Abstract
Myeloid translocation genes (MTGs), originally identified as chromosomal translocations in acute myelogenous leukemia, are transcriptional corepressors that regulate hematopoietic stem cell programs. Analysis of The Cancer Genome Atlas (TCGA) database revealed that MTGs were mutated in epithelial malignancy and suggested that loss of function might promote tumorigenesis. Genetic deletion of MTGR1 and MTG16 in the mouse has revealed unexpected and unique roles within the intestinal epithelium. Mtgr1−/− mice have progressive depletion of all intestinal secretory cells, and Mtg16−/− mice have a decrease in goblet cells. Furthermore, both Mtgr1−/− and Mtg16−/− mice have increased intestinal epithelial cell proliferation. We thus hypothesized that loss of MTGR1 or MTG16 would modify Apc1638/+-dependent intestinal tumorigenesis. Mtgr1−/− mice, but not Mtg16−/− mice, had a 10-fold increase in tumor multiplicity. This was associated with more advanced dysplasia, including progression to invasive adenocarcinoma, and augmented intratumoral proliferation. Analysis of ChIP-seq datasets for MTGR1 and MTG16 targets indicated that MTGR1 can regulate Wnt and Notch signaling. In support of this, immunohistochemistry and gene expression analysis revealed that both Wnt and Notch signaling pathways were hyperactive in Mtgr1−/− tumors. Furthermore, in human colorectal cancer (CRC) samples MTGR1 was downregulated at both the transcript and protein level. Overall our data indicates that MTGR1 has a context dependent effect on intestinal tumorigenesis.
Keywords: MTGR1, Colon cancer, MTG16, Apc1638
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer mortality in the United States1. More than 80% of CRCs feature mutational inactivation of the adenomatous polyposis coli (APC) gene, a tumor suppressor that acts as a regulator of the Wnt/β-catenin signaling pathway2,3. Inactive APC allows β–catenin to accumulate and redistribute to the nucleus activating TCF4-dependent transcriptional programs, promoting tumor development2,4–7. Similar to Wnt signaling, upregulation of the Notch pathway promotes intestinal carcinogenesis8–11. Notch signaling is a critical mediator of intestinal differentiation and is activated when its ligands, Jagged and Delta-like, bind to Notch receptors and induce intracellular proteolytic cleavage by gamma-secretase. This releases the Notch Intracellular Domain (NICD) allowing its translocation to the nucleus, where it binds to the transcription factor CSL (CBF1, Suppressor of Hairless, Lag-1) to block secretory lineage specification and promote stem cell programs11,12. While dysregulation of the Wnt and Notch pathways promotes intestinal tumorigenesis13–15, how each signaling network escapes regulation in this process and becomes activated is incompletely understood.
The Myeloid Translocation Gene (MTG) family consists of three members: MTG8 (ETO), MTGR1 (CBFA2T2), and MTG16 (CBFA2T3)16. MTGs associate with DNA binding proteins and recruit other corepressors and histone deacetylases (HDACs) to form repression complexes that downregulate the transcription of target genes17. MTG8 and MTG16 are pathologically disrupted by chromosomal translocations in acute myeloid leukemia (AML), highlighting their importance in regulating stem cell programs18. Given their prominent role in hematopoietic malignancies and hematopoiesis, and that stem cell programs are frequently activated in tumorigenesis, it was postulated that MTG dysfunction may cooperate with other mutations in driving epithelial tumorigenesis. In support of this hypothesis, MTG8 was identified as a new candidate cancer gene in breast and colorectal cancer19 based on its frequency of mutations. Similarly, our query of The Cancer Genome Atlas (TCGA) database20,21 indicates numerous MTG16 and MTGR1 mutations have been identified. Animal models have revealed unexpected pivotal roles for MTGs in regulating stem cell and differentiation programs in the gut. Genetic deletion of any one of the MTG family members results in striking intestinal phenotypes. A portion of Mtg8−/− mice fail to develop the midgut22, Mtgr1−/− mice have pan-secretory lineage loss17, and Mtg16−/− mice have decreased goblet cells indices23. Moreover, both Mtgr1−/− and Mtg16−/− mice have augmented intestinal epithelial proliferation17,23–25, further suggesting dysregulated stem cell programs. The mechanism underlying their intestinal phenotypes is not deduced, but may reflect alterations in Wnt or Notch signaling levels.
Here we formally tested the roles of MTGs in spontaneous colon tumorigenesis. To accomplish this aim, we employed the Apc1638/+ mouse polyp model and determined that genetic ablation of MTGR1, but not MTG16, increased tumor multiplicity. This was associated with progression to more advanced disease with conversion to high-grade dysplasia and even invasive adenocarcinoma, a feature not observed in this model in wild type mice. Examination of a murine erythroid cell ChIP-seq dataset26 revealed that MTGR1 and MTG16 co-occupy 325 genes, but MTGR1 uniquely occupies an additional 1,063 specific genes. Analysis of these targets predicted MTGR1, but not MTG16, can regulate the Wnt and Notch pathways. Using immunohistochemical and RNA-seq analysis, we determined that both Wnt and Notch signaling were hyperactive in Mtgr1−/− tumors. Lastly, we demonstrate downregulation of MTGR1 in CRC. Our report defines a unique role for MTGR1 as a critical regulator of colorectal cancer programs through dual regulation of Wnt and Notch signaling.
Results
Loss of MTGR1 augments intestinal tumorigenesis
Cancer programs often co-opt normal cellular processes, and we have identified MTGs as regulators of intestinal proliferation, self-renewal and wound healing17,22,25,27,28. MTGs may also play key roles in other non-hematopoietic malignancies; for example, MTG16 has been identified as a putative tumor suppressor in breast cancer29, and mutation of MTG8 is postulated to be a “driver” in breast and colorectal cancer19. Our examination of TCGA data20,21 identified 80 non-synonymous mutations in MTGR1 and 97 in MTG16, some of which were predicted to impair function by MutationAssessor algorithms30, including 6 in MTRG1 and 10 in MTG16 observed in the colon. We postulated that inactivation of MTGR1 or MTG16 would augment tumorigenesis.
