ELECTRONIC ACCESS TO GLOBAL EXPRESSION DATA
We have developed a public Web-based resource to facilitate access to global expression data for Arabidopsis, available at http://mpss.udel.edu/at. Developing an understanding of patterns and levels of transcriptional activity is the starting point for analyzing individual genes or gene families. Genome-wide transcriptional analyses are also revealing the relationship between the structure and organization of a genome and the activity of the genes encoded in that genome. Whole-genome expression data can be obtained from a variety of technologies, including cDNA microarrays (DeRisi et al., 1997), oligonucleotide microarrays (Lockhart et al., 1996), serial analysis of gene expression (SAGE; Velculescu et al., 1995), and massively parallel signature sequencing (MPSS; Brenner et al., 2000a, 2000b). The diversity of microarray platforms, improving densities, relatively low cost per experiment, and the range of technologies (e.g. cDNA, short oligo, and long oligo) have made it easier for labs to obtain their own microarray data than to reanalyze publicly-released data. Tag-based expression data like SAGE and MPSS are more easily utilized by multiple labs because the data format is relatively standard, the genes that are analyzed are not preselected, and the per-library cost discourages unnecessary duplication of experiments. While online resources have been described for SAGE data (Lash et al., 2000; Ball et al., 2001), MPSS data have several unique aspects and must be treated in a different way than SAGE data (Meyers et al., 2004a). None of the existing Web sites for tag-based expression data are customized in a way that links plant genomic and expression data.
We have developed a public, Web-based resource for the analysis of gene expression in the model plant, Arabidopsis. The database and interface is specialized to store and facilitate public access to gene expression data derived by MPSS. Our database currently contains more than 36,991,173 17-base sequence signatures and more than 31,404,553 20-base signatures derived by MPSS from more than 14 Arabidopsis libraries (Meyers et al., 2004a). The MPSS data and Arabidopsis genomic sequence and annotation were used as the basis for the development of publicly-available analysis and comparison tools. Our Web site (http://mpss.udel.edu/at) includes a genome viewer, a set of gene, signature, and library analysis pages, an FTP site for retrieval of the data, and a signature extraction tool to allow specific sequence comparisons to the MPSS data. In this report, we describe the development, organization, and utility of this resource.
THE DATABASE STRUCTURE FOR STORAGE OF MPSS DATA
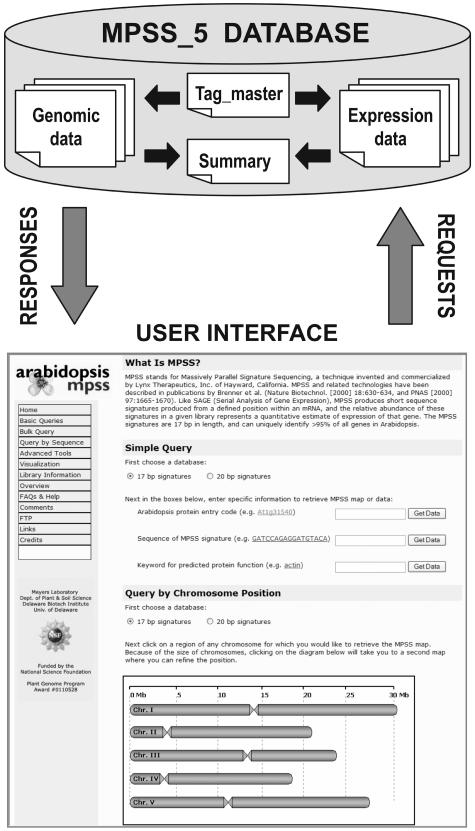
We have designed a relational database for storage and handling of MPSS expression data and genomic sequence information. The general structure for this database is shown in Figure 1; the genomic data are stored separately from the MPSS expression data. A more complete schema for the database is shown in Supplemental Figure 1 (available at www.plantphysiol.org). The database contains 32 tables and includes 13,521,032 records for a total of 14 MPSS libraries plus the Arabidopsis genome annotation. In order to link the MPSS expression data to the Arabidopsis genome, we extract all the potential or genomic signatures from the genomic sequence; these terms are equivalent and simply refer to an occurrence of GATC plus the adjacent 13 or 16 bases of sequence that might be found in MPSS data. This signature extraction procedure is described in more detail in Meyers et al. (2004). Including the additional set of signatures that span exon splice sites, a total of 858,019 genomic signatures are linked via the tag_master table to the 268,132 distinct MPSS signatures that occur in the 14 existing libraries (Meyers et al., 2004a). The genomic tables store physical data for genes and signatures, and the expression tables store information about the MPSS libraries and signature abundances.
Figure 1.
Overview of MPSS database and interface. The database is designed with two major sets of tables, one that contains the genomic annotation and genomic signature information and a second that contains the MPSS expression data. These tables interconnect through additional tables that include a master list of all signature sequences that are utilized in the database. The Web interface connects to the customized database to obtain the data requested by the user and displays the query results in a graphical output.
