Abstract
Collagen is the most abundant protein in animals. The posttranslational hydroxylation of proline residues in collagen contributes greatly to its conformational stability. Deficient hydroxylation is associated with a variety of disease states, including scurvy. The hydroxylation of proline residues in collagen is catalyzed by an Fe(II)- and α-ketoglutarate-dependent dioxygenase, collagen prolyl 4-hydroxylase (CP4H). CP4H has long been known to suffer oxidative inactivation during catalysis, and the cofactor ascorbate (vitamin C) is required to reactivate the enzyme by reducing its iron center from Fe(III) to Fe(II). Herein, we report on the discovery of the first synthetic activators of CP4H. Specifically, we find that 2,2′-bipyridine-4 carboxylate and 2,2′-bipyridine-5-carboxylate serve as ligands for the iron center in human CP4H that enhance the rate of ascorbate-dependent reactivation. This new mode of CP4H activation is available to other biheteroaryl compounds but does not necessarily extend to other prolyl 4 hydroxylases. As collagen is weakened in many indications, analogous activators of CP4H could have therapeutic benefit.
Graphical abstract
Collagen is an abundant protein in all animals. In humans, collagen comprises a third of the total protein and three-quarters of the dry weight of skin, and is the most abundant component of the extracellular matrix.1 The enzyme collagen prolyl 4-hydroxylase (CP4H) is a non-heme Fe(II)- and α-ketoglutarate-dependent dioxygenase (FAKGD) that catalyzes the stereoselective hydroxylation of Cγ of (2S)-proline (Pro) residues in collagen strands.2 The ensuing creation of (2S,4R)-4-hydroxyproline (Hyp) residues is necessary for the formation of stable collagen triple helices.3,4 Molecular oxygen (O2), α-ketoglutarate, and Fe(II) are required for the catalytic activity of CP4H.5 During catalysis, α-ketoglutarate is decarboxylated oxidatively to produce succinate and CO2.6
CP4H can decarboxylate α-ketoglutarate without effecting the hydroxylation of proline residues.7,8 This uncoupled reaction inactivates the enzyme.9 Ascorbate, which is also known as vitamin C (VitCred), rescues the enzyme by reducing the inactive Fe(III) state to the active Fe(II) state.10,11 In humans, a deficiency of ascorbate leads to scurvy,12,13 a disease caused by collagen instability.14 Moreover, nonsyndromic high myopia (which is a leading cause of blindness15) and certain neurological processes16 could arise in part from low CP4H activity. The complete loss of CP4H activity is lethal to Caenorhabditis elegans17,18 and mice.19
Recently, we reported that certain dicarboxylate derivatives of 2,2′-bipyridine (bipy) and other biheteroaryl compounds are specific inhibitors of human CP4H.20,21 These biheteroaryl dicarboxylates compete with α-ketoglutarate for binding to the active-site iron. To our surprise, we discovered two biheteroaryl monocarboxylates that activate CP4H. Activation by 2,2′-bipyridine-4-carboxylate (bipy4C) and 2,2′-bipyridine-5-carboxylate (bipy5C) was unexpected because bipy5C was reported previously to be a weak inhibitor of a related CP4H from chicken.22 Herein, we report on the action of these monocarboxylates and related compounds. Our data suggest that they enhance the rate at which VitCred reduces the iron center. Moreover, we find that under some solution conditions, this mechanism of activation is available to many known inhibitors of human CP4H, including some of the most potent biheteroaryldicarboxylates reported previously.
EXPERIMENTAL PROCEDURES
Chemical Synthesis
Detailed procedures for the synthesis of all compounds used in this study are provided in the Supporting Information.
Instrumentation
Instrumentation for NMR spectrometry and mass spectrometry were as described previously.20
The progress of reactions catalyzed by P4Hs was determined by analytical HPLC as described previously20 or analytical UPLC using an Acquity UPLC® H-Class from Waters (equipped with an Acquity photodiode array detector, Acquity quaternary solvent manager, Acquity sample manager with a flow-through needle, and Empower 3 software). Preparative HPLC was performed as described previously.20 Iron complexes with biheteroaryl ligands were analyzed by spectrophotometry using a Cary 60 UV–Vis Spectrometer from Agilent Technologies (Santa Clara, CA). Protein concentrations were calculated from their absorbance at 280 nm as measured with a NanoVue Plus spectrophotometer from GE Healthcare using an extinction coefficient of 290,000 M−1cm−1 for human CP4H,23 36,900 M−1cm−1 for human PHD2,24 and 44,000 M−1cm−1 for Chlamydomonas reinhardtii P4H (estimated with EXPASY software assuming that all cysteines are in disulfide bonds). EC50-values were calculated from experimental data with the program Prism version 6.0 from GraphPad Software (La Jolla, CA).
