Abstract
The prevalence of enterococci was examined in 280 milk samples collected from dairy cattle diagnosed with mastitis in three provinces of western China. Sixty strains of enterococci were isolated, and the species were determined based on their biochemical characters and 16S rRNA sequences. Resistance to seven antibiotic agents, frequency of seven virulence genes and pathogenicity in Kunming mice were tested to evaluate biological risks. The correlation between the number of virulence genes and pathogenicity in Kunming mice was also evaluated. The 60 isolates were allocated to Enterococcus hirae (68.3%), E. faecium (25.0%), E. mundtii (3.3%) and E. durans (3.3%). A total of 83.3% of the isolates were resistant to penicillin, whereas 15.0% were resistant to ampicillin, 15.0% to vancomycin, 6.7% to tetracycline and 25.0% to ciprofloxacin. Moreover, isolates exhibited 50.0% and 21.7% resistance to high levels of gentamycin and streptomycin, respectively. The gene asa1 was detected in all enterococcal isolates, whereas 66.7% of strains harbored three or more virulence factors and 56.7% were asa1-ccf-gelE-positive. In pathogenicity tests, isolates harboring numerous virulence factors did not show greater invasiveness than isolates harboring fewer virulence traits against Kunming mice. In conclusion, the number of virulence factors does not appear to predict the risk of enterococcal infection. Isolates were commonly resistant to penicillin and sporadically to ampicillin and vancomycin. These results suggest that the use of gentamycin, streptomycin and ciprofloxacin against enterococci should be avoided in mastitic cows. Additionally, the results demonstrate that the majority of isolates are sensitive to tetracycline.
Keywords: antibiotic resistance, pathogenic enterococci, prevalence, virulence
Multidrug-resistant enterococci have become increasingly common in human and veterinary medicine and are causing a worldwide health crisis. Enterococci are commonly found in animal intestines and their feces [10, 11] and were considered commensal organisms until the late 1970s when antibiotics were widely used in both humans and animals [4]. It is thought that oral administration of antibiotic agents eliminated sensitive enterococci and other commensal bacteria, allowing minority strains that survived to become the dominant flora. The dominance of resistant enterococci facilitated their contact with cow teats, increasing the chance of their entry into milk ducts, particularly in cows reared in captivity. Enterococci surviving mucosal immunity can cause clinical or subclinical mastitis [32]. Enterococcus species gained notoriety, because of their dramatic increase in infectious diseases, such as endocarditis, dental surgical infection and urinary tract infection, as well as for their antibiotic resistance and ability to use novel routes of evasion [38, 39]. These organisms are also been widely present in the food processing industry [20, 25, 29]. Because enterococci from various origins facilitate the evolution of resistance, their survival tactics, virulence profile, antibiotic resistance and multiple avenues of evading antibiotic treatment should be closely examined. The prevalence and mechanisms of antibiotic resistance and virulence profiles on enterococci from human medicine have been widely examined [9, 23, 30, 33], whereas reports on enterococci in animal disease are limited, particularly reports concerning enterococcal dairy cow mastitis in China. How an environmental species of bacteria becomes a mastitis-causing pathogen should be further analyzed. Although production of milk and the diversity of milk products continue to increase, the virulence profile of enterococci and their toxins remain unclear. The control of pathogens during dairy production and processing is very important. In the present study, we investigated the virulence profile, antibiotic patterns and pathogenicity of enterococci isolated from mastitic milk and evaluated their effects on dairy cows and public health.
MATERIALS AND METHODS
Bacterial isolates and growth conditions: A total of 280 mastitic milk samples were collected from dairy cows with clinical or subclinical mastitis on six large farms distributed in Gansu province (161), Qinghai province (30) and Ningxia Hui Autonomous Region (89) between November 2011 and May 2012. Briefly, 5–8 ml of milk were collected from each mastitic cow (clinically confirmed mastitis or suspected mastitis with positive Lanzhou mastitis test results) by trained workers immediately after teats were cleaned using towels and disinfected with 2% iodine tincture and 75% ethanol. After sampling, the cows were treated according to individual protocols. All samples were maintained on ice and transported to the laboratory within 6 hr.