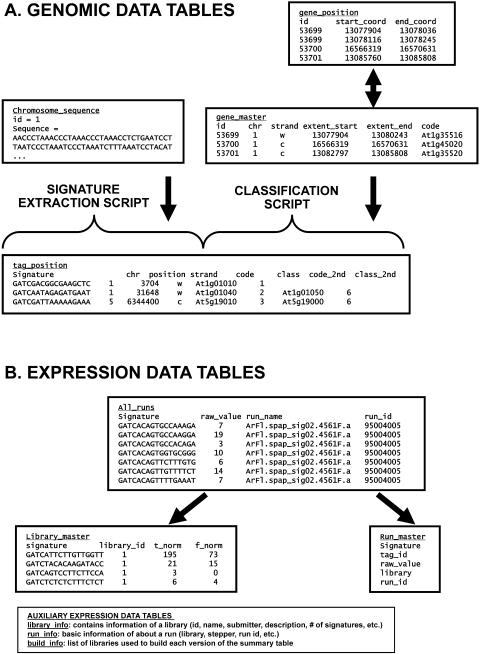
There are two primary tables for storing the annotation data for Arabidopsis genes. This annotation data is parsed directly from the files provided by The Institute for Genomic Research (TIGR; Wortman et al., 2003); the database currently uses TIGR version 3.0, but we anticipate that this will soon be upgraded to TIGR version 5.0. The primary annotation table (gene_master) describes the chromosomal location of the gene, the number of exons, the name and identifier of the gene, and the predicted function of the gene (Fig. 2). The second annotation table (gene_position) stores information regarding the location of each exon of a gene; the position is stored as starting and ending coordinates. A third table, not widely used in version 3.0 of the Arabidopsis annotation, stores the number and exon data for alternate splice variants.
Figure 2.
Genomic and expression data tables. Examples of the primary tables within the genomic and expression datasets in the MPSS database. The table names are underlined and the field names are shown above columns that contain examples of the data found therein. For simplicity, not all tables are shown. The signature sequences are shown in some tables, but the actual sequence of the signatures is stored separately in a master look-up table. A, The tag_position table is constructed using two scripts, one that extracts signatures from the chromosomal sequence and stores the positional information, the other of which compares these positions to the annotated genes to determine the class of the signature, as described in Meyers et al. (2004a). B, The normalized expression level is derived from the raw MPSS data, which is stored in the all_runs table. Two normalized values are obtained for each signature and in each library; these two values correspond to the normalized abundance observed in each MPSS sequencing frame, or stepper, as described in Meyers et al. (2004a), and these data are stored in the library_master table. The final normalized value that is most commonly used for quantitative expression analysis is determined and stored during the construction of the summary table shown in Figure 3A. The run_master table stores information about each MPSS sequencing run.
Because our MPSS expression data includes signatures of both 17 and 20 bases, genomic signatures of these lengths are extracted and stored. The signature extraction script mentioned above is used to identify all occurrences of GATC in the assembled pseudochromosome sequence provided by TIGR (Wortman et al., 2003; Fig. 2); the sequence GATC is derived from the anchoring DpnII site used to generate our MPSS data (Meyers et al., 2004a). We also store 22-base genomic signatures because these data are needed for the analysis of the bad words for the 20-base MPSS signatures, as described in Meyers et al. (2004a). Three tables store the information for each set of signatures of different lengths. The table entitled tag_master stores both the sequence and a unique identifier (pri-mary key) for each distinct signature. Most of the information about the genomic position of a signature is stored in the tag_position table, including the gene with which the signature is associated and the classification of the signature; the class of the signature is determined by the position of the signature relative to annotated genes and exons (Meyers et al., 2004a). Because some signatures are duplicated in the genome (e.g. one signature in tag_master has multiple matches in tag_position), an additional table, z_hits, is used to store the number of occurrences or hits for a given signature. Since the number of hits for a given signature may be in the thousands, the total number of occurrences of a signature that are recorded in the database is much larger than the actual number of stored records.
A set of auxiliary tables stores miscellaneous data about the genome (not shown). These data include the following: the chromosome name; the source and release version of the annotation data; the physical characteristics of each chromosome, such as size and centromere position; and the sequence of the chromosome, although it is not used in routine database queries, and the number of ambiguities found in this sequence.
The remaining tables in the database store information related to the MPSS expression data. There are two primary sets of tables for the expression data. The first set of tables contains the raw data from the individual MPSS sequencing runs. For each library, multiple runs are stored in the run_master table that contains a list of observed signatures and the abundance or expression level of those signatures found in the run (Fig. 2B); this table contains approximately 1,315,827 records for 14 Arabidopsis libraries. There are 4 to 8 runs per library, and each run is sequenced in a particular stepper (Brenner et al., 2000a). However, the expression data represented in the run tables is raw data and requires additional processing to merge the runs and the steppers, and to produce a final normalized value in transcripts per million (TPM) for each signature in the library. The library_master table stores intermediate data in which the runs, but not the steppers, have been merged (Fig. 2B); this table contains approximately 598,891 records for 14 Arabidopsis MPSS libraries.
