SUMMARY
Tropomyosins comprise a large family of actin binding proteins with critical roles in diverse actin-based processes [1], but our understanding of how they mechanistically contribute to actin filament dynamics has been limited. We addressed this question in S. cerevisiae, where tropomyosins (Tpm1 and Tpm2), profilin (Pfy1), and formins (Bni1 and Bnr1) are required for the assembly of an array of actin cables that facilitate polarized vesicle delivery and daughter cell growth. Formins drive cable formation by promoting actin nucleation and by accelerating actin filament elongation together with profilin [2]. In contrast, how tropomyosins contribute mechanistically to cable formation has been unclear, but genetic studies demonstrate that Tpm1 plays a more important role than Tpm2 [3, 4]. Here, we found that loss of TPM1 in strains lacking BNR1 but not BNI1 led to severe defects in cable formation, polarized secretion, and cell growth, suggesting TPM1 function is required for proper Bni1-mediated cable assembly. Further, in vitro TIRF microscopy demonstrated that Tpm1 strongly enhanced Bni1- but not Bnr1-mediated actin nucleation, without affecting filament elongation rate, whereas Tpm2 had no effects on Bni1 or Bnr1. Tpm1 stimulation of Bni1-mediated nucleation also required profilin and its interactions with both G-actin and formins. Together these results demonstrate that yeast Tpm1 works in concert with profilin to promote formin-dependent nucleation of actin cables, thus expanding our understanding of how specific tropomyosin isoforms influence actin dynamics.
Keywords: actin, profilin, formin, tropomyosin, yeast
RESULTS AND DISCUSSION
Tropomyosins (Tpms) are one of the largest families of actin-associated proteins, with over 40 isoforms in mammals generated by alternative splicing from four different genes. Mammalian Tpm isoforms have diverse cellular localization patterns, and perform a wide range of vital roles in actin-based processes, ranging from cell motility and adhesion to intracellular traffic and muscle contraction [1, 5]. Tpms form extended coiled-coil dimers, which in turn polymerize end-to-end to cooperatively decorate the sides of actin filaments. In addition to their canonical roles in regulating myosin activity in both muscle and nonmuscle cells [6, 7], Tpms can govern actin dynamics by influencing the interactions and effects of other actin binding proteins, e.g., by blocking binding of Arp2/3 complex to inhibit branched nucleation [8], and by blocking binding of cofilin to inhibit filament severing and disassembly [9]. Tpms also can relieve formin-mediated deceleration of actin elongation, which occurs in the absence of profilin, enabling filaments to elongate faster, albeit at reduced rates compared to free barbed end growth [10, 11].
We investigated Tpm functions in S. cerevisiae, where only two Tpms (Tpm1 and Tpm2) and two formins (Bni1 and Bnr1) are expressed, compared to over 40 Tpm isoforms and 15 formins in mammals. S. cerevisiae tpm1Δ mutants display a severe loss of actin cables and are highly defective in polarized cell growth [12], whereas tpm2Δ cells display no obvious defects [4]. In a tpm1Δ background, TPM2 is required for viability, suggesting that Tpm1 and Tpm2 share at least some overlapping functions; however, overexpression of Tpm2 fails to rescue tpm1Δ defects [4], indicating that the functions of Tpm1 and Tpm2 are largely distinct. Until now, the mechanistic basis for these differences has gone unexplained. Further, it has remained unclear what specific role(s) Tpm1 plays in promoting cable formation.