Biochemical tests: Based on enterococci’s unique characteristics, 100 µl of each sample was first applied on a Todd-Hewitt selective plate (THBSP) [14, 22, 34], in which isolates able to hydrolyze esculin appeared as black colonies. Plates were incubated at 37°C for 24–48 hr for bacterial enrichment, and then, microbiological procedures were conducted according to standard guidelines [17, 18]. Briefly, 68 gram-positive strains survived THBSP screening, appearing singly, in pairs or as short chains, and were subjected to cAMP and hemolysis reactions before cultivation on plates containing esculin, hippurate salt, 4% bile salt, 6.5% NaCl, plus sorbitol, lactose, trehalose and inulin. From the previous step, 60 isolates showing negative results in the cAMP test, which survived in 6.5% NaCl broth, were further tested for their utilization of pyruvate, l-arabinose, d-raffinose and survival at 45°C and in glucose broth containing 1% potassium tellurite. All suspected “enterococcal” strains were stored at −70°C in Todd-Hewitt broth containing 15% (v/v) glycerol.
Virulence gene analysis and 16S ribosomal DNA sequencing: From 2.5 ml of samples cultivated overnight in Todd-Hewitt broth, genomic DNA was prepared using a commercial DNA isolation kit obtained from Forgene Co., Ltd. (Forgene, Chengdu, China), according to the manufacturer’s protocol. The PCR mixture contained 25 µl 2× PCR master mix (Promega, Madison, WI, U.S.A.), 2 µl (400 nM) of each primer, 2 µl DNA template and 21 µl deionized water, and amplification was conducted in a DNA thermal cycler (Bio-Rad, Hercules, CA, U.S.A.).
All suspected enterococcal isolates were screened for their 16S rRNA and seven virulence genes via PCR with the primers shown in Table 1. The seven virulence genes examined encode enterococcal surface protein (esp), gelatinase (gelE), sex pheromone (ccf), cytolysin activator component A (CylA), aggregation substance (asa1), collagen binding protein (ace) and aggregation protein (agg). Primers were synthesized as previously described and were as follows: CylA [12], gelE, ccf, agg [13], asa1 and esp [35], ace [6] and 16S rRNA [37]. Enterococcus faecalis ATCC 51299 and ATCC 29212 (obtained from Nanjing Bianzhen Biotechnology Ltd., Nanjing, China) were adopted for quality control. Oligo synthesis and PCR product sequencing were conducted by Sangon Co., Ltd. (Sangon, Shanghai, China). Homology of the 16S rRNA and virulence sequences of these isolates were queried on the NCBI nucleotide database using the basic local alignment search tool [24] between September 17, 2014 and September 25, 2014. Enterococci characterization was confirmed according to the biochemical characteristics and 16S rRNA sequences [21, 37].
Table 1. Primers for virulence gene screening and 16S rDNA sequencing.
| Virulence factors | Primer (5´-3´) | Product (bp) length |
|---|---|---|
| CylA | ATGGATGGGACAGATGGAAA | 519 |
| AGCTGCGCTTACTTCTGGAG | ||
| GelE | ACCCCGTATCATTGGTTT | 419 |
| ACGCATTGCTTTTCCATC | ||
| Ccf | GGGAATTGAGTAGTGAAGAAG | 543 |
| AGCCGCTAAAATCGGTAAAAT | ||
| Agg | AAGAAAAAGAAGTAGACCAAC | 1553 |
| AAACGGCAAGACAAGTAAATA | ||
| Asa1 | GCACGCTATTACGAACTATGA | 378 |
| TAAGAAAGAACATCACCACGA | ||
| Esp | AGATTTCATCTTTGATTCTTGG | 510 |
| AATTGATTCTTTAGCATCTGG | ||
| Ace | 6GGAATGACCGAGAACGATGGC | 616 |
| GCTTGATGTTGGCCTGCTTCCG | ||
| 16S rDNA | AGAGTTTGATCCTGGCTCAG | around 1492 |
| TACGGCTACCTTGTTACGACTT |
Pathogenicity test on Kunming mice: Kunming mice were used for pathogenicity tests. These mice were derived from a pair of Swiss mice from Hoffline Institution in 1944 and show strong disease resistance and adaptability as well as high reproduction and survival rates [40]. Isolates showing positive reactions to ≥3 virulence factors were designated as genetically virulent isolates (GVI), whereas those positive to ≤2 virulence factors were designated as genetically non-virulent isolates (NGVI). As shown in Table 2, five GVI (MS 38, MS 39, MS 42, MS 46 and MS 62) harboring four virulence genes and five NGVI isolates (MS 7, MS 10, MS 14, MS 17 and MS 50) harboring no more than two virulence genes were chosen for pathogenicity tests on Kunming mice (detailed information of the virulence screening results are shown in Supplement 1). To confirm the pathogenic, yet non-lethal, dose of overnight enterococcal culture for Kunming mice, an overnight culture of an isolate harboring esp-gelE-asa1 (MS 4) was graded as 0.1 ml, 0.2 ml, 0.3 ml, 0.4 ml and 0.5 ml and inoculated in 15 mouse subjects (three mice in each group). After the pre-experiment, 0.3 ml overnight culture from the five GVI and five NGVI groups was inoculated peritoneally into 30 healthy female mice (three mice in each group, all mice were aged 6–8 weeks). Additional three mice were injected with only 0.3 ml physiological saline and served as controls. The day of inoculation was defined as day 0, after which the mice were observed for 15 days. All experimental mice were obtained from a guaranteed animal experimental center (Gansu University of Chinese Medicine, Gansu, China) and strictly housed and handled ethically in the Animal Center of Gansu Agriculture University, according to the guidelines of the Animal Ethical and Welfare Committee of the College of Veterinary Medicine at Gansu Agricultural University. After 15 days of incubation, living mice were humanely sacrificed according to the center’s guidelines. All livers, hearts and kidneys were anatomically and microbiologically examined; contaminated materials and corpses were handled according to the guidelines of the Animal Ethical and Welfare Committee at Gansu Agricultural University.
