Abstract
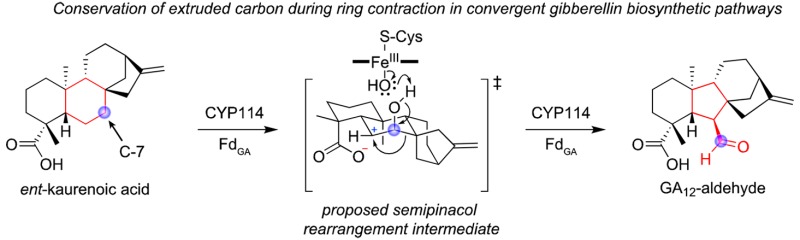
Bacteria have evolved gibberellin phytohormone biosynthesis independently of plants and fungi. Through 13C-labeling and NMR analysis, the mechanistically unusual “B” ring contraction catalyzed by a cytochrome P450 (CYP114), which is the committed step in gibberellin biosynthesis, was shown to occur via oxidative extrusion of carbon-7 from ent-kaurenoic acid in bacteria. This is identical to the convergently evolved chemical transformation in plants and fungi, suggesting a common semipinacol rearrangement mechanism potentially guided by carbon-4α carboxylate proximity.
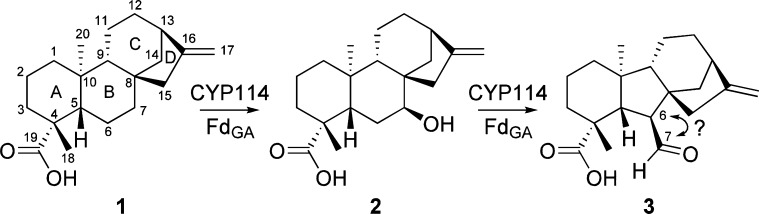
Gibberellins (GAs) are phytohormones that play important roles in plant growth, development, and interactions with microbes.1,2 These diterpenoid-derived compounds are characterized by a 6-5-6–5 fused ring structure, termed the ent-gibberellane carbon skeleton. However, GAs are produced via ent-kaurane precursors, which have a 6-6-6–5 carbon skeleton (see Scheme 1 for numbering and ring nomenclature). Accordingly, the committed step in GA biosynthesis is contraction of the “B” ring from a cyclohexane to cyclopentane. This occurs via oxidation of ent-kaurenoic acid (1; ent-kaur-16-en-19-oic acid), first to 7β-hydroxy-ent-kaurenoic acid (2; ent-7α-hydroxykaurenoic acid), and then to GA12-aldehyde (3), the latter of which involves oxidative extrusion of an endocyclic “B” ring carbon (Scheme 1). The cytochrome P450 mono-oxygenases (CYPs) catalyzing this mechanistically unusual and challenging reaction are termed ent-kaurenoic acid oxidases (KAOs).
Scheme 1. Reaction Catalyzed by CYP114 + FdGA in Bacterial GA Biosynthesis.
1 (ent-kaurenoic acid) is representative of the ent-kaurane backbone, and 3 (GA12-aldehyde) represents the ent-gibberellane backbone.
In addition to their endogenous production by plants, GAs are also produced by certain plant-associated fungi and bacteria, wherein the relevant biosynthetic pathways have independently evolved.3,4 Plant and fungal GA biosynthesis has been extensively studied, and it has been directly demonstrated that the carbon extruded from the “B” ring is C-7 in fungal biosynthesis,5−7 and convincing indirect evidence has been presented that plants also extrude C-7 (i.e., retention of the C-6α proton from 1 during the “B” ring contraction reaction).8,9 Moreover, 2 has been shown to be a bona fide intermediate in both plant and fungal GA biosynthesis. By contrast, 6β,7β-dihydroxy-ent-kaurenoic acid (4; ent-6α,7α-dihydroxy-kaurenoic acid), which is also produced in the KAO-catalyzed oxidation of 1 by plants and fungi, is not,10−14 thus, implicating a mechanism in which C-7, but not C-6, is hydroxylated prior to ring contraction.5,15
The GA biosynthetic pathway in bacteria has only recently been elucidated.4,16 In particular, the role of each enzyme from a CYP-rich gene cluster/operon in symbiotic rhizobia has now been functionally identified, showing that they act to produce GA9 (Scheme S2). While rhizobia only express these enzymes and produce GA after differentiation into their nodule-residing bacteroid form,17 it was possible to observe activity with the individual enzymes upon recombinant expression.4,16,18,19 Notably, “B” ring contraction requires not only a CYP (CYP114) but also the ferredoxin (FdGA) found within the operon, which