Abstract
The epigenome provides a substrate through which environmental exposures can exert their effects on gene expression and disease risk, but the relative importance of epigenetic variation on human disease onset and progression is poorly characterized. Asthma is a heterogeneous disease of the airways, for which both onset and clinical course result from interactions between host genotype and environmental exposures, yet little is known about the molecular mechanisms for these interactions. We assessed genome-wide DNA methylation using the Infinium Human Methylation 450K Bead Chip and characterized the transcriptome by RNA sequencing in primary airway epithelial cells from 74 asthmatic and 41 nonasthmatic adults. Asthma status was based on doctor’s diagnosis and current medication use. Genotyping was performed using various Illumina platforms. Our study revealed a regulatory locus on chromosome 17q12-21 associated with asthma risk and epigenetic signatures of specific asthma endotypes and molecular networks. Overall, these data support a central role for DNA methylation in lung cells, which promotes distinct molecular pathways of asthma pathogenesis and modulates the effects of genetic variation on disease risk and clinical heterogeneity.
Integrated epigenetic, transcriptional, and genetic profiles in airway cells reveal asthma loci and signatures of distinct asthma endotypes.
Introduction
Asthma affects over 235 million people worldwide (1) and is the most common chronic disease in childhood (2). The etiology of asthma is complex, with nearly equal contributions from genes and environment, reflected by heritability estimates of approximately 50% (3). Moreover, asthma is heterogeneous with respect to clinical onset and course, response to therapies, and associated comorbidities, such as allergies (4). Not surprisingly, therefore, variants identified in GWAS have small effect sizes and explain little of the overall risk for asthma. It is likely, therefore, that additional genetic variants remain to be discovered, including those involved in gene-environment interactions that may not reach criteria for significance in GWAS (5–7). Many asthma-promoting environmental exposures alter epigenetic profiles in airway cells (8–10), suggesting that changes in DNA methylation may be a mechanism through which environmental exposures modify asthma risk or contribute to phenotypic heterogeneity. Given the combined importance of environmental exposures and genetic variation on disease risk, a comprehensive understanding of the molecular architecture of asthma requires approaches that integrate genetic, epigenetic, and transcriptional variation with phenotypic measures of specific asthma endotypes (5). Yet no previous study of asthma has included these multiple sources of variation. Here, we characterize the epigenetic, transcriptional, and genetic landscapes in freshly isolated endobronchial airway epithelial cell (AEC) brushings from asthmatic and nonasthmatic subjects (Table 1). All subjects were extensively phenotyped at the University of Chicago Asthma & COPD Center. All asthmatics had a doctor’s diagnosis of asthma and were currently using asthma medications; the nonasthmatic subjects had a negative history of asthma and a normal spirometry and methacholine challenge test (see the Methods for additional details and inclusion/exclusion criteria). Our integrated systems biology approach combining methylation, transcription, and genetic data with pathway analysis revealed a central role for DNA methylation in AECs for modulating the effects of genetic variation on asthma risk and on specific asthma endotypes.
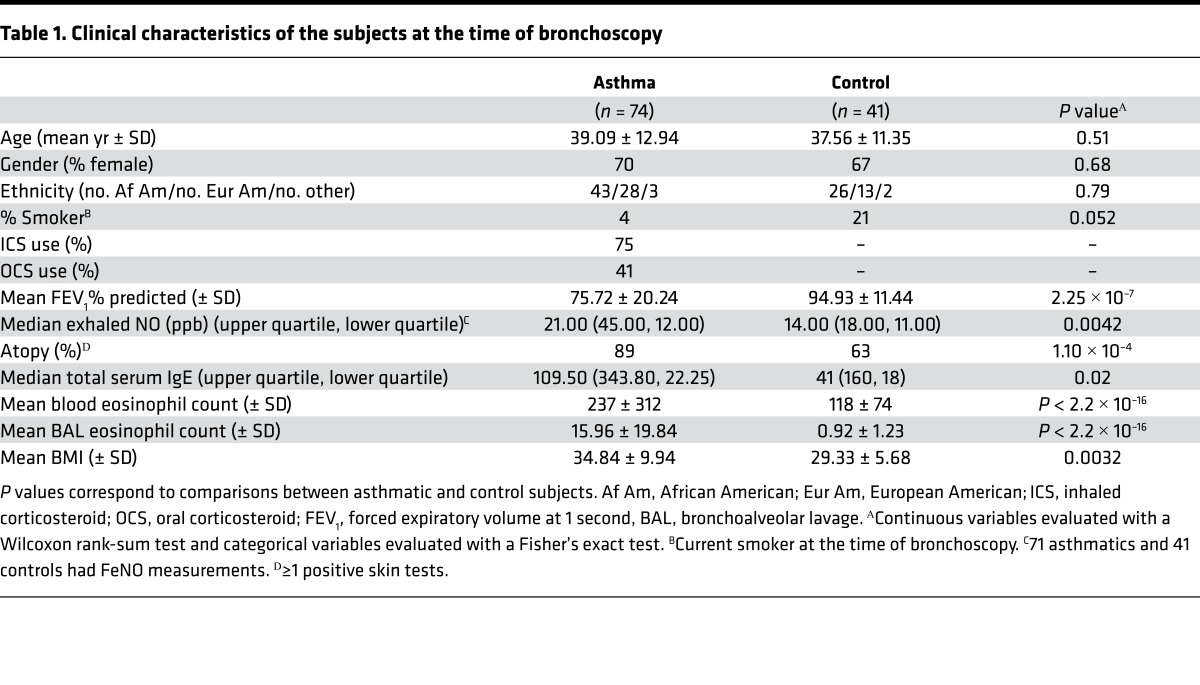
Table 1. Clinical characteristics of the subjects at the time of bronchoscopy.
Results
DNA methylation profiles differ between asthmatic and nonasthmatic subjects.
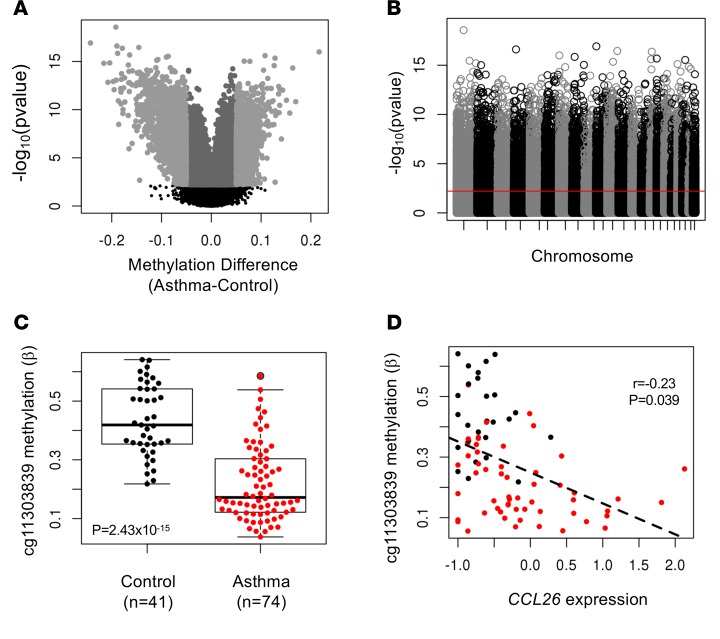
We obtained methylation data from freshly isolated AECs from 115 subjects (74 asthmatics; 41 nonasthmatics) using the Infinium Human Methylation 450K Bead Chip; 327,271 (of >450,000) CpG sites passed quality control checks (see the Methods for details) and were included in our studies. We tested for differences in methylation levels at each of these sites between asthmatic and nonasthmatic subjects using a general linear model framework (11). Overall, 40,892 CpG sites were differentially methylated between these two groups at a FDR of 5% (Figure 1, A and B, and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.90151DS1). The median absolute difference in methylation levels among these differentially methylated CpGs (DMCs) was 2.2% (range, 0.1%–24.3%). Among the DMCs, 22,216 (54%) were more methylated and 18,676 (46%) were less methylated in asthmatics.