Heterologous Production of Human CP4H1
Human CP4H1, which is an α2β2 tetramer containing the α(I) isoform, was produced heterologously in Origami B(DE3) Escherichia coli cells and purified as described previously.23
Assay for the Activation of Human CP4H1
The catalytic activity of human CP4H1 was assayed as described previously.23 Briefly, activity assays were performed at 30 °C in 100 μL Tris–HCl buffer, pH 7.8, containing human CP4H1 (100 nM), activator (0–5 mM), substrate (dansylGlyProProGlyOEt, 500 μM), FeSO4 (50 μM), BSA (1 mg/mL), catalase (0.1 mg/mL), ascorbate (2 mM), DTT (100 μM), and α-ketoglutarate (0.1–10 mM). Unless noted otherwise, reactions were prepared by adding concentrated stock solutions of each component to concentrated assay buffer in the following order: FeSO4, DTT, ascorbate, BSA, catalase, CP4H1, peptide substrate, and activator (or vehicle). Solutions thus prepared were pre-incubated for 2 min at 30 °C, and the reaction was initiated by the addition of α-ketoglutarate. After 15 min, reactions were quenched by boiling for 45 s and subjected to centrifugation at 10,000g. The supernatant (5–10 μL) was injected into an Acquity UPLC® BEH C18 column (2.1 × 50 mm, 1.7-μm particle size) from Waters. The column was eluted at 0.6 mL/min with a linear gradient (2.9 min) of aqueous acetonitrile (20–68% v/v) containing TFA (0.1% v/v). The absorbance of the eluent was monitored at 289 nm. All assays were performed in triplicate. Data are reported as activity (which is the percent conversion of substrate to product) relative to control reactions lacking activator. Control reactions lacking inhibitor typically showed 1–2% conversion of the dansylGlyProProGlyOEt to the dansylGlyProHypGlyOEt product. Dose–response curves were generated for chosen activators by plotting the relative activity versus the log of the inhibitor concentration.
Heterologous Production of Human PHD2
A cDNA encoding human PHD2181–426 with an N-terminal hexahistidine (His6) tag (NHis6-PHD2181–426) was created by using the Gibson method,25 and the encoded protein was produced and purified as described previously.24
Assay for Activation of Human PHD2
The catalytic activity of human PHD2 was assayed as described previously.24 Briefly, activity assays were performed at 30 °C in 100 μL Tris–HCl buffer, pH 7.8, containing human NHis6-PHD2181–426(5 μM), activator (0–10 μM), substrate (HIF-1α peptide556–574, 50 μM), FeSO4 (50 μM), BSA (1 mg/mL), catalase (0.3 mg/mL), ascorbate (2 mM), DTT (1 mM), and α-ketoglutarate (35 μM). Reactions were prepared by adding concentrated stock solutions of each component to concentrated assay buffer in the following order: FeSO4, DTT, ascorbate, BSA, catalase, NHis6-PHD2181–426, peptide substrate, and activator (or vehicle). Solutions thus prepared were pre-incubated for 2 min at 30 °C, and the reaction was initiated by the addition of α-ketoglutarate. After 10 min, reactions were quenched by boiling for 60 s and subjected to centrifugation at 10,000g. The supernatant (50 μL) was injected into a Nucleodur® C18 Gravity reversed-phase column (4.6 × 250 mm, 5-μm particle size) from Macherey–Nagel (Bethlehem, PA). The column was eluted at 1 mL/min with a linear gradient of aqueous acetonitrile (5–56% v/v over 34 min) containing TFA (0.1% v/v). The absorbance of the eluent was monitored at 218 nm. All assays were performed in triplicate. Data are reported as activity (which is the percent conversion of substrate to product) relative to control reactions lacking activator.