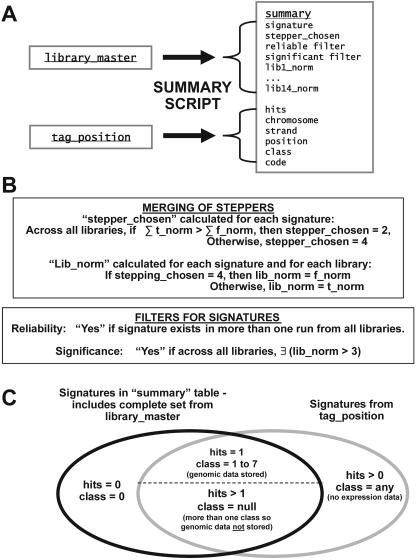
The normalized MPSS expression data for all of the libraries is stored in a single, large table (the summary table, which has 31 columns; Fig. 3A). This is the largest table in our database and contains 4,597,590 entries. The steps are complex to process the raw data from different sequencing reactions into normalized abundances for each signature, and these steps are described in more detail elsewhere (Meyers et al., 2004a). The summary table includes the results of two filtering steps and the merged sequencing runs, summarized in Figure 3B and detailed in Meyers et al. (2004a). In addition to the expression data (in TPM), the summary table contains relational data that associate signatures with the genomic sequence and annotation. Much of these data are redundant with information stored in the other tables described above, but genomic data for signatures duplicated in the genome is not stored in this table, nor are signatures found in the genome but not in the MPSS expression data (Fig. 3C). Although the large size and redundancy of the summary table is contrary to common practice in database design, this table was developed because the queries generated by the Web page, particularly those from the advanced tools page (described below), required data that is found in multiple tables. The queries joined the data for each signature across numerous tables and repeated this process for thousands of signatures, dramatically slowing the access time. By creating a table that stores all of the required data, the disadvantages of data redundancy are outweighed by the improved functionality and enhanced performance of the database. Since the underlying data does not change once it is loaded into the database, this table does not need to be regenerated to maintain the integrity of the database. In general, the database was designed specifically to store and analyze MPSS expression data, and this design was improved based on extensive testing with the goal of rapidly processing the queries generated from our Web site.
Figure 3.
The summary table contains preprocessed data about all of the signatures for each library. A, The summary table is a derived table that contains data extracted from other tables within the database. A set of scripts calculates a final normalized expression level for each signature observed in each library. In the same summary table, information about the genomic position of the expressed signatures is stored; this minimizes the number of different tables that need to be joined when a query requires data about tens of thousands of signatures. B, For each signature, a single normalized value is chosen from the two steppers sequenced in each library, and this value is stored as the final normalized expression level. During the construction of the summary table, the reliability and significance filters are applied to the MPSS data. The normalization step and the filters are briefly described here, but more details about these steps may be found in Meyers et al. (2004a). C, The data in the summary table are completely redundant with the library_master table, but only partially redundant with the tag_position table. The summary table contains only expressed signatures, whether or not the expressed signatures have a genomic match, while the tag_position table includes only signatures found in the genome, independent of the expression data associated with these signatures. Genomic data is stored in the summary for expressed signatures only when the signature is unique in the genome.
THE GRAPHICAL USER INTERFACE FOR ANALYSIS OF MPSS DATA
We developed a graphical interface and analysis tools with which to access our database; this interface is available at our Web site (http://mpss.udel.edu/at). The interface is written in PHP and requires the graphical library, GD. The interface accepts query sequences, Arabidopsis gene identifiers, chromosome position, or MPSS signature sequences to identify a region of interest. The site includes a library browser with information about libraries, tissues from which libraries were derived, and the statistics of signatures derived from each library including success rates and sequencing errors. An advanced analysis tool that permits the direct comparison of entire libraries is available; this tool allows the user to design custom queries across the different libraries and sort the resulting signatures. The main entry page for our Web site (http://mpss.udel.edu/at) provides an access point to all of the pages described in the sections below.
Chromosome Viewer
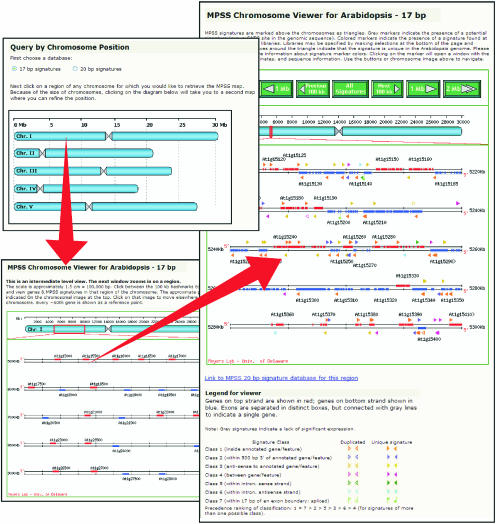
The genomic annotation information combined with the MPSS expression data can be viewed and visually scanned using our customized viewer. This viewer is based on one developed for SAGE data as part of the Saccharomyces Genome Database (SGD; Ball et al., 2001). The code was generously shared by the SGD programmers, and it was rewritten in PHP and extensively modified to include the added features implemented in our database. We call this tool the chromosome viewer (CV), and it is accessed via an image of the five Arabidopsis chromosomes on the main entry page (Fig. 4). From the main entry page, the user is taken to a secondary viewer with a scale of one megabase (Mb) per line, demarcated into 100-kb segments with every fiftieth gene on the chromosome indicated (Fig. 4). This secondary viewer allows the user to more accurately target the window displayed in the primary CV window. The primary viewer has a fixed scale of 20 kb/line, demarcated in 5-kb segments; a standard window shows 100 kb (Fig. 4), but the script permits the display of variable lengths so that entire bacterial artificial chromosome clones may be shown, for example. In both the secondary and primary viewers, the chromosomal location is indicated above the window, and this schematic of the chromosome can be used to navigate. A set of buttons shown above the primary viewer also facilitates movement up or down the chromosome. The primary viewer shows the annotated exons on the top strand of the chromosome in red and the bottom strand in blue, with the set of exons comprising a gene boxed in gray (Fig. 4). The Arabidopsis identifier is indicated above or below the gene.