Figure 1. Differential methylation in airway epithelial cells from asthmatic and control subjects.
(A) Volcano plot showing methylation differences between asthmatic (n = 74) and nonasthmatic (n = 41) individuals. Mean differences in β values are shown on the x axis. Dark gray indicates CpGs that differ between asthmatics and nonasthmatics at q ≤ 0.05; light gray indicates CpGs with a fold change of >5%; The black dots indicate non-significant sites at a q > 0.05. (B) Manhattan plot of 327,271 CpGs in our analysis of asthma-associated differentially methylated CpG sites. P values (y axis) correspond to the differences in methylation between asthmatic and control subjects. The red line corresponds to the q value threshold (FDR 5%). (C) Box plot showing methylation levels at a CpG site (cg11303839) upstream of the transcription start site of CCL26 (encoding eotaxin 3), a chemokine elevated in the airways of asthmatic subjects. Box plot displays the median, first and third quartiles, and 95% confidence intervals. (D) Scatter plot showing the correlation between CCL26 transcript abundance and cg11303839 methylation levels for 81 individuals with both gene expression and methylation data. In C and D, black points are nonasthmatics and red are asthmatics.
To assess the biological relevance of the DMCs, we asked whether methylation levels at these sites were more likely to be correlated with the expression level of nearby genes compared with non-DMCs and if correlations were stronger among DMCs with larger differences in methylation levels between asthmatics and nonasthmatics (i.e., larger effect sizes). Gene expression profiles, determined by RNA sequencing (RNAseq), were available for 81 of the 115 individuals (55 asthmatic and 26 controls). We calculated Spearman correlation P values between methylation levels at each CpG site and expression levels of the nearest gene that was detected (242,887 CpGs and 15,935 genes). As expected, the proportion of correlated CpG-gene pairs increased with increasing DMC effect size: 26% of non-DMCs (55,281 of 214,215), 34% of all DMCs (9,814 of 28,672), 42% of DMCs with effect sizes >5% (1,151 of 2,717), and 45% of DMCs with effect sizes >10% (113 of 249) were at least modestly correlated (r > 0.15) with the expression of the nearest gene. Many of the genes whose expression was correlated with a DMC have been previously implicated in asthma (Figure 1, C and D, and Supplemental Figure 1), suggesting that DMCs in the airways may identify additional asthma candidate genes.
Genetic variations correlated with methylation levels are associated with asthma.
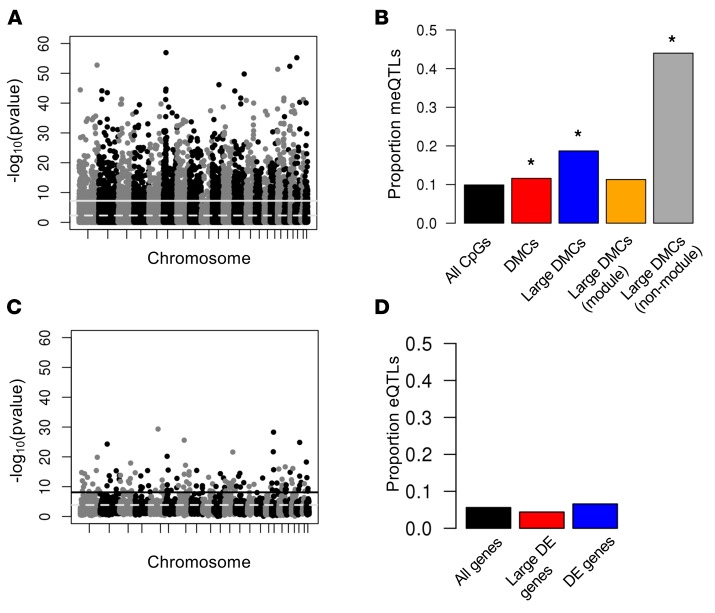
Because DNA methylation levels can be influenced by nearby genetic variation (12, 13), we hypothesized that SNPs correlated with methylation levels in AECs are enriched both among DMCs and among SNPs associated with asthma in GWAS, including those that do not meet stringent thresholds of genome-wide significance. To explore these possibilities, we first examined the influence of local genetic variation on methylation profiles in AECs by mapping methylation quantitative trait loci (meQTLs) in 111 individuals with genotype data, using a linear model framework as implemented in matrixeQTL (Figure 2A and Supplemental Table 2) (14). Among all CpGs within 5 kb of a SNP, 14,325 (9.89%) were associated with at least one meQTL (black bar in Figure 2B), whereas 2,200 (11.96%) DMCs were associated with at least one meQTL (red bar), revealing significantly more meQTLs among DMCs compared with CpG sites with methylation levels that did not differ between asthmatics and controls (i.e., non-DMCs). A parallel study of gene expression detected 16,358 cis expression (e)QTLs (925 unique genes, 14,944 unique SNPs) at an FDR 5% (Figure 2C and Supplemental Table 3) (see the Methods). However, in contrast to meQTLs, cis eQTLs were not enriched among genes that were differentially expressed between asthmatics and controls (Supplemental Table 4), including those with larger expression differences; 5.6% of all genes, 4.4% of differentially expressed genes, and 6.6% of differentially expressed genes with larger differences were associated with an eQTL (Figure 2D).
Figure 2. QTL mapping of methylation (meQTL) and expression (eQTL) levels.
(A) Manhattan plot showing the association P value between each SNP and methylation levels at nearby CpGs. The solid line shows the Bonferroni significance threshold; the dashed line shows the threshold at a FDR of 5%. (B) The proportion of meQTLs present in subsets of CpGs. Only the most significant meQTL per CpG is included. The black bar indicates all CpG sites on the array with a nearby SNP; the red bar indicates differentially methylated CpGs (DMCs) with a nearby SNP; the blue bar indicates DMCs with effect size >5% and with a nearby SNP; the yellow bar indicates DMCs with effect size >5%, a nearby SNP, and in a WGCNA comethylation module; and the gray bar indicates DMCs with effect size >5%, a nearby SNP, and not in a WGCNA comethylation module. *P < 2.2 × 10–16 compared with black bar. (C) Manhattan plot showing the association P value between each SNP and expression levels at nearby genes. The solid line shows the Bonferroni significance threshold; the dashed line shows the threshold at a FDR of 5%. (D) The proportion of eQTLs present in subsets of genes. Only the most significant eQTL per gene is shown. The black bar indicates all genes sites on the array with a nearby SNP; the red bar indicates differentially expressed (DE) genes with a nearby SNP; the blue bar indicates DEs with a >1.2-fold difference and with a nearby SNP.
