ABSTRACT
Background: PD-L1 has been widely reported as immune check points in a range of malignancies as well as some immune-originated diseases. In glioma, the role of PD-L1 remains unclear. We aimed at investigating its role at transcriptome level and relationship with clinical practice.
Method and patients: In total, 976 glioma samples with transcriptome data, including 301 microarray data from Chinese Glioma Genome Atlas (CGGA project) and 675 RNAseq data from TCGA project, were enrolled into our study. Clinical and IDH mutation data were also available. R language was used as the main tool for statistical analysis and graphical work.
Results: PD-L1 expression was found to be positively correlated with WHO grade of glioma. PD-L1 seemed to express more in mesenchymal subtype according to TCGA transcriptional classification scheme and may contribute as a potential marker for mesenchymal subtype in glioblastoma. Pearson correlation test indicated that PD-L1 showed robust correlation with PD1, PD-L2, and CD80 in CGGA dataset. Subsequent gene ontology analysis based on significantly correlated genes of PD-L1 revealed that PD-L1 seemed to be profoundly associated with T cell activation. To further investigate the relationship between PD-L1 expression and immune response, we selected a series of immune signatures, which were then transformed into metagenes, and found that PD-L1 expression was particularly paralleled with T-cells and macrophages-related immune response instead of B cell linage-related immune response. In line with the corresponding biological process, PD-L1 exhibited predictive value for glioma patients: Higher PD-L1 indicated significantly shorter survival, especially in glioblastoma.
Conclusion: PD-L1 is upregulated in glioblastoma, and is synergistic with other check point members. Moreover, PD-L1 is significantly associated with T-cell activation and macrophage-related immune response and predicts much worse survival for patients, warranting clinical trials of PD1/PD-L1 checkpoint inhibitors for potential glioma treatment.
KEYWORDS: Checkpoint inhibitors, glioma, immune response, PD-L1
Introduction
Gliomas are the most prevalent and devastating primary brain tumors in adults. Despite improvements in the standard of care, patients who suffer from glioblastoma, the most aggressive type, only have a median survival time of 15 mo.1,2 A substantial body of work has also pointed to a growing recognition of the interplay between glioma and immunity.3-5
Programmed death-ligand 1 (PD-L1), also termed as cluster of differentiation 274 (CD274) or B7 homolog 1 (B7-H1), is widely reported across a range of malignant tumors,6,7 including breast cancer,8 T-cell lymphoblastic lymphomas,9 non-small cell lung cancer,10,11 primary central nervous system lymphomas,12 non-clear-cell renal cell carcinoma,13 gastrointestinal stromal tumors,14 metastatic bladder cancer,15 etc. PD-L1 is a transmembrane protein that is supposed to play a vital role in blocking the cancer immunity cycle during particular events such as tumorigenesis, pregnancy, autoimmune disease, and other disease states. Many studies sought to determine a correlation between the expression level PD-L1 and survival and have yielded relatively congruent results. In malignant tumors, patients who have over-expression of PD-L1 in their tumors usually live a relatively shorter survival than those with relatively lower PD-L1 expression.10,16,17 Thus, therapies targeting PD1/PD-L1 have been increasing greatly in the past decade,18,19 aiming at restoring the immune activity in tumors, but with inconsistent responding rate across different tumors.9,20
Heretofore, we failed to find a single comprehensive report about PD-L1 expression in whole WHO grade gliomas. Only one study presented by Nduom et al. 21 described PD-L1 expression in glioblastoma, which accounted for about 40% of glioma. To explore the PD-L1 expression status in all glioma, we took advantage of Chinese Glioma Genome Atlas (CGGA) data set, including RNAseq data and mRNA microarray data of whole grade glioma. To further validate what we have revealed in CGGA dataset, we obtained RNAseq data of glioma from TCGA network (http://cancergenome.nih.gov/) and found that the consistency of results between two cohorts was fairly satisfying. This is the first integrative study characterizing PD-L1 expression in whole grade glioma molecularly and clinically.