Tpm1 is required specifically for Bni1-dependent actin cable formation in vivo
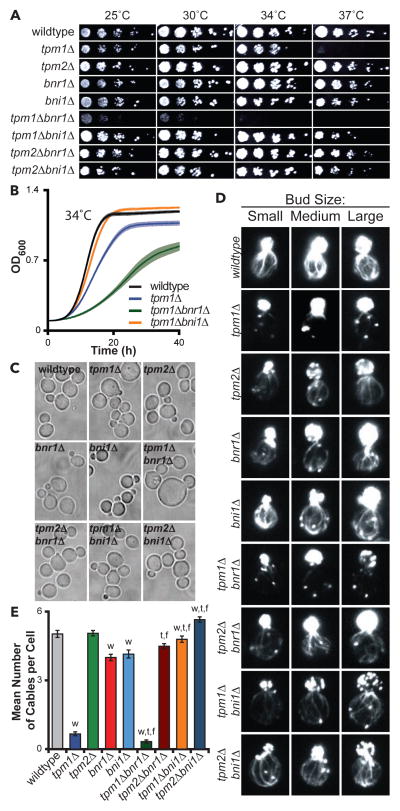
We first tested the importance of Tpm1 and Tpm2 for cell growth separately in bni1Δ and bnr1Δ backgrounds, where we could assess their individual effects on each formin. We generated nine isogenic strains, and directly compared them for growth at different temperatures (Fig 1A). This analysis confirmed that tpm2Δ caused no discernable growth defects [4], and revealed that tpm2Δ does not exacerbate the defects of bni1Δ or bnr1Δ. In contrast, tpm1Δ caused strong growth defects, as previously observed [3], and further exacerbated growth defects specifically in combination with bnr1Δ but not bni1Δ (Fig 1A). These effects were confirmed by analyzing cell doubling rates in liquid cultures grown at 25°C and 34°C (Fig 1B and S1A–S1E). Interestingly, our analysis also showed that tpm1Δ growth defects are suppressed by bni1Δ (Fig 1A, 1B, and S1A). Importantly, this suppression was not due to a spatial redistribution of Bnr1, as Bnr1-GFP localized primarily to the bud neck in wildtype and bni1Δ cells (Fig S1F). Together, these results suggest that Tpm1 plays a critical role specifically in Bni1-dependent processes.
Figure 1. Tpm1 is critical for Bni1-dependent actin cable formation and cell growth.
(A) Five-fold serial dilutions of yeast strains grown on YEPD plates at indicated temperatures. (B) Growth curves for indicated yeast strains grown in YEPD at 34°C with shaking in a microplate absorbance reader with OD600 measured every 5 min. Data averaged from ≥18 replicates/strain. Lighter shading, SEM. (C) Representative DIC images of cells grown to mid-log phase at 25°C in Y EPD and fixed. Scale bar, 5 μm. (D) Representative images of small, medium, and large budded cells grown at 25°C in YEPD, fixed, and stained with Alexafluor-488 phalloidin. Scale bar, 3 μm. (E) Average number of actin cables visible in the mother compartment (n ≥ 50 cells analyzed per strain). Student’s T-test used to determine significant difference (p < 0.05) from wildtype (‘w’), appropriate tropomyosin single mutant (‘t’), or appropriate formin single mutant (‘f’). See also Figure S1.
Further examination showed that tpm1Δ and tpm1Δbnr1Δ cells are enlarged, consistent with impaired polarized growth (Fig 1C and S1G). These morphological defects were more pronounced in tpm1Δbnr1Δ compared to tpm1Δ cells, mirroring the growth analysis above. Defects in actin organization (reduced cable staining and depolarization of patches) also correlated with growth defects (Fig 1D, S1H, and S1I), with the loss of cables being greater in tpm1Δbnr1Δ compared to tpm1Δ (Fig 1E). In contrast, tpm1Δbni1Δ had an almost wildtype number of visible cables, consistent with bni1Δ suppressing tpm1Δ growth defects (Fig 1E); however, cables in tpm1Δbni1Δ cells were noticeably thinner than in wildtype cells (Fig 1D and S1I). Overall, these data indicate a strong correlation between robust cable formation and efficient growth, and reinforce the view that Tpm1 facilitates cable formation by Bni1 but not Bnr1.