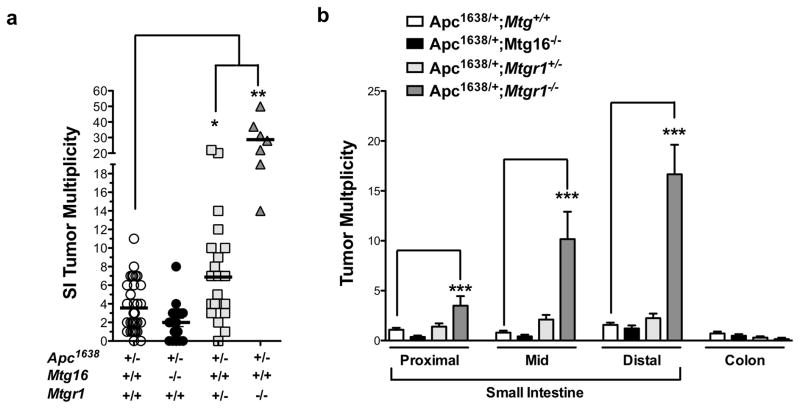
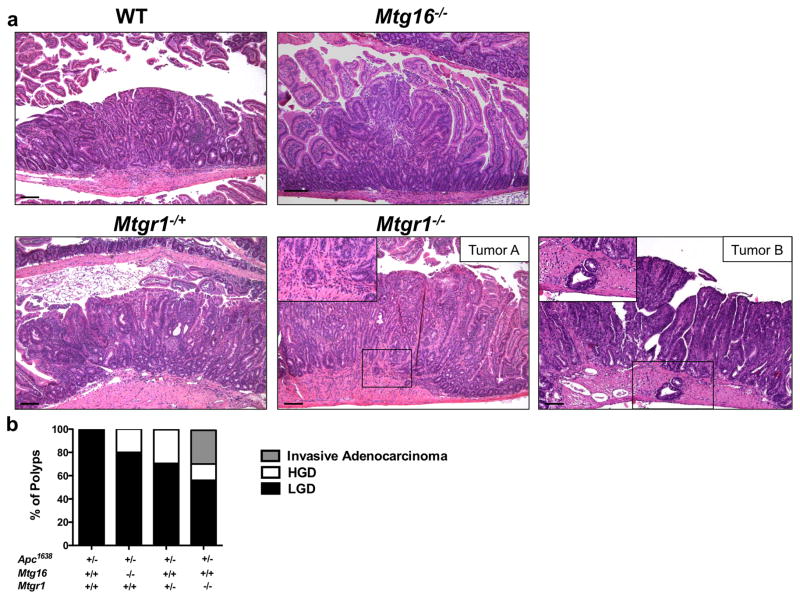
Therefore, we crossed Mtgr1−/− or Mtg16−/− mice with Apc1638/+ polyp-prone mice. Apc1638/+;Mtgr1−/− had decreased survival throughout the duration of the experiment (Supplementary Figure 1), suggesting increased tumor burden and after aging the mice for 36 weeks, we observed increased tumor multiplicity with gene dose-dependent loss of Mtgr1, but surprisingly loss of Mtg16 did not modify tumorigenesis (Apc1638/+;Mtg+/+ 4.3 ± 0.5 vs. Apc1638/+;Mtg16−/− 2.4 ± 0.5 vs. Apc1638/+;Mtgr1−/+ 7.1 ± 1.1 vs. Apc1638/+;Mtgr1−/− 28.9 ± 4.5 tumors per mouse, Figure 1a). Apc1638/+;Mtgr1−/− mice had more tumors in every segment of the small intestine, with the most pronounced effect being in the distal small intestine (Figure 1b). Histopathological analysis indicated tumors from Apc1638/+;Mtgr1−/− and Apc1638/+;Mtgr1−/+ were more dysplastic with progression to invasive adenocarcinoma in some cases (Figure 2a). Indeed, 43% of Apc1638/+;Mtgr1−/− mice showed evidence of invasive adenocarcinoma or high grade dysplasia while Apc1638/+;Mtg+/+ tumors had only low grade dysplastic changes (Figure 2b). Immunostaining for intratumoral B220+ B-cell lymphocytes and CD3+ T-cell lymphocytes revealed no significant differences (Supplementary Figure 2). Overall, our data indicates that loss of MTG16 has no effect on tumorigenesis. Loss of MTGR1, however, substantially augments tumorigenesis in a gene dose-dependent fashion.
Figure 1. Loss of MTGR1, not MTG16, augments intestinal tumorigenesis.
(a) Tumor multiplicity and (b) distribution in Apc1638/+;Mtg+/+, Apc1638/+;Mtg16−/−, Apc1638/+;Mtgr1−/+, Apc1638/+;Mtgr1−/− mice. One way ANOVA test with a Dunnett’s post test was used for statistical analysis. *P < 0.05, **P < 0.01, ***P<0.001
Figure 2. Mtgr1−/− tumors exhibit a higher degree of dysplasia.
(a) Representative H&E images of tumors from Apc1638/+;Mtg+/+, Apc1638/+;Mtg16−/−, Apc1638/+;Mtgr1−/+, Apc1638/+;Mtgr1−/− mice. Two examples (Tumor A and B) of invasive adenocarcinoma from Apc1638/+;Mtgr1−/− mice. Black boxes highlight invasive carcinomas. Size standard is 100 microns. (b) Quantification of degree of dysplasia by histopathological analysis of H&E stained sections.