Table 2. Virulence profile of the inoculated isolates.
| Esp | GelE | Ccf | CylA | Asa1 | Ace | Agg | Virulence count | Identification | ||
|---|---|---|---|---|---|---|---|---|---|---|
| MS7 | - | + | - | - | + | - | - | 2 | E. hirae | NGVI |
| MS10 | - | - | - | - | + | - | - | 1 | E. hirae | NGVI |
| MS14 | - | + | - | - | + | - | - | 2 | E. hirae | NGVI |
| MS17 | - | - | - | - | + | - | - | 1 | E. hirae | NGVI |
| MS50 | - | - | + | - | + | - | - | 2 | E. faecium | NGVI |
| MS38 | - | + | + | - | + | + | - | 4 | E. hirae | GVI |
| MS39 | - | + | + | - | + | + | - | 4 | E. hirae | GVI |
| MS42 | - | + | + | + | + | - | - | 4 | E. hirae | GVI |
| MS46 | + | + | - | - | + | + | - | 4 | E. durans | GVI |
| MS62 | + | + | + | - | + | - | - | 4 | E. faecium | GVI |
“+++”, indicates isolates that were positive to asa1, ccf and gelE. GVI, genetically virulent isolates. NGVI, non-genetically virulent isolates.
Antibiotic resistance tests: Antibiotic resistance tests were performed using the microdilution method according to the National Committee on Clinical Laboratory Standards (NCCLS) [3]. The evaluated antibiotics included penicillin (pen), ampicillin (amp), vancomycin (van), tetracycline (tet), ciprofloxacin (cip), high-level gentamycin (gen) and streptomycin (str). Antibiotics were obtained from Sangon Co., Ltd. (Sangon, Shanghai, China), and all antibiotics were used at United States Pharmacopeia Grade. Supplement 2 shows the explanations of the results, standards and antibiotic gradients. Overnight bacterial culture was adjusted to 0.5 McFarland turbidity with the medium used to dilute the antibiotics, inoculated into gradient antibiotics plates and incubated at 35 ± 2°C for 18–24 hr. The results were assessed as previously described [3], and isolates resistant to ≥3 classes of the antibiotic drugs applied were categorized as multiple-drug-resistant isolates [1].
RESULTS
Biochemical results: Initially, 280 mastitic milk samples were enrolled for this experiment, of which 212 samples were excluded based on the following criteria: (1) gram-negative stained, (2) negative for the THBSP screen, (3) rod bacteria or (4) combined infection. (These excluded strains not described in this study, but subjected to further identification, including 45 Staphylococcus aureus strains, 13 Escherichia coli strains and nine Hafnia alvei strains; data not published.) Eight isolates were identified as Streptococcus spp. based on their biochemical characteristics. The other 60 gram-positive cocci that survived THBSP, tested negative in the cAMP test and survived in 6.5% NaCl were initially suspected to be Enterococcus spp., but their biochemical characteristics could not be used to identify the isolates at the species level. Thus, 16S rRNA and representative virulence gene sequencing were adopted for further characterization [8, 37].