presumably acts as an electron donor.4 This is distinct from plant and fungal KAOs, which simply utilize an archetypical cytochrome P450 reductase for their activity.12,20,21 When expressed alone, CYP114 only converts 1 to 2, while coexpression of CYP114 and FdGA enables full conversion of 1 to 3. This suggests that endogenous ferredoxins from the recombinant host support partial CYP114 activity and indicates a unique role for FdGA in facilitating full activity, presumably through its interaction with CYP114. Although recombinantly coexpressed CYP114 and FdGA are not able to convert 2 to 3, nodule-extracted rhizobial bacteroids can use 2 as a GA precursor, implicating this as an intermediate in bacterial GA biosynthesis as well.4
Though the intermediacy of 2 might be taken to suggest that C-7 also will be extruded during the “B” ring contraction reaction catalyzed by the convergently evolved bacterial enzymes, it is still plausible that C-6 might be extruded instead (e.g., via a pinacol ring rearrangement mechanism).5 The extrusion of C-7 during fungal GA biosynthesis was shown by feeding (2-13C)mevalonolactone (5; the δ-lactone form of mevalonate) to cultures of Fusarium fujikuroi (the anamorph of Gibberella fujikuroi), which leads to specific labeling of C-7 in 1, followed by NMR analysis of the resulting GA3 final product.5,7 This approach was enabled, at least in part, by the high titers of GA3 produced by fungal cultures. By contrast, rhizobia produce only small amounts of GAs.17 Nevertheless, the recombinant coexpression of CYP114 and FdGA, which carry out “B” ring contraction with 1, provides a means to analyze this reaction in more detail (i.e., via incubation with 13C-labeled 1).
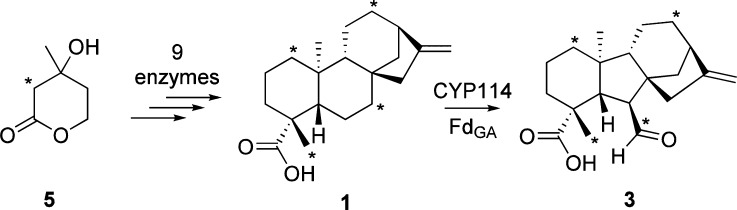
Although bacteria usually produce isoprenoids/terpenoids via the nonmevalonate pathway, Keasling and co-workers have engineered incorporation of the mevalonate-dependent isoprenoid precursor pathway from yeast into E. coli.22 Of particular relevance here, a single plasmid enables production of the universal isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) from 5. In turn, a modular metabolic engineering system has been developed that is compatible with this plasmid.23 This enables the production of diterpenoids via incorporation of a (E,E,E)-geranylgeranyl diphosphate synthase (producing this general diterpene precursor from IPP and DMAPP), subsequently acting diterpene cyclases/synthases, and even further downstream acting CYPs in conjunction with their requisite redox partner.24 Thus, it was possible to produce 13C-labeled 1 by simply feeding (2-13C)-5 to E. coli engineered to produce 1 from 5 (i.e., via coexpression of the necessary nine enzymes; see Scheme S3, Figures S1 and S2). As expected, this enabled isolation of 1 with 13C enrichment at four positions, as initially confirmed by gas chromatography–mass spectroscopy (GC-MS), with comparison to an authentic standard (Figure S2). The expected incorporation of 13C at carbons 1, 7, 12, and 18 (Scheme 2)5,7 was verified by 13C NMR analysis with comparison to unlabeled 1 (Figure S3; Tables S1 and S2).
Scheme 2. 13C Label from (2-13C)-5 Is Specifically Incorporated into 1 via Metabolically Engineered Bacteria.
13C-labeled 1 can then be incubated with bacteria recombinantly co-expressing CYP114 and FdGA to produce 13C-labeled 3.