MTGR1 preferentially associates with Wnt and Notch genes
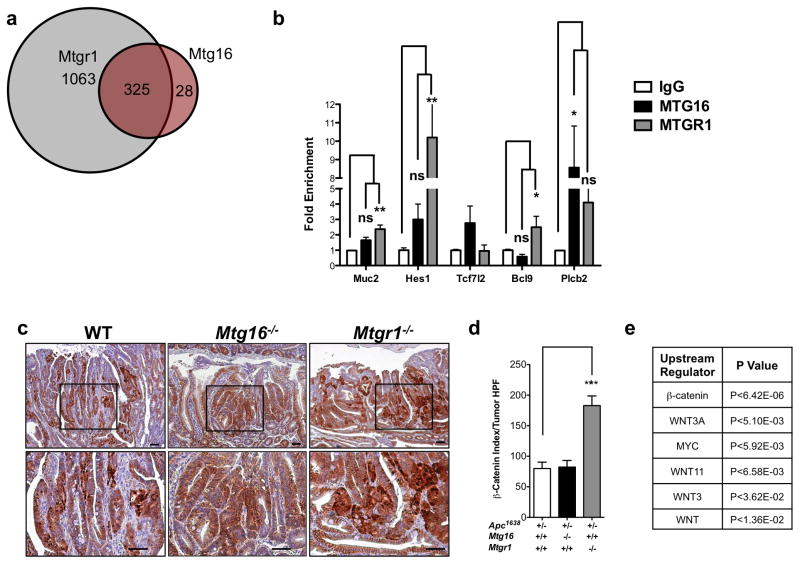
As MTGR1 and MTG16 are transcriptional co-repressors, we reasoned that differential MTG genomic occupancy may underlie these disparate phenotypes. We therefore examined ChIP-seq datasets generated from a murine erythroleukemia cell line (MEL) to identify and compare MTGR1 and MTG16 genomic occupancies26. Using a false discovery rate of 5%, we determined that MTGR1-containing complexes occupy sites proximate to 1,388 specific genes and MTG16-containing complexes occupy 353, of which there was overlap with 325 genes. Thus, there were a large number of unique MTGR1 targets (MTGR1 exclusive: 1,063; MTG16 exclusive: 28, Figure 3a). Protein analysis through evolutionary relationships (PANTHER)31 of the non-overlapping ChIP binding sites predicted that MTGR1, but not MTG16, regulates Wnt and Notch signaling (Supplementary Figure 3). As the ChIP-seq dataset was generated from a murine leukemic cell line using epitope tagged MTGs, we next performed ChIP for endogenous MTGR1 and MTG16 in the colon using the Young Adult Mouse Colon (YAMC) cell line for a subset of the Wnt and Notch targets. In the majority of targets surveyed, MTGR1, but not MTG16, was significantly enriched (Figure 3b). These results suggest that while MTGR1 and MTG16 share occupancy of a subset of targets, the majority of MTGR1 targets are unique to MTGR1.
Figure 3. Mtgr1−/− tumors demonstrate dysregulated Wnt signaling.
(a) Venn diagram of MTGR1 and MTG16 genomic occupancy based on ChIP-seq analysis. (b) Fold enrichment of chromatin immunoprecipitation of endogenous MTGR1 and MTG16 in Young Adult Mouse Cells (YAMC). Data is presented as mean ± SEM. All experiments were performed in triplicate. One-way ANOVA analysis with Dunnett’s post-test. (c) Representative images of β-catenin immunohistochemistry and (d) quantification of β-catenin nuclear localization and intensity. Student’s t-test was used for statistical analysis. (e) RNA-seq analysis of Apc1638/+;Mtgr1+/+ (n=3) and Apc1638/+;Mtgr1−/− (n=3) tumors identifying Wnt-perturbed signaling networks. *P<0.05, **P<0.01, ***P<0.001.
Mtgr1−/− tumors demonstrate hyperactive Wnt and Notch signaling
Because the ChIP-seq data predicted MTGR1 regulation of Wnt targets (Supplementary Figure 3), we used immunohistochemistry to examine the subcellular localization of β-catenin, which is used as a surrogate for Wnt activation. We identified increases in both nuclear β-catenin and extent of its staining, suggesting hyperactive Wnt signaling in the Mtgr1−/− background compared to WT and Mtg16−/− (Figure 3c and 3d). We further evaluated Wnt pathway activation by performing RNA-seq followed by Ingenuity Pathways Analysis (IPA)32 and identified hyperactive Wnt regulatory networks, including β-catenin, Wnt3A, and Myc, supporting the hypothesis that MTGR1 regulates Wnt targets (Figure 3e).
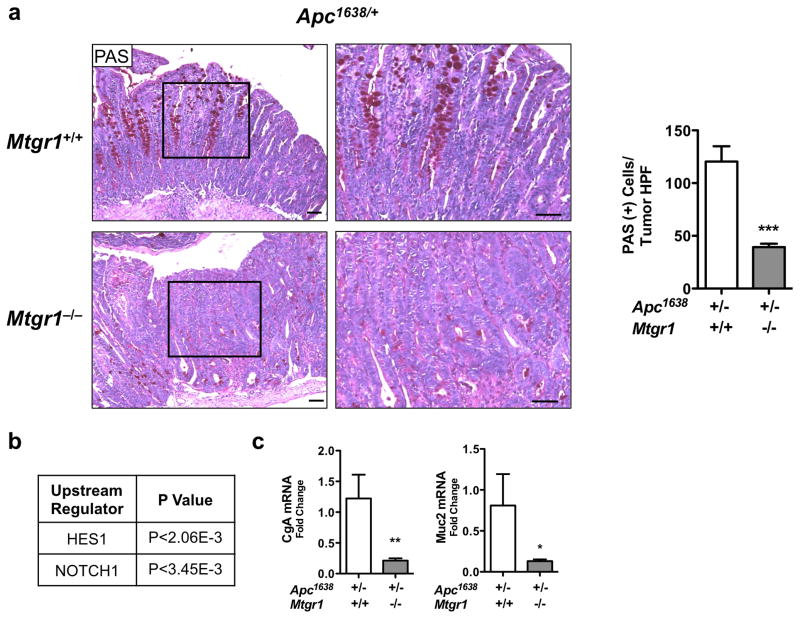
As the ChIP-seq data also suggested preferential Notch regulation by MTGR1, and because we previously determined that the secretory lineage deficiency observed in the Mtgr1−/− intestine was rescued by Notch inhibition24, we next determined if Notch signaling was perturbed in Mtgr1−/− tumors. As active Notch signaling will increase absorptive enterocyte production at the expense of secretory lineages, we stained tumors with periodic acid Schiff (PAS) to identify goblet cells. Mtgr1−/− tumors had dramatically fewer intratumoral goblet cells, consistent with increased Notch signaling Apc1638/+;Mtg+/+ 120 ± 14.5 vs. Apc1638/+;Mtgr1−/− 39.4 ± 3.1, PAS positive cells per tumor HPF, Figure 4a). Supporting this hypothesis, IPA analysis of intratumoral RNA-seq data indicated activation of Notch signaling (Figure 4b). To confirm Notch activation, we performed qPCR for Muc2 and Cga, two Notch repression targets, and observed a 2-fold reduction in both in Mtgr1−/− tumors, further supporting increased Notch tone (Figure 4c). Analysis of Mtg16−/− tumors did not reveal Wnt or Notch hyperactivation compared to WT (Supplementary Figure 4).
