Abstract
MicroRNA-101, a tumor suppressor microRNA (miR), is often downregulated in cancer and is known to target multiple oncogenes. Some of the genes that are negatively regulated by miR-101 expression include histone methyltransferase EZH2 (enhancer of zeste homolog 2), COX2 (cyclooxygenase-2), POMP (proteasome maturation protein), CERS6, STMN1, MCL-1 and ROCK2, among others. In the present study, we show that miR-101 targets transcriptional coactivator SUB1 homolog (Saccharomyces cerevisiae)/PC4 (positive cofactor 4) and regulates its expression. SUB1 is known to have diverse role in vital cell processes such as DNA replication, repair and heterochromatinization. SUB1 is known to modulate transcription and acts as a mediator between the upstream activators and general transcription machinery. Expression profiling in several cancers revealed SUB1 overexpression, suggesting a potential role in tumorigenesis. However, detailed regulation and function of SUB1 has not been elucidated. In this study, we show elevated expression of SUB1 in aggressive prostate cancer. Knockdown of SUB1 in prostate cancer cells resulted in reduced cell proliferation, invasion and migration in vitro, and tumor growth and metastasis in vivo. Gene expression analyses coupled with chromatin immunoprecipitation revealed that SUB1 binds to the promoter regions of several oncogenes such as PLK1 (Polo-like kinase 1), C-MYC, serine-threonine kinase BUB1B and regulates their expression. Additionally, we observed SUB1 downregulated CDKN1B expression. PLK1 knockdown or use of PLK1 inhibitor can mitigate oncogenic function of SUB1 in benign prostate cancer cells. Thus, our study suggests that miR-101 loss results in increased SUB1 expression and subsequent activation of known oncogenes driving prostate cancer progression and metastasis. This study therefore demonstrates functional role of SUB1 in prostate cancer, and identifies its regulation and potential downstream therapeutic targets of SUB1 in prostate cancer.
Introduction
Prostate cancer is the most common malignancy and the second most common cause of cancer death among men in the United States.1 Multiple molecular alterations have been identified in prostate cancer initiation, growth, invasion and metastasis. Further investigations are needed to understand the mechanisms of dysregulation and role in tumorigenesis of many of the dysregulated genes that are implicated in cancer. These studies will enhance the understanding of the disease process and help in developing new therapies. High-throughput gene expression profiling studies and transcriptome analyses have revealed tumor-specific gene signatures and multiple oncogenic drivers in cancers.2, 3, 4, 5, 6, 7, 8, 9, 10
Loss of tumor suppressor microRNAs (miRNAs or miR) is an established mechanism in cancer progression. Our analysis suggested that SUB1 homolog (Saccharomyces cerevisiae) (SUB1) is a target of miR-101. Our previous studies showed that genomic loci encoding miR-101 were deleted in aggressive prostate cancer leading to reduced miR-101 expression, resulting in overexpression of histone methyltransferase enhancer of zeste homolog 2 (EZH2).11 Apart from regulating EZH2, it has been shown that miR-101 can target other critical genes such as COX2 (cyclooxygenase-2), POMP (proteasome maturation protein), CERS6, STMN1, MCL-1 and ROCK2, among others.11, 12, 13, 14, 15, 16
In this study, we characterized SUB1 expression specifically in aggressive prostate cancer. Yeast SUB1 is biochemically identified as a stimulator of in vitro basal transcription that binds to single-strand DNA in the regions of transcription initiation.17, 18 SUB1 is a nuclear protein and is shown to have a role in various cellular processes.19, 20, 21, 22, 23, 24 It is substituted for replication protein A during transcription elongation.25 In addition to its role as transcriptional coactivator, SUB1 has been shown to repress promoter-driven transcription as well as nonspecific transcription in vitro.26, 27 Studies have reported that SUB1 interacts with distinct domains of activators such as VP16, GAL4, AP2, HIV-TAT, P53 and SMYD3 to modulate their functions.19, 22, 28, 29, 30, 31, 32 Previous studies indicated that the overexpression of SUB1 in a population of normal dermal multipotent fibroblasts resulted in the tumorigenic transformation of the cells, indicating its role in tumorigenesis. SUB1 has been recently found to show an upregulated level in different cancers. Expression of SUB1 was correlated with the levels of VEGF-C, VEGF-D and VEGFR-3 during the development of lymphangiogenesis and lymphatic metastasis in lung adenocarcinoma.33 SUB1 is also shown to have a role in non-small-cell lung cancer34 and astrocytoma.35 A recent study demonstrated that SUB1 has a role in SMYD3-mediated transactivation of growth/invasion-stimulatory genes in cancer cells.32
Here, we show increased SUB1 expression in prostate cancer cell lines and tissues. Through gene knockdown studies, we demonstrate that SUB1 has an important role in prostate cancer cell proliferation and invasion both in vitro and in vivo. We investigated the role of miR-101 in regulating SUB1 expression. Furthermore, our studies reveal that SUB1 can regulate several oncogenes including therapeutic targets such as PLK1, BUB1B and C-MYC by directly binding to their promoters. Finally, our studies indicate that SUB1-mediated oncogenic phenotype can be reversed by blocking PLK1 activity using PLK1 inhibitor.
Results
MiR-101 targets SUB1 and regulates its expression
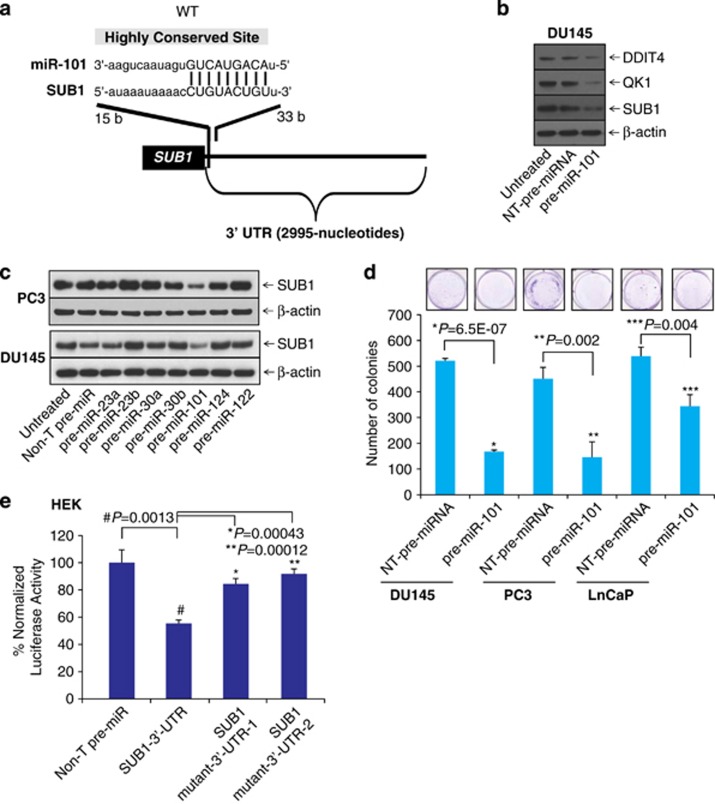
Several studies have revealed that miRNAs are important regulators in cellular processes, such as cell proliferation, invasion and metastasis by repressing several oncogenes.11, 12, 13, 36, 37 To investigate the potential targets of miR-101, we used freely available web-based miR target prediction resources: TargetScan,38 miRanda39 and Diana-microT.40 We identified that miR-101 could potentially target SUB1. The binding site for miR-101 at 3′-UTR (untranslated region) of SUB1 is indicated (Figure 1a). MiR-101 is known to have tumor suppressor function by targeting several oncogenes including EZH2,11 proteasome assembly factor POMP,13 COX2,12 and others, therefore we sought to determine its role in SUB1 regulation. We treated prostate cancer cell line DU145 with precursor miRNA, miR-101 and tested some of the potential targets. We observed downregulation of QK1 and DDIT4 protein levels along with SUB1 (Figure 1b). Quantitative real-time PCR (qPCR) analyses also showed that miR-101 downregulates SUB1, DDIT4 and STC1 mRNA levels (Supplementary Figure S1). Here we show that miR-101 targets other known players in prostate cancer DDIT4, STC1 and QK1. To find the effect on SUB1 protein levels, we treated prostate cancer cells with precursor miRs, miR-101, -23a, -23b, -30a, -30b, -124 and -122 individually and measured SUB1 protein levels. As shown in Figure 1c, miR-101-treated cells showed significant reduction in SUB1 protein levels, whereas the control and other miR precursors did not alter SUB1 expression. Further, we measured the effect of miR-101 on cell growth using a colony growth assay using DU145, PC3 and LnCaP cells (Figure 1d). To determine whether miR-101 directly binds SUB1 3′-UTR and regulates it, HEK-293T cells were co-transfected with miR-101 and pMir-REPORT-SUB1 3′-UTR plasmids. MiR-101 showed substantial reduction in luciferase reporter activity compared with non-targeting (Non-T) control miR (Figure 1e). This effect is reversed by mutating miR-101 target site (Supplementary Figures S2a and b). These results indicate that SUB1 is a direct target of miR-101.