Efficient secretory vesicle traffic requires Tpm1-enhanced Bni1-mediated cable assembly
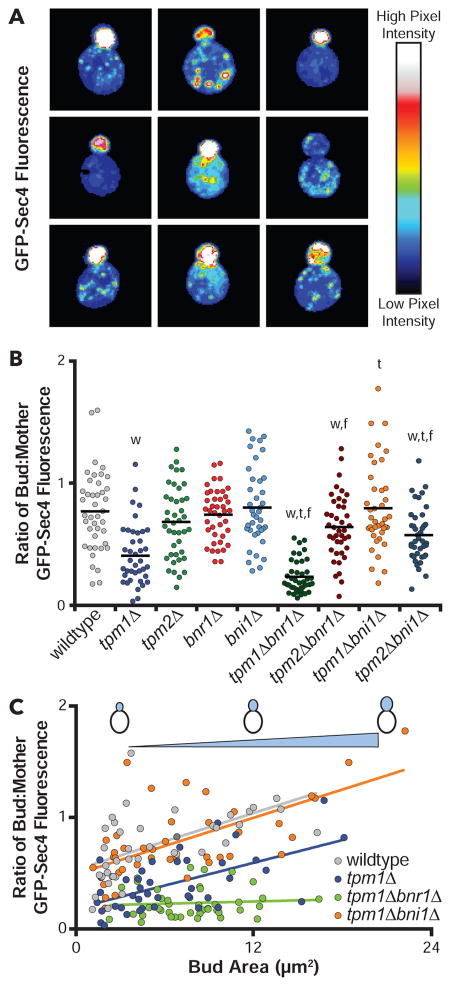
Post-Golgi vesicles are transported by myosin V (Myo2) along actin cables and accumulate at the bud tip, poised for exocytosis. This polarized distribution of secretory vesicles (marked by Myo2 or Sec4) reflects efficient polarized traffic, and is rapidly lost upon acute disruption of formins [13, 14]. We quantified vesicle distribution as a physiological test of cable function in our strains by comparing the ratio of GFP-Sec4 fluorescence in the bud versus mother for small- and medium-budded cells (Fig 2A and 2B), which we refer to as the ‘polarity ratio’. In wildtype cells, the polarity ratio was 0.77, which reflects efficient polarized trafficking (note: this value is not higher because mother cells are much larger on average than buds). Four mutants showed significantly decreased polarity ratios compared to wildtype cells. The most severely impaired was tpm1Δbnr1Δ (0.23), with more modest defects in tpm1Δ (0.41), tpm2Δbni1Δ (0.58), and tpm2Δbnr1Δ (0.64).
Figure 2. Efficient secretory vesicle traffic requires Tpm1 function in Bni1-mediated actin cable assembly.
(A) Representative images of early log phase cells showing distribution of GFP-Sec4 (false colored). Scale bar, 3 μm. (B) Ratio of bud to mother GFP-Sec4 fluorescence. Each dot is the ratio quantified for one cell (n ≥ 40 cells per strain analyzed). Black line, Mean. Student’s T-test used to determine the indicated significant difference (p < 0.05) from wildtype (‘w’), appropriate tropomyosin single mutant (‘t’), or appropriate formin single mutant (‘f’). (C) Ratio of bud to mother GFP-Sec4 fluorescence plotted as a function of bud size (same data as in B). Lines fit by linear regression using standard equations in Prism 6.
Closer examination revealed that tpm1Δ cells have a partial polarization of vesicles at the bud tip, but also an abnormal accumulation of vesicles at the rear of the mother. This phenotype can be explained by the absence of long cables reaching the back of the mother compartment. tpm1Δ cells have short wispy cables near the neck, which may be ineffective in transporting vesicles from the rear of the mother (Fig 2A). This leads to an intermediate polarity ratio for tpm1Δ mutants, between wildtype and tpm1Δbnr1Δ. By comparison, tpm1Δbnr1Δ mutants showed a complete absence of GFP-Sec4 polarization, consistent with its more severe loss of cables compared to tpm1Δ mutants (Fig 1D and 1E). These data suggest that in tpm1Δ cells, most of the visible cables that remain, and which support vesicle transport near the neck, are derived from Bnr1-mediated cable assembly.
For wildtype and key mutant strains, we also plotted the polarity ratios as a function of bud size and performed linear regression analysis (Fig 2C). Since the bud continuously accumulates GFP-Sec4 marked vesicles as it grows, we expect this ratio to scale with bud size. Indeed, this was observed for wildtype cells, and for all but one of the mutants. The exception was tpm1Δbnr1Δ, which showed no correlation between bud size and polarity ratio (Fig 2C) consistent with tpm1Δbnr1Δ having severe defects in vesicle transport.
