Abstract
Gene therapy applications depend on vector delivery and gene expression in the appropriate target cell. Vector infection relies on the distribution of natural virus receptors that may either not be present on the desired target cell or distributed in a manner to give off-target gene expression. Some viruses display a very limited host range, while others, including herpes simplex virus (HSV), can infect almost every cell within the human body. It is often an advantage to retarget virus infectivity to achieve selective target cell infection. Retargeting can be achieved by (i) the inclusion of glycoproteins from other viruses that have a different host-range, (ii) modification of existing viral glycoproteins or coat proteins to incorporate peptide ligands or single-chain antibodies (scFvs) that bind to the desired receptor, or (iii) employing soluble adapters that recognize both the virus and a specific receptor on the target cell. This review summarizes efforts to target HSV using these three strategies.
Introduction
With the approval of Glybera (alipogene tiparvovec) in Europe [1,2] for the treatment of lipoprotein lipase deficiency and IMLYGIC (T-VEC, talimogene laherparepvec) [3,4] in the US [5] for the treatment of malignant melanoma, gene therapy is beginning to show promise as an approved alternative to drug and radiation therapies in the treatment of human disease. However, further progress will require the refinement of gene therapy approaches before it becomes a true everyday therapeutic. Current gene therapy applications, such as Glybera, often rely on tissue-specific promoters to limit therapeutic gene expression to specific cells. While this strategy helps minimize off-target effects, it does not ensure delivery of the viral vector to the intended cell or tissue. One strategy being employed to refine gene therapy is the use of transductional targeting to limit viral vector infection to only the desired target cell, a strategy called “retargeting”.
Limiting vector transduction to specific cell types can benefit many gene therapy applications, including those for chronic pain and cancer. For example, in pain gene therapy, most standard approaches fail to limit infection to specific peripheral nerve fiber subtypes. Ideally, one only wants to silence the activity of C-fiber neurons that are the nociceptors involved in pain sensation, while avoiding nerve fibers that regulate proprioception, pressure or itch so that they will function normally. Cancer gene therapy approaches have recently drawn interest due to the success of T-VEC. However, even that oncolytic herpesvirus (oHSV) is not restricted to which type of cells it can transduce, but is designed to selectively replicate in rapidly dividing tumor cells based on the absence of the viral ICP34.5 gene. As an alternative approach to limiting oHSV infection and lysis to tumor cells, our group and the groups of Campadelli-Fiume and Roizman have designed retargeted oHSV that preferentially infect breast or brain tumors. In this strategy, viral envelope glycoproteins that bind to widespread cellular receptors for HSV, are modified to ablate natural receptor binding and incorporate ligands or single-chain antibodies (scFv) that recognize the human epidermal growth factor (EGF) receptor 2 (HER2) [6,7], the interleukin-13 receptor IL-13Rα2 [8], or the EGF receptor (EGFR) [9,10] that are often over-expressed in these and other tumors.
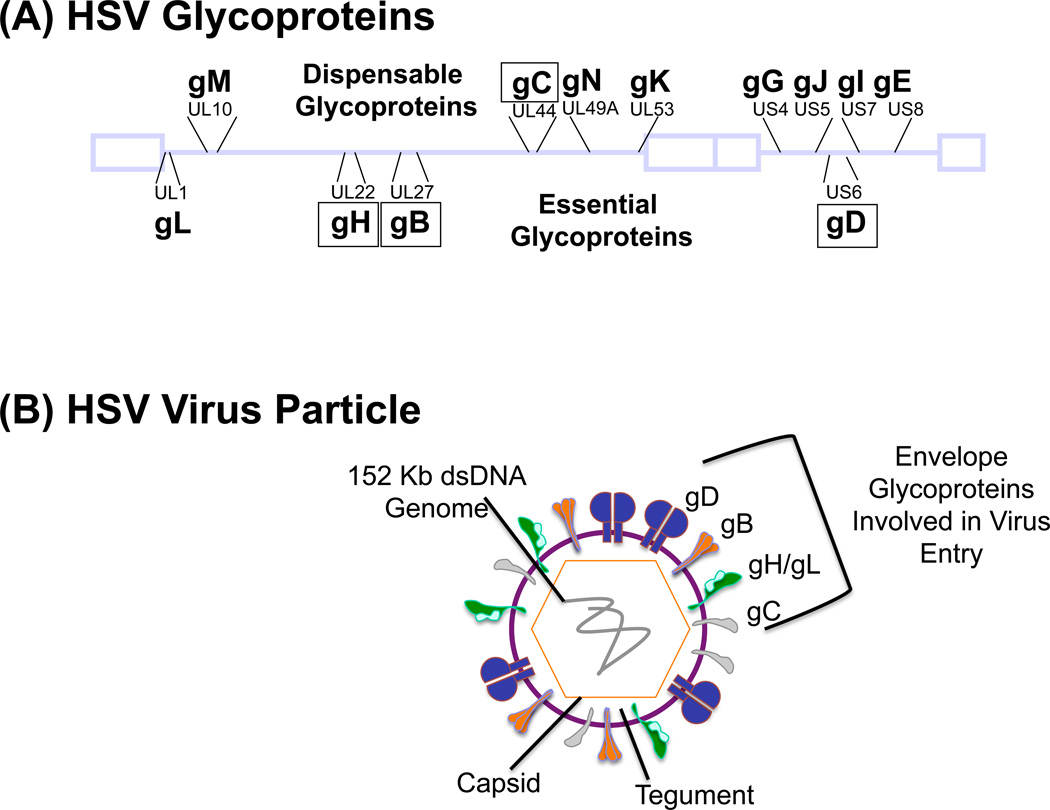
All viruses display a natural tropism for specific cell types, tissues and organs within the body. Tropism-determining factors include how the virus (i) encounters the host, (ii) attaches to host cell receptors that enable entry, (iii) establishes itself within the host, (iv) influences pathogenesis and disease, and (v) counteracts the host immune response. Retargeting can be performed to either expand the tropism of viruses that infect only a very limited number of cell types or, more commonly, to restrict the tropism of viruses that infect many cells of the host. Non-enveloped viruses employ one or more viral capsid proteins to interact with the host cell, while enveloped viruses use one or more glycoproteins to bind to and enter host cells. While many enveloped viruses, such as measles, influenza or HIV, employ a single or two glycoproteins to achieve cell binding and entry, members of the herpesvirus family rely on an array of glycoproteins to enter cells and spread from infected to uninfected cells. Herpes simplex virus (HSV) encodes 12 different glycoproteins (Figure 1A) and uses glycoproteins B, C, D, E, H, I, K and L for entry and cell-to-cell spread within the host (reviewed in [11–13]). Most of these glycoproteins contribute to viral tropism, making retargeting of HSV a distinct challenge. HSV retargeting is a worthwhile pursuit, however, given the attractive features HSV offers as a gene therapy platform, including a capacity to accommodate very large or multiple transgenes and infection without integration into host chromosomes.