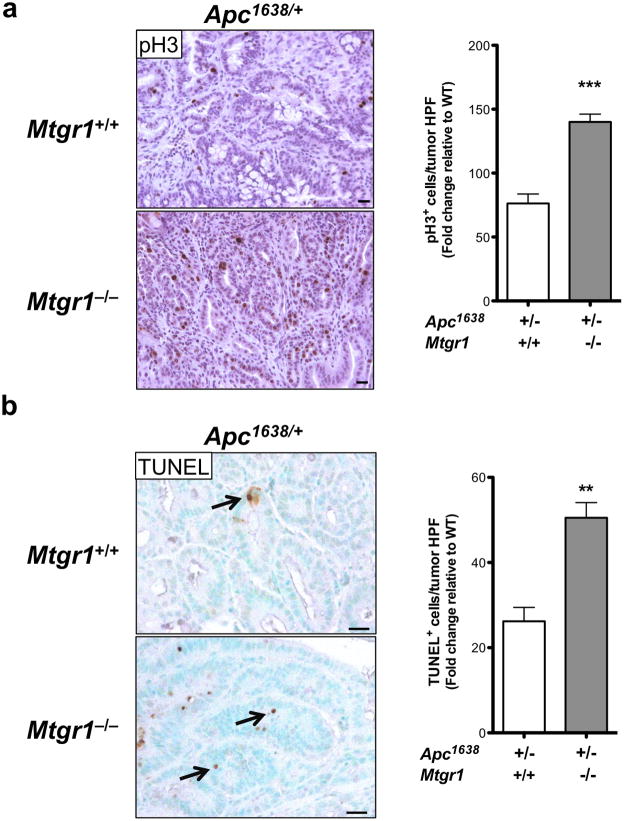
Figure 4. Intratumoral Notch signaling is hyperactive upon MTGR1 inactivation.
(a) Periodic Acid Schiff (PAS) staining for goblet cells in tumors from Apc1638/+;Mtgr1+/+ and Apc1638/+;Mtgr1−/− and quantification of PAS-positive cells per tumor HPF. Size standard is 50 microns. (b) RNA-seq analysis of Apc1638/+;Mtgr1+/+ (n=3) and Apc1638/+;Mtgr1−/− (n=3) tumors identifying perturbed Notch signaling networks. (c) qPCR for Cga and Muc2 in Apc1638/+;Mtgr1+/+ and Apc1638/+;Mtgr1−/− tumors. Student’s t test was used. *P<0.05, **P<0.01, ***P<0.001.
We next reasoned that if Wnt and Notch signaling were increased in Mtgr1−/− tumors, then cellular processes such as proliferation and apoptosis should be affected. We measured intratumoral proliferation by immunohistochemical staining for phosphohistone H3 and apoptosis by TUNEL staining and observed increases in both indices in Mtgr1−/− tumors (pH3: Apc1638/+;Mtg+/+ 76 ± 7.4 vs. Apc1638/+;Mtgr1−/− 139 ± 10.3 positive cells per tumor HPF and TUNEL: Apc1638/+;Mtg+/+ 26 ± 3.3 vs. Apc1638/+;Mtgr1−/− 50 ± 3.5 positive cells per tumor HPF) (Figure 5). These data support the hypothesis that MTGR1 is a coregulator of Notch and Wnt signaling.
Figure 5. Increased intratumoral proliferation and apoptosis in Mtgr1−/− animals.
(a) Proliferation (phospho-histone H3) and (b) apoptosis (TUNEL) immunohistochemical assessment per tumor high power field. Size standard is 20 microns, Student’s t test was used. **P<0.01, ***P<0.001.
MTGR1 is underexpressed early in CRC development
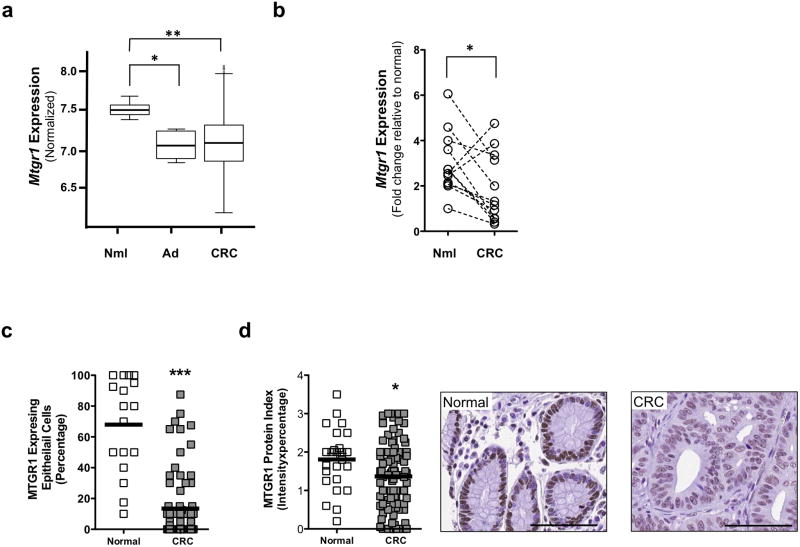
Because our data implicates MTGR1 as a tumor suppressor, we reasoned that its levels may be reduced in CRC. Therefore, we assessed MTGR1 levels in CRC in the Moffitt/Vanderbilt-Ingram Cancer Center expression array dataset consisting of 10 normal controls, 6 adenomas, and 250 carcinomas33,34. MTGR1 mRNA expression was significantly reduced in both the adenoma and carcinoma stages (Figure 6a). We subsequently collected 12 matched samples of normal colon tissue and colorectal carcinoma. qPCR for MTGR1 revealed a decrease in the matched samples (Figure 6b). To corroborate this decrease in MTGR1 mRNA, we used high-resolution in situ hybridization (RNAscope) to measure its expression in a separate CRC tissue microarray consisting of 25 normal colon controls and 102 carcinomas. We observed that MTGR1 RNA was also reduced in carcinomas compared to normal colons (normal colons: 68% ± 7.5% vs. carcinomas: 13% ± 2.5% MTGR1 expressing epithelial cells per core, Figure 6c; Supplementary Figure 5). Similarly, we detected less MTGR1 protein in carcinomas compared to normal colons by immunohistochemistry (normal colons: 1.8 ± 0.15 vs. carcinomas: 1.3 ± 0.09 MTGR1 staining protein index, Figure 6d). While MTGR1 expression was decreased in carcinomas, MTGR1 expression did not correlate with disease outcome, survival, or grade (data not shown). Furthermore, we identified statistically significant inverse correlations between MTGR1 expression and Wnt and Notch activation in the Vanderbilt/Moffitt Cancer Center expression array. MTGR1 negatively correlated with Hes1 and β-catenin and positively correlated with secretory markers Atoh1 and Sox9 (Supplementary Figure 6). Thus, our data demonstrates that MTGR1 is underexpressed at an early stage in CRC and that its transcript levels are inversely related to the expression of genes pivotal to Wnt and Notch activation.