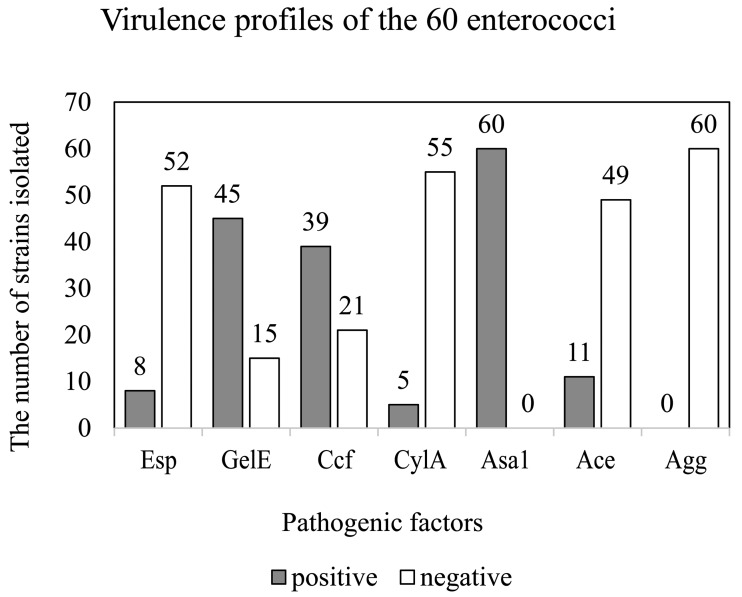
16S rRNA sequencing and virulence factors: In combined biochemical tests, 16S rRNA and virulence gene sequencing confirmed the survival of 60 gram-positive strains under THBSP selection, comprising Enterococcus hirae (n=41), E. faecium (n=15), E. mundtii (n=2) and E. durans (n=2). Table 3 shows the six virulence genes detected in each species, whereas Fig. 1 shows the frequency of positive and negative enterococcal isolate ratios to the six virulence genes.
Table 3. Frequencies of virulence factor of the 60 enterococcal isolates.
| Species | Esp | GelE | Ccf | CylA | Asa1 | Ace | Agg |
|---|---|---|---|---|---|---|---|
| E. hirae | 5 | 31 | 28 | 5 | 41 | 9 | 0 |
| E. faecium | 2 | 11 | 9 | 0 | 15 | 1 | 0 |
| E. mundtii | 0 | 2 | 2 | 0 | 2 | 0 | 0 |
| E. durans | 1 | 1 | 0 | 0 | 2 | 1 | 0 |
| Total | 8 | 45 | 39 | 5 | 60 | 11 | 0 |
| Positive rate | 13.30% | 75.00% | 65.00% | 8.30% | 100.00% | 18.30% | 0 |
Fig. 1.
Virulence patterns of the 60 enterococci isolates from mastitic dairy cows in Gansu province and Ningxia Hui autonomous region. Esp, enterococcus surface protein; gelE, gelatinase; ccf, sex pheromone; CylA, cytolysin activator component A; asa1, aggregation substance; ace, collagen binding protein; agg, aggregation protein.
All 60 isolates were positive for asa1, whereas the rates of positive reaction for the other virulence genes were as follows: gelE (45, 75%), ccf (39, 65%), ace (11, 18.3%), esp (8, 13.3%) and CylA (5, 8.3%). Agg was not detected in our isolates. Of the 60 (66.7%) isolates, 40 were GVI [1], and 34 of the GVI were asa1-gelE-ccf-positive. The other 20 (33.3%) isolates were NGVI.
Pathogenicity: The health status of GVI- and NGVI-inoculated mice declined rapidly in the 24 hr after inoculation, although most mice gradually recovered in 72 hr. The five mice in the quality control group remained healthy throughout the experiment. The peak of loss among subject mice occurred on days 4 and 5. On day 4, one mouse died in the GVI group, and seven died in the NGVI group. On day 5, one mouse died in the GVI group, and two mice died in the NGVI group. On day 8, one mouse died in the NGVI group. There were no further losses on days 0–3, 6, 7 and 9–14. Under examination, gram-positive cocci of common morphology with the inoculated enterococcal isolates were detected in the blood, kidney and liver samples of the deceased subjects. No combined infections were detected. For the virulence genes, five NGVI were all negative to esp, ace and agg (Table 2). Isolates in the GVI group harbored all virulence factors that were positive in the NGVI isolates (asa1, gelE and ccf), but GVI did not cause a greater loss of Kunming mice than NGVI. In contrast, NGVI caused a greater loss of Kunming mice than GVI. However, the clinical signs and pathological changes of the two groups were similar.