Figure 1.
MiR-101 targets and downregulates SUB1 expression. (a) The predicted miR-101 binding site at the 3′-UTR of SUB1. (b) Immunoblot analysis showing SUB1, QK1 and DDIT4 in pre-miR-101- and control pre-miR-treated DU145 cell lysates. (c) Immunoblot analysis showing SUB1 protein expression in DU145 and PC3 cells treated with a panel of miRNAs. (d) Images of colony growth of cells either treated with non-T pre-miRNA or pre-miR-101. Quantitative data were presented in the histogram. (e) Luciferase reporter assay of SUB1 3′-UTR. HEK-293T cells were transfected either with pre-miR-101 or non-T pre-miR along with either SUB1 3′-UTR wild-type, mutant-1 or mutant-2 luciferase constructs. pRL-TK vector was used as an internal control.
Transcriptional coactivator SUB1 expression in prostate cancer
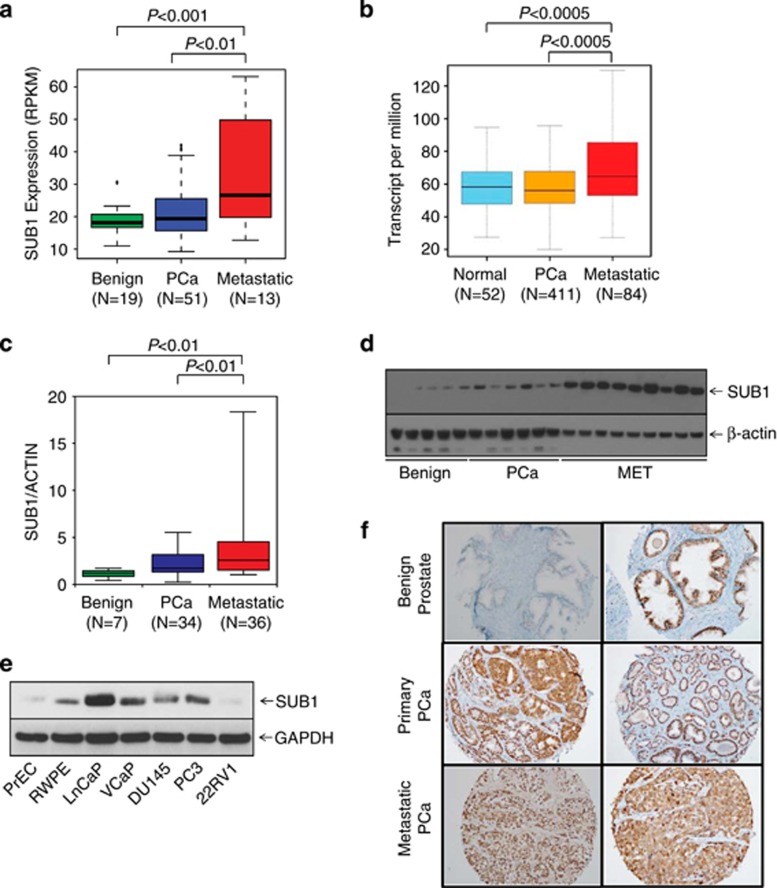
The expression profiling and transcriptome sequence analysis showed upregulation of SUB1 in metastatic prostate cancer (Figure 2a). Moreover, The Cancer Genome Atlas (TCGA) data show that SUB1 is overexpressed in metastatic prostate adenocarcinoma (Figure 2b). To validate this observation, we performed qPCR using RNA from multiple prostate cancer and benign tissues and confirmed increased expression of SUB1 in metastatic prostate cancer tissues relative to benign prostate samples (Figure 2c). Immunoblot analysis using SUB1-specific antibody (Figure 2d) indicated SUB1 protein overexpression. Similarly, elevated levels of SUB1 protein was observed in metastatic prostate cancer cell lines relative to benign cell lines (Figure 2e). Additionally, we investigated SUB1 protein expression in a large number of prostate cancer samples by immunohistochemical analysis, which showed weak or no reactivity in many benign tissues but stronger staining in aggressive prostate cancer tissue and metastatic prostate tumors (Figure 2f).
Figure 2.
SUB1 is overexpressed in aggressive prostate cancer. (a) SUB1 gene expression from next-generation RNA sequencing (RPKM (log 2)) data from benign, prostate carcinoma (PCa) and metastatic prostate cancer (MET) tissues. (b) Expression of SUB1 in normal prostate, primary tumor and metastatic tumor samples from TCGA. (c) qPCR of SUB1 using RNA from benign, PCa and MET tissues. (d) SUB1 protein expression by immunoblot analysis of prostate tissue extracts using SUB1 antibody. β-Actin was used as a loading control. (e) Immunoblot analysis of SUB1 in prostate cancer cell lines. GAPDH was used as a loading control. (f) Immunohistochemical analysis of SUB1 in benign prostate epithelia (top), primary PCa (middle) and metastatic PCa (bottom).
SUB1 expression is essential for prostate cancer cell proliferation and invasion
To determine the functional significance of SUB1 expression in prostate cancer, we perturbed SUB1 levels in prostate cells and investigated the effect of this modulation on cell proliferation, migration and invasion. We used both transient RNA interference and stable knockdown strategies targeting SUB1 in aggressive prostate cancer cell lines DU145 and PC3 and hormone-responsive LnCaP and VCaP cells. The efficiency of SUB1 knockdowns were confirmed by immunoblot (Figure 3a) and qPCR (Supplementary Figures S3a–g) analyses. We observed significant decrease in cell proliferation upon transient knockdown of SUB1 compared with control cells transfected with non-T small interfering RNAs (siRNAs) (Figure 3a). Next, we tested cell motility after stable SUB1 knockdown in prostate cancer cells using wound healing assay. The efficiency of SUB1 stable knockdowns were confirmed by immunoblot (Supplementary Figure S4a). SUB1 knockdown showed a wider wound area 24 h post wound generation relative to control cells, the delayed time to heal indicating an inability of SUB1 knockdown cells to migrate (Supplementary Figures S4b and c). Additionally, SUB1 knockdown in DU145 and PC3 reduced the invasive potential of these cells as assessed by Boyden chamber Matrigel invasion assay (Figure 3b). Further we tested for colony formation after transient knockdown of SUB1 and colonies were quantified (Figures 3c and d). Taken together, these observations demonstrate the involvement of SUB1 in the proliferation, migration, invasion and colony formation of prostate cancer cells in vitro.