Figure 6. MTGR1 is underexpressed in Human CRC.
(a) Mtgr1 mRNA expression in Moffitt/Vanderbilt Ingram Cancer Center expression array (10 normal controls, 6 adenomas, and 250 carcinomas). A Wilcoxon rank sum test was used to compare expression in normal, adenoma, and cancer tissue. (b) qPCR for Mtgr1 expression in 12 samples consisting of matched normal and colorectal cancer specimen. Paired t test was used for statistical analysis. (c) MTGR1 RNA expression and (d) MTGR1 protein expression (size standard is 100 microns) in Vanderbilt Tissue Microarray of CRC (25 normal colon controls and 102 carcinomas). Student’s t test was used for statistical analysis. *P<0.05, **P<0.01, ***P<0.001.
Discussion
In this report, we show that while MTGR1 and MTG16 are 65% homologous at the protein level, genetic inactivation of MTGR1, but not MTG16, increased tumorigenesis ~10-fold in the Apc1638/+ mouse model of CRC. Moreover, Mtgr1−/− tumors had a higher degree of dysplasia with increased proliferation and apoptosis. Immunohistochemical and RNA-seq analysis of Mtgr1−/− tumors indicated Wnt and Notch pathway activation. We also determined that in sporadic CRC, MTGR1 was underexpressed in the majority of CRC samples analyzed. Overall, our data demonstrate that loss of MTGR1 augments tumorigenesis with associated dysregulation of Wnt and Notch signaling.
The Myeloid Translocation Gene family consists of three members: Mtg8, Mtgr1, and Mtg1616. MT8 was initially discovered as a foundational translocation in AML, and while MTGs have since been shown to regulate hematopoietic stem cell processes, their genetic deletion in mice has revealed a unique intestinal role for each family member. Mtg8−/− mice demonstrate a severe mid-gut deletion phenotype22; Mtgr1−/− mice have pan-secretory cell loss and are smaller than wild-type mice17; and Mtg16−/− mice have decreased goblet cell indices23. The mechanisms underlying these various phenotypes remain unclear, although there are reports that MTGR1 directly binds to TCF4, competing for β-catenin occupancy and that it interacts with CSL in repressing Notch signaling24,35. However, evidence that MTGs occupy Wnt/Notch targets in the gut has been lacking. Our analysis of previously published ChIP-seq data discovered that MTGR1, but not MTG16, uniquely occupies Wnt and Notch targets. Thus, a possible explanation for their differential effects on tumor phenotypes is that MTGR1 and MTG16 occupy different genomic loci and have non-overlapping regulatory roles. As MTGs lack the ability to bind DNA directly, their target specificity is determined by the trans-acting DNA binding factors with which they associate16. MTGR1 and MTG16 are 65% homologous and share four highly conserved domains termed Nervy Homology Domain16. The regions between these domains are variable and likely are important factors in explaining the unique genomic associations of each family member.
Our report identifies MTGR1 as a Modifier of Min (MOM) similar to Muc2, Mom2, and Rassf1a36–38. Where inactivation of MTGR1 fits in the multistep model of CRC mutations is unclear, but our data suggests that loss of MTGR1 plays an important role in both tumor initiation and progression. We propose that because Mtgr1 loss in mice leads to basal elevation of Wnt and Notch signaling, the threshold to trigger tumorigenesis is reduced. This is underscored by the fact that Mtgr1 haploinsufficiency increased tumor multiplicity and progression (Figure 1a and Figure 2). Moreover, that Mtgr1−/+ mice show increased tumor multiplicity suggests that MTGR1 levels are tightly regulated to restrict Wnt and Notch signaling. Future work would test if pharmacologic inhibition of Wnt and/or Notch signaling could reduce tumor burden in Mtgr1−/− mice. Although we are not aware of a specific Wnt signaling inhibitor, previous groups have shown Notch inhibition using a gamma secretase inhibitor converts adenomas into non-proliferating goblet cells14,39.
MTGR1 plays an important role in protecting the gut from injury. Mtgr1−/− mice are extremely sensitive to colitis-induced injury, exhibiting severe weight loss, mucosal injury, and increased inflammatory infiltrates27. In our prior report, we showed that Mtgr1−/− mice had fewer polyps in the AOM/DSS inflammatory carcinogenesis model, indicating that MTGR1 is required in colitis-driven dysplasia28. This was unexpected given that MTGR1 negatively regulates Wnt signaling through an interaction with TCF435. In the current report we analyzed the contribution of MTGR1 in Wnt-initiated tumorigenesis in the absence of epithelial injury and inflammation. We show that loss of MTGR1 augments sporadic tumorigenesis. Taken together, we propose a model in which Mtgr1−/− cells, in the setting of injury, are predisposed to undergo apoptosis, thus resulting in clearance of initiated cells during inflammation mediated injury. In the absence of damage, however, elevated Wnt and Notch signaling in Mtgr1−/− cells synergizes with Apc loss and accelerates tumorigenesis. Collectively, our work demonstrates that MTGR1 can function as a context dependent tumor modifier.