Methods
Sample and data collection
In CGGA data set, we have collected transcriptome data of 301 samples, ranging from WHO grade II to grade IV, generated by Agilent Whole Human Genome Array platform.22 In TCGA dataset, RNAseq data (level 3, RSEM normalized data) of 675 samples of whole WHO grades, ranging from WHO grade II to grade IV, were available. Upon analyzing with TCGA RNAseq data, RSEM value was log transformed. Thus, in total, PD-L1 transcriptional expression data of 976 samples were evaluated. Written informed consents were obtained from the patients (or their families) for CGGA project. This study was approved by the Ethics Committee of Capital Medical University, Beijing, China.
Detection of biomarkers of glioma
IDH1/2 mutations are the most common mutations in glioma, especially in lower grade glioma and secondary glioblastoma, which developed from a less malignant lesion. For CGGA cohort, IDH1/2 mutations were detected by pyrosequencing which has been described in our previous study.23 For TCGA cohort, IDH mutation data were downloaded from TCGA website, which were called from whole exon sequencing data or pyrosequencing.
Statistical analysis
R language was used as the main tool for the statistical analysis and generating figures.24 Multiple variates cox proportional hazard model was performed using coxph function provided in survival package.25 Other figures were generated by several R packages, such as pheatmap, pROC,26 circlize,27 and corrgram. Gaussian test was performed before data analysis which required Gaussian distribution. A p value less than 0.05 is considered to be statistically significant. All statistical tests were two-sided.
Results
PD-L1 expression was upregulated in glioblastoma and downregulated in IDH mutant glioma
Due to prominent heterogeneity of molecular nature across different glioma, PD-L1 expression of 301 samples with mRNA microarray data was analyzed according to the WHO grade system and IDH mutation status. In CGGA cohort, glioblastoma showed the highest PD-L1 expression when compared to grade II and grade III glioma (Student t test, p value = 3.71 × 10−8 and 0.0024, respectively; Fig. S1A). This result was well validated in TCGA RNA sequencing data (Fig. S1B), which further suggested that higher PE-L1 expression was accompanied by higher malignancy in glioma in line with other malignant tumors reported previously. Moreover, when IDH mutation status was added as a sub-classifier, we found that in IDH wild-type glioblastoma showed significantly distinct pattern of PD-L1 expression from IDH mutant glioblastoma of both CGGA and TCGA data set (Figs. 1A and B). Furthermore, IDH mutant-type showed universally lower expression of PD-L1 than that of IDH mutant glioma, across different grades, though no statistical significance was observed in some groups. This indicated that PD-L1 check-point-related immune response were more prevalent in IDH wild-type glioma which further reflected different biological pattern between these two kinds of tumors.
Figure 1.
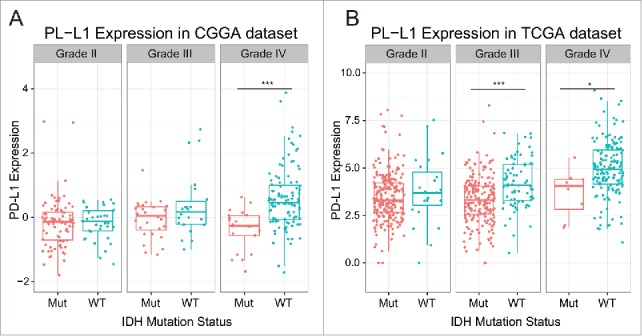
PD-L1 expression in CGGA (A) dataset and TCGA (B) data set according to IDH status. *** indicates p value< 0.001, * indicates p value < 0.05.
PD-L1 was a potential marker for mesenchymal molecular subtype
To investigate the molecular relevance between PD-L1 and glioma, we asked the distribution of PD-L1 expression in different molecular subtypes defined by TCGA network. As shown in Figs. 2A and B PD-L1 was significantly upregulated in mesenchymal subtype than other subtypes in both CGGA and TCGA dataset, except for classical subtype in CGGA data set, which also showed apparent trend although not significant. This result enlightened us that PD-L1 may serve as a biomarker for mesenchymal subtype. ROC curves for PD-L1 expression and mesenchymal subtype in all glioma were performed and under curve area is 80.1% and 80% in CGGA and TCGA dataset, respectively (Figs. 2C and D).
