis; Ubi, ubiquitous expression; No, no detectable signal; ND = not determined). NS: p>0.01.
(DOCX)
P0 Lfng-GFP cochlear cultures were maintained for 24 hours in DAPT or DMSO, followed by FACS sorting for GFP fluorescence. DESeq was used to identify transcripts enriched in cultures treated with either DAPT or DMSO. The table shows a list of 2088 genes enriched in Lfng-GFP+ cells that were significantly altered in DAPT-treated cultures compared to DMSO controls. Up-regulated transcripts after DAPT treatment are highlighted in red; down-regulated transcripts are highlighted in green. Duplicate samples of LfngGFP+ and GFP- sorted cells were prepared for postnatal day 0 (P0) and postnatal day 5 (P5) cultured for 24 hours in DMSO or DAPT (10uM). 10,000–20,000 sorted cells were used as starting material to generate approximately 100–600 ng RNA (measured by Nanodrop spectrophotometer). cDNA received an initial amplification using the NuGen Ovation Kit. cDNA libraries for RNAseq were generated using RNA Seq Truseq RNA sample preparation kit v2 (Illumina) following the low sample protocol for RNA extraction, cDNA synthesis, indexing and amplification. The quality and integrity of RNA samples and the final quality of the sequencing libraries was checked by electrophenogram in an Agilent Bioanalyzer. Paired-end sequencing was performed in HiSeq2000 sequencing platform (Illumina). Fastq files of paired end reads have been deposited in the NCBI GEO database, Accession No. GSE83357. Reads were aligned to the Mus musculus Ensembl mm9 iGenome (Ilumina) using TopHat 1.4.1 software and the number of reads per gene and per library was obtained using DESeq program. After comparing the level of expression of each gene within each pair of related libraries, the most significant differentially expressed genes (DEG) were annotated and analyzed separately for both approaches. In order to find genes significantly changing in isolated Lfng-EGFP+ cells obtained from cultured explants (treated or not with DAPT), the level of DEG significance was a FC higher than 2 and a p value and FDR < 0.01.
(XLS)
Data Availability Statement
Fastq files of paired end reads for all RNA-seq data have been deposited in the NCBI GEO database, Accession No. GSE83357.