Tpm1 stimulates Bni1-mediated actin filament nucleation in vitro
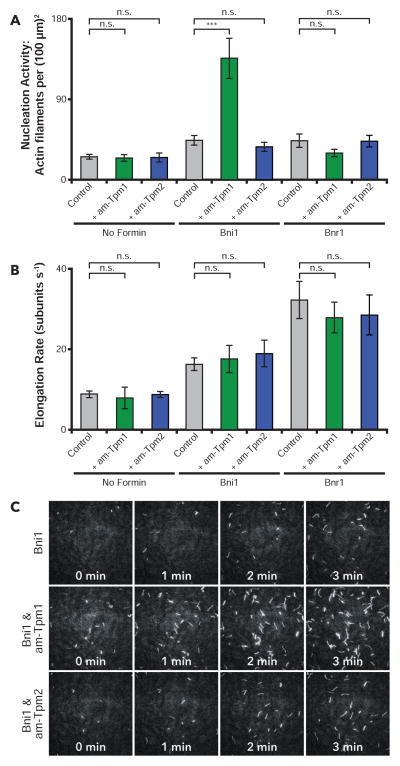
Our in vivo observations above suggest that Tpm1 may have a role in promoting Bni1-rather than Bnr1-mediated cable assembly. To test this model biochemically, we used total internal reflection fluorescence (TIRF) microscopy to directly visualize the formation of actin filaments in real time, and separately quantify the effects of each Tpm on actin filament nucleation and elongation with each formin. TIRF reactions contained actin monomers and yeast profilin (Pfy1) with different combinations of Tpm1 or Tpm2 and Bni1 or Bnr1 (active FH1-FH2-C fragments). Tpm1 and Tpm2 effects were tested at 2.5 μM, a concentration in their physiological range (B.G., unpublished observations).
Acetylation of S. cerevisiae Tpm1 and Tpm2 is critical for their normal biochemical interactions with actin [15], and Tpm1 acetylation is required in vivo for actin cable formation and polarized cell growth [16]. Therefore, we extended our analysis to compare non-acetylated (na) and acetylation-mimetic (am) forms of Tpm1 and Tpm2; am-Tpm forms carry an N-terminal Met-Ala-Ser extension that gives them similar actin binding properties to natively acetylated Tpm1 and Tpm2 [17, 18]. In the absence of formins, am-Tpm1 and am-Tpm2 showed no significant effects on filament nucleation or elongation (Fig 3A and 3B), and similar results were observed for na-Tpm1 and na-Tpm2 (Fig S2A and S2B). In reactions lacking Tpms, Bni1 and Bnr1 each stimulated actin nucleation and increased the rate of filament elongation (Fig 3A-C). Addition of am-Tpm1 to Bni1-containing reactions resulted in a substantial increase in nucleation activity (approximately 3-fold), with no significant effect on rate of filament elongation (Fig 3A-C; movie S1), and the nucleation-promoting effects of am-Tpm1 were concentration-dependent (Fig S2C). In contrast, am-Tpm1 had no effects on Bnr1-mediated nucleation or elongation (Fig 3A and 3B; and movie S2). Moreover, am-Tpm2 had no effects on Bni1- or Bnr1-mediated actin nucleation or elongation (Fig 3A; movie S1 and S2), and the non-acetylated forms of Tpm1 and Tpm2 likewise showed no effects on actin nucleation or elongation in the presence of Bni1 or Bnr1 (Fig S2A and S2B). Thus, the nucleation-promoting effects of am-Tpm1 on Bni1 were highly specific. These stimulatory effects were also confirmed using yeast actin (Fig S2D), and closely mirrored our in vivo observations, where Tpm1 makes an important contribution to Bni1- but not Bnr1-mediated actin cable formation.
Figure 3. Tpm1 and Tpm2 effects on formin-mediated actin filament nucleation and elongation.
(A) Actin nucleation effects determined by in vitro TIRF microscopy. Reactions contained 1 μM monomeric actin (20% Oregon Green-labeled), 3 μM profilin, and variable components: 250 pM Bni1, 200 pM Bnr1, 2.5 μM am-Tpm1, and 2.5 μM am-Tpm2. Number of filaments per field of view (FOV) was quantified 5 min after initiation of actin assembly from four independent experiments (16 FOV per condition). (B) Actin filament elongation rates determined from the same TIRF reactions as A. Elongation rates were calculated from the slopes of individual filament traces of length versus time (n ≥ 30 filaments per condition). (C) Representative time-lapse images from TIRF reactions. Scale bar, 20 μm. Error bars, SEM. Student’s T-test used to determine significant differences: n.s. – not significant, * < 0.05, ** < 0.01, *** < 0.001. See also Figure S2 and Movies S1 & S2.