Figure 2.
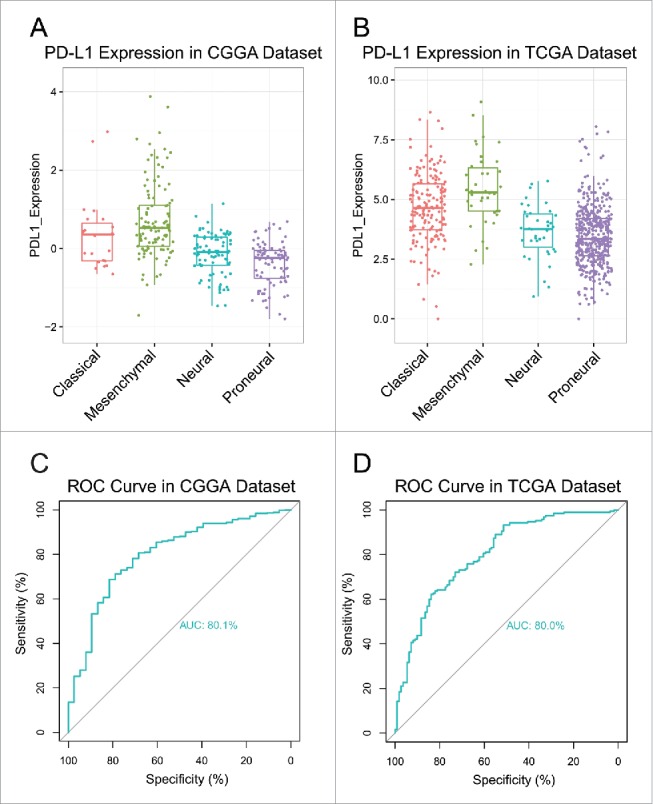
PD-L1 expression in molecular subtypes (A, B) and predictive value for mesenchymal subtype (C, D).
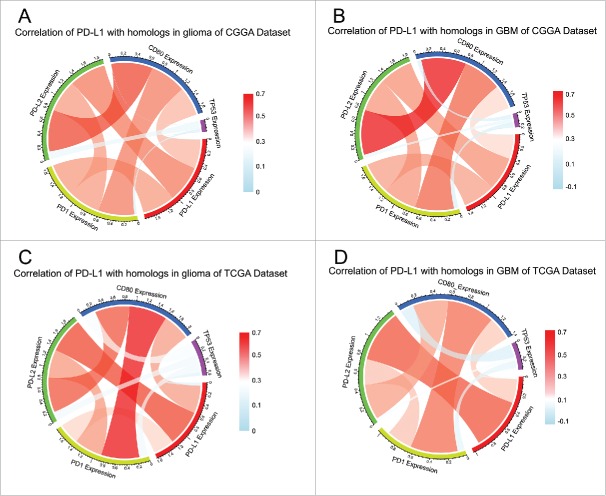
PD-L1 was synergistic with other check point members in tumor-induced immune response
PD-L1 is one of the ligands of PD1 and the conjugation of two molecules can suppress immune response by suppressing T-cells functioning. Cortez et al.28 identified a novel mechanism that TP53 could regulate the expression of PD-L1 through microRNA-34 which contributed to immune evasion of tumor. Thus, we put p53 into analysis together with PD-L1, PD1, CD80, and PD-L2 expression. Pearson correlation was performed with these five factors in both CGGA and TCGA data set. In whole grade glioma of CGGA, PD-L1 showed high concordance with PD1, PD-L2, and CD80, indicating active PD1/PD-L1 pathway (Fig. 3A). To our knowledge, higher inflammation and immune response are induced more in glioblastoma than lower grade glioma. To examine the relationship among these immune check point members in glioblastoma, Pearson correlation was performed additionally. As Fig. 3B indicated, these check point members showed even higher correlation with each other. In line with CGGA dataset, the correlation among check point markers was also very robust in TCGA data set (Figs. 3C and D), suggesting synergistic effects of these markers. Moreover, only a very weak correlation between p53 expression and PD-L1 expression was revealed, which may be accounted for by noise.