Figure 1. HSV glycoproteins.
(a) The location of the HSV-1 encoded viral glycoproteins within the HSV-1 genome is depicted. Those listed below the viral genome are essential for virus replication, while those above it are non-essential and their deletion does not block virus replication in culture or in vivo. However, these dispensable glycoproteins can contribute to virus host range, pathogenesis and the host response to viral infection in vivo. Glycoprotein K (gK) is not found in the envelope of mature virus particles. Glycoproteins in boxes have been altered in efforts to retarget HSV. (b) The HSV particle is composed of an icosahedral-shaped capsid containing a 152-kb double-stranded linear DNA genome. The viral nucleocapsid is surrounded by an amorphous tegument layer consisting of both viral and cellular proteins. The virus envelope contains 12 viral glycoproteins, those essential for entry (gD, gB, gH/gL) and modified for vector retargeting (gD, gB, gC, gH) are depicted.
Years ago we performed some of the first retargeting studies with HSV by genetic fusion of full-length erythropoetin (EPO) to HSV glycoprotein C (gC) that had been N-terminally truncated to eliminate attachment of the virus to cellular heparan sulfate proteoglycans (HS or HSPG) in the background of a virus lacking the HS-binding region of gB [14]. Although this resulted in entry of the gC-EPO virus into EPO receptor-bearing cells, the virus was still able to enter cells that possess the natural receptors for HSV entry. Thus, we had expanded HSV’s tropism rather than restricting it to EPO receptor-bearing cells. In order to achieve full retargeting of HSV, virus interaction with the canonical HSV entry receptors must be blocked or eliminated and functionally replaced with alternate ligand-receptor interactions. The choice of alternate receptors is limited to candidates that are markers for the target cell and are recognized by peptide ligands or single-chain antibodies (scFvs) that preferentially do not activate the normal physiological function of the receptor. As summarized below, studies by a number of laboratories over the past 20 years have greatly enhanced out understanding of HSV entry, ultimately allowing the development of a first generation of fully retargeted HSV vectors.
HSV Attachment and Entry
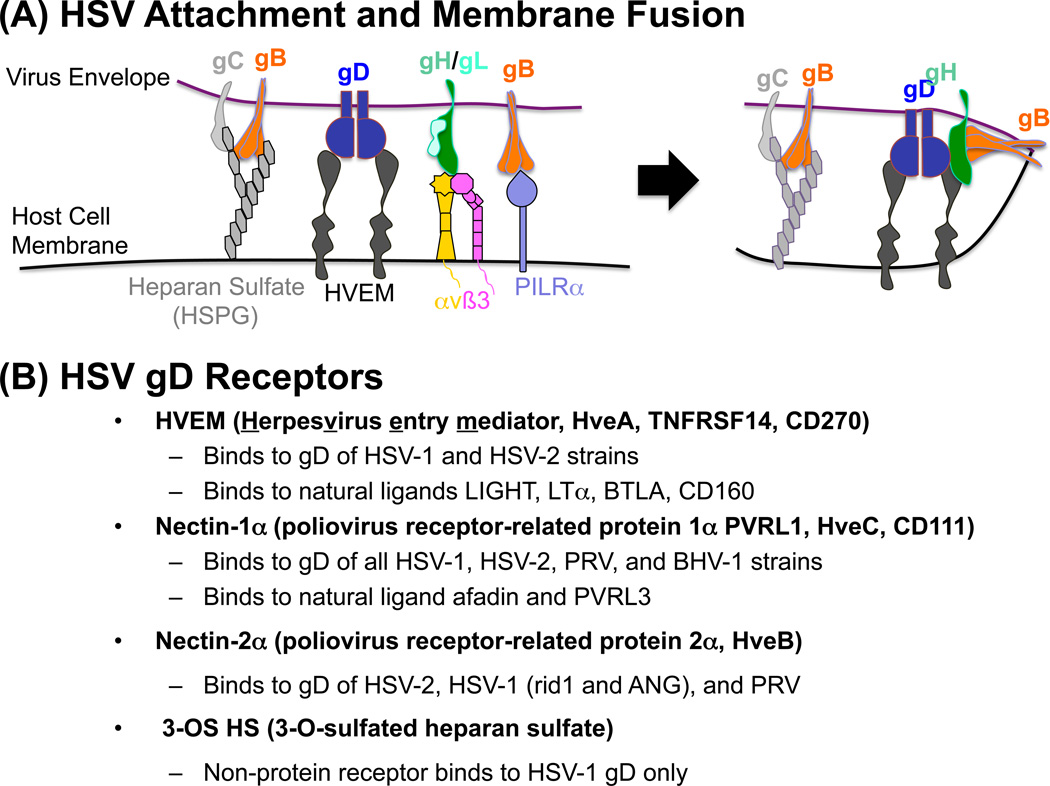
The HSV particle (Figure 1B) is composed of an icosahedral-shaped nucleocapsid containing the 152-kb double-stranded DNA genome (Figure 1A), an amorphous tegument layer consisting of viral and cellular proteins, and a lipid envelope acquired from the host cell that contains the 12 viral glycoproteins. The glycoproteins are grouped according to whether they are essential or non-essential for HSV entry [15–18]. Another group of important players in the HSV entry process are the cellular receptors that HSV normally engages to achieve attachment and entry. Initially, HSV gB and gC bind via arginine- and lysine-rich regions to negatively-charged HSPG present on almost all cells of the body (Figure 2A) [19–21]. This interaction enables virus surfing of the cell surface membrane [22] until HSV gD encounters one of its cognate receptors that are more restricted in their expression than HSPG, yet are present on numerous cell types (Figure 2A).