Figure 4.
Accessing specific chromosomal regions using the MPSS viewer. The chromosome viewer is launched by clicking on the image of the Arabidopsis chromosomes located on the main entry page. Red arrows in this figure indicate the direction of navigation from the main page to the secondary chromosomal viewer page, as described in the text. The magnification on the secondary page is five times less than that of the primary page, and this secondary page allows the user to focus on a particular subgenomic region for display in the primary CV window. The primary chromosomal viewer focuses on a selected approximately 100-kb region of an Arabidopsis chromosome, and displays the annotated genes and exons, with identifiers, on both strands of the DNA. Above and below the genes, the viewer displays the significantly expressed signatures. The genes and signatures are linked to the gene analysis and signature analysis Web pages.
In CV, unexpressed genomic signatures are shown in gray and signatures significantly expressed in any library (e.g. greater than 3 TPM) are shown in color. The colors of the expressed signatures indicate the class; the seven classes are determined based on the position of each signature relative to annotated genes and exons, and a legend is shown below the viewer to explain these classes (Fig. 4). The classification system is modeled after a similar system used in the SGD (Ball et al., 2001), but we have added three additional classes. These modifications are described in greater detail in Meyers et al. (2004a), and the interpretation of expressed signatures of different classes is detailed below. Within CV, the unexpressed genomic signatures can be hidden or revealed by selecting the button “signatures on”, whereas the expressed signatures are always shown. If the viewer is accessed from the signature or gene analysis pages described below, the signature or gene that referred to the chromosome viewer is indicated in red as a reminder to the user.
Future modifications to CV will include access to the mitochondrial and chloroplast genomes, for which some matching signatures have been identified in our MPSS libraries (Meyers et al., 2004a). Because the latest versions of the Arabidopsis annotation incorporate full-length cDNAs that include 5′ and 3′ untranslated regions (UTRs), future versions of CV will differentiate UTRs from coding regions.
Gene Analysis and Bulk Query Tool
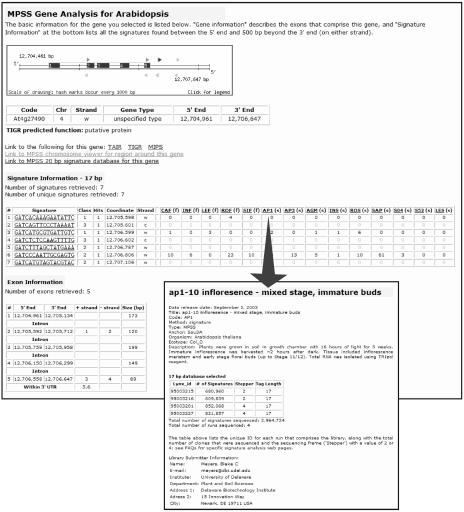
Analysis of specific genes can be performed by entering an Arabidopsis identifier from the main entry page, or by selecting a gene from the CV window. Either of these options takes the user to the gene analysis (GA) page. This page shows all of the genomic signatures derived from the sequence of the gene and the downstream region that may contain the 3′ UTR. The expression data derived from these signatures can be used to investigate the level and pattern of transcriptional activity. This represents an electronic or virtual northern blot for the gene, although an absence of MPSS expression data does not necessarily mean that the gene is not expressed, due to sequence-specific effects that reduce or eliminate MPSS data for certain signatures (Meyers et al., 2004a). GA includes a smaller version of the viewer described above that uses an expanded scale and is specific for the gene of interest (Fig. 5). The function of the gene may be determined based on the TIGR Arabidopsis annotation that contains known functions from published studies or protein homologies. The GA pages include direct links to the specific Web pages for each gene at TIGR (http://www.tigr.org), TAIR (http://www.arabidopsis.org), and MIPS (http://mips.gsf.de). GA also links to the chromosome viewer to display the larger genomic context of the gene of interest. The MPSS expression data in the GA output page is summarized in a table that contains the normalized abundance in each library for the signatures derived from the gene. These data, with values in TPM, are displayed in a table that also contains the chromosomal position and the number of hits or occurrences of each signature in the genome. The libraries are indicated by a three-letter code and either “f” or “s” that indicates if the MPSS method used was either full (also known as classic) or signature; the variation in the method is minor and is briefly discussed in Meyers et al. (2004a). The libraries are listed in an arbitrary order (the order in which they were received and entered into our database), and the three-letter code is linked to a pop-up window that summarizes the relevant information about the tissue, RNA and MPSS sequencing runs (Fig. 5; see description of “Library Information Pages” below).