Figure 3.
SUB1 is essential for prostate cancer cell proliferation and invasion. (a) Transient knockdown of SUB1 in prostate cancer cell lines reduces prostate cancer cell proliferation. Immunoblot analysis of protein using lysates from prostate cancer cell lines DU145, PC3, LnCaP and VCaP treated with two specific and independent SUB1 siRNA duplexes. β-Actin was used as a loading control. Cell proliferation assay of these cells transfected with either SUB1 siRNA duplex or Non-T siRNA control. (b) Knockdown of SUB1 reduces DU145 and PC3 cell invasion. Boyden chamber Matrigel invasion assay was performed using DU145 or PC3 cells in which SUB1 was transiently knocked down using two independent SUB1 siRNA duplexes. Non-T siRNA-treated cells served as control. Invaded cells were stained with crystal violet and the absorbance was measured at 560 nm. (c) Representative images of colony formation assay. (d) Quantification of colonies formed. The colony formation efficiency was significantly reduced in both SUB1 siRNA1- and 2-treated cells as compared with untreated and Non-T siRNA controls.
SUB1 modulates gene expression in prostate cancer
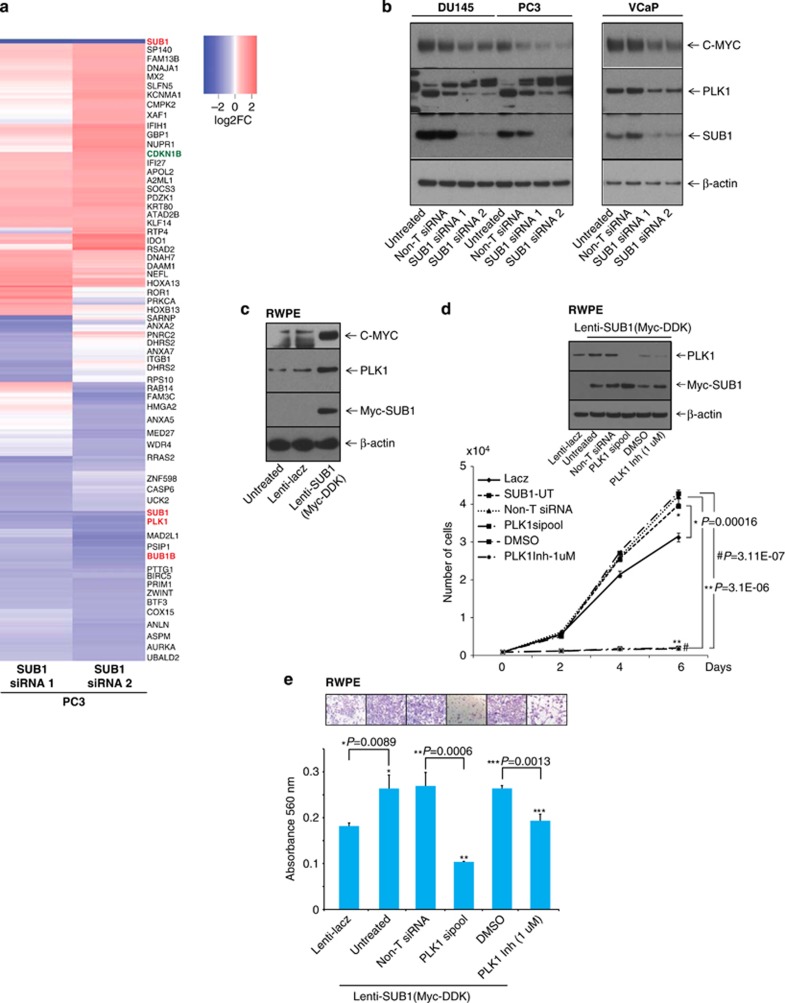
To evaluate SUB1-mediated effects in prostate cancer progression, we performed gene expression analysis using RNA from SUB1 knockdown prostate cell lines. We identified multiple genes that were modulated upon SUB1 knockdown including PLK1, C-MYC, BUB1B and CDKN1B, among others (Figure 4a). PLK1 and C-MYC are known to have a role in cell proliferation, invasion and metastasis.41, 42 We validated the activation of CDKN1B and the downregulation of PLK1, C-MYC and BUB1B, both at the mRNA and protein levels upon SUB1 knockdowns (Supplementary Figures S5a–h, Figure 4b and Supplementary Figures S6a–c) and observed induction of these genes upon SUB1 overexpression in RWPE cells (Figure 4c). TCGA data show that PLK1 and BUB1B are overexpressed in metastatic prostate adenocarcinoma (Supplementary Figures S7a and b). Additionally, we validated both PLK1 mRNA and protein levels across benign, prostrate carcinoma and metastatic prostate cancer tissues by qPCR and immunoblot analysis, respectively (Supplementary Figures S7c and d), which confirmed the direct correlation between SUB1 and PLK1. Additionally, SUB1 and PLK1 mRNA levels are correlated in prostate cancer cell lines (Supplementary Figures S7e and f). Further, we generated stable SUB1-expressing RWPE cells using lentivirus (Supplementary Figure S8). Overexpression of SUB1 resulted in increased cell proliferation (Supplementary Figure S8 and Figure 4d) and invasion (Figure 4e). Furthermore, SUB1 overexpression elevated PLK1 and C-MYC expression, and reduced CDKN1B expression (Figure 4c and Supplementary Figure S6c), showing SUB1 dysregulation triggers alterations in critical oncogenes and tumor suppressors in prostate cancer. To verify the role of PLK1 in cell proliferation and invasion, we treated stable SUB1-overexpressing RWPE cells with PLK1 siRNA SMARTpool or PLK1 inhibitor (volasertib (BI6727)) and analyzed Myc-DDK-tagged SUB1 and PLK1 (Figure 4d inset). Both PLK1 knockdown and PLK1 inhibitor decreased cell proliferation (Figure 4d) and Matrigel invasion (Figure 4e). Thus, these consolidated observations underscore a downstream role for PLK1, C-MYC and CDKN1B in SUB1-mediated prostate cancer cell proliferation and invasion.
Figure 4.
SUB1 modulates PLK1 expression in prostate cancer cells. (a) Heatmap of select genes in stable SUB1 knockdown PC3 cells. (b) Immunoblot analysis showing SUB1, PLK1 and C-MYC in prostate cancer cells after transient knockdown of SUB1. (c) Immunoblot analysis showing Myc-DDK-tagged SUB1, PLK1 and C-MYC in stable RWPE-SUB1- (Myc-DDK-tagged) overexpressing cells. (d and e) Immunoblot, cell proliferation and Matrigel invasion assays were performed using lenti-lacZ or lenti-SUB1 cells. Untreated cells, non-T siRNA, PLK1-specific siRNA SMARTpool-treated cells or in the presence of PLK1 inhibitor (volasertib (BI6727)) were used. Invaded cells were stained with crystal violet and the absorbance was measured at 560 nm. Inset, photomicrographs of invaded cells.