We next asked whether SNPs that are meQTLs or eQTLs in AECs are also associated with asthma in published GWAS. For this analysis, we classified the most significant SNP (meQTL or eQTL) associated with each CpG site or gene as the QTL for that CpG site or gene (15). We then extracted association P values for SNPs included in two of the largest asthma GWAS to date, the EVE (16) and GABRIEL (17) consortia, and retained the overlapping SNPs between our study and each GWAS. We stratified each set of SNPs by their GWAS P values (< 0.01 vs. ≥0.01) and by those with and without QTLs (meQTLs and eQTLs separately, FDR ≤ 5% vs. > 5%), and tested for nonrandom distributions using a Fisher’s exact test. Indeed, SNPs that were meQTLs or eQTLs in AECs were more likely to be associated with asthma than SNPs that were not QTLs (meta-analysis of EVE + GABRIEL: PmeQTL = 0.0017, PeQTL = 0.0015; Supplemental Table 5). Previous studies of many complex diseases have shown that disease-associated SNPs in GWAS are enriched for eQTLs (18–21) and meQTLs (13, 22). Our data show that meQTLs (in airway cells) are also enriched for asthma-associated SNPs.
An integrated omics approach identifies an asthma locus.
The combined analysis of gene expression, methylation, and genetic variation can both facilitate the identification of novel disease loci that did not reach statistical significance in GWAS and provide an understanding of regulatory mechanisms underlying associations. To explore this further, we considered the 35 SNPs that were both meQTLs and eQTLs in AECs and associated with asthma in either the EVE (16) or GABRIEL (17) studies at a P value of less than 0.01. These 35 SNPs were meQTLs for 25 CpG sites and eQTLs for 13 genes, including potentially novel asthma loci (Supplemental Table 6). Many of the overlapping SNPs were correlated with the expression of genes on chromosome 17q12-21, the most significant and most replicated asthma locus (discussed in ref. 5). Asthma-associated SNPs at this locus span an approximately 200-kb block of linkage disequilibrium (LD) and are eQTLs for two coregulated genes, ORMDL3 and GSDMB, in blood cells (19, 23, 24) and whole lung tissue (19, 25). As a result of the extensive LD in this region and the coregulatory effects of associated SNPs on the expression of these two genes, it has been difficult to localize the causal SNP(s) and disentangle the relative roles of ORMDL3 and GSDMB in asthma risk.
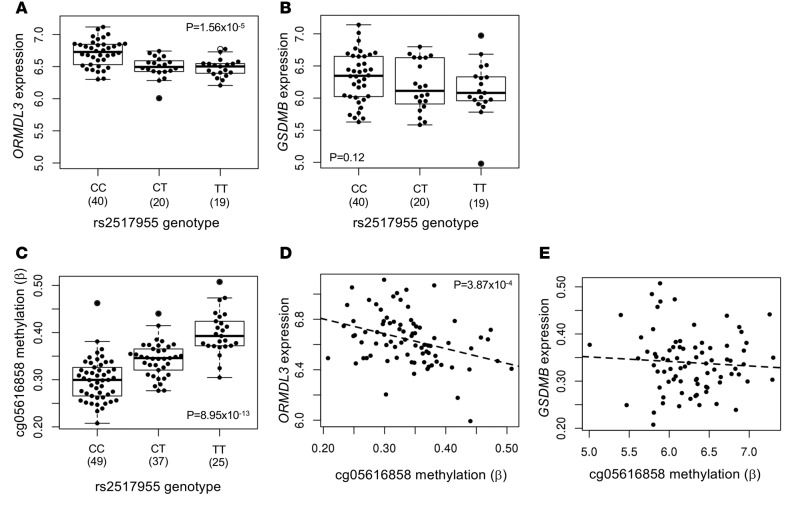
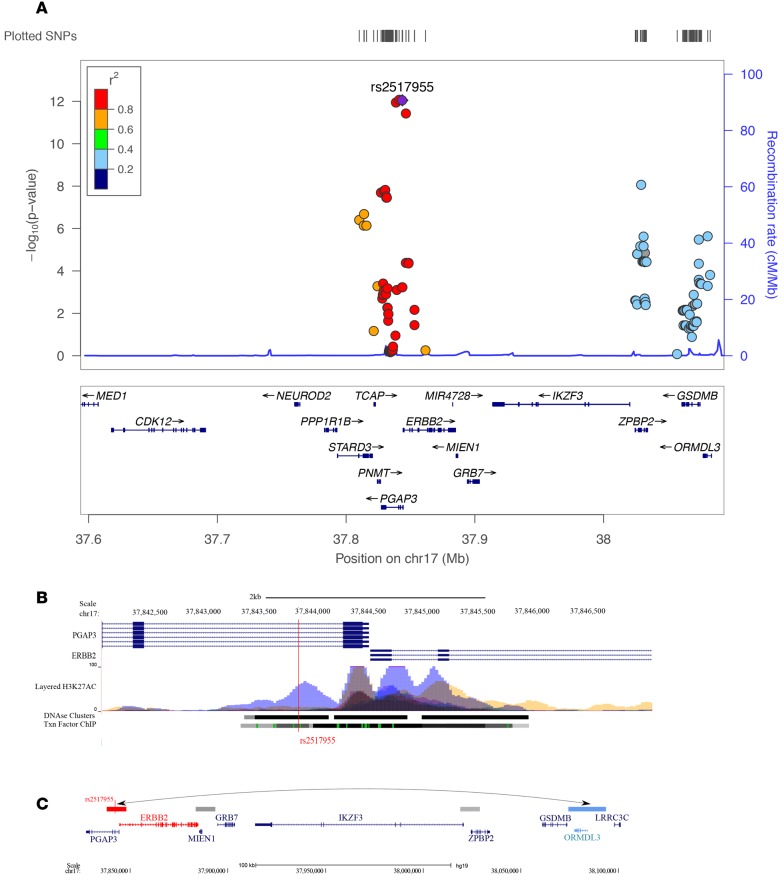
In our study, the most significant eQTL for ORMDL3 in AECs is located ~240 kb upstream of its transcription start site at a 17q locus outside of the LD block at the established 17q locus: LD r2 between rs2517955 at the locus and rs12936321 at the established locus was 0.27 (Figure 3A). To confirm that the effect of genotype at rs2517955 on ORMDL3 expression was independent of SNPs at the established locus, we repeated the eQTL analysis, including genotype at rs1293631 as a covariate. In this conditional analysis, the eQTL P value for rs2517955 and ORMDL3 changed from 2.56 × 10–5 to 8.81 × 10–4, indicating that the two loci are indeed independent. Moreover, unlike the eQTLs at the established locus, this SNP is not associated with expression of GSDMB in AECs in our study (Figure 3B), consistent with results in whole lung tissue in the Gene-Tissue Expression (GTEx) consortium studies (19). Although SNPs at this locus reached genome-wide significance in the GABRIEL study (P = 1.2 × 10–9) and were nearly genome-wide significant in the EVE study (P = 2.2 × 10–7), they were considered to be part of the association at the established 17q locus and not recognized as an independent asthma locus in either report. The allele that is associated with asthma in GWAS is associated with increased expression of ORMDL3 in airway epithelial cells in our study, suggesting that elevated ORMDL3 in the airways is associated with asthma risk.
Figure 3. SNPs at a novel locus are eQTLs for ORMDL3.
Box plots showing the association of genotype at rs2517955 with expression levels of ORDML3 (A) and GSDMB (B) and methylation levels at cg05616858 (C). Numbers in parentheses represent the number of individuals with the specified genotype. Scatter plots showing the correlation of methylation levels at cg05616858 with expression levels of ORMDL3 (D) and GSDMB (E) for the 81 individuals with gene expression, methylation data, and genotypes. Box plots display the median, first and third quartiles, and 95% confidence intervals.