That MTGR1 can promote inflammatory—yet suppress sporadic—tumorigenesis is not necessarily surprising. While inflammatory tumorigenesis and sporadic tumorigenesis share several pathologic and molecular features, including alterations in the Wnt signaling pathway40, the development of inflammation driven tumorigenesis has a unique molecular pathogenesis. Loss of APC, for example, is observed at a later stage in colitis-associated cancer progression compared to sporadic CRC40. Further, recent work has identified several genes that exhibit context dependent phenotypes in tumorigenesis modeling. For example, loss of Mmp9 or Myd88 promotes inflammatory carcinogenesis in the AOM/DSS model41,42, but loss of either inhibits tumorigenesis in the ApcMin model of sporadic colorectal cancer43,44. Mice deficient for Tlr4 had reduced inflammatory carcinogenesis45, suggesting Tlr4 is also required for inflammatory carcinogenesis. Constitutive intestinal epithelial expression of Tlr4, however, reduced tumor burden in the ApcMin model of sporadic colon cancer, indicating that high levels of Tlr4 suppress tumorigenesis when Apc is inactivated46. Our work now adds MTGR1 to the list of genes that exhibiting context dependent phenotypes in inflammatory versus sporadic carcinogenesis.
In this report, we have identified that loss of MTGR1 augments Wnt dependent tumorigenesis. The clinical importance of this observation is underscored by our finding that MTGR1 is downregulated at both the transcript and protein level in the majority of human CRC. Patients who have reduced MTGR1 expression may be at risk for progression from precancerous to cancerous colon tumors. Indeed, given that even partial loss of MTGR1 promoted tumor formation, MTGR1 could serve as a valuable biomarker for patients at risk for CRC.
Materials and methods
Mouse experiments and analysis
Mice were housed, maintained, and then euthanized using isoflurane and cervical dislocation according to a protocol approved by the Vanderbilt Institutional Animal Care and Use Committee. The mice are on the C57BL6 background and both male and female mice were used in these studies (Apc1638/+;Mtg+/+ n=36; Apc1638/+;Mtg16−/− n=19; Apc1638/+;Mtgr1−/+ n=25; Apc1638/+;Mtgr1−/− n=7). The small intestine was removed and divided into equal thirds. Each segment, along with the large intestine, was then bisected longitudinally. Tumor number was counted grossly. Tumor samples and normal, non-malignant tissue were collected for RNA and stored in RNAlater (Invitrogen). The remaining intestinal segments were “Swiss rolled” so that the distal most segment was innermost28. Microscopic analysis was performed by a gastrointestinal pathologist (MKW) for dysplasia on haematoxylin and eosin (H&E) stained sections (processed by the Vanderbilt Translational Pathology Shared Resource core). All in vivo procedures were carried out in accordance with protocols approved by the Vanderbilt Institutional Animal Care and Use Committee.
Chromatin immunoprecipitation
A qChIP assay was performed as described47. Briefly, cells were cross-linked with 1% formaldehyde, quenched with 125 mM glycine, washed twice with ice cold PBS, and lysed for 5 min in lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, protease inhibitor mixture, 1 mM PMSF). Lysate was sonicated on ice into chromatin fragments with an average length of 500 bp. Protein A/G magnetic beads (Millipore, Cat no. 16-663) were washed twice in RIPA buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 140 mM NaCl) and resuspended in 250 ul of RIPA buffer with 2.5 ug of respective antibodies (IgG Cell Signaling #3900; MTG16 Abcam #33072; MTGR1 Proteintech #11336-1AP). Diluted chromatin (1:10 in RIPA buffer) was added to antibody-bead complexes and immunoprecipitated overnight at 4°C. Precipitated immune complexes were washed five times, and cross-links were reversed by incubating samples with 150 ml of elution buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl) containing 1% SDS and 50 mg/ml proteinase K for 2h at 68 °C. DNA was recovered by capturing beads, and the ChIP material was reincubated in 150 ml of elution buffer/SDS/proteinase K for 5 min, and both supernatants were pooled. DNA was isolated using DNA clean and concentrator (Zymo Research Cat No. ZRC 162780). Real time PCR was performed using primers described below.
Primer sequences were generated based on the publicly available ChIP-seq database26:
MUC2F: GGGAACACCACTCACCAACT
MUC2R: AACATCCTGGCCTCGATAAA
Hes1F: CAGAGGAGAGGATTCTAAACTGC
Hes1R: CCTGCCAAGCCACTATTCC
TCF712F: TAGGGTGTGACACGGCATAA
TCF712R: GGGGTTCTGACAAAGAACGA
Pclb2F: GCACTCTACCCAGGTGTTGC
Pclb2R: TTGGTGATGCTTTGCCTACAT
Bcl9F: GTCACAAGGCCTCTATTAGGAAAA
Bcl9R: AGGAGGAGGAGGAGGAGGAG
qPCR mRNA analysis
RNA was made from tumor tissue stored in RNAlater using the RNeasy Mini kit (Qiagen), according to the manufacturer’s directions. cDNA was then made using the SuperScript cDNA kit (Invitrogen). qPCR was then performed using SYBR Green (Biorad) with primers for Gapdh (Realtimeprimers). Primers for CgA and Muc2 were previously described24. Primers for Ascl2, Axin2, c-Myc, and Klf4 were purchased from RealtimePrimers. Reactions were performed according to the manufacturer’s recommendations.