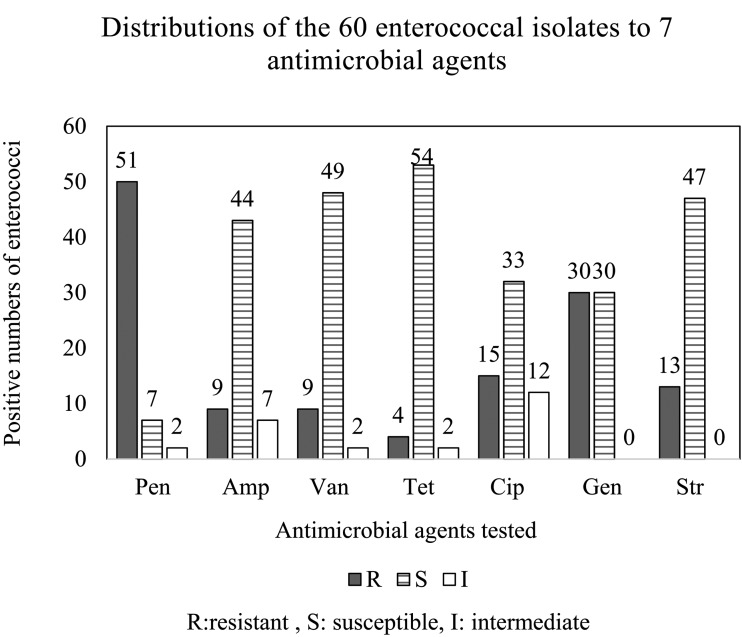
Antibiotic resistance tests: Antibiotic results were evaluated according to the NCCLS [3]. As shown in Fig. 2 and Table 4, the most prevalent resistance was to β-lactams, with 51 of 60 (85.0%) isolates resistant to penicillin and nine of 60 (15.0%) to ampicillin. Considerable resistance to glycosides was also observed, with nine of 60 (15.0%) isolates resistant to vancomycin, which is considered a drug of last resort for enterococcal infection. Among the isolates, 15 of 60 (25.0%) were resistant to ciprofloxacin in the quinolone class, and four of 60 (6.7%) isolates were resistant to tetracycline. For aminoglycosides, ten isolates were resistant to both high-level gentamycin and streptomycin. In addition, 30 (50.0%) and 13 (21.7%) of 60 isolates were respectively resistant to gentamycin and streptomycin. In analysis of antibiotic, the susceptibility results revealed 17 multiple-drug-resistant isolates.
Fig. 2.
Frequency of the 60 enterococcal isolates to seven antibiotic agents. Pen: penicillin. Amp: ampicillin. Van: vancomycin. Tet: tetracycline. Cip: ciprofloxacin. Gen: gentamicin. Str: streptomycin.
Table 4. Resistance of the four enterococcal species (n=60) to seven antibiotic agents.
| E. hirae | E. faecium | E. mundtii | E. durans | Total R | |||||
|---|---|---|---|---|---|---|---|---|---|
| n=45 | n=15 | n=2 | n=2 | ||||||
| I | R | I | R | I | R | I | R | ||
| Pen | 2 | 38 | 0 | 11 | 0 | 1 | 0 | 1 | 51 (85%) |
| Amp | 6 | 6 | 1 | 2 | 0 | 0 | 0 | 1 | 9 (15%) |
| Van | 2 | 7 | 0 | 2 | 0 | 0 | 0 | 0 | 9 (15%) |
| Tet | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 4 (6.7%) |
| Cip | 10 | 11 | 2 | 3 | 0 | 0 | 0 | 1 | 15 (25%) |
| Gena) | 0 | 25 | 0 | 4 | 0 | 0 | 0 | 1 | 30 (50%) |
| Strb) | 0 | 12 | 0 | 1 | 0 | 0 | 0 | 0 | 13 (21.7%) |
a) high-level gentamycin resistance (500 µg/ml). b) high-level streptomycin resistance (1,000 µg/ ml).
DISCUSSIONS
In human clinics, E. faecalis is the most commonly reported species and is present in many mammals at similar frequency [19]. However, E. hirae has rarely been reported as one of the main bacterial species in clinical dairy cow mastitis or contaminated milk products. How this species evolved as to become a dominant pathogen causing bovine mastitis has remained unclear.