The SNP that is the eQTL for ORMDL3, rs2517955, is also among the most significant meQTLs for a nearby CpG site (cg05616858, Figure 3C and Figure 4A), and methylation levels at this site were correlated with expression of ORMDL3 (Figure 3D) but not with expression of GSDMB (Figure 3E). To elucidate the causal relationship among genotype, methylation level, and gene expression, we performed Mendelian randomization (26). We tested for an effect of methylation at cg05616858 on ORMDL3 transcript abundance, using rs2517955 as the instrumental variable. The randomization P value was 0.001, signifying that methylation at cg05616858 contributes to ORMDL3 gene expression independent of genotype at rs2517955. Thus, methylation at this locus is likely the underlying molecular mechanism for the observed eQTL. Overall, these data show that rs2517955 at an asthma locus is associated with the expression of ORMDL3, but not GSDMB, in AECs through its effect on methylation at cg056168858.
Figure 4. A 17q locus is within a putative enhancer.
(A) Locus zoom plot of meQTL results for the 17q12-21 region. Only SNPs included in the meQTL study (those within 5 kb pairs of a CpG) are shown. The meQTL P values are shown on the left y axis for each CpG-SNP pair; only the smallest meQTL P value is shown when there is more than one SNP within 5 kb of a CpG site. LD (r2) is shown between the most significant meQTL (rs2517955) and all other SNPs. Note that rs2517955 shows little LD (r2 = 0.27 based on 1,000 genomes CEU) with rs12936231, a SNP at the established 17q21 locus, which spans from ZPBP2 to ORMDL3. (B) ENCODE H3K27ac histone marks align to the region flanking rs2517955. The vertical red line shows the position of rs2517955. (C) Results of in situ Hi-C studies. Interaction between the putative distal enhancer locus (red bar) and a region including the ORMDL3 promoter (blue bar) is shown by an arrow. Gray bars show additional sites of interaction with the enhancer locus. The vertical red line shows the position of rs2517955.
We next examined the regulatory architecture around rs2517955 using the ENCODE (27) data on histone marks of enhancers. The associated SNPs at this locus are within a strong H3K27ac histone mark (Figure 4B). H3K27ac is a mark of poised or active enhancers in mammalian cells (28, 29). Moreover, capture Hi-C studies in lymphoblastoid cell lines showed looping and physical interaction between the 17q locus and the promoter of ORMDL3, 240 kb away at the established 17q locus (Figure 4C). Thus, integration over multiple omic platforms revealed further complexity at the 17q asthma locus and identified SNPs at a regulatory locus that are associated with asthma and correlated specifically with the expression of ORMDL3 in AECs.
Epigenetic variations define asthma endotypes and molecular pathways of pathogenesis.
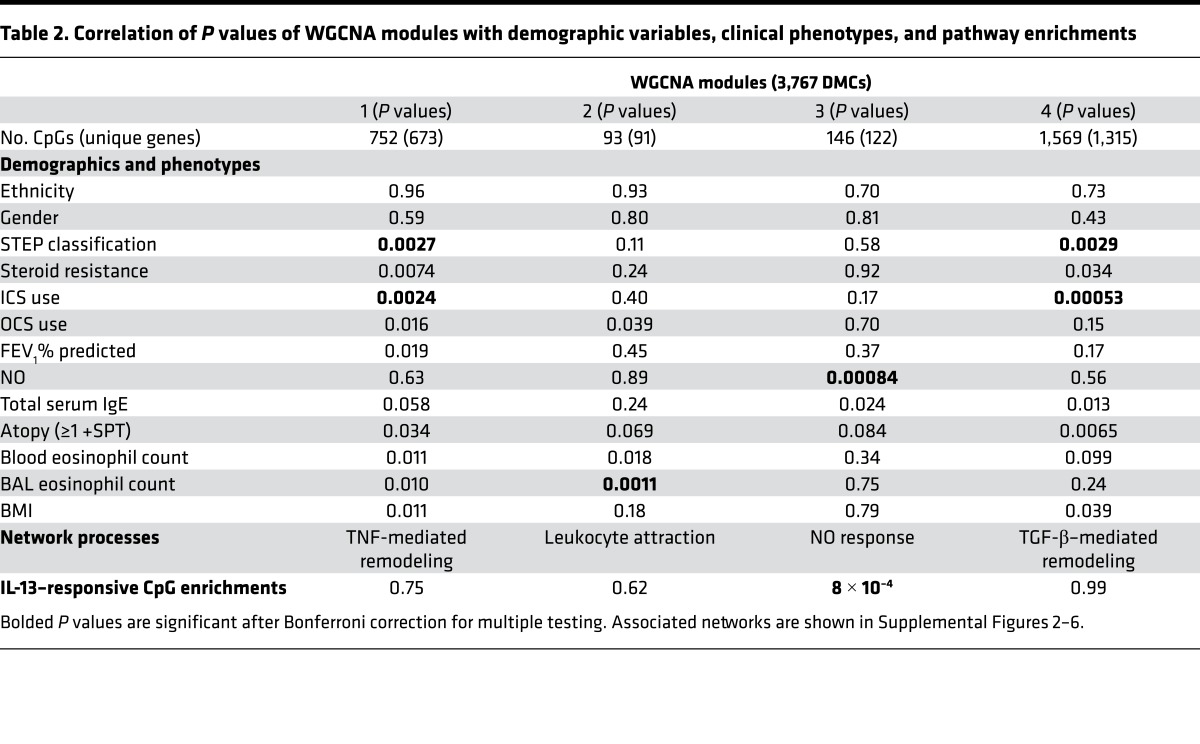
Both the epigenetic and genetic signatures in AECs described above provide rich sources of variation that may influence asthma endotypes. To examine this more closely, we focused on the subset of high confidence DMCs that were differentially methylated between asthmatics and nonasthmatics with an effect size of at least 5% (n = 3,767), and used a systems biology approach, as implemented in weighted gene coexpression network analysis (WGCNA) (30), to examine the correlation structure of these DMCs. WGCNA grouped 68% of the DMCs into four comethylation modules (Table 2). To evaluate the clinical relevance of these modules, we tested for correlations between each of the modules and asthma-relevant phenotypes. For this analysis, we calculated the trajectory of average methylation change for each module (eigengene) using WGCNA and correlated the module eigengenes with phenotypes in the 74 asthmatic subjects. The modules were correlated with three distinct phenotypic signatures or endotypes. Modules 1 and 4 were both associated with a classifier of asthma severity (STEP classification) and inhaled corticosteroid (ICS) usage, an indicator of severity. Module 2 was associated with eosinophilia in bronchial alveolar lavage (BAL) fluid, and module 3 was associated with fractional exhaled NO. Both BAL eosinophilia and exhaled NO are markers of airway inflammation. Module 3 was also enriched for CpGs that were IL-13 responsive in an AEC culture model (31). Because 75% of the asthmatics in this study used ICS and long-acting β agonists (LABA) to control their symptoms (Table 1), and ICS usage is associated with WGCNA modules 1 and 4, we examined the effects of this commonly used combination therapy on methylation changes in cultured primary AECs. To test the effects of these pharmacologics on methylation patterns, we treated AEC cultures established from primary cells with vehicle or with a combination of 10–5 M dexamethasone and 10–7 M fluticasone for 6, 24, and 48 hours. None of the 2,560 CpGs included in the four comethylation modules changed after exposure to ICS/LABA at any of the time points assessed (FDR > 5%). These studies revealed that the correlated DMCs within the modules do not change in response to this therapy in cell culture and suggest that the methylation differences between asthmatic and nonasthmatic individuals in our study are not due to inhaled therapy use in the asthmatics.
Table 2. Correlation of P values of WGCNA modules with demographic variables, clinical phenotypes, and pathway enrichments.