Figure 5.
Output of gene analysis and library information pages. The gene analysis Web page is reached by entering a specific Arabidopsis identifier on the main entry page, by selecting a gene from a list of identifiers generated by a keyword search or the bulk query page, by selecting a gene shown in the viewer, or from a direct link in the signature analysis page. The small viewer window at the top of the gene analysis page shows the annotated exons for the gene and the associated genomic signatures. The exons are numbered from 5′ to 3′. The signatures are indicated as triangles and listed in the table below; these signatures are linked to the signature analysis Web page. The table in the center contains the normalized expression data and a summary of the genomic information for signatures located in or immediately 3′ of the gene of interest. The small table at the bottom contains the genomic coordinates for the exons and introns and indicates the signatures contained therein. The library information page (inset, and indicated by arrow) is a JavaScript pop-up window that is activated by selecting the three-letter library code, and it contains the basic information about the tissue, RNA, and MPSS sequencing runs. The library information is also accessible from elsewhere in the MPSS Web site.
For users interested in a large family or set of genes, the page entitled Bulk Query allows the user to quickly view all the expression data for sets of genes. The genes are listed using Arabidopsis gene identifiers, and the list is entered on a separate page that is accessible from the left frame of the main entry page. The BQ page produces a summary of the genes along with only the subset of genomic signatures for which significant expression data exist in the database. As an alternative way to sort through large numbers of genes, we have implemented a keyword search that generates a list of the genes with a match in the TIGR annotation and description to a user-entered keyword. The keyword search is accessible from the main entry page, and the output is a series of Arabidopsis identifiers linked to the GA pages. As an example, entering “kinase” as the argument to the “keyword for predicted protein” function produces a list of 1,124 genes. While the keyword search does not permit wildcard entries, partial word matches are allowed (for example, “kina” produces the same list of 1,124 genes).
Future modifications to GA may include a way to restrict or order the list of libraries that are shown on a single screen; the current set of 14 libraries fills most of the page, and a larger set of libraries would require that the user scroll far to the right to find the library of interest.
Signature Analysis
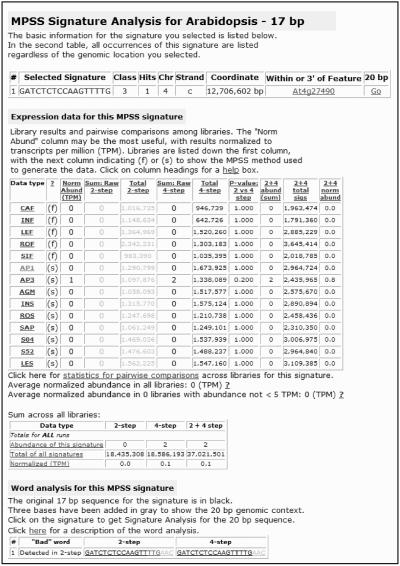
From the table in GA, from the viewer, or from the main entry page, specific signatures can be selected for analysis. An MPSS signature is a string of 17 or 20 nucleotides that start with GATC, for example GATCCAGAGGATGTACA. The signature analysis (SA) page contains all of the genomic data and the normalized and raw expression data for each signature (Fig. 6). The uppermost table lists all positions in the genome for the specific signature and contains links between the 17- and 20-base genomic signatures. If a signature is among the MPSS data but does not match the genome (a Class 0 signature), this is indicated in the upper table. The raw and normalized expression data for the Arabidopsis libraries are listed below this genomic information. The table of expression data identifies the library with a three-letter code, the MPSS method used for each library (as mentioned above for GA), the normalized abundance (calculated as described in Meyers et al., 2004a), and the raw abundances for the 2- and 4-step sequencing reactions along with the total number of signatures sequenced in either stepper for each library. The raw abundances are shown in gray when the stepper is not chosen during the normalization steps described in Meyers et al. (2004a). We also provide a series of comparisons and sums of the 2-step and 4-step raw abundances (Fig. 6). For each library, the SA page uses a Z-test to assess the difference and calculate a P-value for the 2-step and 4-step abundances, and the page reports the sum of the 2- and 4-step sequencing reactions for each library and for all libraries. These calculations provide different ways for users to evaluate the MPSS expression data for a given signature.
Figure 6.
Output of signature analysis page. The signature analysis page is reached by clicking on signatures from the GA, CV, and AT pages, or by entering the exact sequence of a signature from the main page. The upper table lists all genomic matches for a specific signature. This table also contains links between the 17- and 20-base genomic signatures. The raw and normalized expression data are listed in the large central table; the smaller table below this contains the summed expression data from all libraries. At the bottom of the page is an analysis of the MPSS sequencing frames for each signature. This analysis is used to screen for the presence of the 20 bad words (Meyers et al., 2004a). The presence of one or more bad words in frame with the MPSS sequencing reaction can result in under-representation of the signature in the expression data.