Figure 2. HSV-1 attachment and entry into host cells and the glycoprotein D cell surface receptors.
(a) Virus attachment to susceptible cells involves the movement of a virus particle along the host cell membrane surface until gC and gB encounter and bind to heparan sulfate proteoglycan (HSPG) moieties present on various glycosylated proteins on the cell surface that brings the virus into close proximity with the cell membrane in order for the virus to initiate binding to specific entry receptors. Diffusion of the virus along the cell surface results in contact of gD with either the TNF receptor superfamily member HVEM (HveA, TNFRSF14, CD270), the immunoglobulin superfamily member nectin-1 (PVRL1, HveC, CD111 receptor), or 3-O sulfated heparan sulfate. At this point gB can bind to PILRα while the gH/gL heterodimer engages integrins αvβ3, αvβ6 or αvβ6. Binding of gD to its cognate receptors leads to a conformational change in gD resulting in exposure of the C-terminal profusion or fusion-activating domain of gD. The exposed profusion domain of gD initiates fusion of the virus envelope with the cell membrane through signal transfer via gH to gB with concomitant gL release from the gH/gL heterodimer. Fusion results in release of the virus capsid into the cytoplasm of the cell. (b) Host cell receptors involved in the entry of alpha herpesviruses HSV-1, HSV-2, bovine herpes virus-1 (BHV-1), pseudorabies virus (PRV) as well as the HSV-1 rid1 and ANG mutants. HVEM, nectin-1 and -2 are present on almost all cell types of the human body as well as on most vertebrate cells except those of the testes and ovaries. The alpha herpesviruses that bind these receptors are indicated, as well as the natural ligands for these receptors.
HSV gD can bind to either (i) HVEM (TNFRSF14, HveA, CD270), a member of the TNF/NGF superfamily of receptors whose natural ligands include LIGHT, LTα, BTLA and CD160 [23], (ii) nectin-1 (PVRL1, HveC, CD111), a member of the immunoglobulin superfamily of receptors whose natural ligands include afadin and PVRL3 (nectin-3) [24], and (iii) 3-O-sulfated heparan sulfate (3-OS HS) generated by specific sulfotransferase family members, including 3-OST-3A/B [25,26] (Figure 2B). The region of gD containing the binding sites for these receptors has been mapped to the first ~230 residues of the protein and mutants with alterations in this region have enabled the identification of residues that are crucial to receptor binding [27–30]. In addition, mutation and deletion studies have defined a profusion domain beyond residue 260 that is required for virus entry [31,32], supporting the idea that gD represents the fusion trigger [33]. Refined knowledge of the interactions between gD and nectin-1 [34] or HVEM [35] has come from crystallography studies compared to un-liganded gD [36] and are consistent with the model that the gD-receptor interaction results in a conformational change that displaces the N-terminus of gD and exposes the gD pro-fusion domain [37,38]. The next step in the entry process involves transmission of the fusion signal from the fusion trigger (gD) to the fusion regulator (Figure 2A), the gH/gL heterodimer [12,33,39], whose structure has been defined by crystallography [40]. Additionally, it has been shown that gH can bind to integrins like αvβ3 [41] and αvβ6/αvβ8 [42], aiding in the formation of the fusion complex (Figure 2A). During the process of fusion complex formation, gL is displaced from the gH/gL heterodimer [33,39]. The final member of the fusion complex is gB (Figure 2A), which has been shown to bind to PILRα [71]. The crystal structure of gB [43,44] confirms that it is the true HSV fusogen, being the only HSV glycoprotein with sequence similarity to viral class III fusion proteins, in particular the vesicular stomatitis virus (VSV) G glycoprotein [45,46]. These final interactions within the fusion complex lead to exposure of the gB fusion loops [47], which enable fusion of the HSV envelope with the cell surface or endosomal membrane [33,39].
Retargeting of HSV Attachment/Entry
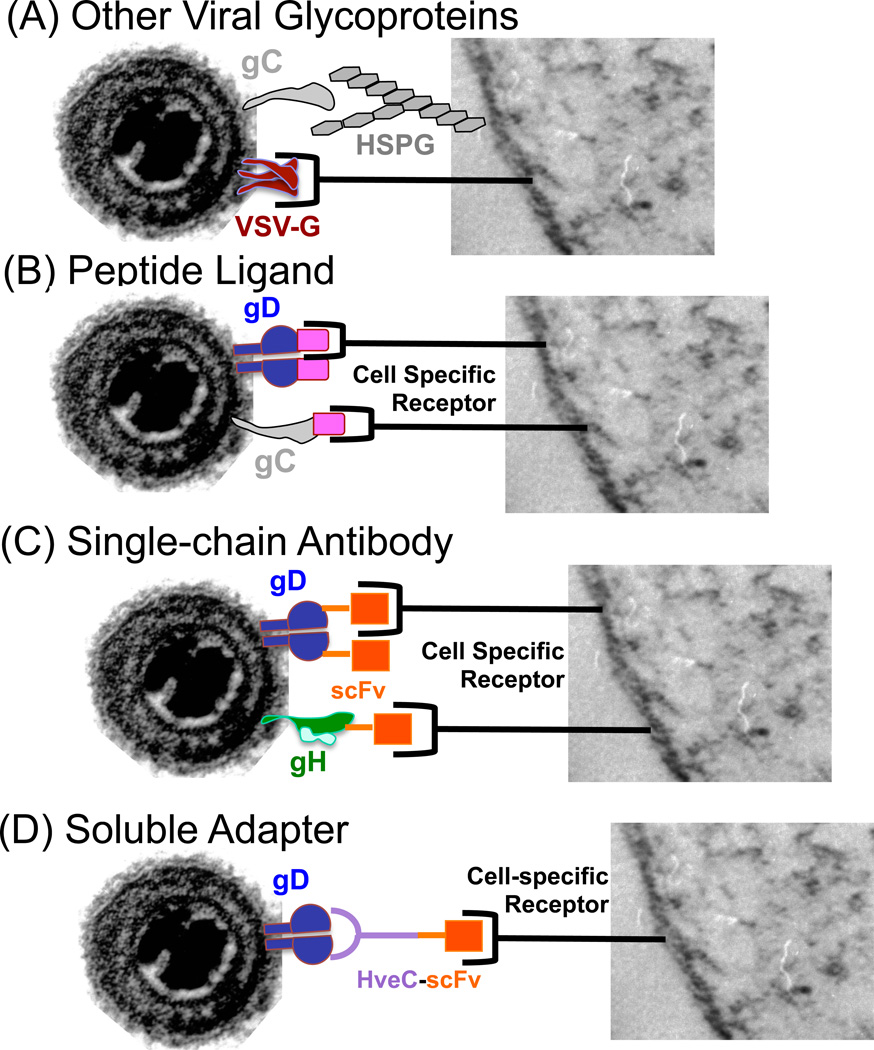
Because of the complex involvement of various HSV glycoproteins in the attachment and entry process, it has been a difficult task to completely retarget HSV, including the elimination of entry via the native HSV gD receptors while achieving levels of infection similar to those obtained with wild-type virus on susceptible cells. Multiple strategies have been employed (Figure 3 and Table 1) including (a) the use of pseudotyped virus replacing one or more HSV glycoproteins with those of another virus that has a different natural tropism, (b) the incorporation of peptide ligands into HSV gD or gC, (c) the incorporation of scFvs into gD, gC or gH, and (d) the use of adapter molecules that are capable of binding to both HSV and the desired target receptor.