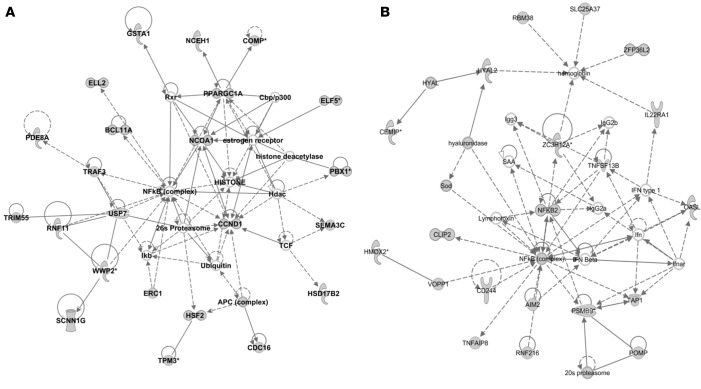
To further evaluate the potential biological relevance of the modules, we used ingenuity pathway analysis (IPA) to identify protein-protein interaction networks and upstream regulators of proteins whose genes were associated with each module. For this analysis, we included the closest gene to each DMC if methylation and expression levels of the CpG-gene pair were at least modestly correlated (r > 0.15). Each module was significantly enriched for at least one network (IPA network score ≥ 25) (Table 2, Figure 5, and Supplemental Figures 4–7). Genes correlated with CpG methylation levels in modules 1 and 4, both of which were associated with asthma severity, are enriched in many networks with protein hubs implicated in a broad number of remodeling, cell growth, and inflammatory pathways, such as ERK1/2, NF-κB, and Ras/Raf kinase. However, despite having similar hubs and similar phenotype associations, the CpGs within these modules are largely correlated with different genes (only 9 genes overlap between modules, 2% of module 1 and 4% of module 4). The overlapping network hubs, therefore, likely represent different signaling pathways that act through the same intermediate molecules (Figure 5). Consistent with this is that modules 1 and 4 are also enriched for different upstream regulator molecules. Module 1 was enriched for the upstream regulator TNF (P = 4.81 × 10–4), while module 4 was enriched for the upstream regulator TGFB1 (P = 7.74 × 10–9) (Supplemental Figure 6). These analyses suggest that the genes in modules 1 and 4 alter different pathways that influence asthma severity in the airways. Module 2, which was specifically associated with BAL eosinophil count, is enriched for a single network, with hubs that include VEGF, SELE, and SMAD2/TGFB1, all genes involved in processes related to eosinophilia (32) or eosinophil migration across epithelium (33, 34) (Supplemental Figure 3). Module 3 was associated with exhaled NO levels, and the one network associated with this module is centered on induced NO synthetase (NOS2) (35) as well as additional components of induced NO response (Supplemental Figure 6). Collectively, these data suggest that each module comprises methylation changes associated with central yet distinct components of asthma pathogenesis: airway remodeling (modules 1 and 4), leukocyte attraction (module 2), and NO response (module 3). Notably, parallel studies on gene expression did not yield coexpression modules
Figure 5. IPA networks of genes correlated with CpGs in modules 1 and 4 are both centered on NF-κB but have few overlapping genes.
(A) Module 1 network (score 42). (B) Module 4 network (score 29). The expression of the genes included in these networks was correlated with methylation levels at nearby differentially methylated CpGs (DMCs) (r > 0.15). Genes that were associated with a DMC are shown in gray. Description of molecule shapes can be found at http://ingenuity.force.com/ipa/articles/Feature_Description/Legend
meQTLs are enriched among uncorrelated DMCs with large effect sizes.
Finally, we explored the relationship between meQTLs and the WGCNA modules to ask whether genetic variation contributes to specific asthma endotypes. Although meQTLs were enriched among DMCs with large effect sizes, they were not equally distributed among the large effect DMCs assigned to comethylation modules and those that were not: 11.3% of the comethylated DMCs were associated with a meQTL compared with 39.3% of DMCs that were not comethylated (yellow and gray bars, respectively, in Figure 2B). The latter group of non-comethylated DMCs also showed stronger associations overall (i.e., smaller meQTL P values) compared with comethylated DMCs in the modules (Supplemental Figure 7). These observations of both more meQTLs and more significant meQTL P values among the uncorrelated large effect DMCs suggest that the correlated methylation profiles of DMCs within modules may be more influenced by nongenetic factors, such as environmental exposures or downstream effects of the disease process itself.
Discussion
Our findings indicate that DNA methylation in the airway plays a central role in mediating the effects of genetic variation on asthma risk and clinical course. Using a systems biology approach that integrated genome-wide genetic, epigenetic, and transcriptomic data in freshly isolated airway epithelial cells with publicly available genomic and GWAS data led to the discovery and mechanistic understanding of a regulatory locus for asthma and the identification of epigenetic signatures of distinct endotypes and molecular pathways. These observations would not have been revealed if we had focused on individual CpG sites or on a single omics readout, such as gene expression. In fact, it was unexpected, and notable, that epigenetic profiling in the asthmatic airways revealed insights into asthma pathogenesis that were not observable in the gene expression data. Differentially expressed genes between asthmatics and nonasthmatic subjects did not cluster into coexpression modules or show enrichment for eQTLs, as was observed for DMCs. These findings suggest that asthma-associated epigenetic changes in the airways may be a more stable marker of disease and therefore a more relevant focus for biomedical discovery.
Our study also revealed a relative depletion of meQTLs for large effect DMCs within correlation modules that are associated with specific endotypes and molecular pathways compared with uncorrelated large effect DMCs. We suggest that large coordinated shifts in DNA methylation levels at CpGs within the modules reflect functional connectivity, possibly in response to environmental exposures or even downstream effects of the disease process. The paucity of meQTLs for these sites may be due to biological constraints on these interconnected pathways. In contrast, uncorrelated CpGs in the asthmatic airways may be under less constraint and therefore may be more likely to be influenced by nearby genetic variation. Overall, this systems level analysis revealed a partitioning between correlated and uncorrelated DMCs with large effect sizes, potentially reflecting environmental and genetic influences on asthma endotypes and susceptibility, respectively.
Finally, our study identified an asthma locus outside of the LD block at the previously characterized 17q12-21 locus. Unlike previous studies of this locus, we show that methylation levels at a CpG site (cg056168858) within a putative enhancer locus are associated with the expression of ORMDL3 but not GSDMB in freshly isolated AECs and that a SNP (rs2517955) at the enhancer locus is associated with ORMDL3 expression via its primary effects on methylation at cg056168858. These results indicate that the regulation of expression of ORMDL3 differs between airway and blood cells and further highlights the importance of focusing omic studies on specific cell populations from disease-relevant tissues (19, 36). Using a chromatin configuration assay, we demonstrated physical interaction between the putative enhancer and the promoter of ORMDL3, supporting a direct effect of this locus on ORMDL3 expression. Integration with published GWAS data supports an association between rs2517955 and asthma and suggests that expression of ORMDL3 is increased in the airways of asthmatic individuals.