Tpm1 enhancement of Bni1-mediated actin nucleation requires profilin and its interactions with G-actin and formins
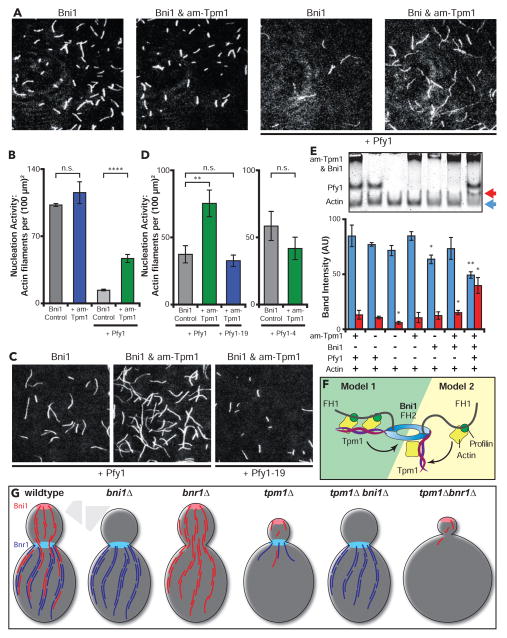
We included profilin in our TIRF experiments above because it enables formins to accelerate actin filament elongation [19]. However, profilin also suppresses both spontaneous and formin-mediated actin nucleation, except when specific formin nucleation co-factors such as APC and Bud6 are present [20, 21]. When yeast profilin was excluded from reactions, am-Tpm1 failed to enhance actin nucleation by Bni1 (Fig 4A and 4B; movie S3). Thus, profilin is required along with am-Tpm1 for enhanced Bni1-mediated nucleation. To dissect this mechanism, we repeated the assays using two profilin mutants: Pfy1-19 (Y120D), which is impaired in binding the poly-L-proline stretches in formin FH1 domains, and Pfy1-4 (K66A), which is impaired in G-actin binding [22, 23]. These experiments revealed that profilin interactions with the formin and G-actin are both critical for am-Tpm1 enhancement of Bni1 nucleation activity (Fig 4C and 4D; movie S4 and S5).
Figure 4. Tpm1 stimulation of Bni1-mediated actin nucleation requires profilin and its interactions with G-actin and formins.
(A) Representative FOVs from TIRF reactions containing 1.0 μM monomeric actin (20% Oregon Green-labeled) polymerized in the presence of 250 pM Bni1 and variable components: 3.0 μM yeast profilin and 2.5 μM am-Tpm1. Scale bar, 20 μm. (B) Actin nucleation effects determined from the reactions in A. Number of filaments per FOV was quantified as in Figure 3A from two independent experiments (12 FOV per condition). (C) Representative FOVs from TIRF reactions; conditions as in A, except using 3 μM Pfy1 and Pfy1-19 as indicated. Scale bar, 20 μm. (D) Actin nucleation effects determined by TIRF microscopy; conditions as in A, except using 3 μM Pfy1, Pfy1-4, or Pfy1-19 as indicated. The number of filaments per FOV was quantified as in Figure 3A from ≥ 2 separate experiments (≥ 12 FOVs per condition). (E) Native gel shift analysis of G-actin. Reactions contained 1.0 μM latrunculin bound G-actin with one or more variable components: 0.5 μM Bni1, 5.0 μM am-Tpm1, and 3 μM yeast profilin (Pfy1). Reactions fractionated by native PAGE, stained with Coomassie Blue and actin band intensity was quantified at the unshifted (blue arrow) and shifted (red arrow) positions. Data averaged from three independent experiments (blue and red bars, as above). Significant differences are compared to actin & profilin control (Lane 2). (F) Two possible models for how Tpm1 and profilin works together to enhance Bni1-mediated actin filament nucleation. (G) Graphical summary of actin cable pheontypes in indicated strains studied. Error bars, SEM. Student’s T-test used to determine significant differences: n.s. – not significant, * < 0.05, ** < 0.01, **** < 0.0001. See also Figure S3 and Movies S3, S4, and S5.