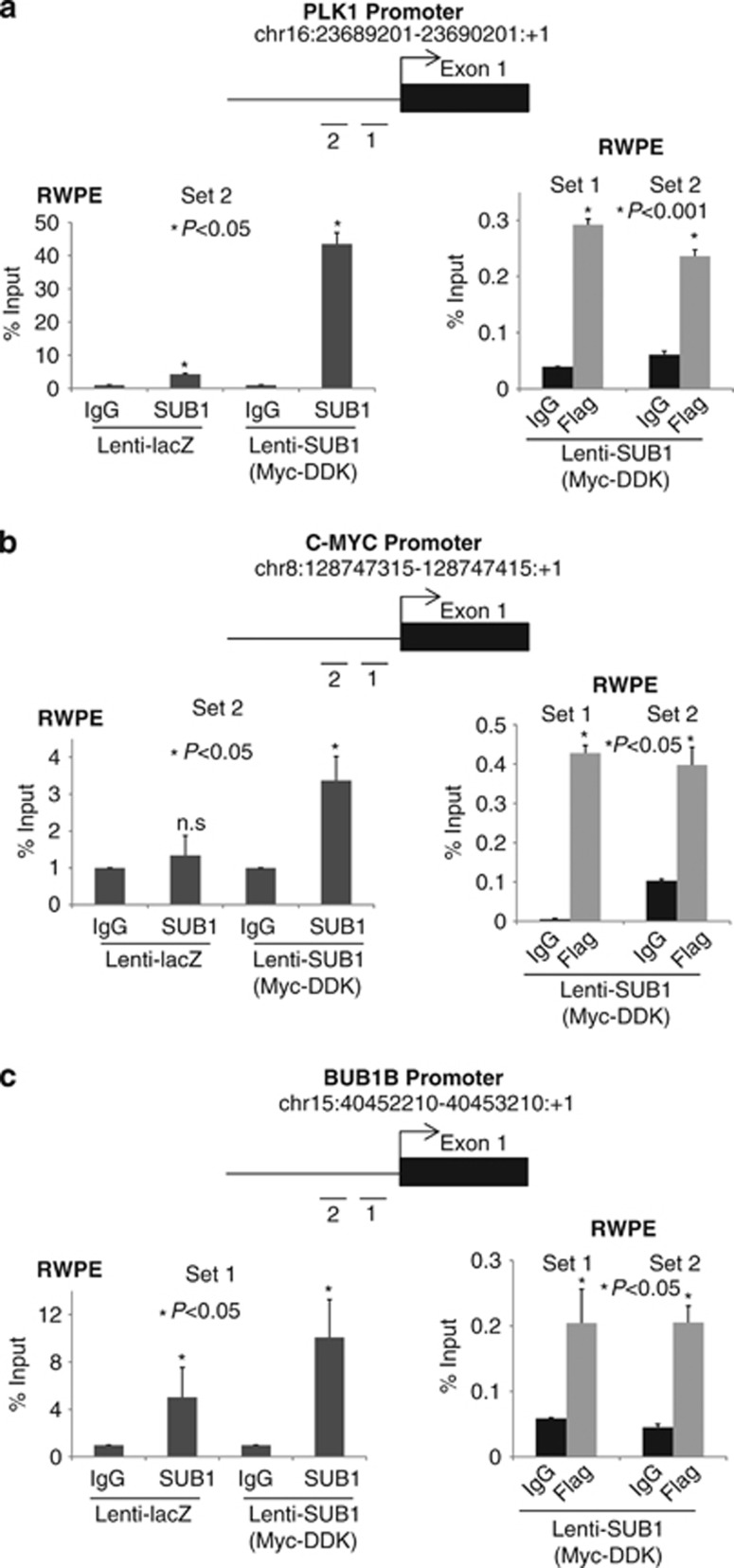
Earlier studies suggest the importance of SUB1 in regulating transcription in vivo. For example, SUB1 enhances transcriptional activation by the activators GCN5 and HAP4 in yeast,43 and stimulates transcription in vitro with diverse kinds of activators, including SMYD3,32 VP16,24, 44 BRCA-145 and octomer transcription factor-1,46 possibly by facilitating assembly of the preinitiation complex. First, we compared motif occurrences within 500 bp upstream regions of downregulated genes and undifferentially expressed genes (when SUB1 is knocked down) using a computer program called CLOVER.47 Next, to validate the binding of SUB1 to PLK1, BUB1B and C-MYC promoters, we conducted chromatin immunoprecipitation (ChIP) assays using commercially available anti-SUB1 or -DDK antibodies in stable RWPE cells overexpressing lacz or Myc-DDK-tagged SUB1. As expected, SUB1 is enriched at PLK1, C-MYC and BUB1B promoter regions (Figures 5a–c). These data demonstrate that PLK1, C-MYC and BUB1B promoters are transcriptionally activated by SUB1 in prostate cancer.
Figure 5.
SUB1 transactivates gene expression by directly binding to specific motifs of their promoter regions. ChIP-PCR analysis for the SUB1 occupancy on (a) PLK1, (b) C-MYC and (c) BUB1B promoters in SUB1- or lacZ-overexpressing RWPE cells. ChIP was performed using antibodies against SUB1-, Myc-DDK-tagged SUB1 and a control IgG. Inset: Schematic representation of the respective genomic regions showing gene and amplicon positions.
SUB1 has a role in prostate tumor growth and metastasis
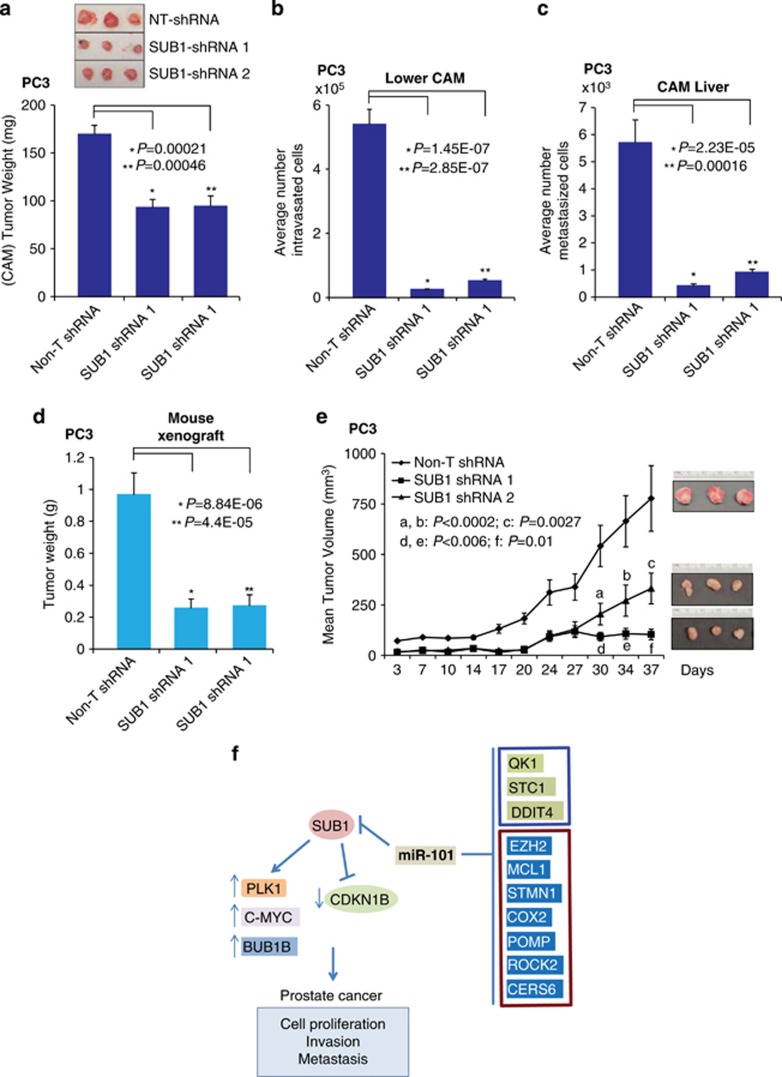
To demonstrate the role of SUB1 on tumor growth in vivo, we used a chick chorioallantoic membrane (CAM) assay and measured spontaneous metastasis, including local invasion, intravasation and metastasis to distant organs. CAM assay was performed as described previously,37, 48 using prostate cancer PC3-SUB1 knockdown cells. Depletion of SUB1 resulted in significantly reduced tumor weight compared with non-target short hairpin RNA (shRNA)-transfected control cells (Figure 6a). SUB1 knockdown in PC3 cells impaired their ability to invade the CAM basement membrane and resulted in a significantly decreased number of intravasated cells in the lower CAM compared with control cells (Figure 6b). Furthermore, there was attenuation of tumor metastasis in the SUB1 knockdown group compared with the control group (Figure 6c). Next, we examined SUB1-mediated tumorigenesis in a murine PC3 xenograft model using Non-T shRNA or two independent SUB1 stable knockdown PC3 cells. Both SUB1-shRNA 1 and 2 cells showed significantly reduced tumor growth and tumor weight in mice (Figures 6d and e) relative to control animals, demonstrating that SUB1 inhibition attenuates tumor growth (CAM assay and murine xenografts) and metastasis (CAM assay) in vivo. These observations show that SUB1 has a role in prostate tumor growth in vivo.