The SA page also performs an analysis of the MPSS sequencing frames for each signature to identify the potential bad words; 20 of the 256 possible 4-base words that occur in the MPSS sequencing frames are under-represented in the MPSS expression data (Meyers et al., 2004a). As previously described by Meyers et al. (2004a), a complete analysis of the words in the 17-base signatures requires an additional 3 bases of sequence extracted from the genome (20 bases total), and analysis of the 20-base signatures requires an additional 2 bases of sequence extracted from the genome (22 bases total). For each genomic location, the database stores the 17-, 20-, and 22-base signatures; the SA Web page performs the analysis of the words that occur in the MPSS sequencing frames in these signatures. This analysis is useful to determine if the measured abundance of one or both of the two MPSS steppers might be diminished by sequence-specific effects.
Advanced Tools
The Advanced Tools (AT) page builds complex queries using a set of pull-down combo boxes (lists), radio buttons and check-boxes for selection, and textboxes for entering data provided by the user. Signatures can be retrieved that match a series of criteria set by the user; these criteria may be based on the genomic location or characteristics of expression in one or more libraries. This page bypasses the graphical maps described above and can generate sorted lists of signatures. Because AT reports results based on the normalized expression data (in TPM), it required the development of the precomputed summary table described above. Thus, the summary table is the basis for all queries submitted via this page.
The large number of libraries for which the queries could be designed necessitated an entry page that allows the user to select a subset of libraries for analysis (Fig. 7). The selected libraries are then listed in a second page on which the complex queries can be designed to sort through MPSS signatures found in the library. This second page of AT offers a range of search criteria for creating complex queries. On this second page, adjacent to the list of libraries selected from the entry page, the “select” option will follow the defined criteria to generate a list of signatures; the “view” option will retrieve and show expression data for that library but does not allow the user to define the expression data criteria; and the “ignore” option hides the expression data in the final output page. The primary function of the AT page is to screen the expression data for signatures of interest; the user is required to pick at least one library as part of the criteria or for viewing.
Figure 7.
“Advanced tools” permits complex query building to access the MPSS data. The advanced tools page permits users to custom-design queries. Arrows in this figure indicate the direction of navigation from a library selection page, to the criteria definition page, and to the output page. The user chooses libraries on the first page, and the selected libraries appear on a second page along with a series of pull-down windows, selectable options, and entry boxes to define specific genomic locations or characteristics of expression that are of interest to the user. The output can be sorted according to expression levels, genomic matches, or other criteria.
The main criteria for sorting and selecting signatures in AT is the normalized expression level. The first setting that the user chooses is whether the settings for different libraries are compared using the “AND” or “OR” Boolean operators. The AND option will return a list of signatures precisely matching the abundance levels for all the selected libraries, while the OR operator will return signatures that match at least one of the selected abundance levels for one of the libraries. In this regard, the AND operator is exclusive while the OR operator is less restrictive and more inclusive. To define the expression level for each selected library, the user has a choice of setting a predefined range, a specific range, or a single, exact value. If the user does not define an abundance level at this point, the library is ignored during the final analysis. After setting the expression level, the user must decide whether to retrieve 17- or 20-base MPSS signatures. The relative merits of 17- and 20-base signatures are described elsewhere (Meyers et al., 2004a).
We have implemented a tool within the AT page for the identification of signatures that are differentially expressed when two libraries are compared (Fig. 7). When this option is selected, a pair of libraries is selected for comparison, and the user defines either a percentage or fold difference for signatures found in one library as compared to the other. The comparison will identify signatures for which the expression is within a range of differences, above or below a certain difference, or exactly matching a given difference. This tool also allows the comparison to be made in only one direction or in both directions; for example, the user may chose to identify signatures for which the abundance is 2-fold higher in the callus versus silique library, or by selecting “both directions,” the same comparison can identify signatures found 2-fold higher in either the callus or silique library in the pairwise comparison. When this section of AT is activated, a JavaScript program defines the formula that the user is building for the pairwise comparison.
The next set of options in the primary query page of AT restricts the signatures to those that lie in a certain region of genes or the genome, or that have a certain number of hits (occurrences in the genome). The default for this section is an unrestricted search. The user may restrict the query to signatures with a defined position in the gene based on the classification system described above and detailed in Meyers et al. (2004a); essentially, seven classes of genomic signatures have been defined based on comparisons to the genome annotation and depending on the position and strand relative to the exons of annotated genes. AT is also one of two pages in the Web site that can directly access signatures that do not match to the genome (Class 0 signatures); the signature extraction page described below is the other way to access these signatures. In addition to the class of the signature, the search can be limited to signatures that match within a defined set of coordinates or positions on one of the chromosomes. The user can also limit the search to signatures that are unique in the genome (hits = 1), or to the “duplicated” signatures with more than one occurrence in the genome (hits > 1).
Finally, the signatures can be sorted that match the queries defined above for AT. The user can select from numerous criteria in defining the sorting steps, including the number of hits, the class, chromosome, position, strand, or the most commonly used criterion is the abundance level in one of the libraries. After selecting the basis for the sort, it is then further defined as ascending or descending. There are three sorting steps that can occur sequentially; all sorting is turned off by default, and one, two, or all three sorts can be utilized for organizing the data and output of AT. The combinations of query options and criteria make the advanced tools page a powerful tool for comparing libraries and designing specific searches.