Asa1 and ccf, both coded by sex-pheromone plasmid pAD1, were originally reported early only in connection with E. faecalis [15], whereas later studies confirmed that E. faecium also harbors the asa1 sequence [26, 35], suggesting that pAD1 and the sex pheromone-plasmid-exchanging system is not an exclusive trait of E. faecalis; other enterococcal species may also function in this manner. The gelE gene is necessary, but not sufficient for gelatinase activity. Gelatinase activity was reported to be co-controlled by the genes gelE and fsr, and a lack of fsr was inadequate to produce gelatinase [16, 28, 36]. Additionally, the high level of gelatinase detected in milk samples in cows with mastitis supports the hypothesis that gelatinase promotes Enterococcus survival and invasion in host tissues by facilitating Enterococcus migration. Because 40 of the 60 enterococcal isolates harbor more than three virulence genes, we propose that multiple virulence properties confer enterococci with greater survival probability against the innate bovine immune system.
The seven virulence genes tested in this study were reported to contribute to enterococci virulence, and the NGVI were positive for esp (0), gelE (2), ccf (1), cylA (0) and ace (0). The pathogenicity to Kunming mice showed that the NGVI still caused a greater loss of Kunming mice than the GVI, which were positive for esp (2), gelE (4), ccf (4), cylA (1) and ace (3). Asa1 was positive in all in these isolates, whereas agg was negative in all isolates. The described virulence factors are insufficient for determining the prognosis of an enterococcal infection in our mouse model, suggesting that unknown mechanisms or virulence factors participated in their pathogenicity in Kunming mice. No obvious differences were found in hemolysis properties; there was one strain with β-hemolysis and two stains with α and γ in both the GVI and NGVI, respectively. However, these enterococcal strains were all isolated from mastitic cows, indicating that the strains had become well adapted to survive in the mammary tissue of dairy cows. The lethal effect in mice appeared to occur through different mechanisms.
Enterococcal isolates in this study showed a high frequency of resistance to penicillin, as well as a relatively high ratio of resistance to large doses of gentamycin and streptomycin. Resistance to penicillin (85.0%) was greater than 64.8% (68/105), vancomycin resistance was 15.0% to 0, ampicillin 15.0% to 0.9% (1/105), and high-level gentamycin resistance was 50.0 to 30.5% (32/105) as reported by Cortés, et al. [5]. Notably, vancomycin had never been used at the source farms, yet resistant strains were detected. Vancomycin-resistant enterococci were also reported in dust from pig breeding facilities (15%, 26/171) [36], wild Eurasian otter (17.2%, 5/29) [37], and poultry feed and feed ingredients (1.9%, 8/414) [7]. These results agree with our findings. Resistance to tetracycline was 6.7% (4/60), which was lower than that reported by Cortés et al. and da Costa et al., who respectively reported 60% (63/105) and 18% (31/171) resistance [5, 7]. Resistance to ciprofloxacin was 25% (15/60), higher than that found in broiler feed at 3.9% (16/414) [7]. Ciprofloxacin is considered to have only modest antibacterial activity against enterococci [27] and thus is not the first choice for treating enterococcal infection. Schaberg also reported that an increasing number of ciprofloxacin-resistant cases were accompanied by high-level gentamycin resistance [31], which is similar to our findings. High-level aminoglycoside resistance should be further examined, as aminoglycoside resistance reduces the synergetic action of combined penicillin-aminoglycoside treatment, which would result in a selective elimination of anaerobic bacteria parallel to an increase in enterococci [2].
Although virulence genes and antibiotic resistance have been detected in milk samples, Enterococcus spp. have been safely present in milk products for centuries. However, when considering the safety of enterococci in food or probiotic use, strict monitoring mechanisms are necessary to guarantee consistent safety for consumers.
In conclusion, enterococci are emerging as one of the main species causing dairy cow mastitis in Gansu province and the surrounding region. This is the first report of enterococcal mastitis in western China and the first report of a virulence factor combination, asa1-ccf-gelE, found in mastitis originating from enterococcal strains. Based on our results, the seven examined virulence genes (asa1, ccf, gelE, esp, CylA, ace and agg) are inadequate for determining the prognosis of enterococcal infection. However, more work is needed to reveal the mechanism of enterococci pathogenesis. Tetracycline remains one of the most effective drugs for treating these isolates, followed by glycopeptides and new generations of β-lactams. Although few reports have demonstrated the threat of enterococcal to public health, strict and guaranteed levels of enterococci in the food and probiotic industries are necessary.