Finally, we asked whether any stable complexes form between Latrunculin-bound G-actin and different combinations of the proteins required for enhanced nucleation (Bni1, profilin, and am-Tpm1). In native gels, a shift in G-actin migration was detected only when Bni1, profilin, and am-Tpm1 were present (blue and red arrows, lower panel Fig 4E). Note that the interaction of profilin alone with G-actin is too transient to shift G-actin migration in these assays. By comparison, no G-actin shift was detected with am-Tpm2 and Bni1 in the presence or absence of profilin, or if Bnr1 was substituted for Bni1 (Fig S3B). Further, in pull-down assays lacking G-actin no direct interactions were observed between Bni1 and am-Tpm1 or am-Tpm2 with or without profilin (Fig S3C and S3D). Thus, we detect stable interactions only when Bni1, am-Tpm1, profilin, and G-actin are all present. While Tpm interactions with G-actin alone have not been observed before, our results indicate that Tpm1 interacts with profilin- and/or Bni1-bound actin monomers. Consistent with this possibility, the profilin- and Tpm-binding surfaces on actin are distinct, permitting simultaneous binding with actin [24, 25]. Given these observations, and that Tpm1 dimers have multiple actin binding sites, one model for enhanced nucleation is that am-Tpm1 stabilizes/organizes arrays of profilin-G-actin complexes on the FH1 domains of Bni1 (Model 1, Fig 4F). An alternative model is that profilin-bound actin monomers are recruited by the FH1 domains and dynamically handed off to Tpm1 molecules transiently associated with Bni1 (Model 2, Fig 4F). In both models, the FH2 domain is likely to participate in formation of the initial F-actin seed, capturing its barbed end to initiate processive elongation.
Taken together, our findings fill a long-standing gap in our understanding of how Tpm1 contributes to actin cable formation. Specifically, we have shown that Tpm1 works in concert with profilin in vitro to enhance Bni1- but not Bnr1-mediated actin nucleation. Further, tpm1Δ diminished actin cables in cells solely expressing Bni1, i.e., a bnr1Δ background. This begs the question, why Bni1 and not Bnr1 would evolve to rely on Tpm1 enhancement in nucleating actin assembly? One possibility is the weaker nucleation activity of Bni1 compared to Bnr1 [26]. However, there are also key differences in the environments of the bud and mother compartments, where Bni1 and Bnr1 assemble cables, respectively [27]. During polarized growth, cortical actin patches nucleated by Arp2/3 complex are found primarily in the bud, and Arp2/3 complex and formins are known to compete for available actin monomers [28–30]. Thus, Tpm1 could be required to enhance Bni1-mediated nucleation in the bud due to increased local competition for actin monomers.
Our findings also offer new insights into why tpm1Δ, tpm1Δbni1Δ, and tpm1Δbnr1Δ cells display such different cable phenotypes. In wild type cells, there is a robust cable arrays generated by the combined activities of Bni1 at the bud cortex (red cables, Fig 4G) and Bnr1 at the neck (blue cables, Fig 4G). Bni1-derived cables are released from the bud cortex [31, 32] and at the bud neck may be captured by Bnr1 and incorporated into the thicker, more organized cables filling the mother cell (Fig 4G) [33]. Accordingly, bni1Δ mutants have no cables in the bud and thinner cables in the mother, and bnr1Δ mutants have normal cables in the bud and thinner cables in the mother. Further, tpm1Δbni1Δ mutants resemble bni1Δ mutants, because Tpm1 does not contribute to Bnr1-mediated actin assembly. This is also consistent with tpm1Δbnr1Δ mutants showing a more severe loss of cables than bnr1Δ mutants, because the one formin expressed, Bni1, has compromised nucleation activity without Tpm1. What seems paradoxical is that tpm1Δ mutants exhibit a more severe loss of cables than bni1Δ mutants (Fig 4G). However, this may be due to additional roles of Tpm1 in stabilizing Bnr1-generated cables from cofilin-mediated disassembly. Alternatively, tpm1Δ may not only compromise Bni1 nucleation activity, but also leave Bnr1 competing with Bni1 for available profilin-actin monomers, resulting in Bni1 and Bnr1 each assembling very short cables that are rapidly disassembled by cofilin and co-factors. This is supported by bni1Δ suppressing tpm1Δ cable defects (Fig 4G). Further, both models are supported by aip1Δ and srv2Δ suppressing tpm1Δ defects [34, 35].