Figure 6.
SUB1 is required for prostate tumor growth in vivo. (a) Tumor growth of stable SUB1 knockdown PC3 prostate cancer cells or control non-T shRNA PC3 cells in the chick CAM tumor assay. Extraembryonic tumors were harvested and weighed after 72 h of postimplantation of cells. Inset: Photomicrographs of CAM tumors (b) and (c) SUB1 knockdown reduces metastasis of PC3 cells in the CAM models. Cells that metastasized to the lower CAM and liver of chicken embryos were quantified using human Alu-specific PCR. (d) SUB1 knockdown in PC3 cells inhibits tumor growth in a mouse xenograft model. Plot of mean tumor volume at indicated time points for mice inoculated with non-T shRNA (solid line with filled diamonds) or two independent SUB1 stable knockdown shRNA 1 (solid line with filled squares) and 2 (solid line with filled triangles) cells. Inset: photomicrographs of xenograft tumors (e) Tumor weights of corresponding mouse xenograft models. n=8 mice per group. (f) Proposed model of SUB1 and miR-101 regulation in prostate cancer progression. SUB1 has a role in cell proliferation, invasion, metastasis and tumor growth, and is regulated by miR-101. Additionally, our study showed that miR-101 downregulates STC1, DDIT4 and QK1 expression (blue box). Additional known targets of miR-101 from published literature are also shown (brown box).11, 12, 13, 14, 15, 16
Discussion
In this study, we show that miR-101 regulates SUB1 expression and SUB1 has a role in prostate cancer growth. We and others have earlier shown that reduced expression of miR-101 leads to overexpression of oncogenic histone methyltransferase EZH2 in multiple tumors.11, 13, 36, 49 A genomic loss of miR-101 or epigenetic silencing leads to reduction in miR-101 expression in multiple cancers.50, 51 These observations suggest that attenuation of miR-101 expression is an important event in oncogenesis. Our investigations also demonstrate the role of SUB1 in prostate cancer cell proliferation and invasion.
SUB1 protein has important roles in various cellular processes including transcription, replication, chromatin organization, cell cycle progression, DNA damage repair and apoptosis.20, 21, 34, 52 Earlier, it was shown that SUB1 is amplified and overexpressed in non-small-cell lung cancer cell lines,53 invasive intraductal papillary mucinous neoplasm of the pancreas54 and in carcinomatoses.55 Additionally, it has been shown that exogenous overexpression of SUB1 could induce the transformation of a population of normal dermal multipotent fibroblasts to acquire malignant characteristics of anchorage-independent growth in vitro and tumorigenicity in nude mice.56 Additionally, it was reported that SUB1 is upregulated in esophageal squamous cell carcinoma and its absence increased radiosensitivity of esophageal squamous cell carcinoma cells, and suppressed non-homologous end-joining activity via downregulation of XLF.52 Targeting coactivators and transcription factors through chemical inhibitors has been challenging.57, 58, 59, 60
Although earlier studies showed the role of SUB1 in cancer, its mechanistic insight in oncogenesis is not fully understood. The present study shows an increased expression of SUB1 in prostate cancer. Moreover, it is involved in tumorigenesis through the activation of PLK1, BUB1B and C-MYC, and repression of CDKN1B during cancer progression. These proteins are involved in several cellular processes including cell proliferation, cell cycle and tumorigenesis.41, 42, 61, 62 The oncoprotein PLK1 is known to have a role in critical cell cycle events and acts in concert with cyclin-dependent kinase 1-cyclin B1 and Aurora kinases.63 Moreover, in cancer these kinases are often dysregulated, promoting uncontrolled cell proliferation and aberrant cell division.63, 64 The PLK family members have been associated with poor prognosis, which lead to enhanced interest as promising targets for anticancer drug development.65 In our study, we demonstrated that SUB1 positively regulated PLK1 expression at the transcriptional level. Furthermore, we investigated the potential role of SUB1-induced PLK1 in prostate cancer invasion by using PLK1-specific siRNA pool or inhibitor volasertib. The PLK1 inhibitor volasertib attenuated stable RWPE-SUB1 cells ability to proliferate as well as to invade through Matrigel in vitro. Moreover, it is currently the most clinically advanced inhibitor.66 Studies in various cancer cell lines (prostate, lung, colon, melanoma, hematopoietic malignancies and urothelial tumors) demonstrated that volasertib inhibits cell division leading to cell death.67, 68, 69, 70 Additionally, we also observed that SUB1 also regulates BUB1B.
In summary, here we show that reduced miR-101 expression results in transcriptional coactivator SUB1 overexpression (Figure 6f). SUB1 triggers increased cell proliferation, invasion, metastasis and modulates expression of several including PLK1, BUB1B, C-MYC and CDKN1B. Finally, SUB1-mediated oncogenic event can be alleviated using PLK1 siRNA or inhibitor. This study shows SUB1 overexpression in aggressive prostate cancer and reveals therapeutic options to block SUB1-mediated oncogenesis.
Materials and methods
Cell lines
Prostate cancer cell lines DU145, PC3 and LnCaP were grown in RPMI-1640 (Life Technologies, Carlsbad, CA, USA), whereas VCaP was grown in Dulbecco's modified Eagle's medium with penicillin–streptomycin (100 U/ml) and 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) in 5% CO2 cell culture incubator. The HEK293 (ATCC), RWPE-1 (henceforth referred as RWPE; ATCC, Manassas, VA, USA) cells were grown in their respective medium as specified by the suppliers. Lentiviruses were generated by the University of Michigan Vector Core (Ann Arbor, MI, USA). Prostate cancer cells were infected with lentiviruses expressing SUB1 shRNA or Non-T shRNA controls and stable cell lines were generated by selection with 1 μg/ml puromycin (Life Technologies).