Supplementary Material
REFERENCES
- 1.Barigye R., Gautam A., Piche L. M., Schaan L. P., Krogh D. F., Olet S.2012. Prevalence and antimicrobial susceptibility of virulent and avirulent multidrug-resistant Escherichia coli isolated from diarrheic neonatal calves. Am. J. Vet. Res. 73: 1944–1950. doi: 10.2460/ajvr.73.12.1944 [DOI] [PubMed] [Google Scholar]
- 2.Chow J. W.2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31: 586–589. doi: 10.1086/313949 [DOI] [PubMed] [Google Scholar]
- 3.Cockerill F. R.2012. Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement; [... provides updated tables for... M02-A11 and M07-A9]. National Committee for Clinical Laboratory Standards. [Google Scholar]
- 4.Cohen M. L.1992. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science 257: 1050–1055. doi: 10.1126/science.257.5073.1050 [DOI] [PubMed] [Google Scholar]
- 5.Cortés C., De la Fuente R., Contreras A., Sánchez A., Corrales J. C., Ruiz-Santa-Quiteria J. A., Orden J. A.2006. Occurrence and preliminary study of antimicrobial resistance of enterococci isolated from dairy goats in Spain. Int. J. Food Microbiol. 110: 100–103. doi: 10.1016/j.ijfoodmicro.2006.01.033 [DOI] [PubMed] [Google Scholar]
- 6.Creti R., Imperi M., Bertuccini L., Fabretti F., Orefici G., Di Rosa R., Baldassarri L.2004. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 53: 13–20. doi: 10.1099/jmm.0.05353-0 [DOI] [PubMed] [Google Scholar]
- 7.da Costa P. M., Oliveira M., Bica A., Vaz-Pires P., Bernardo F.2007. Antimicrobial resistance in Enterococcus spp. and Escherichia coli isolated from poultry feed and feed ingredients. Vet. Microbiol. 120: 122–131. doi: 10.1016/j.vetmic.2006.10.005 [DOI] [PubMed] [Google Scholar]
- 8.David J.1991. 16S/23S rRNA sequencing. pp 115–144. In: Nucleic acid Techniques in Bacterial Systematics, Wiley, New York. [Google Scholar]
- 9.De Angelis G., Cataldo M. A., De Waure C., Venturiello S., La Torre G., Cauda R., Carmeli Y., Tacconelli E.2014. Infection control and prevention measures to reduce the spread of vancomycin-resistant enterococci in hospitalized patients: a systematic review and meta-analysis. J. Antimicrob. Chemother. 69: 1185–1192. doi: 10.1093/jac/dkt525 [DOI] [PubMed] [Google Scholar]
- 10.Devriese L. A., Hommez J., Laevens H., Pot B., Vandamme P., Haesebrouck F.1999. Identification of aesculin-hydrolyzing streptococci, lactococci, aerococci and enterococci from subclinical intramammary infections in dairy cows. Vet. Microbiol. 70: 87–94. doi: 10.1016/S0378-1135(99)00124-8 [DOI] [PubMed] [Google Scholar]
- 11.Devriese L., Van de Kerckhove A., Kilpper-Bälz R., Schleifer K.1987. Characterization and identification of Enterococcus species isolated from the intestines of animals. Int. J. Syst. Bacteriol. 37: 257–259. doi: 10.1099/00207713-37-3-257 [DOI] [Google Scholar]
- 12.Drahovská H., Slobodníková L., Kocíncová D., Seman M., Konceková R., Trupl J., Turňa J.2004. Antibiotic resistance and virulence factors among clinical and food enterococci isolated in Slovakia. Folia Microbiol. (Praha) 49: 763–768. doi: 10.1007/BF02931562 [DOI] [PubMed] [Google Scholar]
- 13.Eaton T. J., Gasson M. J.2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67: 1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facklam R. R.1973. Comparison of several laboratory media for presumptive identification of enterococci and group D streptococci. Appl. Microbiol. 26: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galli D., Lottspeich F., Wirth R.1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4: 895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x [DOI] [PubMed] [Google Scholar]
- 16.Galloway-Peña J. R., Bourgogne A., Qin X., Murray B. E.2011. Diversity of the fsr-gelE region of the Enterococcus faecalis genome but conservation in strains with partial deletions of the fsr operon. Appl. Environ. Microbiol. 77: 442–451. doi: 10.1128/AEM.00756-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia L. S.2013. Clinical microbiology procedures handbook, 3rd ed., American Society for Microbiology Press, California. [Google Scholar]
- 18.Huochun Y.2006. Veterinary Microbiology Experiment Manual. 2nd ed., Agricultural Press, Beijing. [Google Scholar]