Figure 3.
Correlation of PD-L1 homologs and family members in glioma (A, C) and GBM (B, D).
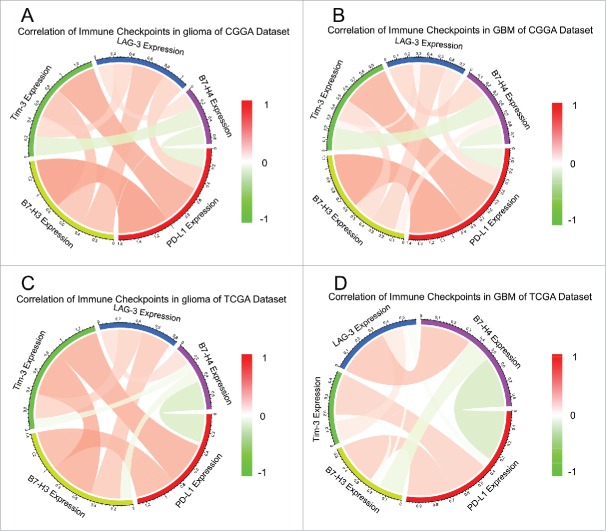
Many other check point members that have been assessed as therapeutic targets in clinical or preclinical trials, additional immune genes or immune checkpoints were enrolled into the analysis, such as B7-H3, B7-H4, LAG-3, and TIM-3,29,30 summarized by Pardoll.6
TIM-3 was reported to be upregulated in higher grade glioma than lower grade lesions.31 Circos plots demonstrated that PD-L1 expression was tightly associated with Tim-3 and B7-H3 (Fig. 4). These results remind us that when glioma acquire resistance to PD-L1 inhibitors, we need to be aware of the arising Tim-3 or B7-H3.
Figure 4.
Correlation of check point members in glioma (A, C) and GBM (B, D).
PD-L1-related biological process
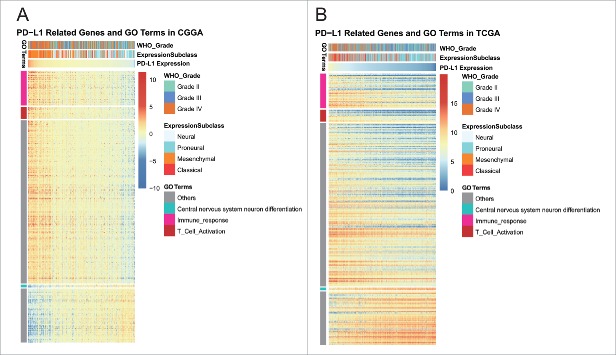
PD-L1 expression was heterogeneous across different grade glioma. To investigate the biological feature of glioma with different PD-L1 expression, we asked the genes that strongly correlated with PD-L1 expression (Pearson |R| > 0.5) in each dataset. Totally, 989 and 775 genes in CGGA and TCGA data set were eligible for subsequent analysis, respectively. To yield an accurate result, significantly related genes that were shared by two dataset were chosen for Gene Ontology analysis with online methods (DAVID, https://david.ncifcrf.gov/). We found that genes that positively correlated with PD-L1 expression were more involved in immune response, especially T cell activation (Figs. 5A and B). While genes that negatively correlated with PD-L1 expression were more involved in normal biological process, such as central nervous system neuron differentiation, chordate embryonic development, etc. This suggested that PD-L1, together with PD1, CD80 were induced more as immune suppressors in the tumor environment in which inflammatory and immune response were more active. As aforementioned, PD-L1 expressed more in GBM or mesenchymal molecular subtype, taking GBM as a distinct group of glioma, we expanded our knowledge of GBM by evaluating associated biological process. Significantly correlated genes shared by GBM of two data sets were further analyzed, and it turned out that, other than higher immune response, PD-L1 were positively associated with angiogenesis (Figs. S2A and B). These findings suggested that PD-L1 pathway activation was paralleled by vascularization of GBM, which indicated a more aggressive nature of glioma.