Benign and tumor tissues
In this study, we used tissues from clinically localized prostate cancer patients who underwent radical prostatectomy. Samples were also obtained from androgen-independent metastatic prostate cancer patients from a rapid autopsy program through the University of Michigan Prostate SPORE Tissue Core as described previously.71, 72 The detailed clinical and pathological data are maintained in a secure relational database. The Institutional Review Board at the University of Michigan Medical School approved this study. Both radical prostatectomy series and the rapid autopsy program are part of the University of Michigan Prostate SPORE Tissue Core.
Gene expression from TCGA
The patients clinical data for prostate adenocarcinoma were downloaded using TCGA assembler.73 However, downloaded data comprised of only tumor pathologic and node pathologic information. Thus, based on tumor pathologic and node pathologic data as per 'https://cancerstaging.org/references-tools/quickreferences/Documents/ProstateSmall.pdf', samples were categorized into primary and metastatic tumor. Afterwards, level3 TCGA RNA-seq data (including raw_read_count and scaled_estimate for each sample) for all primary tumor, metastatic tumor and matched normal samples were downloaded using TCGA assembler. Transcript per million values for each gene was obtained by multiplying scaled_estimate by 1 000 000. Boxplot was generated using R (https://cran.r-project.org/).
Immunohistochemistry
Benign and prostate cancer tissues were obtained from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program, both part of the University of Michigan Prostate SPORE programs, through appropriate informed consent. Institutional Review Board approval was obtained to procure and analyze the tissues used in this study. Immunohistochemistry was carried out to evaluate SUB1 expression using rabbit polyclonal antibody against SUB1 (Novus Biologicals, Littleton, CO, USA; cat. no. NBP1-82454). Immunohistochemistry was performed using an automated protocol developed for the DISCOVERY XT automated slide staining system (Ventana Medical Systems Inc., Tucson, AZ, USA) using Ultramap anti-rabbit HRP (cat. no. 760-4315; Ventana Medical Systems Inc.) and was detected using ChromoMap DAB (cat. no. 760-159; Ventana Medical Systems Inc.). Hematoxylin II (cat. no. 790-2208; Ventana-Roche, Tucson, AZ, USA) was used as the counterstain. The study pathologist Dr Kunju (PK) evaluated the immunohistochemical staining.
Immunoblot analyses
Antibodies used in the study are listed in Supplementary Table S1. All antibodies were used at dilutions optimized in our laboratory. For immunoblot analysis, 10 μg protein samples were separated on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto Immobilon-P PVDF membrane (EMD Millipore, Billerica, MA, USA). The membrane was incubated for 1 h in blocking buffer (Tris-buffered saline, 0.1% Tween (TBS-T), 5% nonfat dry milk), followed by incubation overnight at 4 °C with the primary antibody. After a wash with TBS-T, the blot was incubated with horseradish peroxidase-conjugated secondary antibody and signals were visualized by Luminata Crescendo chemiluminescence western blotting substrate as per the manufacturer's protocol (EMD Millipore).
Gene expression analysis
Global gene expression data was generated using RNA isolated from SUB1 siRNA knockdown PC3 and non-target control cells in profiling analysis as well as in transcriptome sequencing analysis.6 Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Agilent, Santa Clara, CA, USA) and the analysis was performed according to the manufacturer's protocol. A bioconductor package ‘agilp'74 was used to extract and normalize raw data from two-channel experiment arrays. Loess normalization was applied on each array. The gene expression profiling data has been deposited at gene expression omnibus (GSE74895). The differential expression of each gene was estimated by subtracting loess normalized log-2-transformed signal intensity of control sample from that of knockdown sample. Genes with absolute fold change of 2 were selected as differentially expressed genes. The heatmap.2 function of R package ‘gplots' was used to create the heat map. To measure mRNA expression levels, total RNA was isolated from prostate cancer cell lines and prostate cancer tissue samples using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). qPCR was performed as described.37 All primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA) and PCR reactions were performed in triplicate. Primer sequences are listed in Supplementary Table S2.
RNA interference and miRNA transfection
The siRNA duplexes (duplex 1, cat. no. SI04174835 and duplex 2, cat. no. SI00678580) used to inhibit SUB1 expression were purchased from Qiagen, and PLK1 SiGenome SMARTpool (cat. no. M-003290-01-0005) was purchased from GE Dharmacon (Lafayette, CO, USA). Precursors of respective microRNAs and negative controls were purchased from Ambion (Life Technologies). Transfections were performed either with oligofectamine or Lipofectamine RNAiMAX (Life Technologies). SUB1 shRNA (pGipz SUB1-shRNA 1 (V2LHS_85556) and SUB1-shRNA2 (V3LHS_331788)) were purchased from GE Dharmacon. Lentiviruses for these stable knockdowns were generated by the University of Michigan Vector Core. For miRNA transfection or RNA inference, we plated prostate cancer cells at 1 × 105 cells per well in a 6-well plate, and after 12 h, the cells were transfected either with siRNA duplexes or miRNAs. A second identical transfection was performed 24 h later. Seventy-two hours after the first transfection, cells were harvested for RNA isolation or immunoblot analysis.
In vitro overexpression
SUB1 cDNA (Origene Technologies, Rockville, MD, USA; cat. no. RC204999; Myc-DDK tagged) was cloned into Gateway expression system (Life Technologies). To generate lentiviral constructs, PCR8-SUB1 (Myc-DDK tagged) was recombined with pLenti6/V5-Dest (Life Technologies) using LR Clonase II (Life Technologies). Lentiviruses were generated by the University of Michigan Vector Core. Benign immortalized prostate cells (RWPE) were infected with lentiviruses expressing SUB1 or lacZ, and stable clones were selected with 3.5 μg/ml blasticidin (Santa Cruz Biotechnology Inc., Dallas, TX, USA).
Cell proliferation assays
Cell proliferation was measured by cell counting. For this, transient SUB1 and PLK1 knockdown and stable cells overexpressing SUB1 were used. After 72 h of transfection using specific siRNA, the cells were trypsinized and seeded at a density of 10 000 cells per well in 24-well plates (n=3). Non-T siRNA-treated cells were served as controls. The stable RWPE lacZ- or SUB1-overexpressing cells were plated at the same density as mentioned earlier, trypsinized and counted at specified time points by Z2 Coulter particle counter (Beckman Coulter, Brea, CA, USA). Each experiment has been performed with three replicates per sample.
Wound healing assay
DU145, PC3 scramble shRNA or SUB1 stable knockdown cells were seeded in 6-well plates in growth medium containing 10% fetal bovine serum and puromycin (10 μg/ml) for DU145 and PC3, and then allowed to grow to confluent monolayer. For DU145 and PC3, the cells were serum starved for 12 h and replenished with 10% fetal bovine serum-RPMI medium. The wound-induced migration was triggered by scraping the cells with a 200 ul pipette tip, washed with Dulbecco's phosphate-buffered saline and replenished with respective medium. The wound was imaged immediately (0 h) and at 24 h with an inverted phase-contrast microscope under × 4 objective.
Matrigel invasion assay
Matrigel invasion assays were performed as described earlier.37, 75 Various test cells were seeded onto BD BioCoat Matrigel matrix (Corning Life Sciences, Tewksbury, MA, USA) in the upper chamber of a 24-well culture plate. The lower chamber containing respective medium was supplemented with 10% serum as a chemoattractant. After 48 h, the noninvading cells and Matrigel matrix were gently removed with a cotton swab. Invasive cells located on the lower side of the chamber were stained with 0.2% crystal violet in methanol, air-dried and photographed using an inverted microscope (× 4). Invasion was quantified by colorimetric assay. For colorimetric assays, the inserts were treated with 150 μl of 10% acetic acid and the absorbance measured at 560 nm.