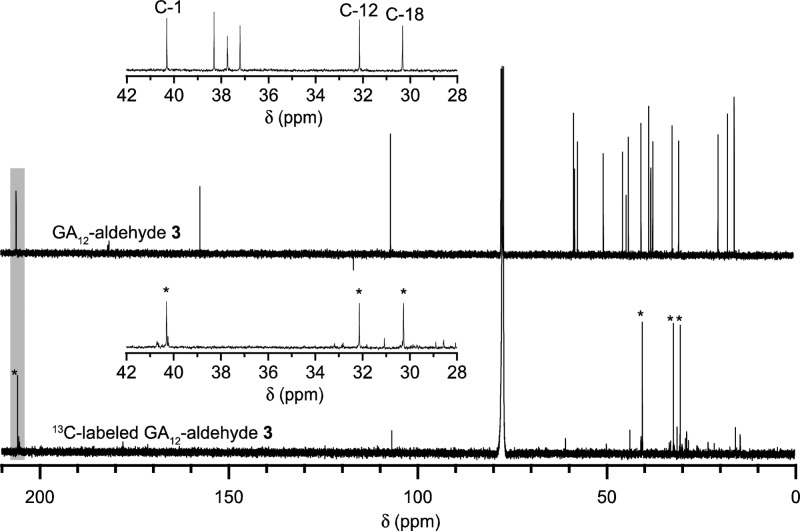
To investigate the origin of the extruded carbon, 13C-enriched 1 was fed to bacterial cultures recombinantly coexpressing CYP114 and FdGA. This allowed isolation of 3 enriched at four positions, as confirmed by GC-MS comparison to an authentic standard (Figure S4). Following purification, 13C NMR analysis showed three enriched carbons with chemical shifts between 30 and 50 ppm, indicative of alkyls, while the fourth had a chemical shift of over 200 ppm, representing a carbonyl carbon (Figure 1; Tables S3 and S4). These shifts were further verified by comparison to those measured for unlabeled 3. Thus, it was demonstrated that C-7 of 1 was extruded and oxidized to the aldehyde of 3.
Figure 1.
Comparison of the 13C-labeled 313C NMR spectrum to that of the unlabeled standard (800 MHz, CDCl3 for each) reveals that C-7 is extruded during the ring contraction from ent-kaurenoic acid 1 to GA12-aldehyde 3. The 13C-enriched carbons in the labeled substrate are indicated with asterisks (*).
As with plant and fungal GA biosynthesis, 2 is observed and seems to serve as an intermediate in bacteria as well,4 implying C-7β hydroxylation prior to ring contraction. It is known for plants and fungi that the 6β hydrogen of 1 is removed prior to rearrangement/ring contraction, although 4 does not serve as an intermediate,8,9,25 and seems to be a side product of the corresponding CYPs in both kingdoms. Interestingly, closer analysis of incubations of 1 in cells coexpressing CYP114 and FdGA showed that a trace amount of 4 is produced (Figure S5). However, feeding 4 to bacterial cultures recombinantly coexpressing CYP114 and FdGA does not result in further conversion (Figure S5), suggesting that 4 is a side product of bacterial KAO activity. Thus, the KAOs from all three biological kingdoms not only extrude C-7 in “B” ring contraction but also exhibit a conserved order of chemical transformations, with conversion of 1 to 3 via 2, but apparently not 4.
Although it is possible that the conversion of 1 to 3 could proceed via transient formation of a C-19,6-γ-lactone ring, which is used to achieve “B” ring contraction in the chemical synthesis of GAs,26 this corresponds to the known kaurenolide side products of GA biosynthesis in both plants and fungi.27 Notably, these compounds were not observed here, nor have these previously been reported in other investigations of bacterial GA biosynthesis.4,16 To further evaluate the possibility of the “B” ring contraction reaction proceeding through this type of intermediate, kaurenolide and 7β-hydroxykaurenolide (see Figure S6 for chemical structures) were fed to bacterial cultures recombinantly coexpressing CYP114 and FdGA. However, these were not converted in this system (Figure S6) and likely are not intermediates in bacterial GA biosynthesis, similar to what has been reported for plants and fungi.27
All three biological kingdoms have convergently evolved KAOs that carry out this committed step in GA biosynthesis.4 In each case the KAO is a member of the CYP superfamily, but falls within phylogenetically distinct families specific to each biological kingdom, as bacterial KAOs come from the CYP114 family, the fungal KAOs from the CYP68 family, and those from plants from the CYP88 family.14,28 However, the results reported here show that these convergently evolved KAOs all catalyze extrusion of the same carbon, using a conserved order of central chemical transformations (i.e., from 1 to 3 via 2) in each case.4 This suggests a physical/chemical restraint for the “B” ring contraction reaction.