We note several limitations of this study. First, it was performed in cells from asthmatics with established disease and nonasthmatic controls. Thus, although we identified global epigenetic signatures of distinct endotypes among the asthmatics, the design of our study does not allow us to imply causality. Whether the observed profiles are a result of the disease process itself or whether they reflect responses to the environment that preceded disease cannot be distinguished within the context of this study. In contrast, genetic variants that are meQTLs and associated with asthma in GWAS are more likely to be causally related to the observed differences in methylation levels and potentially to asthma inception or progression, as we show for the 17q SNP. Second, due to the relatively small size of our sample, we could not formally test for interactions among genotype, methylation levels, and asthma, which may identify potential sites of gene-environment interactions. Nonetheless, enrichments of meQTLs for CpG sites with methylation levels that differ between asthmatics and controls (DMCs) suggest that such interactions exist and could potentially be identified in larger studies. Finally, studies of airway cells in asthmatics with established disease are always potentially confounded by medication usage. Indeed, 75% of the asthmatics were using a standard combination therapy of ICSs and LABAs at the time of our studies. It is possible therefore that some of the DNA methylation and gene expression differences observed between asthmatic and nonasthmatic individuals was due to exposure to these therapies. We addressed this by measuring DNA methylation and gene expression responses to these compounds in an airway epithelial cell model. Although the results of those studies suggest that the correlated DMCs within the comethylation modules are not responsive to the effects of this treatment in vitro, it is possible that some proportion of the observed DMCs are responsive to treatment in vivo.
The integrated approach described in this paper represents a step toward unraveling the genetic and environmental components of asthma, as well as the underlying molecular structure of asthma endotypes, and should be applicable to complex diseases in general. Expanding these studies to include other relevant cell types, additional epigenetic marks, and further modeling of disease-promoting or -protective exposures in cell culture models will continue the process of fleshing out the underlying regulatory architecture of asthma. Elucidating how both genetic and epigenetic variation, individually and as interactions, contribute to this architecture and ultimately to disease onset and progression are important steps toward the ultimate goal of personalized therapeutics and prevention strategies for asthma.
Methods
Studies in freshly isolated cells.
One hundred twenty-three adult subjects (76 with asthma, 47 without asthma) underwent bronchoscopy between March 2010 and March 2014 at the University of Chicago. Endobronchial brushings were obtained during bronchoscopy, as previously described (37). The asthmatic subjects had a current doctor’s diagnosis of asthma, no conflicting pulmonary diagnoses, and were using asthma medications. Controls were subjects that had no current or previous diagnosis of asthma and had normal spirometry and methacholine challenge tests. In 5 subjects (1 asthmatic, 4 nonasthmatics), we were unable to obtain sufficient quantity or quality of DNA from the brushings for methylation studies, and methylation data from 3 additional subjects (1 asthmatic, 2 nonasthmatics) failed quality control checks. The remaining 115 subjects (74 asthmatics, 41 nonasthmatics) were included in the methylation studies (Table 1). For studies of differential expression, we obtained RNAseq read depths of >10 million mapped reads in 85 individuals who were used in studies of differential expression (58 asthmatics, 27 nonasthmatics); 81 of the 85 also had methylation data. We obtained genome-wide genotypes for 116 individuals. One hundred and eleven individuals also had methylation data available and were used for meQTL studies, and 79 had expression data and were used for eQTL studies.
DNA and RNA were isolated from epithelial cell brushings using the QIAzol lysis reagent (QIAgen). DNA was concentrated using the Millipore Amicon Ultra centrifugal filters, 0.5 ml 30K membrane (Millipore), according to the manufacturer’s instructions.
Cell culture studies.
To assess the effects of asthma medications on methylation levels in AECs, we cultured primary AECs from 7 human donor lungs that were not suitable for transplantation and obtained through the Gift of Hope. Primary AEC cultures were established from these lungs at the University of Chicago Lung Biospecimen Core; all donors were European American, 4 were female, and the median age was 44 years (range 37–48 years). Cells were cultured as previously described (38) and then treated with a combination of 10–5 M dexamethasone and 10–7 M fluticasone or with vehicle for 6, 24, and 48 hours. DNA for methylation was extracted from vehicle and treated samples using the QIAgen AllPrep kit. Samples with sufficient amounts of DNA after extraction were analyzed for methylation as described below. After 6 hours, 7 treated and untreated samples provided sufficient amounts of DNA for methylation studies; after 24 hours, 6 treated and 5 untreated samples provided sufficient amounts of DNA; and after 48 hours, 3 treated and 4 untreated samples provided sufficient amounts of DNA. RNA for expression studies and DNA for methylation studies were extracted from treated and untreated cultures using the QIAgen AllPrep kit as described previously (38).
Methylation studies.
Methylation was assessed in freshly isolated and cultured AEC samples using the Infinium Human Methylation 450K Bead Chip (39). Probes located on the sex chromosomes and those that had a detection P value of greater than 0.01 in 75% of samples were removed. We also excluded probes that mapped to more than one location in a bisulfite-converted genome or overlapped with the location of known SNPs (13), leaving 327,271 CpGs. Methylation data were processed using the minfi package (40); Infinium type I and type II probe bias was corrected for using the SWAN algorithm (41). We corrected raw probe values for color imbalance and background by controls normalization. At each CpG site, the methylation level was reported as a β value, which is the fraction of signal obtained from the methylated beads over the sum of methylated and unmethylated bead signals.
Principal component analysis was used to determine the effects of known confounding variables on global methylation profiles. In the freshly isolated cells, chip, gender, ethnicity, age, and BMI were significantly correlated with principal components. The effects of chip were regressed out using COMBAT. Residual methylation β values after regression were used for all analyses in which gender, ethnicity, and age were included as covariates in the model to identify DMCs. Because BMI was strongly correlated with age and no longer significant after age was included in the model, BMI was not included as a covariate. Smoking status was not significantly associated with principal components, but we included it in the model due to its known effect on methylation profiles (42). Methylation array data were deposited into Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession GSE85568. In the cell culture model, to assess the effects of inhaled combination therapy on methylation, DNA concentration, age of individual, and gender were associated with methylation levels. In analyses of those data, DNA concentration was regressed out and age of individual and gender were retained as covariates in linear models.
To assess associations of asthma on methylation levels at each CpG site in the freshly isolated cells, we used the R package limma, with gender, age, smoking status, and ethnicity included as covariates. Analysis of methylation changes in the cell culture model was performed using a paired mixed-effects linear regression analysis of treated versus untreated conditions with individual ID coded as a random effect for 6-hour samples. Due to sample loss in the 24- and 48-hour samples, we did not have enough paired samples in those treatments and performed analyses of 24- and 48-hour samples using standard linear regression.
Gene expression studies.
cDNA libraries were constructed using the TruSeq RNA Sample Preparation v2 according to the manufacturer’s instructions (Illumina) and run on the Illumina HiSeq 2000 platform. RNAseq data were mapped to the transcriptome using BWA (43). Sequences that overlapped with protein-coding regions were determined using BEDTools (44). The median number of mapped reads was 19,210,000 mapped reads per individual, with a range of 10,100,000–51,150,000 mapped reads. The gene counts were adjusted for gene length and variation in sample read depth. Expression estimates were obtained for 18,931 autosomal ENSEMBL genes and estimates were filtered to genes that had at least 5 reads in 2 individuals and were annotated in biomart reducing the number of genes to 16,535. RNAseq counts were processed using RUVSeq (45). We estimated the factors of unwanted variation using RUVSeq by selecting 10% of the least variable genes in our sample (1,653 genes). These genes were subsequently excluded from analyses of differential expression but were retained for eQTL analyses. Two unwanted sources of variation (RUVs) were identified and included in the negative binomial generalized linear model, as implemented in RUVSeq. To better understand the variables associated with these RUVs, we correlated the RUVs to clinical phenotypes of interest. One RUV was significantly correlated with RIN score (P = 1.1 × 10–6), and one was significantly correlated with ethnicity (P = 8.1 × 10–8), average base pair read length (P = 1.4 × 10–7), and flow cell (P = 1.6 × 10–12). After regressing out these two RUVs, principal component analysis was performed on log-transformed residuals of counts per million. No additional correlations with sources of variation were detected. RNAseq data have been deposited into GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession GSE85568.
Genotyping and imputation.