These findings have broad implications for isoform-specific Tpm functions in other systems. Recent studies in fission yeast show that two distinct populations of the same tropomyosin (acetylated and non-acetylated Cdc8) decorate distinct actin structures nucleated by different formins (Cdc12 and For3) [5]. In budding yeast, we cannot presently address whether Tpm1 and Tpm2 differentially decorate cables made by Bni1 and Bnr1 until new tools become available (e.g., isoform-specific antibodies and/or functional GFP tags on Tpm1 and Tpm2). However, our data expand on the more general theme of Tpm isoform-specific functions by demonstrating a pairing between one tropomyosin and one formin to promote actin nucleation. Related tropomyosin-formin pairings to stimulate actin assembly may occur in other systems, and will be particularly intriguing to examine in mammalian systems, where there are more than 40 different tropomyosin isoforms and 15 different formins implicated in assembling a range of different cellular actin structures [1, 36].
Supplementary Material
Acknowledgments
We are grateful D. Mulvihill and A. Bretscher for providing reagents, to J. Eskin for creating graphics, J. Henty-Ridilla for scientific advice throughout the project, and J. Eskin, S. Guo, J. Henty-Ridilla, S. Jansen, and A. Johnston for helping edit the manuscript. This project was supported by the Brandeis MRSEC grant NSF MRSEC-1420382, and by a grant from NIH (GM083137) to B.G.
Footnotes
Supplemental information includes three figures, five movies, and experimental procedures.
AUTHOR CONTRIBUTIONS
Conceptualization, S.A. and B.L.G.; Methodology, S.A.; Investigation, S.A., M.V.G., and D.B.; Formal Analysis, S.A. and M.V.G.; Writing – Original Draft, S.A. and B.L.G.; Writing – Review & Editing, S.A. and B.L.G.; Funding Acquisition & Supervision, B.L.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J Cell Sci. 2015;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- 2.Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- 4.Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995;128:383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson M, East DA, Mulvihill DP. Formins determine the functional properties of actin filaments in yeast. Curr Biol. 2014;24:1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 6.Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hundt N, Steffen W, Pathan-Chhatbar S, Taft MH, Manstein DJ. Load-dependent modulation of non-muscle myosin-2A function by tropomyosin 4.2. Sci Rep. 2016;6:20554. doi: 10.1038/srep20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 9.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46:8146–8155. doi: 10.1021/bi700686p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HP, Bretscher A. Purification of tropomyosin from Saccharomyces cerevisiae and identification of related proteins in Schizosaccharomyces and Physarum. Proc Natl Acad Sci U S A. 1989;86:90–93. doi: 10.1073/pnas.86.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- 14.Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- 15.Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269:24310–24315. [PubMed] [Google Scholar]
- 16.Singer JM, Shaw JM. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc Natl Acad Sci U S A. 2003;100:7644–7649. doi: 10.1073/pnas.1232343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maytum R, Geeves MA, Konrad M. Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry. 2000;39:11913–11920. doi: 10.1021/bi000977g. [DOI] [PubMed] [Google Scholar]
- 18.Maytum R, Konrad M, Lehrer SS, Geeves MA. Regulatory properties of tropomyosin effects of length, isoform, and N-terminal sequence. Biochemistry. 2001;40:7334–7341. doi: 10.1021/bi010072i. [DOI] [PubMed] [Google Scholar]
- 19.Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 20.Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013;201:595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG. In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol. 2000;150:895–904. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 25.von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 27.Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015;32:43–53. doi: 10.1016/j.devcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell. 2015;32:54–67. doi: 10.1016/j.devcel.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buttery SM, Yoshida S, Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol Biol Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 33.Yu JH, Crevenna AH, Bettenbuhl M, Freisinger T, Wedlich-Soldner R. Cortical actin dynamics driven by formins and myosin V. J Cell Sci. 2011;124:1533–1541. doi: 10.1242/jcs.079038. [DOI] [PubMed] [Google Scholar]
- 34.Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol Biol Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, Goode BL. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell. 2013;24:31–41. doi: 10.1091/mbc.E12-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.