Colony formation assay
After 72 h of transfection, untreated, Non-T siRNA/miRNA- or miR-101-treated cells were counted and seeded 800 cells per one well of 6-well plates (triplicate) and incubated at 37 °C with 5% CO2 for 7–10 days. Colonies were fixed with 10% (v/v) ethanol for 30 min and stained with crystal violet (Sigma-Aldrich, St Louis, MO, USA) for 20 min. Then, the photographs of the colonies were taken using Amersham Imager 600RGB (GE Healthcare Life Sciences, Pittsburgh, PA, USA). Colony quantification was carried out using ImageQuant TL Colony v.8.1 software (GE Healthcare Life Sciences).
miR reporter luciferase assays
Wild-type or mutant 3′-UTR of SUB1 were cloned into the pMIR-REPORT miRNA Expression Reporter Vector (Life Technologies). HEK293 cells were co-transfected with pre-miR-101 or controls and wild-type or mutant 3′-UTR-luc, as well as pRL-TK vector as an internal control for luciferase activity. Forty eight hours post-transfection, the cells were lysed and luciferase assays were conducted using the dual luciferase assay system (Promega, Madison, WI, USA). Each experiment was performed in triplicate.
ChIP assays
ChIP assays were carried out with respective antibodies (Supplementary Table S1) using the EZ-Magna ChIP Kit (EMD Millipore, Billerica, MA, USA) as described.37 The primer sequences for the promoters analyzed are provided in Supplementary Table S3.
Chicken CAM assay
The CAM assay for local cell invasion, intravasation, metastasis and tumor (or xenograft) formation was performed as described previously.37, 48, 75 After 3 days of implanting the cells in each egg, lower CAM was harvested and extraembryonic tumors were isolated and weighed. For metastasis assay, the embryonic livers were harvested on day 18 of embryonic growth and analyzed for the presence of tumor cells by quantitative human Alu-specific PCR. Genomic DNA from lower CAM and livers were prepared using Puregene DNA purification system (Qiagen) and quantification of human Alu was performed as described.37, 48, 75 An average of eight eggs per group was used in each experiment.
Tumor xenograft model
All procedures involving mice were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan and conform to all regulatory standards. To evaluate the role of SUB1 in tumor formation in vivo, we propagated stable SUB1 knockdown PC3 cells using two independent shRNAs and Non-T shRNA control cells, and inoculated 1 × 106 cells subcutaneously into the dorsal flank of 5-week-old male athymic nude mice (n=8 for each group; Harlan Laboratories, Evigo Indianapolis, IN, USA). The tumor data obtained using scramble cells is the same as that used in an earlier study as SUB1 tumor xenograft study was conducted simultaneously using common control animals.37 Tumor size was measured biweekly, and tumor volumes were calculated using the formula (π/6) (L × W2), where L is the length and W is the width of the tumor. After 5 weeks, mice from different groups were killed, and then the tumors were photographed, weighed and plotted.
Statistical analysis
To determine significant differences between two groups, Student's two-tailed t-test was used for all experiments except for microarray. P-values <0.05 were considered significant.
Acknowledgments
We thank Twani Dhar, Sherin John, Sivakumar Nallasivam, Samuel Lee and Sai Akshaya Hodigere Balasubramanya for their assistance. We also thank the University of Michigan Vector Core for generating lentivirus. This work is supported, in part, by National Institutes of Health R01CA157845. SV is also supported by R01CA154980 and the University of Michigan Prostate Cancer SPORE Career Development award (to SV). Gene expression data has been deposited in the GEO database under accession number GSE74895
Author contributions
BVSKC and SV designed the experiments. BVSKC, MTG, SSP, ADR, SA, JS, SLC, XJ, NP and SV performed the experimental work. LPK and SD, DSC and MC performed statistical analysis. AMC was involved in fruitful discussions and suggestions. BVSKC and SV wrote the paper. All authors discussed the results and commented on the manuscript when needed.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002; 30: 41–47. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K et al. Delineation of prognostic biomarkers in prostate cancer. Nature 2001; 412: 822–826. [DOI] [PubMed] [Google Scholar]
- Ferreira PG, Jares P, Rico D, Gomez-Lopez G, Martinez-Trillos A, Villamor N et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res 2014; 24: 212–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ et al. Combinatorial microRNA target predictions. Nat Genet 2005; 37: 495–500. [DOI] [PubMed] [Google Scholar]
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 2010; 16: 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999–2009. [DOI] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530–536. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322: 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet 2015; 11: e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schulz R, Edmunds S, Kruger E, Markert E, Gaedcke J et al. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol Cell 2015; 59: 243–257. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Cao K, Kato S, Komizu Y, Mizutani N, Tanaka K et al. Targeting ceramide synthase 6-dependent metastasis-prone phenotype in lung cancer cells. J Clin Invest 2016; 126: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang R, Wang HB, Hao CJ, Cui Y, Han XC, Hu Y et al. MiR-101 is involved in human breast carcinogenesis by targeting Stathmin1. PLoS One 2012; 7: e46173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Zhang T, Liu H, Lv T, Yuan D, Yao Y et al. MiR-101 and Mcl-1 in non-small-cell lung cancer: expression profile and clinical significance. Med Oncol 2012; 29: 1681–1686. [DOI] [PubMed] [Google Scholar]
- Henry NL, Bushnell DA, Kornberg RD. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem 1996; 271: 21842–21847. [DOI] [PubMed] [Google Scholar]
- Lada AG, Kliver SF, Dhar A, Polev DE, Masharsky AE, Rogozin IB et al. Disruption of transcriptional coactivator sub1 leads to genome-wide re-distribution of clustered mutations induced by APOBEC in active yeast genes. PLoS Genet 2015; 11: e1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta K, Kundu TK. Activation of p53 function by human transcriptional coactivator PC4: role of protein–protein interaction, DNA bending, and posttranslational modifications. Mol Cell Biol 2007; 27: 7603–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C, Acker J. Sub1/PC4 a chromatin associated protein with multiple functions in transcription. RNA Biol 2010; 7: 287–290. [DOI] [PubMed] [Google Scholar]
- Das C, Gadad SS, Kundu TK. Human positive coactivator 4 controls heterochromatinization and silencing of neural gene expression by interacting with REST/NRSF and CoREST. J Mol Biol 2010; 397: 1–12. [DOI] [PubMed] [Google Scholar]
- Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 1994; 78: 513–523. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Ge H, Amin AA, Hurwitz J. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J Biol Chem 1996; 271: 22111–22116. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 1994; 78: 525–534. [DOI] [PubMed] [Google Scholar]
- Sikorski TW, Ficarro SB, Holik J, Kim T, Rando OJ, Marto JA et al. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol Cell 2011; 44: 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershnar E, Wu SY, Chiang CM. Immunoaffinity purification and functional characterization of human transcription factor IIH and RNA polymerase II from clonal cell lines that conditionally express epitope-tagged subunits of the multiprotein complexes. J Biol Chem 1998; 273: 34444–34453. [DOI] [PubMed] [Google Scholar]