Whole blood–derived DNA was genotyped on the Illumina Omni2.5-8v1A (n = 35), Omni1MDuo (n = 37), or Human Core (n = 44) arrays. SNPs on each array were oriented to the plus strand; SNPs were excluded if they had call rates <99% within each platform, and individuals were excluded if they were missing genotypes at >90% of SNPs. Genotypes within each platform and each ethnicity (European American, African American) were phased using MACH (46) and imputed with minimac3 (47) using the 1,000 genomes phase 3 reference panel. SNPs with an imputation efficiency >0.7 within each ethnicity/platform analysis were retained, resulting in a set of 1,046,454 common SNPs across all platforms. Biallelic SNPs with a MAF >10% in both the African American and European American samples were used in all subsequent studies (n = 988,004).
Correlation of methylation levels with the nearest gene.
We calculated the Spearman correlation coefficient for 81 individuals with both gene expression (of 85 individuals with RNAseq data) and methylation (of 115 subjects with methylation data) data using R. Each CpG site (locations annotated by Illumina) was mapped to the closest gene transcription start site (according to ENSEMBL). Spearman correlation r and P values were recorded for each CpG-gene pair in which the nearest gene was detected as expressed in our study (n = 242,877 CpG-gene pairs and 15,935 genes).
WGCNA comethylation analyses.
Methylation levels for the 3,767 DMCs with effect sizes >5% were clustered into comethylation modules using WGCNA (48). The soft thresholding power was determined to be 12; all other settings were kept at the default values. Genes that were differentially expressed between asthmatics and controls were also clustered using WGCNA. The soft thresholding power for the gene expression studies was 5. Eigengenes for each comethylation module were derived through WGCNA and correlated with clinical phenotypes among the asthmatic subjects.
IPA networks.
Network analyses were performed separately for the set of genes correlated with CpGs within each of the WGCNA comethylation modules. For these analyses, we used the gene nearest to each CpG that was correlated with the expression of the nearest gene (correlation r > 0.15). Network scores are based on the network hypergeometric distribution and are calculated with the right-tailed Fisher’s exact test to identify enrichment of those genes that were associated with WGCNA modules in the network relative to the IPA database. Networks with a score ≥25, corresponding to a Fisher exact P value of 10–25, were considered significantly enriched for the input genes. Ingenuity Upstream Regulator analysis was performed using the same module-associated gene lists. Upstream Regulator analysis identifies genes in each list that are enriched for common upstream regulators, i.e., molecules known to influence the expression of those genes.
QTL analyses.
meQTL and eQTL mapping were performed using matrix eQTL (14). We used windows of 500 kb pairs from each transcription start site (1-Mb window) and 5 kb from each CpG (10-kb window) for eQTL and meQTL mapping, respectively. In the meQTL studies, the significant covariates identified by principal component analysis and included as covariates were gender, smoking, age, and ethnicity. Ethnicity was estimated using the first 5 principal components of a principal component analysis for ancestry. Ancestry informative markers and the procedure used to estimate ancestry were described previously (49). No covariates were included in the eQTL analyses, because they were performed on the log-transformed counts per million RUV-adjusted count data, which was already adjusted for known covariates and RUVS.
GWAS enrichment analyses.
Publicly available lists of P values for each SNP were obtained for two asthma GWAS, EVE (16) and GABRIEL (17). GWAS SNPs from each study were subset to those that were present in both the eQTL and meQTL studies, (i.e., we excluded nonoverlapping SNPs). To reduce LD from multiple SNP associations with the same gene or CpG site, overlapping SNPs were further subset to include only the most significant eQTL or meQTL per gene or CpG, respectively. A Fisher’s exact test was used to assess if distributions were different between GWAS SNPs with low P values (P < 0.01) and meQTLs or eQTLs (FDR < 5%). A meta-analysis of the asthma GWAS (EVE and GABRIEL) was performed by combining the odds ratios and standard errors derived from the count data in the R package meta (50).
Promoter capture in situ Hi-C.
In situ Hi-C was performed as described previously (51). Human lymphoblast cells (LCL; GEO accession GM19141; https://www.ncbi.nlm.nih.gov/geo/) were cultured according to Battle et al. (52). Five million LCLs were treated with formaldehyde 1% to cross-link interacting DNA loci. Cross-linked chromatin was treated with lysis buffer (10 mM Tris-HCl pH 8.0, 10 mM NaCl, 0.2% Igepal CA630 [Sigma-Aldrich, I8896], 1X protease inhibitor cocktail [cOmplete, Roche, 11697498001]) and digested with MboI endonuclease (New England Biolabs, R0147). Subsequently, the restriction fragment overhangs were filled in and the DNA ends were marked with biotin-14-dATP (Life Technologies, 19524-016). The biotinylated DNA was ligated, cross-link reversed, and sheared to a size of 300–500 bp, using a Covaris S2 instrument (duty cycle, 5; intensity, 5; cycles/burst, 200; time, 60 seconds for 2 cycles). The biotin-labeled DNA was pulled down using Dynabeads MyOne Stretavidin T1 beads (Life Technologies, 65602) and prepared for Illumina paired-end sequencing. The in situ Hi-C library was amplified directly off of the T1 beads with 9 cycles of PCR, using Illumina primers and protocol (Illumina, 2007). Promoter capture was performed as described previously (53) with the following changes: the in situ Hi-C library was hybridyzed to 81,735 biotinylated 120-bp custom RNA oligomers (Custom Array, Supplemental Table 7) targeting promoter regions. Oligomers were selected by Agilent’s SureDesign software with default parameters to target regions at the ends of MboI restriction fragments longer than 200 bp (hg19) (54). The 4 nearest targets, 2 on the left and 3 on the right of RefSeq transcription start sites (> 1,000 kb apart), were submitted to SureDesign. Postcapture PCR (12 amplification cycles) was performed on the DNA bound to the beads via biotinylated RNA.
Promoter capture in situ Hi-C data analysis.
Alignment of 100-bp paired-end reads to hg19 was performed independently for each mate using bowtie2-2.2.3 with the --local option. Reads with mapping quality lower than 10 were discarded. Mates were paired by name with a custom script. HOMER v4.7.2 (55) was used to call interactions with P < 1 × 10–5 using -res 2000 and -superRes 10000 to bin reads. Interactions were mapped to RefSeq (56) mRNA transcription start sites based on alignment to hg19 provided by the UCSC genome browser. Reads were deposited at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/)under accession GSE79718.
Data and materials availability.
The methylation and gene expression data for freshly isolated airway epithelial cells have been deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession GSE85568.
Statistics.
Data were analyzed with R software (version 3.1.2). CpG-gene pair correlations were determined using Spearman correlations between β values and the log-transformed RUV-adjusted residual counts per million. The Kolmogorov-Smirnov test was used to compare the distributions of Spearman correlation P values. A P value less than 0.05 was considered significant. meQTL enrichments among CpG subsets were performed by filtering all CpG sites in our study to those that are within 5 kb of a SNP and subset further to the most significant meQTL per CpG. Subsequent distributions were compared using a χ2 test in which the “all CpG” group included the set of CpGs complementary to each comparison group. A P value less than 0.05 was considered significant. Mendelian randomization was performed using the ivreg2 function for R (http://www.r-bloggers.com/an-ivreg2-function-for-r/), using genotype as the instrumental variable. An instrumental variable is a variable that is associated with variation in an intermediate factor (methylation in this case) but not associated with other factors that are known to influence the causal effect of methylation on gene expression (i.e., confounding variables that may affect both gene expression and methylation). A P value less than 0.05 was considered significant. We used the associations between genotype (at rs2517955) and both methylation (cg05616858) and gene expression (ORMDL3) to assess the causal effect of methylation on ORMDL3 expression.