Figure 5.
Gene ontology analysis for PD-L1 in glioma.
PD-L1-related inflammatory activities
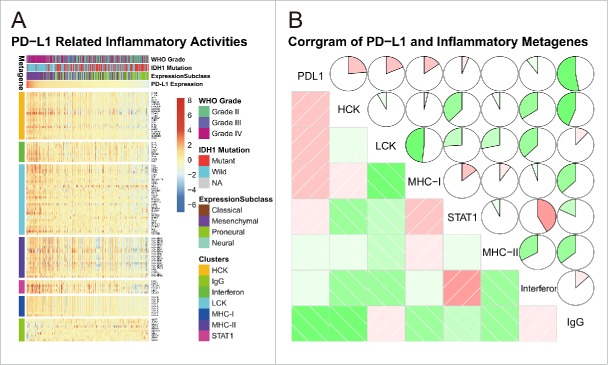
To get further understanding of PD-L1-related inflammatory activities, we chose seven clusters of 104 genes in total which were subsequently defined as metagenes8 (Table S1), representing different types of inflammation and immune response (Table S1). As shown in Fig. 6A, in CGGA dataset, most clusters were positively associated with PD-L1 expression except for IgG, which was mainly associated with activities of B lymphocytes. To validate what we found in clusters, seven metagenes were generated with results of Gene Sets Variation Analysis (GSVA)32 of corresponding clusters of genes. Corrgrams were derived according to Pearson r value between PD-L1 and seven metagenes (Fig. 6B). PD-L1 was positively associated with HCK, LCK, and MHC-I, but was negatively associated with IgG, in consistent with what we observed in Fig. 6A. We observed similar pattern in glioblastoma of CGGA data set (Fig. S3). These results suggested that PD-L1 was upregulated as an immune suppressor when macrophages and T cells signaling transduction were activated in glioma. While B linage-related immune responses were not much involved in glioma, in contrast to frequent interaction between PD1/PD-L1 pathways and T-cell-related gene expression signatures.
Figure 6.
PD-L1-related immune response.
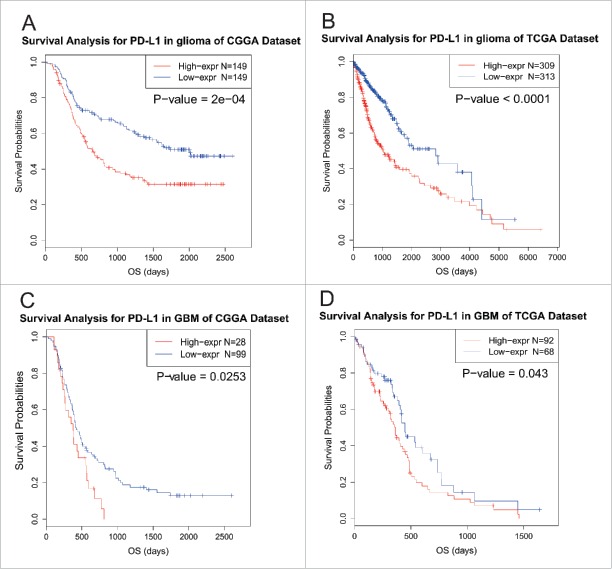
PD-L1 predicts worse survival in glioma
To explore the influence on survival, we tested the prognostic value of PD-L1. In two datasets, survival data were available for 298 patients and 622 in CGGA and TCGA data set, respectively. As shown in Figs. 7A and B, when taking all types of glioma into account, patients who had higher PD-L1 expression in their tumors lived a significantly shorter survival than the counterparts. Due to heterogeneity across different grade of glioma, we additionally investigated the prognostic value of PD-L1 expression in GBM of two datasets (Figs. 7C and D). When comparing the two groups defined by PD-L1 expression, we observed a similar pattern of Kaplan-Meier curves as Figs. 7A and B. Those findings indicated that PD-L1 was a negative prognosticator in glioma due to suppressive effect on T-cell-related immune response.