Immunohistochemistry and immunofluorescence
Five micrometer sections were cut, dewaxed, hydrated and endogenous peroxidase activity quenched with 0.03% hydrogen peroxide in MeOH. Antigen retrieval was performed using the boiling sodium citrate method in a microwave (20 mmol sodium citrate pH 6.5) for 16 minutes at 30% power. After blocking, primary antibodies were incubated overnight at 4°C at the following concentrations: α-phosphohistone-H3 (Millipore #06-570), 1:150, α-β-catenin (BD Transduction Laboratories #610153), 1:1000, or α-CD3 (Serotec #145-2C11) 1:500. The Vectastain ABC Elite System (Vector Labs) was used to visualize staining for immunohistochemistry. Identification of intratumoral and crypt apoptotic cells was performed using the ApopTag Plus Peroxidase in situ Apoptosis Kit (Chemicon) according to the manufacturer's protocol. Control stains were obtained by omitting the terminal transferase (TnT) enzyme. Apoptosis and proliferation indices were generated by counting the number of positive cells per high-powered field (HPF; 40× objective) within each tumor by a blinded observer. A β-catenin index was employed, as previously reported28. This index is generated by multiplying the staining intensity (on a scale of 1–5) by percentage of the cells demonstrating nuclear staining.
For B220 immunofluorescence, antigen retrieval was performed using a microwave and 10 mM Na Citrate pH 6.0. Samples were incubated with primary antibody (B220/CD45r 1:100, BD Pharmingen #550286 and E-cadherin 1:500, BD Transduction Laboratories #610181) at 4°C overnight. Goat anti-Rat 568 (1:500, Life Technologies #11077) and Goat anti-IgG2A 488 (1:500, Life Technologies #21131) were applied and slides were mounted with ProLong Gold antifade reagent with DAPI (Life Technologies). CD3 and B220 quantification was performed by counting the number of intratumoral positive cells per HPF (x40 objective).
RNA Sequencing and Analysis
Tumor RNA from Mtgr1−/− (n=3) and WT (n=3) mice was sequenced by the Vanderbilt Sequencing Core Facility. Initial raw sequencing data was aligned to a reference mouse genome (mm9) using TopHat (version 1.3.1) software48. The transcript of mouse genome (mm9) was downloaded from UCSC as implemented in the Bioconductor package GenomicFeatures. The Bioconductor packages Rsamtools and DESeq were then used to estimate the read count for expression of each gene and to detect differentially expressed (DE) genes. For count based gene expression data, DESeq uses a model based on the negative binomial distribution which includes a dispersion parameter to better estimate variance49. The p-values from DESeq were adjusted by Benjamini and Hochberg’s method to control false discovery rate (FDR)50.
Tissue Microarrays
All tissue samples were collected, coded, and de-identified in advance, and their use in this work was approved by the Institutional Review Board. Tissues were stained with H&E and representative regions were selected for inclusion in a tissue array. Tissue cores with a diameter of 0.6 mm were retrieved from the selected regions of the donor blocks and punched to the recipient block using a manual tissue array instrument (Beecher Instruments); samples were punched in duplicate. Control samples from normal epithelial specimens were punched in each sample row. Five micrometer sections were transferred to polylysine-coated slides (Menzel-Glaser) and incubated at 37°C for 2 hours. The resulting tumor tissue array was used for immunohistochemical analysis. Further clinical information regarding samples is described (Supplementary Table 1).
Antigen retrieval was conducted by boiling in citrate pH 6.0 at 104°C for 20 min. Slides were then cooled down at room temperature for 10 min before being quenched with 0.03% H202 with sodium azide for 5 min. Serum-free protein was used to block for 20 min. Primary antibody (MTGR1/CBFA2T2 Proteintech CAT# 11336-1-AP) was used at a 1:200 dilution and incubated for 60 min. Envision HRP Labeled Polymer was applied for 30 minutes for detection. DAB was used as a chromogen after incubation for 5 min. Cores were scored for the proportion of epithelial cells that stained positive and for the intensity of the stain using an index from 1–4. The index was generated by multiplying the two scores together.
Moffitt/Vanderbilt-Ingram Cancer Center Expression Array
The array was previously described33. The source of the data: GSE17538.
RNA Scope
RNA in situ probes for MTGR1 were ordered from Advanced Cell Diagnostics and sequences are available on their online database. Tissue microarrays were processed and stained exactly according to manufacturer’s protocol. Staining was scored as percentage of positive cells per core. All cores were also stained with a positive control probe for housekeeping gene Peptidylprolyl Isomerase B (Cyclophilin B, PPIB). Cores that did not stain robustly with positive control were omitted.
Statistical Methods
For human studies, all available samples were analyzed and absolute numbers are given in figure legends. For mouse experiments, number of mice were determined based on previous experience with the Apc1638 model. Where sample size varied between Mtgr1+/− and Mtgr1−/−, this was based on the availability of mice from Mtgr1+/− x Mtgr1+/− breedings. There were no excluded cases and all available mice were analyzed from the breedings hence there was no randomization required. Due to differences in coat color between the WT, Mtgr1+/− (black), vs. the Mtgr1−/− (brown) the investigators could not be blinded to genotype, however all aspects of microscopic analysis the pathologist was blinded. A student’s t-test was used when comparing two groups such as apoptosis, proliferation, and tumor counts. In Figure 1a, a one-way ANOVA test was used with a post-hoc Dunnett’s analysis to compare multiple groups. In Figure 6a, a Wilcoxon rank sum test was used to compare MTGR1 expression in normals, adenomas, and carcinomas. Data is presented as the mean +/− the standard error of the mean (SEM) in bar graphs and a line identifying the mean is shown when all data points are plotted. All of these analyses were performed using GraphPad Prism®5.0c.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants K08DK080221, R01DK099204 (CSW), P50CA095103 (MKW), 1F30DK096718 (BP), 1F31 CA167920 (CWB), T32 GM07347 (NIH/NIGMS) (BP), Merit Review Grant from the Office of Medical Research, Department of Veterans Affairs 1I01BX001426 (CSW), ACS-RSG 116552 (CSW), R01AT004821 (KTW), P30DK058404 (Vanderbilt Digestive Disease Research Center), NIH grant UL1TR000445 (Vanderbilt CTSA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank the members of the Williams lab, and Karie Ballard for help in revising and proofreading the manuscript. We would also like to thank Drs. R. Daniel Beauchamp, David Bader, and Barbara Fingleton for their advice and counsel in preparing the manuscript.