Study approval.
These studies were approved by the University of Chicago Institutional Review Board. Written informed consent was obtained from all research subjects.
Author contributions
JNJ and CO conceptualized the study and wrote the manuscript. JNJ cultured the primary cells, processed nucleic acids, performed genotype imputation, and processed and analyzed the methylation, genotype, and expression data. RAM performed RNAseq quality control and mapping. NJS, DRS, and MAN performed the Hi-C studies. ETN, AIS, JS, SRW, MAN, DLN, and YG contributed to study design and interpretation of the data. DKH, ETN, and SRW provided primary AECs and clinical data. JS arranged the Gift of Hope/Regional Organ and Tissue Donor Network lung procurement at the University of Chicago.
Supplementary Material
Acknowledgments
The authors thank Katherine Naughton, Christine Billstrand, and Jyotsna Sudi for sample processing and Kyung Won Kim and Catherine Stanhope for statistical support. Human lung tissue for cell culture studies was provided by the Gift of Hope/Regional Organ and Tissue Donor Network through the generosity of donor families. This research was supported by National Institute of Allergy and Infectious Diseases grant U19 AI095230 (to CO and SRW), the Office of Research on Women’s Health (to CO and SRW), and NIH grants P01 HL070831 (to CO) and U01 AI106683 (to CO). JNJ was supported by an American Heart Association postdoctoral fellowship and by NIH grant T32 HL07605.
Footnotes
JNJ’s present address is: Research and Development, USANA Health Sciences Inc., Salt Lake City, Utah, USA.
RAM’s present address is: Duke Center for Applied Genomics and Precision Medicine and Duke Department of Medicine, Duke University, Durham, North Carolina, USA.
Conflict of interest: J. Solway has been a scientific advisor for and has a financial interest in PulmOne Advanced Medical Devices Ltd., Israel, and received reimbursement for expenses. He served on the Respiratory Therapy Clinical Advisory Board for Hollister Inc., and for this received honoraria and was reimbursed for travel and meal expenses incurred during meetings. He has received a research grant from AstraZeneca Inc., from 2006 to 2014, that was administered through the University of Chicago. He has multiple patents concerning a smooth muscle gene promoter and one pending concerning a method to determine respiratory physiological parameters (6090618; 6114311; 6284743; 6291211; 6297221; 6331527; 7169764). He has consulted for Novartis Institute for Biomedical Research, for which he received an honorarium and travel reimbursement. He was a member of the scientific advisory board for Cytokinetics Inc., for which he received honoraria and travel reimbursement.
Reference information:JCI Insight. 2016;1(20):e90151. doi:10.1172/jci.insight.90151.
Contributor Information
Debora R. Sobreira, Email: dsobreira@bsd.uchicago.edu.
Julian Solway, Email: jsolway@medicine.bsd.uchicago.edu.
Yoav Gilad, Email: gilad@uchicago.edu.
Carole Ober, Email: c-ober@bsd.uchicago.edu.
References
- 1. Asthma Fact Sheet. World Health Organization. http://www.who.int/mediacentre/factsheets/fs307/en/ Accessed October 28, 2016.
- 2.Bisgaard H, Szefler S. Prevalence of asthma-like symptoms in young children. Pediatr Pulmonol. 2007;42(8):723–728. doi: 10.1002/ppul.20644. [DOI] [PubMed] [Google Scholar]
- 3.Polderman TJ, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47(7):702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- 4.Wesolowska-Andersen A, Seibold MA. Airway molecular endotypes of asthma: dissecting the heterogeneity. Curr Opin Allergy Clin Immunol. 2015;15(2):163–168. doi: 10.1097/ACI.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bønnelykke K, Ober C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J Allergy Clin Immunol. 2016;137(3):667–679. doi: 10.1016/j.jaci.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27(3):107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furlong LI. Human diseases through the lens of network biology. Trends Genet. 2013;29(3):150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep. 2012;12(3):211–220. doi: 10.1007/s11882-012-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North ML, Ellis AK. The role of epigenetics in the developmental origins of allergic disease. Ann Allergy Asthma Immunol. 2011;106(5):355–61; quiz 362. doi: 10.1016/j.anai.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Liang L, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520(7549):670–674. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 12.Bell JT, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12(1):R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banovich NE, et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10(9):e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flutre T, Wen X, Pritchard J, Stephens M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 2013;9(5):e1003486. doi: 10.1371/journal.pgen.1003486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torgerson DG, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Genetic associations with expression for genes implicated in GWAS studies for atherosclerotic cardiovascular disease and blood phenotypes. Hum Mol Genet. 2014;23(3):782–795. doi: 10.1093/hmg/ddt461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong H, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6(5):e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamazon ER, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18(3):340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calışkan M, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368(15):1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy A, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19(23):4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy. 2015;70(10):1309–1318. doi: 10.1111/all.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith GD. Mendelian Randomization for Strengthening Causal Inference in Observational Studies: Application to Gene × Environment Interactions. Perspect Psychol Sci. 2010;5(5):527–545. doi: 10.1177/1745691610383505. [DOI] [PubMed] [Google Scholar]
- 27.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicodemus-Johnson J, et al. Genome-Wide Methylation Study Identifies an IL-13-induced Epigenetic Signature in Asthmatic Airways. Am J Respir Crit Care Med. 2016;193(4):376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gagliardo R, et al. The role of transforming growth factor-β1 in airway inflammation of childhood asthma. Int J Immunopathol Pharmacol. 2013;26(3):725–738. doi: 10.1177/039463201302600316. [DOI] [PubMed] [Google Scholar]
- 33.Makinde T, Murphy RF, Agrawal DK. Immunomodulatory role of vascular endothelial growth factor and angiopoietin-1 in airway remodeling. Curr Mol Med. 2006;6(8):831–841. doi: 10.2174/156652406779010795. [DOI] [PubMed] [Google Scholar]
- 34.Michail S, Mezoff E, Abernathy F. Role of selectins in the intestinal epithelial migration of eosinophils. Pediatr Res. 2005;58(4):644–647. doi: 10.1203/01.PDR.0000180572.65751.F4. [DOI] [PubMed] [Google Scholar]
- 35.Zheng S, et al. Impaired innate host defense causes susceptibility to respiratory virus infections in cystic fibrosis. Immunity. 2003;18(5):619–630. doi: 10.1016/S1074-7613(03)00114-6. [DOI] [PubMed] [Google Scholar]
- 36.Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicodemus-Johnson J, et al. Maternal asthma and microRNA regulation of soluble HLA-G in the airway. J Allergy Clin Immunol. 2013;131(6):1496–1503. doi: 10.1016/j.jaci.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicodemus-Johnson J, et al. Genome-Wide Methylation Study Identifies an IL-13-induced Epigenetic Signature in Asthmatic Airways. Am J Respir Crit Care Med. 2016;193(4):376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3(6):771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 40.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee KW, Pausova Z. Cigarette smoking and DNA methylation. Front Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28(14):1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32(9):896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 49.Tandon A, Patterson N, Reich D. Ancestry informative marker panels for African Americans based on subsets of commercially available SNP arrays. Genet Epidemiol. 2011;35(1):80–83. doi: 10.1002/gepi.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartung J, Knapp G. On tests of the overall treatment effect in meta-analysis with normally distributed responses. Stat Med. 2001;20(12):1771–1782. doi: 10.1002/sim.791. [DOI] [PubMed] [Google Scholar]
- 51.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battle A, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347(6222):664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.