Visualization of Library Comparisons
We have precalculated and displayed the pairwise comparisons of the callus, flower, leaf, root, and silique libraries. In the visualization Web pages at our site, Arabidopsis genes are represented by 8 × 8 pixel squares colored accordingly with their level of expression; these data are mirrored at http://allometra.com/mpss.shtml. For each gene in the two libraries in the comparison, gene expression in library A is reflected by the portion of the red component in the final color, and expression in library B is reflected by the portion of the green component in the final color. The position of each gene in the image reflects its position in the Arabidopsis genome and the gene annotation appears on mouse-over. Each square can be selected and is linked to the Gene Analysis page for the selected gene. The library comparison and Web images were generated using PyMood, a genomics data visualization and Web-publishing program (Allometra, Davis, CA).
Library Information Pages
We have stored all of the information about each MPSS library in tables that are accessible as pop-up windows throughout the SA, GA, and AT output pages (Fig. 5). The set of libraries currently stored in our database is summarized on the library information Web page, and this list is linked to the same set of pop-up information windows. The information stored about each library includes the public release date for each library, a brief title for the library, the three-letter code used throughout the Web site, the MPSS method (signature or full/classic), the type (MPSS, although SAGE data is an un-implemented option), the anchoring restriction enzyme (DpnII or its isoschizomer Sau3A), the organism and ecotype from which the tissue was obtained, and a short yet detailed description of how the plant material was grown and the RNA obtained. The library information also includes summary statistics for the MPSS sequencing runs that were obtained for each library, along with the name and contact information of the individual who submitted the sample.
We developed a tool to assess the sensitivity of the MPSS data in our database. This tool is available on the library information Web page. This tool will determine the likelihood of detecting a transcript known to be expressed at a particular level, based on the number of signatures in a library or set of libraries. This calculation is based on a Poisson distribution and can be used to determine the threshold of sensitivity for a given library size. The user selects a confidence interval for detection in a set of libraries that they are analyzing and then selects the libraries or specific number of signatures that they are analyzing. The number of MPSS signatures (N) that was obtained for the selected libraries is entered into the algorithm. The script then calculates the lowest level of expression (in normalized units of TPM) that is detectable at the selected confidence level in the N-sized population of signatures. This calculation is useful, because for a weakly expressed transcript it is possible that the transcript was present but not detected due to sampling error. Although this calculation ignores the biological differences among libraries that may result in an absence of a particular transcript, it does allow the user to determine the level of expression that should be detected within a given confidence level.
Signature Extraction and Query by Sequence
Some users may have specific sequences that they would like to compare against the MPSS expression data. For example, these sequences could include cDNA sequences that have not been included in the genomic data used in our site. The signature extraction (SE) tool, located under the “query by sequence” menu option on our Web site, allows the user to enter one or more sequences in FASTA format. The SE algorithm locates all occurrences of the anchoring site for the enzyme used in the MPSS reactions, which is GATC (from DpnII) for the libraries in our database. The potential signatures are then obtained from the flanking sequence; these potential signatures include either 13 or 16 bases 3′ of the GATC, depending on whether the user is querying the 17- or 20-base expression data, respectively. The signatures are extracted from 5′ to 3′ and then, depending on the user's choice, the reverse complement of the strand is generated and signatures extracted in the same way. The user selects the set of libraries to analyze and chooses whether to show only signatures with expression data or every potential signature extracted from the sequence. SE, like the advanced tool page described above, allows the user to access MPSS data that has not been mapped to the genome (Class 0 signatures).
Help Pages and Frequently Asked Questions (FAQs)
We have linked help pages and a frequently asked questions (FAQs) page throughout our site. This page is launched through JavaScript as a pop-up box and contains basic information to clarify some of the more common difficulties that users encounter in understanding the MPSS data.
THE USE AND INTERPRETATION OF DATA FROM THE ARABIDOPSIS MPSS WEBSITE
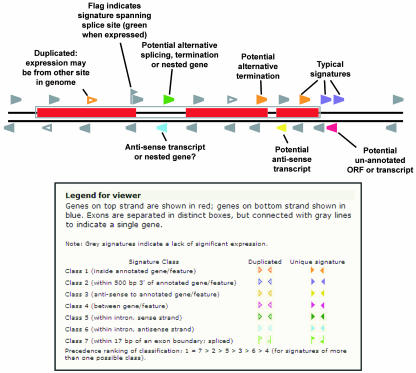
Analyses of the Arabidopsis MPSS expression data indicate that a significant number of novel transcripts are produced from this genome. While transcripts matching to sense-strand expression of annotated genes are predominant, the MPSS data also identify alternatively terminated or polyadenylated transcripts, antisense transcripts, and transcripts mapping to intergenic regions (Meyers et al., 2004a, 2004b). These novel transcripts are easily recognized in our viewer. In Figure 8, we provide a guide for the interpretation of the expressed signatures that map to regions of the gene other than those expected for normal transcripts that would map to the 3′ end of the sense strand. The white triangles indicate signatures that are duplicated in the genome; if expression data is found for a duplicated 17-base signature, the 20-base MPSS data may provide validation of the source of the expression data by providing a higher level of specificity. The quantitative and qualitative data that can be obtained from the MPSS Web site should serve as the starting point for additional laboratory-based experiments to confirm these results. Measurements of gene expression performed using different technology platforms are likely to vary, and in some cases this variation may be substantial.