Figure 3. Strategies employed for HSV vector retargeting.
(a) Introduction of glycoproteins from other viruses in place of HSV glycoproteins. The G-glycoprotein from Vesicular stomatitis virus (VSV-G) has been used to pseudotype various viruses and has been introduced into genomic and amplicon-based vectors lacking either gD or gB. VSV-G retargeted viruses could be neutralized by antibodies to VSV-G or VSV. (b) Peptide ligands for naturally occurring receptors have been introduced into HSV gD or gC, in most instances replacing canonical receptor-binding sequences of those glycoproteins. (c) Similar to the introduction of ligands, sequences encoding single-chain antibodies (scFvs) have been introduced into the HSV gD and recently gH genes to achieve retargeting. Introduction of either peptide ligands or scFvs into HSV gD with a combination of specific deletions and/or mutations have enabled complete virus retargeting producing entry of cells solely via the novel receptor target. (d) Soluble adapter molecules have also been employed and are composed of a dual interactive component, one capable of binding to HSV gD and the other to the cell surface receptor target. Virus must be mixed with the adapter prior to infection in order to attain entry via the specific target receptor.
Table 1.
HSV Retargeting Studies
| (A) Other Virus Glycoproteins | ||||||
|---|---|---|---|---|---|---|
| Virus | gD | gB | gC | gH/L | Ligand-Location | References |
| ΔUs3–8 | HveA−/HveC− * | Wt | Wt | Wt | VSV-G (gD) | [48] |
| Amplicon VSV-G | Wt | ΔgB | Wt | Wt | VSV-G (gB) | [49] |
| (B) Peptide Ligands | ||||||
|---|---|---|---|---|---|---|
| Virus | gD | gB | gC | gH/L | Ligand-Location | References |
| KgBpK:-gC-EPO | Wt | HS− | HS− * | Wt | EPO (gC) | [14] |
| KgBpK-:gC-preS1 | Wt | HS− | HS− * | Wt | HBV- sAg (gC) | [65] |
| R5111 | HveA+/HveC+ * | HS− | HS− * | Wt | IL--13 (gD) | [8,53,54] |
| R5141 | HveA−/HveC− * | HS− | HS− * | Wt | IL--13 (gD) | [8] |
| R5181 | HveA+/HveC+ * | HS− | HS− * | Wt | IL--13 (gC): uPA (gD) | [53] |
| Amplicon pCONGA-H | Wt | Wt | HS− * | Wt | HIS-tag (gC) | [66] |
| Amplicon pCONGA-MG11 | Wt | Wt | HS− * | Wt | MG11 peptide (gC) | [67] |
| Amplicon gC-BDNF | Wt | HS− | HS− * | Wt | pre-pro BDNF (gC) | [63,64] |
| Amplicon gC-GDNF | Wt | HS− | HS− * | Wt | pre-pro GDNF (gC) | [63,64] |
| Amplicon NMDA NR2A/2B | Wt | HS− | HS− * | Wt | NMDA NR2A/B Ab (gC) | [68,69] |
| (C) Single Chain Antibodies (scFvs) | ||||||
|---|---|---|---|---|---|---|
| Virus | gD | gB | gC | gH/L | Ligand-Location | References |
| KGNEp | HveA−/HveC− * | N/T | Wt | Wt | scFv EpCAM (gD) | [9] |
| KGNE | HveA−/HveC− * | N/T | Wt | Wt | scFv EGFR (gD) | [9,10] |
| KGNC | HveA−/HveC− * | Wt | Wt | Wt | scFv CEA (gD) | [10] |
| R-LM113 | HveA−/HveC+ * | Wt | Wt | Wt | scFv HER2 (gD) | [55,57,58,60] |
| R-LM249 | HveA−/HveC− * | Wt | Wt | Wt | scFv HER2 (gD) | [7,58,59] |
| R-VG809 | HveA−/HveC− | Wt | Wt | Wt * | scFv HER2 (gH) | [6] |
| R-LM31 | Wt * | Wt | Wt | Wt | scFv HER2 (gD) | [57] |
| R-LM11 | HveA−/HveC+ * | Wt | Wt | Wt | scFv HER2 (gD) | [56] |
| R-LM39 | HveA−/HveC+ * | Wt | Wt | Wt | scFv HER2 (gD) | [57] |
| HSV1716 scFv CD55 | HveA−/HveC− * | Wt | Wt | Wt | scFv CD55 (gD) | [70] |
| Amplicon pCONGA-MR1-1 | Wt | HS− | HS− * | Wt | scFv EGFR MR1-1 (gC) | [67] |
| (D) Adapters | ||||||
|---|---|---|---|---|---|---|
| Virus | gD | gB | gC | gH/L | Ligand-Location | References |
| HVEM:CEA Adapter | Wt | Wt | Wt | Wt | scFv CEA | [52] |
| Nectin1:EGFR Adapter | Wt | Wt | Wt | Wt | scFv EGFR | [51] |
| Nectin1 Adapter | Wt | Wt | Wt | Wt | nectin1-HveC | [50] |
denotes the presence of the retargeting molecule within the virus
Two early studies employed the VSV-G glycoprotein for “pseudotyping” HSV (Figure 3a and Table 1a), replacing HSV-1 gD [48] or gB [49]. Interestingly, when VSV-G was substituted for HSV gB, entry was seen at levels approaching that of wild-type HSV in rat striatum. These data are consistent with the structural similarity between VSV-G and HSV gB, apparently allowing VSV-G to act as the fusogen for HSV-1 entry. However, it is unknown whether the presence of gD in these particles still allows virus entry via the canonical gD receptors. In the gD replacement study, the pseudotyped viruses showed reduced entry efficiency and appeared to enter cells solely via the endocytic pathway, with the majority of virions being trapped in and degraded by the low pH of the endosomal pathway. Importantly, these studies illustrated that the HSV-1 entry machinery could be manipulated, replacing individual glycoproteins with functional alternatives, and infectious virions could be produced with the alternate proteins mediating entry into cells bearing the appropriate receptors.