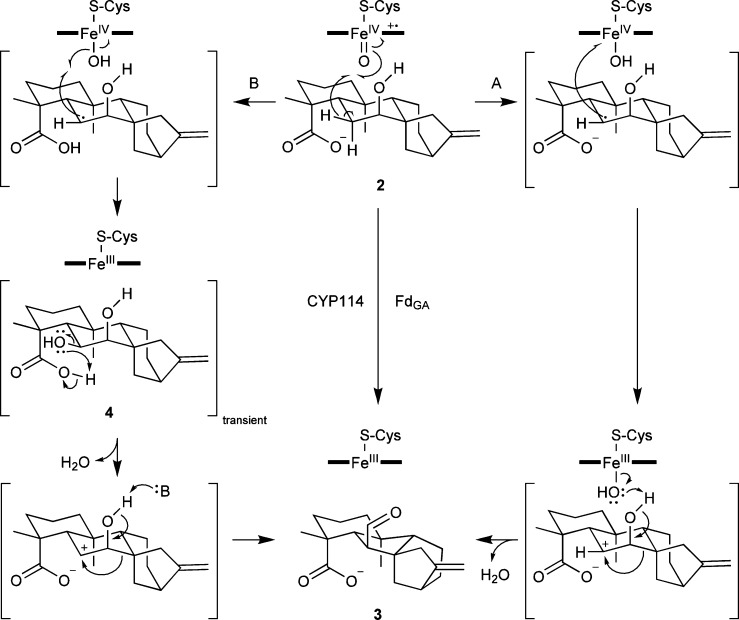
Intriguingly, there appears to always be a free C-4α carboxylate (C-19) present. While CYPs typically catalyze radical based reactions, it has been previously suggested that this “B” ring contraction reaction might proceed via a carbocation based mechanism instead, with transfer of the unpaired electron from the initially formed C-6 radical of 2 to the heme-iron.29,30 Notably, the C-4α carboxylate is ideally positioned to offer anchimeric assistance to formation of this putative C-6 carbocation, potentially guiding the observed oxidative extrusion of C-7 via a semipinacol rearrangement mechanism (Scheme 3, path A).31 Alternatively, hydroxylation of 2 to form 4 as a transient intermediate would enable rearrangement via a classical pinacol mechanism. If protonated in the CYP114 active site (pKa ≈ 4.6), the C-4α moiety might then provide anchimeric assistance by acting as an acid to protonate the 6β-hydroxyl group, leading to specific extrusion of C-7 (Scheme 3, path B). While the lack of enzymatic conversion of 4 may argue against the classic pinacol mechanism, it is possible that suboptimal expression of CYP114 and/or FdGA prevented turnover here.
Scheme 3. Proposed Reaction Mechanisms for “B” Ring Contraction during GA Biosynthesis That Proceed Either through a Semipinacol (Path A) or Classic Pinacol (Path B) Rearrangement.
Consistent with a role for anchimeric assistance by the free C-4α carboxylate, neither the methyl ester of 1 nor ent-kaurenal (which in the predominant diol form sterically resembles 1) is further transformed by recombinantly expressed CYP114 (either with or without coexpression of FdGA).4 Additional support for the proposed mechanism stems from the use of pinacol-like intermediates to achieve “B” ring contraction in the chemical synthesis of GA.26 Thus, it seems likely that the independently evolved CYPs catalyzing the characteristic “B” ring contraction in gibberellin biosynthesis in all three biological kingdoms may have converged on a common (semi)pinacol rearrangement mechanism to selectively carry out this unusual and challenging reaction.
Acknowledgments
We thank D. Bruce Fulton (Iowa State University) for expert assistance with the NMR analysis and Dr. Peter Hedden (Rothamsted Research, U.K.) for insightful discussion and for the 6β,7β-dihydroxy-ent-kaurenoic acid (4), kaurenolide, and 7β-hydroxykaurenolide substrates. This work was supported by grants from the National Institutes of Health (GM109773) and National Science Foundation (CHE-1609917) to R.J.P. and by the DFG Collaborative Research Center 813 “Chemistry at Spin Centers” (J.S.D.).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.orglett.6b02569.