Figure 7.
Survival analysis for PD-L1.
Conclusion
PD-L1 is upregulated in glioblastoma, and is synergistic with other check point members. Moreover, PD-L1 is significantly associated with T-cell activation and macrophage-related immune response and predicts much worse survival for patients, warranting clinical trials of PD1/PD-L1 checkpoint inhibitors for potential glioma treatment.
Discussion
Gliomas are the most common primary malignant brain tumors in adults. Limited improvement of treatment has been made despite combination of surgery, chemotherapy and radiotherapy after diagnosed. Gliomas develop within the relatively immune-privileged central nervous system and thrive in a microenvironment of relative immunosuppression. Immune therapy becomes an increasingly eye-catching method for glioma,33,34 especially in GBM, shedding a shimmering light on glioma treatment.
We also have noticed that a study about PD-L1 expression in glioblastoma which accounted for about 40% of glioma published in Neuro-oncology as we finished this manuscript. To further address the role of PD-L1 expression and distribution in glioma, we extended our analysis to whole grade glioma as an integrative research. As immuno-therapy is becoming more and more important to malignant tumors, a primitive investigation of PD-L1 in glioma is of great necessity. In this study, we took advantage of CGGA and TCGA data set and totally 976 samples were enrolled into the analysis. We found that PD-L1 expression was upregulated in higher grade gliomas and mesenchymal subtypes, indicating that PD-L1 expression was associated with more malignant biological process. Additionally, PD-L1 may serve as an indicator of mesenchymal subtype. Lower expression of PD-L1 usually was companied by IDH mutation, suggesting IDH mutant gliomas were involved in less tumor-induced immune response than that IDH wild-type gliomas.
PD-L1 were expressed synergistically with PD1, PD-L2, and CD80, with were also involved in PD1/PD-L1 pathway. Through seven immune-related clusters, we revealed that PD-L1 expression were positively correlated with T cell activation, macrophage-related immune response and probabilities of activation of interferon. This result is in line with the primary function of PD1/PD-L1 pathway, suppressing T cell functioning in tumors. Immune response, especially in relatively immune privileged environment, is induced by tumor-related pathological process in glioma. Lymphatic vessels have been discovered in central nervous system by Louveau,35 providing solid proof and explanation to high level of T cell activities in glioma. Furthermore, high expression of macrophages-related gene signatures may derive from microglia cells, which are considered to be resident macrophages of the brain and spinal cord and function as immune defender in central nervous system.
Moreover, PD-L1 expression predicted significantly worse survival for glioma patients. This prognostic value was also observed when taking GBM as a separated type of glioma. Interestingly, PD-L1 seemed to be correlated with angiogenesis, corresponding to highly vascularization of GBM in contrast to lower grade glioma, which further validated the relationship between PD-L1 expression and malignant process.
Many pre-clinical and clinical trials have been carried out to investigate the effects of blocking PD1/PD-L1 pathway,7,20 and observed durable antitumor effect in a range of malignancies. In this study, we confirmed PD-L1 expression was highly activated in glioma, especially in GBM. A clinical study with specific immune checkpoint inhibitors seems to be warranted in glioma. Meanwhile, evaluation of the composition of the tumor microenvironment should be included in clinical trials. Thus, we propose immune-therapy using PD-L1 blockade as an avenue toward new therapy against human glioma.
About neuro-oncology, they have done a good job, focusing on PD-L1 expression and prognostic value on glioblastoma. We extended the research to all grades of glioma, ranging from WHO grade II to WHO grade IV, which profoundly extended the spectrum of research on PD-L1.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate the generosity of TCGA network for sharing the huge amount of data.
Funding
This work was supported by The Research Special Fund For Public Welfare Industry of Heath (No. 201402008), National Key Research and Development Program Focused on Precision Medicine Research Projects (2016YFC0902500) and a Grant from the biological medicine department, Beijing Municipal Science & Technology Commission (No. Z141100000214009).