Our group was the first to explore the use of adapter molecules to enable HSV retargeting (Figure 3d). The adapter protein consists of a portion that can bind to an HSV glycoprotein such as gD and a second component that is able to bind to a suitable target receptor. This adapter, or bridging protein, can be produced in bacteria or mammalian cells, purified using protein tags, and then added to virus just prior to infection of the target cells. Although this approach produced reasonable efficiency of retargeted cell transduction in vitro [50,51], it required meticulous purification of the adapter and precise determination of the timing of adapter addition to the virus prior to infection. A recent study by Baek and colleagues [52] employed a nectin-1:scFv anti-CEA adapter that was shown to increase the transduction efficiency of MKN45 flank tumors in nude mice compared to the no adapter control animals and produced a 3-fold reduction in tumor volume. While adapters that rely on intact nectin-1- or HVEM-binding sites in gD can mediate targeting to alternate receptors, detargeting from nectin-1 or HVEM can be sub-optimal due to incomplete occupancy of these binding sites by the adapter, allowing background entry through nectin-1 or HVEM. Furthermore, spread from initially infected cells requires either the presence of natural HSV-1 receptors on the uninfected acceptor cells or continued availability of the adapter.
The retargeting methods that have proven most efficacious have been the introduction of peptide ligands (Figure 3b and Table 1b) or scFvs (Figure 3c and Table 1c) into gD, or more recently gH. The majority of these studies have been performed to limit oHSV infection to only specific tumor cell types. The Roizman group has shown success in rendering virus entry uniquely dependent on the expression of IL-13Rα2 or the urokinase plasminogen activator receptor (uPAR) on target cells [8,53,54] by genetic incorporation of peptide ligands into both gC and gD. Retargeting of gC was aimed at increasing the selectivity of virus attachment while gD retargeting was expected to limit entry to target receptor-expressing cells. Despite promising results in vitro, the potential increased ability of these viruses to infect and kill glial tumor targets in animals has yet to be confirmed. The Campadelli-Fiume group has employed recombinant oHSV containing an anti-HER2 scFv in gD to target HER2-expressing tumor cells [7,55–60] in culture and in vivo. Early studies employed gD recombinants that could still enter cells by binding to the canonical gD receptor nectin-1 [55–57,60] and although they displayed good efficacies in ovarian and glioma tumor models in mice [55,57,60], exclusive targeting of the HER2 tumor receptor was not achieved. Subsequent reports from this group [7,58,59] employed oHSV that were mutationally inactivated for binding to nectin-1 and HVEM, resulting in sole entry into HER2-bearing tumor cells. These vectors displayed reduced toxicity when injected into the brains of nude mice, suggesting decreased off-target infection, and entry could be completely blocked using the Herceptin HER2 antibody, supporting the conclusion that these oHSV gained entry into tumor cells exclusively by retargeted gD interaction with cell-surface HER2.
Recent work from the Campadelli-Fiume group described an alternative approach to vector retargeting by the introduction of the anti-HER2 scFv into gH [6], either in a virus that retained the ability to enter via the canonical gD receptors or one lacking this ability. The results not only demonstrated HER2-specific entry of the anti-HER2 scFv-gH virus, but also established the possibility of virus entry via receptor engagement by a gH-based ligand in the absence of functional gD. This finding steps away from the established view of HSV entry, where gD binding to its receptor triggers signaling through gH to the fusogen gB to achieve fusion between the virus envelope and the cell membrane. Instead, it suggests that modifying gH to incorporate an alternative receptor binding function can circumvent the requirement for gD-receptor interactions. This finding opens up a range of possibilities in HSV targeting, allowing modification of not only gD, but also gH as complete retargeting strategies.
Studies in our own lab [10] and with collaborators [9] have succeeded in scFv-mediated oHSV retargeting to EGFR and its uniquely tumor-specific variant EGFRvIII [10], the carcinoembryonic antigen (CEA) [10] present on gastric cancer tumor cells and to epithelial cell adhesion molecule (EpCAM) found in abundance on colon cancer cells [9]. In these instances, the targeting scFv was introduced into gD modified to disrupt the binding sites for its natural receptors. Both recombinants included modifications in gB (gB:N/T) previously shown in our lab to dramatically increase the rate of HSV entry [61,62]. The inclusion of these mutations enabled target receptor-dependent entry of the EGFR, CEA and the EpCAM retargeted oHSV at levels approaching those of virus bearing wild-type gD into cells bearing a canonical gD receptor. The EGFR/EVFRvIII retargeted vector was shown to be effective in the treatment of an orthotopic glioblastoma multiforme (GBM) model employing primary human GBM cells [10].
Conclusions
Over the past two decades, the field of HSV retargeting has made tremendous progress. Much of our current ability to target infection to different receptors and to prevent infection of nectin-1 and HVEM expressing cells has come from a greater understanding of the contributions of individual amino acids and functional domains of gD, gC, gB, and gH/gL to the processes of attachment and entry. This knowledge has been substantially refined by resolution of the crystal structures of gD, nectin-1-bound gD, HVEM-bound gD, gB, and the gH/gL heterodimer. The field has also benefited from studies defining novel receptor-ligand pairs as well as the availability of scFvs that do not activate the target receptor, a concern that is most significant when using HSV as a vector for therapeutic gene delivery. While all completely retargeted HSV vectors reported to date are replication-competent oHSVs, the development of complementing cell lines expressing suitable target receptors will make this technology transferable to replication-defective HSV vectors that offer a large payload capacity for gene therapy. In recent years, the use of HSV BACs to rapidly generate novel recombinant viruses has dramatically affected HSV vector design, and CRISPR technology will further facilitate the creation and propagation of future generations of retargeted HSV vectors. Such progress will allow for more rapid evaluation of retargeting receptor-ligand pairs, including scFvs and similar targeting molecules, facilitate the identification of glycoprotein mutations that further enhance retargeted virus entry, and open new approaches toward optimization of HSV cell-to-cell spread. In the coming years, these and other efforts can be expected to bring retargeted HSV vectors closer to clinical applicability.