Figure 8.
Interpretation of expressed signatures of different classes. A diagram of a gene is shown as it would appear in our chromosome viewer or gene analysis page. The gray triangles indicate genomic signatures for which no MPSS expression data has been identified, while the colored triangles indicate different classes of signatures for which significant expression data exist. The legend, below, is taken from our Web page, and explains the relationship between the color of the expressed signatures and our classification system. The text in the figure indicates the interpretation of expressed signatures that uniquely map to introns, internal exons, anti-sense positions, or positions outside of currently annotated genes. These transcripts, and the associated signature classes, are discussed in more detail in Meyers et al. (2004a).
The interface has been designed to display additional libraries. Each of the pages dynamically responds to the contents of the database. As new libraries are added to the Web page, these appear automatically on each of the pages described above. We will take advantage of this design for future MPSS projects; for example, we anticipate that our site will soon host data for a new project that is generating approximately 65 MPSS libraries from rice (Oryza sativa; B.C. Meyers and G.L. Wang, unpublished data), as well as data for the rice blast pathogen, Magnaporthe grisea (G.L. Wang, C.D. Haudenschild, and B.C. Meyers, unpublished data), and grape (Vitis vinifera; A. Iandolino and B.C. Meyers, unpublished data). If more than 20 MPSS libraries are obtained for any single organism, it may become unwieldy to display these data, as the tables would become much wider or taller than would fit within a single Web page. Additional modifications to the interface may become necessary to show selected subsets of libraries in which a user has a specific interest. As more eukaryotic genomes become available in draft and finished forms, and MPSS or SAGE data are more widely used to annotate and explore transcription in these genomes, it may be advantageous to adapt the database and interface described here to the genomes of these organisms.
The database and Web interface described above are continually refined to improve the accuracy of the data and to facilitate different types of analyses. For example, analysis of the unmatched Class 0 signatures is continuing, and many of these signatures are likely to be matched with the impending release of the TIGR version 5.0 annotation; this newer version includes many more full-length cDNA sequences than the version (3.0) that we are currently using (Haas et al., 2003; Wortman et al., 2003 and C.D. Town, personal communication). The higher-abundance Class 0 signatures may be derived from as-yet uncharacterized transcripts or splicing events, so additional full-length cDNA sequences may be matched at a high rate by these signatures (Meyers et al., 2004a). The customized database and set of data-handling scripts make this a unique repository for MPSS data; apart from the libraries in our database, very little of these data are available in the public domain, and those that are available are extremely difficult to access or work with in the absence of any bioinformatics tools (Jongeneel et al., 2003). However, public access to the large amounts of transcriptional data such as those generated by MPSS is extremely important. Our goal was to build a simple and clearly-designed interface that permits biologists to review and assess both the raw and processed MPSS data and to build complex queries to compare libraries. This is a novel public resource for gene expression analysis, and for plant biologists it offers a powerful means to analyze specific genes and gene families.
Supplementary Material
Acknowledgments
We thank Michael Cherry, Shuai Weng, and Kara Dolinski of Stanford University for supplying the code to the SAGE viewer. We appreciate the advice and suggestions of Christian Haudenschild of Lynx Therapeutics during the construction and implementation of the Web site.
This research was supported by the National Science Foundation Plant Genome Research Program (award no. DBI 0110528) and by an NSF Research Experience for Undergraduate (REU) supplement for D.K.L.
The online version of this article contains Web-only data.
References
- Ball CA, Jin H, Sherlock G, Weng S, Matese JC, Andrada R, Binkley G, Dolinski K, Dwight SS, Harris MA, et al (2001) Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res 29: 80–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000. a) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18: 630–634 [DOI] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, Mao JI, Luo S, Kirchner JJ, Eletr S, et al (2000. b) In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc Natl Acad Sci USA 97: 1665–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel CV, Iseli C, Stevenson BJ, Riggins GJ, Lal A, Mackay A, Harris RA, O'Hare MJ, Neville AM, Simpson AJ, et al (2003) Comprehensive sampling of gene expression in human cell lines with massively parallel signature sequencing. Proc Natl Acad Sci USA 100: 4702–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF (2000) SAGEmap: a public gene expression resource. Genome Res 10: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild C, Agrawal V, Edberg SB, Ghazal H, Decola S (2004. a) The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res (in press) [DOI] [PMC free article] [PubMed]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild C (2004. b) Analysis of the transcriptional complexity of Arabidopsis by massively parallel signature sequencing. Nat Biotechnol (in press) [DOI] [PubMed]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484–487 [DOI] [PubMed] [Google Scholar]
- Wortman JR, Haas BJ, Hannick LI, Smith RK Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al (2003) Annotation of the Arabidopsis genome. Plant Physiol 132: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.