References
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA 2010; 60:166-93; PMID:20445000; http://dx.doi.org/ 10.3322/caac.20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang P, Zhang W, Wang Y, Peng X, Chen B, Qiu X, Li G, Li S, Wu C, Yao K et al.. IDH mutation and MGMT promoter methylation in glioblastoma: Results of a prospective registry. Oncotarget 2015; 6:40896-906; PMID:26503470; http://dx.doi.org/ 10.18632/oncotarget.5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain SF, Yang D, Suki D, Aldape K, Grimm E, Heimberger AB. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol 2006; 8:261-79; PMID:16775224; http://dx.doi.org/ 10.1215/15228517-2006-008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garofalo S, D'Alessandro G, Chece G, Brau F, Maggi L, Rosa A, Porzia A, Mainiero F, Esposito V, Lauro C et al.. Enriched environment reduces glioma growth through immune and non-immune mechanisms in mice. Nat Commun 2015; 6:6623; PMID:25818172; http://dx.doi.org/ 10.1038/ncomms7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rolle CE, Sengupta S, Lesniak MS. Mechanisms of immune evasion by gliomas. Adv Exp Med Biol 2012; 746:53-76; PMID:22639159; http://dx.doi.org/ 10.1007/978-1-4614-3146-6_5 [DOI] [PubMed] [Google Scholar]
- 6.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12:252-64; PMID:22437870; http://dx.doi.org/ 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K et al.. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Eng J Med 2012; 366:2455-65; PMID:22658128; http://dx.doi.org/ 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D et al.. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res 2009; 11:R15; PMID:19272155; http://dx.doi.org/ 10.1186/bcr2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos J, Gonzalez-Sanchez L, Villa-Morales M, Ors I, Lopez-Nieva P, Vaquero C, González-Gugel E, Fernández-Navarro P, Roncero AM, Guenet JL et al.. The stromal gene encoding the CD274 antigen as a genetic modifier controlling survival of mice with gamma-radiation-induced T-cell lymphoblastic lymphomas. Oncogene 2010; 29:5265-73; PMID:20639904; http://dx.doi.org/ 10.1038/onc.2010.280 [DOI] [PubMed] [Google Scholar]
- 10.Azuma K, Ota K, Kawahara A, Hattori S, Iwama E, Harada T, Matsumoto K, Takayama K, Takamori S, Kage M et al.. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014; 25:1935-40; PMID:25009014; http://dx.doi.org/ 10.1093/annonc/mdu242 [DOI] [PubMed] [Google Scholar]
- 11.He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment for Non-Small Cell Lung Cancer. Scientific Reports 2015; 5:13110; PMID:26279307; http://dx.doi.org/ 10.1038/srep13110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berghoff AS, Ricken G, Widhalm G, Rajky O, Hainfellner JA, Birner P, Raderer M, Preusser M. PD1 (CD279) and PD-L1 (CD274, B7H1) expression in primary central nervous system lymphomas (PCNSL). Clin Neuropathol 2014; 33:42-9; PMID:24359606; http://dx.doi.org/ 10.5414/NP300698 [DOI] [PubMed] [Google Scholar]
- 13.Choueiri TK, Fay AP, Gray KP, Callea M, Ho TH, Albiges L, Bellmunt J, Song J, Carvo I, Lampron M et al.. PD-L1 expression in nonclear-cell renal cell carcinoma. Ann Oncol 2014; 25:2178-84; PMID:25193987; http://dx.doi.org/ 10.1093/annonc/mdu445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertucci F, Finetti P, Mamessier E, Pantaleo MA, Astolfi A, Ostrowski J, Birnbaum D. PDL1 expression is an independent prognostic factor in localized GIST. Oncoimmunology 2015; 4:e1002729; PMID:26155391; http://dx.doi.org/ 10.1080/2162402X.2014.1002729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brower V. Anti-PD-L1 antibody active in metastatic bladder cancer. The Lancet Oncology 2015; 16:e11; PMID:25638546; http://dx.doi.org/ 10.1016/S1470-2045(14)71167-2 [DOI] [PubMed] [Google Scholar]