Highlights.
Understanding the contributions of HSV gD, gC, gB, and gH/gL in attachment and entry
Complete elimination of HSV attachment and entry via natural receptors
Selection of glycoprotein mutations increasing entry of retargeted HSV
Efficient retargeting using single chain antibodies or ligands in HSV gD and gH
Acknowledgments
The work on HSV retargeting in the Glorioso group was sponsored by grants CA119298, HL66949, NS544323, CA163205, DK44935, and AR50733 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests
JBC and JCG are co-inventors of intellectual property licensed to Oncorus, Inc. (US patent applications 13/641,649s and 15/032,958). JCG is a founder of Oncorus, Inc. and Chairman of the Scientific Advisory Board. WFG is a consultant for Oncorus, Inc.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Salmon F, Grosios K, Petry H. Safety profile of recombinant adeno-associated viral vectors: Focus on alipogene tiparvovec (Glybera(R)) Expert Rev Clin Pharmacol. 2014;7(1):53–65. doi: 10.1586/17512433.2014.852065. [DOI] [PubMed] [Google Scholar]
- 2.Yla-Herttuala S. Endgame: Glybera finally recommended for approval as the first gene therapy drug in the european union. Mol Ther. 2012;20(10):1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, Milhem M, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 4.Andtbacka RH, Ross M, Puzanov I, Milhem M, Collichio F, Delman KA, Amatruda T, Zager JS, Cranmer L, Hsueh E, Chen L, et al. Patterns of clinical response with talimogene laherparepvec (T-VEC) in patients with melanoma treated in the optim phase III clinical trial. Ann Surg Oncol. 2016 doi: 10.1245/s10434-016-5286-0. {Epub ahead of print} [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pol J, Kroemer G, Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. Oncoimmunology. 2016;5(1):e1115641. doi: 10.1080/2162402X.2015.1115641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gatta V, Petrovic B, Campadelli-Fiume G. The engineering of a novel ligand in gH confers to HSV an expanded tropism independent of gD activation by its receptors. PLoS Pathog. 2015;11(5):e1004907. doi: 10.1371/journal.ppat.1004907. •• This paper was the first report of insertion of the retargeting sequences, the HER2 scFv, into gH both in the background of wild-type and detargeted gD. Infection of SK-OV-3 ovarian tumor cells in culture could be completely blocked using herceptin antibody against HER2. This suggests that gD-receptor interaction can be abrogated as long as specific binding still occurs via gH.
- 7. Leoni V, Gatta V, Palladini A, Nicoletti G, Ranieri D, Dall'Ora M, Grosso V, Rossi M, Alviano F, Bonsi L, Nanni P, et al. Systemic delivery of HER2-retargeted oncolytic-HSV by mesenchymal stromal cells protects from lung and brain metastases. Oncotarget. 2015;6(33):34774–34787. doi: 10.18632/oncotarget.5793. •• This work employed a completely detargeted/retargeted oHSV with the scFv to HER2 inserted in the 60–218 amino acid deleted region of gD. In animal tumor studies the virus inhibited lung and brain metastases when the virus was delivered in mesenchymal stem cell (MSCs) as carriers.
- 8. Zhou G, Roizman B. Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13alpha2 receptor. Proc Natl Acad Sci U S A. 2006;103(14):5508–5513. doi: 10.1073/pnas.0601258103. • This work details the retargeting of the IL-13 receptor using an HSV recombinant virus in which the IL-13 ligand was introduced into the HS-binding region of gC as well as the N-terminus of gD that had been completely detargeted from the native gD receptors. The same ligand inserted into two different HSV glycoproteins could help aid in retargeting.
- 9. Shibata T, Uchida H, Shiroyama T, Okubo Y, Suzuki T, Ikeda H, Yamaguchi M, Miyagawa Y, Fukuhara T, Cohen JB, Glorioso JC, et al. Development of an oncolytic HSV vector fully retargeted specifically to cellular EpCAM for virus entry and cell-to-cell spread. Gene Ther. 2016;23(6):479–488. doi: 10.1038/gt.2016.17. •• This manuscript highlights the use of a completely gD receptor detargeted oHSV possessing the gB:N/T ROE mutations that enable efficient retargeting to EpCAM- or EGFR-bearing tumor cells depending on the scFv employed. The EpCAM-retargeted oHSV will not infect and kill glioma tumors (EGFR+) while the EGFR-retargeted virus is unable to infect and lyse the colon cancer tumors (EpCAM+) further enforcing the specifcity of the retargeting.
- 10. Uchida H, Marzulli M, Nakano K, Goins WF, Chan J, Hong CS, Mazzacurati L, Yoo JY, Haseley A, Nakashima H, Baek H, et al. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol Ther. 2013;21(3):561–569. doi: 10.1038/mt.2012.211. •• This was the first report using the gB:N/T (D285N/A549T) compensatory ROE mutations in combination of a fully detargeted/retargeted gD to achieve efficient killing of human GBM30 glioma cell tumors in nude mice even assessed by MRI. Biodistribution analysis using this detargeted/EGFR-retargeted oHSV demonstrated the homing of the virus to the tumor volume in tumor-bearing compared to normal mice when administered by intravenious tail vein injection further attesting to specificity.