- 19.Klein G.2003. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88: 123–131. doi: 10.1016/S0168-1605(03)00175-2 [DOI] [PubMed] [Google Scholar]
- 20.Lauková A., Marcináková M., Strompfová V., Ouwehand A. C.2008. Probiotic potential of enterococci isolated from canine feed. Folia Microbiol. (Praha) 53: 84–88. doi: 10.1007/s12223-008-0012-3 [DOI] [PubMed] [Google Scholar]
- 21.Lee I. M., Hammond R. W., Davis R. E., Gundersen D. E.1993. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology 83: 834–842. doi: 10.1094/Phyto-83-834 [DOI] [Google Scholar]
- 22.Litsky W., Mallmann W. L., Fifield C. W.1953. A new medium for the detection of enterococci in water. Am. J. Public Health Nations Health 43: 873–879. doi: 10.2105/AJPH.43.7.873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupis F., Giordano S., Pampinella D., Scarlata F., Romano A.2009. [Infective endocarditis: review of 36 cases]. Infez. Med. 17: 159–163. [PubMed] [Google Scholar]
- 24.Mount D. W.2007. Using the basic local alignment search tool (BLAST). Cold Spring Harbor Protocols 2007:pdb. top17. [DOI] [PubMed]
- 25.Nueno-Palop C., Narbad A.2011. Probiotic assessment of Enterococcus faecalis CP58 isolated from human gut. Int. J. Food Microbiol. 145: 390–394. doi: 10.1016/j.ijfoodmicro.2010.12.029 [DOI] [PubMed] [Google Scholar]
- 26.Padmasini E., Divya G., Karkuzhali M., Padmaraj R., Ramesh S. S.2014. Distribution of cylA, esp, asa1, hyl and gelE virulence genes among clinical isolates of Enterococcus faecium and Entrococcus faecalis. BMC Infect. Dis. 14suppl 3: 32. doi: 10.1186/1471-2334-14-s3-p3224428855 [DOI] [Google Scholar]
- 27.Perry J. D., Ford M., Gould F. K.1994. Susceptibility of enterococci to ciprofloxacin. J. Antimicrob. Chemother. 34: 297–298. doi: 10.1093/jac/34.2.297 [DOI] [PubMed] [Google Scholar]
- 28.Qin X., Singh K. V., Weinstock G. M., Murray B. E.2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183: 3372–3382. doi: 10.1128/JB.183.11.3372-3382.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saavedra L., Taranto M. P., Sesma F., de Valdez G. F.2003. Homemade traditional cheeses for the isolation of probiotic Enterococcus faecium strains. Int. J. Food Microbiol. 88: 241–245. doi: 10.1016/S0168-1605(03)00186-7 [DOI] [PubMed] [Google Scholar]
- 30.Savini V., Gherardi G., Astolfi D., Polilli E., Dicuonzo G., D’Amario C., D’Antonio D.2012. Insights into airway infections by enterococci: a review. Recent Pat. Anti-Infect. Drug Discovry 7: 36–44. [DOI] [PubMed] [Google Scholar]
- 31.Schaberg D. R., Dillon W. I., Terpenning M. S., Robinson K. A., Bradley S. F., Kauffman C. A.1992. Increasing resistance of enterococci to ciprofloxacin. Antimicrob. Agents Chemother. 36: 2533–2535. doi: 10.1128/AAC.36.11.2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder J. W.1997. Bovine mastitis and milking management [accessed 08 November 2012]. https://www.ag.ndsu.edu/pubs/ansci/dairy/as1129.pdf.
- 33.Sujatha S., Praharaj I.2012. Glycopeptide resistance in gram-positive cocci: a review. Interdiscip. Perspect. Infect. Dis. 2012: 781679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Switzer R. E., Evans J. B.1974. Evaluation of selective media for enumeration of group D streptococci in bovine feces. Appl. Microbiol. 28: 1086–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vankerckhoven V., Van Autgaerden T., Vael C., Lammens C., Chapelle S., Rossi R., Jabes D., Goossens H.2004. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42: 4473–4479. doi: 10.1128/JCM.42.10.4473-4479.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waters C. M., Antiporta M. H., Murray B. E., Dunny G. M.2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185: 3613–3623. doi: 10.1128/JB.185.12.3613-3623.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J.1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wirth R.1995. The sex pheromone system of Enterococcus faecalis. EJB Reviews 1994: 117–128. [DOI] [PubMed] [Google Scholar]
- 39.Wirth R.2000. Sex pheromones and gene transfer in Enterococcus faecalis. Res. Microbiol. 151: 493–496. doi: 10.1016/S0923-2508(00)00163-7 [DOI] [PubMed] [Google Scholar]
- 40.Zhang G. M., Yao G. H.1997. A survey on the genetic background document of Chinese Kunming mouse (KM mouse). Chin. J. Lab. Anim. Sci. 7: 246–251. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.