Footnotes
There are no conflicts of interest to disclose
References
- 1.Cancer Facts & Figures. Health Policy (New York) 2010 [Google Scholar]
- 2.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 3.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacDonald BT, Tamai K, He X. Wnt/b-Catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 6.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 7.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 8.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 2005;15:1686–1691. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 10.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 11.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh M, Katoh M. Notch signaling in gastrointestinal tract (Review) Int J Oncol. 2007;30:247–251. [PubMed] [Google Scholar]
- 13.Peignon G, Durand A, Cacheux W, Ayrault O, Terris B, Laurent-Puig P, et al. Complex interplay between β-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 15.Fre S, Pallavi SK, Huyghe M, Laé M, Janssen K-P, Robine S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- 17.Amann JM, Chyla BJI, Ellis TC, Martinez A, Moore AC, Franklin JL, et al. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Mol Cell Biol. 2005;21:9576–9585. doi: 10.1128/MCB.25.21.9576-9585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chyla BJ, Moreno-Miralles I, Steapleton Ma, Thompson MA, Bhaskara S, Engel M, et al. Deletion of Mtg16, a target of t(16;21), alters hematopoietic progenitor cell proliferation and lineage allocation. Mol Cell Biol. 2008;28:6234–6247. doi: 10.1128/MCB.00404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjoblom T, Jones S, Wood L, Parons D, Lin J, Barber T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabi F, Pannell R, Pavloska G. Gene targeting reveals a crucial role for MTG8 in the gut. Mol Cell Biol. 2001;21:5658–5666. doi: 10.1128/MCB.21.16.5658-5666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poindexter SV, Reddy VK, Mittal MK, Williams AM, Washington MK, Harris E, et al. Transcriptional co-repressor MTG16 regulates small intestinal crypt proliferation and crypt regeneration after radiation-induced injury. Am J Physiol - Gastrointest Liver Physiol. 2015;308:562–571. doi: 10.1152/ajpgi.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parang B, Rosenblatt D, Williams A, Washington MK, Revetta F, Short SP, et al. The transcriptional corepressor MTGR1 regulates intestinal secretory lineage allocation. FASEB J. 2014;29:786–795. doi: 10.1096/fj.14-254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams CS, Bradley AM, Chaturvedi R, Singh K, Piazuelo MB, Chen X, et al. MTG16 contributes to colonic epithelial integrity in experimental colitis. Gut. 2013;62:1446–1455. doi: 10.1136/gutjnl-2011-301439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soler E, Andrieu-Soler C, De Boer E, Bryne JC, Thongjuea S, Stadhouders R, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez JA, Williams CS, Amann JM, Ellis TC, Moreno-Miralles I, Washington MK, et al. Deletion of Mtgr1 sensitizes the colonic epithelium to dextran sodium sulfate-induced colitis. Gastroenterology. 2006;131:579–588. doi: 10.1053/j.gastro.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Barrett CW, Fingleton B, Williams A, Ning W, Fischer MA, Washington MK, et al. MTGR1 is required for tumorigenesis in the murine AOM/DSS colitis-associated carcinoma model. Cancer Res. 2011;71:1302–1312. doi: 10.1158/0008-5472.CAN-10-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochetkova M, Mckenzie OL, Bais AJ, Martin JM, Secker GA, Seshadri R, et al. CBFA2T3 ( MTG16 ) is a putative breast tumor suppressor gene from the breast xancer loss of heterozygosity region at 16q24. 3. Cancer Res. 2002;16:4599–4604. [PubMed] [Google Scholar]
- 30.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;4:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams CS, Zhang B, Smith JJ, Jayagopal A, Barrett CW, Pino C, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest. 2011;121:4056–4069. doi: 10.1172/JCI44228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore AC, Amann JM, Williams CS, Tahinci E, Farmer TE, Martinez JA, et al. Myeloid translocation gene family members associate with T-cell factors (TCFs) and influence TCF-dependent transcription. Mol Cell Biol. 2008;28:977–987. doi: 10.1128/MCB.01242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K, Popova NV, Wan CY, Lozonschi I, Tadesse S, Kent S, et al. Interaction of Muc2 and Apc on Wnt signaling and in intestinal tumorigenesis: potential role of chronic inflammation. Cancer Res. 2008;68:7313–7322. doi: 10.1158/0008-5472.CAN-08-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverman KA, Koratkar R, Siracusa LD, Buchberg AM. Identification of the modifier of Min 2 (Mom2) locus, a new mutation that influences Apc-induced intestinal neoplasia. Genome Res. 2002;12:88–97. doi: 10.1101/gr.206002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Der Weyden L, Arends MJ, Dovey OM, Harrison HL, Lefebvre G. Loss of Rassf1a co-operates with Apc Min to accelerate intestinal tumourigenesis. Oncogene. 2008;27:4503–4508. doi: 10.1038/onc.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of Notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918–928. doi: 10.1053/j.gastro.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 41.Garg P, Sarma D, Jeppsson S, Patel NR, Gewirtz AT, Merlin D, et al. Matrix metalloproteinase-9 functions as a tumor suppressor in colitis-associated cancer. Cancer Res. 2010;70:792–801. doi: 10.1158/0008-5472.CAN-09-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai R-M, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207:1625–1636. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinnamon MJ, Carter KJ, Fingleton B, Matrisian LM. Matrix metalloproteinase-9 contributes to intestinal tumourigenesis in the adenomatous polyposis coli multiple intestinal neoplasia mouse. Int J Exp Pathol. 2008;89:466–475. doi: 10.1111/j.1365-2613.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SH, Hu L-L, Gonzalez-Navajas J, Seo GS, Shen C, Brick J, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 2010;16:665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, Krishnareddy S, et al. Toll-Like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133:1869–81. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Teo WL, Low MJ, Meijer L, Sanderson I, Pettersson S, et al. Constitutive TLR4 signalling in intestinal epithelium reduces tumor load by increasing apoptosis in APC(Min/+) mice. Oncogene. 2013:1–9. doi: 10.1038/onc.2012.581. [DOI] [PubMed] [Google Scholar]
- 47.Dahl JA, Collas P. A quick and quantitative chromatin immunoprecipitation assay for small cell samples. Front Biosci. 2007;12:4925–4931. doi: 10.2741/2438. [DOI] [PubMed] [Google Scholar]
- 48.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.