- 11.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: A story with many characters. Viruses. 2012;4(5):800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karasneh GA, Shukla D. Herpes simplex virus infects most cell types in vitro: Clues to its success. Virol J. 2011;8(481) doi: 10.1186/1743-422X-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spear PG. Herpes simplex virus: Receptors and ligands for cell entry. Cell Microbiol. 2004;6(5):401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- 14.Laquerre S, Anderson DB, Stolz DB, Glorioso JC. Recombinant herpes simplex virus type 1 engineered for targeted binding to erythropoietin receptor-bearing cells. J Virol. 1998;72(12):9683–9697. doi: 10.1128/jvi.72.12.9683-9697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai WH, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62(8):2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai PJ, Schaffer PA, Minson AC. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: Evidence that gH is essential for virion infectivity. J Gen Virol. 1988;69(Pt 6):1147–1156. doi: 10.1099/0022-1317-69-6-1147. [DOI] [PubMed] [Google Scholar]
- 17.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66(4):2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligas MW, Johnson DC. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62(5):1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold BC, Visalli RJ, Susmarski N, Brandt CR, Spear PG. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol. 1994;75(Pt 6):1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 20.Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72(7):6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trybala E, Bergstrom T, Svennerholm B, Jeansson S, Glorioso JC, Olofsson S. Localization of a functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulphate. J Gen Virol. 1994;75(Pt 4):743–752. doi: 10.1099/0022-1317-75-4-743. [DOI] [PubMed] [Google Scholar]
- 22.Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing. Biochem Biophys Res Commun. 2010;391(1):176–181. doi: 10.1016/j.bbrc.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87(3):427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 24.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280(5369):1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 25.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99(1):13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari V, O'Donnell C, Copeland RJ, Scarlett T, Liu J, Shukla D. Soluble 3-O-sulfated heparan sulfate can trigger herpes simplex virus type 1 entry into resistant chinese hamster ovary (CHO-K1) cells. J Gen Virol. 2007;88(Pt 4):1075–1079. doi: 10.1099/vir.0.82476-0. [DOI] [PubMed] [Google Scholar]
- 27.Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci U S A. 2007;104(47):18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchida H, Shah WA, Ozuer A, Frampton AR, Jr, Goins WF, Grandi P, Cohen JB, Glorioso JC. Generation of herpesvirus entry mediator (HVEM)-restricted herpes simplex virus type 1 mutant viruses: Resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J Virol. 2009;83(7):2951–2961. doi: 10.1128/JVI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce de Leon M, Peng T, Nicola AV, Montgomery RI, Warner MS, Soulika AM, et al. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71(8):6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon M, Zago A, Shukla D, Spear PG. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J Virol. 2003;77(17):9221–9231. doi: 10.1128/JVI.77.17.9221-9231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc Natl Acad Sci U S A. 2004;101(19):7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou G, Roizman B. Separation of receptor-binding and profusogenic domains of glycoprotein D of herpes simplex virus 1 into distinct interacting proteins. Proc Natl Acad Sci U S A. 2007;104(10):4142–4146. doi: 10.1073/pnas.0611565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: A structural view of the herpesvirus entry machinery. Nat Rev Microbiol. 2011;9(5):369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 2011;7(9):e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8(1):169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 36.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 2005;24(23):4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gallagher JR, Saw WT, Atanasiu D, Lou H, Eisenberg RJ, Cohen GH. Displacement of the C terminus of herpes simplex virus gD is sufficient to expose the fusion-activating interfaces on gD. J Virol. 2013;87(23):12656–12666. doi: 10.1128/JVI.01727-13. • This paper details the overall change in gD structure post receptor binding to the gD amino terminus that leads to the displacement of the C-terminus as a way of communicating the fusion signal to the gB-gH/gL complex.
- 38. Lazear E, Whitbeck JC, Zuo Y, Carfi A, Cohen GH, Eisenberg RJ, Krummenacher C. Induction of conformational changes at the N-terminus of herpes simplex virus glycoprotein D upon binding to HVEM and nectin-1. Virology. 2014;448:185–195. doi: 10.1016/j.virol.2013.10.019. • This paper details a comparison of the mutational analyses of the HVEM and Nectin-1 binding domains of gD with the actual crystal structure of receptor-bound and free gD. It provides a greater understanding of the conformational change in gD upon engaging the two different cellular gD rreceptors.
- 39.Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, Eisenberg RJ. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. 2010;84(8):3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17(7):882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parry C, Bell S, Minson T, Browne H. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol. 2005;86(Pt 1):7–10. doi: 10.1099/vir.0.80567-0. [DOI] [PubMed] [Google Scholar]
- 42.Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM, Campadelli-Fiume G. Alphavbeta6-and alphavbeta8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog. 2013;9(12):e1003806. doi: 10.1371/journal.ppat.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313(5784):217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 44. Zeev-Ben-Mordehai T, Vasishtan D, Hernandez Duran A, Vollmer B, White P, Prasad Pandurangan A, Siebert CA, Topf M, Grunewald K. Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc Natl Acad Sci U S A. 2016;113(15):4176–4181. doi: 10.1073/pnas.1523234113. • This work employs electron cryotomography to examine the differences in pre- and post-fusion crystallographic forms of gB helping to detail the conformational change in gB enabling fusion of the viral envelop with the host cell or endosomal membrane.