- Werten S, Stelzer G, Goppelt A, Langen FM, Gros P, Timmers HT et al. Interaction of PC4 with melted DNA inhibits transcription. EMBO J 1998; 17: 5103–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway AF, Occhiodoro F, Mittler G, Meisterernst M, Shannon MF. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J Biol Chem 2000; 275: 21668–21677. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Stelzer G, Meisterernst M. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J 1995; 14: 3520–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan P, Tainsky MA. Coactivator PC4 mediates AP-2 transcriptional activity and suppresses ras-induced transformation dependent on AP-2 transcriptional interference. Mol Cell Biol 1999; 19: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Wang Y, Kannan P, Tainsky MA. Functional characterization of the interacting domains of the positive coactivator PC4 with the transcription factor AP-2alpha. Gene 2003; 320: 155–164. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kim K, Schmidt T, Punj V, Tucker H, Rice JC et al. Cooperation between SMYD3 and PC4 drives a distinct transcriptional program in cancer cells. Nucleic Acids Res 2015; 43: 8868–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S, Yu J, Xu Y, Deng B, Sun T, Hu P et al. PC4 induces lymphangiogenesis dependent VEGF-C/VEGF-D/VEGFR-3 axis activation in lung adenocarcinoma. Am J Cancer Res 2015; 5: 1878–1889. [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang J, Zhang E, Sun H, Wang Q, Wang T et al. Human positive coactivator 4 is a potential novel therapeutic target in non-small cell lung cancer. Cancer Gene Ther 2012; 19: 690–696. [DOI] [PubMed] [Google Scholar]
- Chen L, Du C, Wang L, Yang C, Zhang JR, Li N et al. Human positive coactivator 4 (PC4) is involved in the progression and prognosis of astrocytoma. J Neurol Sci 2014; 346: 293–298. [DOI] [PubMed] [Google Scholar]
- Yan F, Shen N, Pang J, Xie D, Deng B, Molina JR et al. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis 2014; 5: e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi BV, Pathi SS, Goswami MT, Cieslik M, Zheng H, Nallasivam S et al. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget 2014; 5: 6654–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015. 4e05005. [DOI] [PMC free article] [PubMed]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008; 36: D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 2009; 37: W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezuk JA, Brassesco MS, Morales AG, de Oliveira JC, de Paula Queiroz RG, Machado HR et al. Polo-like kinase 1 inhibition causes decreased proliferation by cell cycle arrest, leading to cell death in glioblastoma. Cancer Gene Ther 2013; 20: 499–506. [DOI] [PubMed] [Google Scholar]
- Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res 2011; 71: 2034–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J 1996; 15: 1933–1940. [PMC free article] [PubMed] [Google Scholar]
- Ge H, Zhao Y, Chait BT, Roeder RG. Phosphorylation negatively regulates the function of coactivator PC4. Proc Natl Acad Sci USA 1994; 91: 12691–12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haile DT, Parvin JD. Activation of transcription in vitro by the BRCA1 carboxyl-terminal domain. J Biol Chem 1999; 274: 2113–2117. [DOI] [PubMed] [Google Scholar]
- Luo Y, Ge H, Stevens S, Xiao H, Roeder RG. Coactivation by OCA-B: definition of critical regions and synergism with general cofactors. Mol Cell Biol 1998; 18: 3803–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 2004; 32: 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami MT, Chen G, Chakravarthi BV, Pathi SS, Anand SK, Carskadon SL et al. Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 2015; 6: 23445–23461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Song Q, Cai Y, Wang P, Wang M, Zhang D. RLIP76-dependent suppression of PI3K/AKT/Bcl-2 pathway by miR-101 induces apoptosis in prostate cancer. Biochem Biophys Res Commun 2015; 463: 900–906. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol cell 2011; 43: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology 2014; 59: 1850–1863. [DOI] [PubMed] [Google Scholar]
- Qian D, Zhang B, Zeng XL, Le Blanc JM, Guo YH, Xue C et al. Inhibition of human positive cofactor 4 radiosensitizes human esophageal squmaous cell carcinoma cells by suppressing XLF-mediated nonhomologous end joining. Cell Death Dis 2014; 5: e1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Yasui K, Saito-Ohara F, Koshikawa K, Iizasa T, Fujisawa T et al. A novel target gene, SKP2, within the 5p13 amplicon that is frequently detected in small cell lung cancers. Am J Pathol 2002; 161: 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Maitra A, Iacobuzio-Donahue CA, van Heek NT, Cameron JL et al. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol 2004; 164: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleivi K, Lind GE, Diep CB, Meling GI, Brandal LT, Nesland JM et al. Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer 2007; 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Zhu Y, Chung LWK, Su Y, Cheng T. PC4 is a novel oncogenic gene for mesenchymal stem cell transformation and mediates the reciprocal actions between mesenchymal stem cells and prostate cancer cells. Exp Hematol 2008; 36: S82. [Google Scholar]
- Darnell JE Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2002; 2: 740–749. [DOI] [PubMed] [Google Scholar]
- Hammitzsch A, Tallant C, Fedorov O, O'Mahony A, Brennan PE, Hay DA et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc Natl Acad Sci USA 2015; 112: 10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Park HS, Park SA, Ryu SH, Meng W, Jurgensmeier JM et al. A novel small-molecule inhibitor targeting CREB-CBP complex possesses anti-cancer effects along with cell cycle regulation, autophagy suppression and endoplasmic reticulum stress. PLoS One 2015; 10: e0122628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu Y, Chow DC, Yan F, Hsu CC, Stossi F et al. Characterization of a steroid receptor coactivator small molecule stimulator that overstimulates cancer cells and leads to cell stress and death. Cancer Cell 2015; 28: 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiela-Iparraguirre J, Prado-Marchal L, Pajuelo-Lozano N, Jimenez B, Perona R, Sanchez-Perez I. Mad2 and BubR1 modulates tumourigenesis and paclitaxel response in MKN45 gastric cancer cells. Cell Cycle 2014; 13: 3590–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet 2001; 27: 222–224. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9: 153–166. [DOI] [PubMed] [Google Scholar]
- Liu X. Targeting Polo-like kinases: a promising therapeutic approach for cancer treatment. Transl Oncol 2015; 8: 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov 2010; 9: 643–660. [DOI] [PubMed] [Google Scholar]
- Gjertsen BT, Schoffski P. Discovery and development of the Polo-like kinase inhibitor volasertib in cancer therapy. Leukemia 2015; 29: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res 2009; 15: 3094–3102. [DOI] [PubMed] [Google Scholar]
- Wissing MD, Mendonca J, Kortenhorst MS, Kaelber NS, Gonzalez M, Kim E et al. Targeting prostate cancer cell lines with polo-like kinase 1 inhibitors as a single agent and in combination with histone deacetylase inhibitors. FASEB J 2013; 27: 4279–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-C, Lin Y-M, Tsai Y-C, Pu Y-S, Cheng A-L, Yang JC. Abstract 3725: Combined cytotoxic effects of volasertib and cisplatin in urothelial carcinoma cells. AACR 103rd Annual Meeting, vol. 72 (8 Suppl), Cancer Research: Chicago, IL, USA, 2012. [Google Scholar]
- Smith M, Keir S, Maris J, Kolb A, Reynolds P, Kang M et al. Abstract LB-317: Pediatric Preclinical Testing Program (PPTP) evaluation of volasertib (BI 6727), a Polo-like kinase (PLK) inhibitor. AACR 103rd Annual Meeting, vol. 72 (8 Suppl). Cancer Research: Chicago, IL, USA, 2012. [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 2007; 448: 595–599. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005; 310: 644–648. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods 2014; 11: 599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain B, Bowen H, Hammond J, Posch W, Rasaiyaah J, Tsang J et al. Error, reproducibility and sensitivity: a pipeline for data processing of Agilent oligonucleotide expression arrays. BMC Bioinform 2010; 11: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q et al. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia 2012; 14: 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.