- 16.Sabatier R, Finetti P, Mamessier E, Adelaide J, Chaffanet M, Ali HR, Viens P, Caldas C, Birnbaum D, Bertucci F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 2015; 6:5449-64; PMID:25669979; http://dx.doi.org/ 10.18632/oncotarget.3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura T, Ohira M, Tanaka H, Muguruma K, Toyokawa T, Kubo N, Sakurai K, Amano R, Kimura K, Shibutani M et al.. Programmed Death-1 Ligand-1 (PDL1) Expression Is Associated with the Prognosis of Patients with Stage II/III Gastric Cancer. Anticancer research 2015; 35:5369-76; PMID:26408698 [PubMed] [Google Scholar]
- 18.Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology 2012; 1:1223-5; PMID:23243584; http://dx.doi.org/ 10.4161/onci.21335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JW, Eder JP. Prospects for targeting PD-1 and PD-L1 in various tumor types. Oncology (Williston Park) 2014; 28(Suppl 3):15-28; PMID:25387682 [PubMed] [Google Scholar]
- 20.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN et al.. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515:563-7; PMID:25428504; http://dx.doi.org/ 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nduom EK, Wei J, Yaghi NK, Huang N, Kong L-Y, Gabrusiewicz K, Ling X, Zhou S, Ivan C, Chen JQ et al.. PD-L1 expression and prognostic impact in glioblastoma. Neuro-Oncol 2016; 18:195-205; PMID:26323609; http://dx.doi.org/ 10.1093/neuonc/nov172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan W, Zhang W, You G, Zhang J, Han L, Bao Z, Wang Y, Liu Y, Jiang C, Kang C et al.. Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol 2012; 14:1432-40; PMID:23090983; http://dx.doi.org/ 10.1093/neuonc/nos263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W, Zhang W, You G, Bao Z, Wang Y, Liu Y, Kang C, You Y, Wang L, Jiang T. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One 2012; 7:e30339; PMID:22291938; http://dx.doi.org/ 10.1371/journal.pone.0030339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.Rproject.org/. [Google Scholar]
- 25.T T. A Package for Survival Analysis in S. version 2.38 2015. [Google Scholar]
- 26.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformat 2011; 12:77; PMID:21414208; http://dx.doi.org/ 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014; 30:2811-2; PMID:24930139; http://dx.doi.org/ 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- 28.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK et al.. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2016; 108; PMID:26577528; http://dx.doi.org/ 10.1093/jnci/djv303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero D. Immunotherapy: PD-1 says goodbye, TIM-3 says hello. Nat Rev Clin Oncol 2016; 13:202-3; PMID:26977783; http://dx.doi.org/ 10.1038/nrclinonc.2016.40 [DOI] [PubMed] [Google Scholar]
- 30.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, Gandhi L, Redig AJ, Rodig SJ, Asahina H et al.. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 2016; 7:10501; PMID:26883990; http://dx.doi.org/ 10.1038/ncomms10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Han H, He X, Li S, Wu C, Yu C, Wang S. Expression of the galectin-9-Tim-3 pathway in glioma tissues is associated with the clinical manifestations of glioma. Oncol Lett 2016; 11:1829-34; PMID:26998085; http://dx.doi.org/ 10.3892/ol.2016.4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14:7; PMID:23323831; http://dx.doi.org/ 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bovenberg MSS, Degeling MH, Tannous BA. Cell-based Immunotherapy Against Gliomas: From Bench to Bedside. Mol Therapy 2013; 21:1297-305; PMID:23648695; http://dx.doi.org/21149252 10.1038/mt.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heimberger AB, Sampson JH. Immunotherapy coming of age: what will it take to make it standard of care for glioblastoma? Neuro Oncol 2011; 13:3-13; PMID:21149252; http://dx.doi.org/ 10.1093/neuonc/noq169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS et al.. Structural and functional features of central nervous system lymphatic vessels. Nature 2015; 523:337-41; PMID:26030524; http://dx.doi.org/ 10.1038/nature14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.