- 45.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313(5784):187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 46.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315(5813):843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 47.Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81(9):4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson DB, Laquerre S, Ghosh K, Ghosh HP, Goins WF, Cohen JB, Glorioso JC. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables mutant virus attachment and entry. J Virol. 2000;74(5):2481–2487. doi: 10.1128/jvi.74.5.2481-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang J, Yang T, Ghosh HP, Geller AI. Helper virus-free HSV-1 vectors packaged both in the presence of vsv g protein and in the absence of HSV-1 glycoprotein B support gene transfer into neurons in the rat striatum. J Neurovirol. 2001;7(6):548–555. doi: 10.1080/135502801753248132. [DOI] [PubMed] [Google Scholar]
- 50.Kwon H, Bai Q, Baek HJ, Felmet K, Burton EA, Goins WF, Cohen JB, Glorioso JC. Soluble V domain of nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J Virol. 2006;80(1):138–148. doi: 10.1128/JVI.80.1.138-148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakano K, Asano R, Tsumoto K, Kwon H, Goins WF, Kumagai I, Cohen JB, Glorioso JC. Herpes simplex virus targeting to the EGF receptor by a gD-specific soluble bridging molecule. Mol Ther. 2005;11(4):617–626. doi: 10.1016/j.ymthe.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Baek H, Uchida H, Jun K, Kim JH, Kuroki M, Cohen JB, Glorioso JC, Kwon H. Bispecific adapter-mediated retargeting of a receptor-restricted HSV-1 vector to CEA-bearing tumor cells. Mol Ther. 2011;19(3):507–514. doi: 10.1038/mt.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamiyama H, Zhou G, Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13(7):621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 54.Zhou G, Roizman B. Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13ralpha2 receptor of malignant glioma cells. J Virol. 2005;79(9):5272–5277. doi: 10.1128/JVI.79.9.5272-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gambini E, Reisoli E, Appolloni I, Gatta V, Campadelli-Fiume G, Menotti L, Malatesta P. Replication-competent herpes simplex virus retargeted to HER2 as therapy for high-grade glioma. Mol Ther. 2012;20(5):994–1001. doi: 10.1038/mt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menotti L, Cerretani A, Campadelli-Fiume G. A herpes simplex virus recombinant that exhibits a single-chain antibody to HER2/neu enters cells through the mammary tumor receptor, independently of the gD receptors. J Virol. 2006;80(11):5531–5539. doi: 10.1128/JVI.02725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Menotti L, Cerretani A, Hengel H, Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J Virol. 2008;82(20):10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Menotti L, Nicoletti G, Gatta V, Croci S, Landuzzi L, De Giovanni C, Nanni P, Lollini PL, Campadelli-Fiume G. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc Natl Acad Sci U S A. 2009;106(22):9039–9044. doi: 10.1073/pnas.0812268106. • This paper employs a gD-detargeted HER2-retargeted oHSV with the scFv to HER2 inserted into the Ig fold of gD (amino acids 61–218). Tumor studies in nude mice showed 60% survival of SK-OV-3 ovarian tumor-bearing mice injected with the virus out to seven months. Moreover, the oHSV proved to be non-toxic in normal nude mice.
- 59. Nanni P, Gatta V, Menotti L, De Giovanni C, Ianzano M, Palladini A, Grosso V, Dall'ora M, Croci S, Nicoletti G, Landuzzi L, et al. Preclinical therapy of disseminated HER-2(+) ovarian and breast carcinomas with a HER-2-retargeted oncolytic herpesvirus. PLoS Pathog. 2013;9(1):e1003155. doi: 10.1371/journal.ppat.1003155. • Interperitoneal injection of the HER2 retargeted virus produced an increase in the medium survival time of time of nude mice bearing SK-OV-3 ovarian tumors and 20% survival of mice possessing breast cancer metastases out to 100 days post-injection.
- 60.Reisoli E, Gambini E, Appolloni I, Gatta V, Barilari M, Menotti L, Malatesta P. Efficacy of HER2 retargeted herpes simplex virus as therapy for high-grade glioma in immunocompetent mice. Cancer Gene Ther. 2012;19(11):788–795. doi: 10.1038/cgt.2012.62. [DOI] [PubMed] [Google Scholar]
- 61.Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, Cohen JB, Glorioso JC. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol. 2010;84(23):12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uchida H, Chan J, Shrivastava I, Reinhart B, Grandi P, Glorioso JC, Cohen JB. Novel mutations in gB and gH circumvent the requirement for known gD receptors in herpes simplex virus 1 entry and cell-to-cell spread. J Virol. 2013;87(3):1430–1442. doi: 10.1128/JVI.02804-12. •• Using a novel selection methodology, this work detailed the isolation of viral mutations in gB fusogenic molecule and gH fusion effector gene product that compensated for weak signaling via gD. Thus dependence on a gD-receptor interaction to provide the fusion signal to the gB-gH/gL complex can be compensated by alterations of the amino acid sequences of gB and gH.
- 63.Cao H, Zhang GR, Wang X, Kong L, Geller AI. Enhanced nigrostriatal neuron-specific, long-term expression by using neural-specific promoters in combination with targeted gene transfer by modified helper virus-free HSV-1 vector particles. BMC Neurosci. 2008;9(37) doi: 10.1186/1471-2202-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Kong L, Zhang GR, Sun M, Geller AI. Targeted gene transfer to nigrostriatal neurons in the rat brain by helper virus-free HSV-1 vector particles that contain either a chimeric HSV-1 glycoprotein C-GDNF or a gC-BDNF protein. Brain Res Mol Brain Res. 2005;139(1):88–102. doi: 10.1016/j.molbrainres.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Argnani R, Boccafogli L, Marconi PC, Manservigi R. Specific targeted binding of herpes simplex virus type 1 to hepatocytes via the human hepatitis B virus preS1 peptide. Gene Ther. 2004;11(13):1087–1098. doi: 10.1038/sj.gt.3302266. [DOI] [PubMed] [Google Scholar]
- 66.Grandi P, Wang S, Schuback D, Krasnykh V, Spear M, Curiel DT, Manservigi R, Breakefield XO. HSV-1 virions engineered for specific binding to cell surface receptors. Mol Ther. 2004;9(3):419–427. doi: 10.1016/j.ymthe.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 67.Grandi P, Fernandez J, Szentirmai O, Carter R, Gianni D, Sena-Esteves M, Breakefield XO. Targeting HSV-1 virions for specific binding to epidermal growth factor receptor-vIII-bearing tumor cells. Cancer Gene Ther. 2010;17(9):655–663. doi: 10.1038/cgt.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao H, Zhang GR, Geller AI. Antibody-mediated targeted gene transfer to NMDA NR1-containing neurons in rat neocortex by helper virus-free HSV-1 vector particles containing a chimeric HSV-1 glycoprotein C-staphylococcus A protein. Brain Res. 2010;1351:1–12. doi: 10.1016/j.brainres.2010.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao H, Zhang GR, Geller AI. Antibody-mediated targeted gene transfer of helper virus-free HSV-1 vectors to rat neocortical neurons that contain either NMDA receptor 2B or 2A subunits. Brain Res. 2011;1415:127–135. doi: 10.1016/j.brainres.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conner J, Braidwood L, Brown SM. A strategy for systemic delivery of the oncolytic herpes virus HSV1716: Redirected tropism by antibody-binding sites incorporated on the virion surface as a glycoprotein D fusion protein. Gene Ther. 2008;15(24):1579–1592. doi: 10.1038/gt.2008.121. [DOI] [PubMed] [Google Scholar]
- 71.Saitoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear PG, Lanier LL, Arase H. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132(6):935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]