Experimental methods, Supplemental figures and tables (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hedden P.; Thomas S. G. Biochem. J. 2012, 444, 11–25. 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- De Bruyne L.; Höfte M.; De Vleesschauwer D. Mol. Plant 2014, 7, 943–959. 10.1093/mp/ssu050. [DOI] [PubMed] [Google Scholar]
- Hedden P.; Sponsel V. J. Plant Growth Regul. 2015, 34, 740–760. 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett R. S.; Montanares M.; Marcassa A.; Lu X.; Nagel R.; Charles T. C.; Hedden P.; Rojas M. C.; Peters R. J.. Nat. Chem. Biol. [Online early access]. 10.1038/nchembio.2232. Published Online: November 14, 2016. http://dx.doi.org/1038/nchembio.2232 (accessed November 14, 2016). [DOI] [Google Scholar]
- Citron C. A.; Brock N. L.; Tudzynski B.; Dickschat J. S. Chem. Commun. (Cambridge, U. K.) 2014, 50, 5224–5226. 10.1039/C3CC45982A. [DOI] [PubMed] [Google Scholar]
- Birch A. J.; Richards R. W.; Smith H.; Harris A.; Whalley W. B. Tetrahedron 1959, 7, 241–251. 10.1016/S0040-4020(01)93192-8. [DOI] [Google Scholar]
- Evans B. R.; Hanson J. R.; Siverns M. J. Chem. Soc., Perkin Trans. 1 1975, (15), 1514–1517. 10.1039/p19750001514. [DOI] [PubMed] [Google Scholar]
- Graebe J. E.; Hedden P.; MacMillan J. J. Chem. Soc., Chem. Commun. 1975, (5), 161. 10.1039/c39750000161. [DOI] [Google Scholar]
- Castellaro S. J.; Dolan S. C.; Hedden P.; Gaskin P.; MacMillan J. Phytochemistry 1990, 29, 1833–1839. 10.1016/0031-9422(90)85024-A. [DOI] [Google Scholar]
- Hanson B. J. K.; White A. F. Chem. Commun. 1969, 4, 7–8. [Google Scholar]
- Graebe J. E.; Hedden P.; Gaskin P.; MacMillan J. Phytochemistry 1974, 13, 1433–1440. 10.1016/0031-9422(74)80304-3. [DOI] [Google Scholar]
- Helliwell C. A.; Chandler P. M.; Poole A.; Dennis E. S.; Peacock W. J. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 2065–2070. 10.1073/pnas.98.4.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S. E.; Elliott R. C.; Helliwell C. A.; Poole A. T.; Reid J. B. Plant Physiol. 2003, 131, 335–344. 10.1104/pp.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M. C.; Hedden P.; Gaskin P.; Tudzynski B. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 5838–5843. 10.1073/pnas.091096298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan J.; Ward D. A.; Phillips A. L.; Sánchez-Beltrán M. J.; Gaskin P.; Lange T.; Hedden P. Plant Physiol. 1997, 113, 1369–1377. 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsukami Y.; Ueda M. Sci. Rep. 2016, 6, 27998. 10.1038/srep27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C.; Baginsky C.; Hedden P.; Gong F.; Carú M.; Rojas M. C. Phytochemistry 2014, 98, 101–109. 10.1016/j.phytochem.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Morrone D.; Chambers J.; Lowry L.; Kim G.; Anterola A.; Bender K.; Peters R. J. FEBS Lett. 2009, 583, 475–480. 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- Hershey D. M.; Lu X.; Zi J.; Peters R. J. J. Bacteriol. 2014, 196, 100–106. 10.1128/JB.01031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek S.; Rojas M. C.; Hedden P.; Gaskin P.; Hopkins P.; Tudzynski B. J. Biol. Chem. 2004, 279, 25075–25084. 10.1074/jbc.M308517200. [DOI] [PubMed] [Google Scholar]
- Troncoso C.; Cárcamo J.; Hedden P.; Tudzynski B.; Rojas M. C. Phytochemistry 2008, 69, 672–683. 10.1016/j.phytochem.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Martin V. J. J.; Pitera D. J.; Withers S. T.; Newman J. D.; Keasling J. D. Nat. Biotechnol. 2003, 21, 796–802. 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Morrone D.; Lowry L.; Determan M. K.; Hershey D. M.; Xu M.; Peters R. J. Appl. Microbiol. Biotechnol. 2010, 85, 1893–1906. 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka N.; Lu X.; Yang B.; Peters R. J. Mol. Plant 2015, 8, 6–16. 10.1016/j.molp.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J. R.; Hawker J.; White A. F. J. Chem. Soc., Perkin Trans. 1 1972, 1, 1892–1895. 10.1039/P19720001892. [DOI] [Google Scholar]
- Mander L. N. Chem. Rev. 1992, 92, 573–612. 10.1021/cr00012a005. [DOI] [Google Scholar]
- Macmillan J. Nat. Prod. Rep. 1997, 14, 221–243. 10.1039/np9971400221. [DOI] [Google Scholar]
- Bömke C.; Tudzynski B. Phytochemistry 2009, 70, 1876–1893. 10.1016/j.phytochem.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano P. R.Substrate Oxidation by Cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Ortiz De Montellano P. R., Ed.; Springer International Publishing: Switzerland, 2015; Vo1. 1, pp 111–176. [Google Scholar]
- Ortiz de Montellano P. R.; Nelson S. D. Arch. Biochem. Biophys. 2011, 507, 95–110. 10.1016/j.abb.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. L.; Fan C. A.; Tu Y. Q. Chem. Rev. 2011, 111, 7523–7556. 10.1021/cr200055g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.