Heterologous Production of C. reinhardtii P4H-1
A cDNA encoding C. reinhardtii P4H 130-245 with an N-terminal hexahistidine (His6) tag (NHis6-CrP4H-1) under the control of the T7 promoter was inserted into a pET-22b(+) vector using the Gibson method.25 The expression vector was transformed into Origami B(DE3) cells by electroporation, and transformants were grown on LB agar containing kanamycin (15 μg/mL), ampicillin (100 μg/mL), and tetracycline (12.5 μg/mL). A starter culture of TB medium containing antibiotics as described above was inoculated with a fresh colony, and the cells were grown overnight at 37 °C with shaking at 200 rpm. The starter culture was used to inoculate 4 L of TB medium supplemented with antibiotics as described above to OD600 = 0.02. The culture was incubated at 37 °C with shaking at 200 rpm until reaching an OD600 = 0.9. Protein expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG, 500 μM), and the culture was grown for 18 h at 21 °C with shaking at 200 rpm. Cells were harvested by centrifugation, and the cell pellet (6.0 g) was resuspended in 30 mL of lysis buffer, which was 50 mM sodium phosphate buffer, pH 7.4, containing NaCl (300 mM) and imidazole (2.5 mM). The cells were lysed at 22 kPSI in a T series Cell Disrupter 2.2 kW from Constant Systems Limited (Northants, UK). Insoluble material was cleared from the lysate by centrifugation at 30,000g for 45 min, and NHis6-CrP4H-1 was purified from the soluble fraction by nickel-affinity chromatography using a 5-mL HisTrap FF column (GE Healthcare, Piscataway, NJ). The column was equilibrated with lysis buffer, the supernatant was injected, and the A280 of the flowthrough was monitored until its return to baseline. NHis6-CrP4H-1 was eluted from the column with a linear gradient of aqueous imidazole (2.5–500 mM), with most of the NHis6-CrP4H-1 eluting between 50–250 mM imidazole. Fractions were subjected to SDS–PAGE on a 12% w/v gel, and those containing purified NHis6-CrP4H-1 were combined, dialyzed against 10 mM Tris–HCl, 100 mM glycine, 100 mM NaCl, pH 7.8, and concentrated to 3 mg/mL using a Vivaspin concentrator with a molecular weight cut off of 8,000 Da. Aliquots were flash frozen in liquid N2 and stored at −80 °C until use in P4H activity assays. This protocol yielded NHis6-CrP4H-1 at ~24 mg per gram of cell pellet.
Assay of C. reinhardtii P4H-1 Activity in the Presence of Activators
The catalytic activity of NHis6-CrP4H-1 was assayed in a manner similar to that for human CP4H1 with minor modifications. Briefly, activity assays were performed at 30 °C in 100 μL HEPES–HCl buffer (50 mM), pH 7.0, containing NHis6-CrP4H-1 (1.2 μM), activator (0–10 μM), substrate (dansylGlyProProGlyOEt, 500 μM), FeSO4 (200 μM), BSA (1 mg/mL), catalase (0.1 mg/mL), ascorbate (2 mM), DTT (100 μM), and α-ketoglutarate (150 μM). Assay mixtures were prepared by adding concentrated stock solutions of each component to concentrated assay buffer in the following order: FeSO4, DTT, ascorbate, BSA, catalase, NHis6-CrP4H-1, peptide substrate, and activator (or vehicle). Solutions thus prepared were pre-incubated for 2 min at 30 °C, and the reaction was initiated by the addition of α-ketoglutarate. After 1 h, reactions were quenched by adding EDTA (1 μL of a 0.5 M solution) and boiling for 45 s. Quenched reactions were subjected to centrifugation at 10,000g for 5 min, after which the supernatant (5–10 μL) was injected into an Acquity UPLC® BEH C18 column (2.1 × 50 mm, 1.7-μm particle size) from Waters. The column was eluted at 0.6 mL/min with a linear gradient of aqueous acetonitrile (20–68% v/v over 2.9 min) containing TFA (0.1% v/v). The absorbance of the eluent was monitored at 289 nm. All assays were performed in triplicate. Data are reported as activity (which is the percent conversion of substrate to product) relative to control reactions lacking activator.
Assay of Fe(II)-Affinity for Biheteroaryl Ligands
The affinity of biheteroaryl ligands for Fe(II) was determined comparatively by measuring the half-maximal concentration (EC50) required for binding 20 μM Fe(II) (Fe20-EC50) in sodium phosphate buffer, pH 7.0, as described previously.20
Determination of Fe(II) Complex Stoichiometry
The stoichiometry of biheteroaryl complexes with Fe(II) was estimated with Job’s method,26,27 as described previously.20
RESULTS AND DISCUSSION
Discovery of activators of CP4H1 and requirement for vitamin C
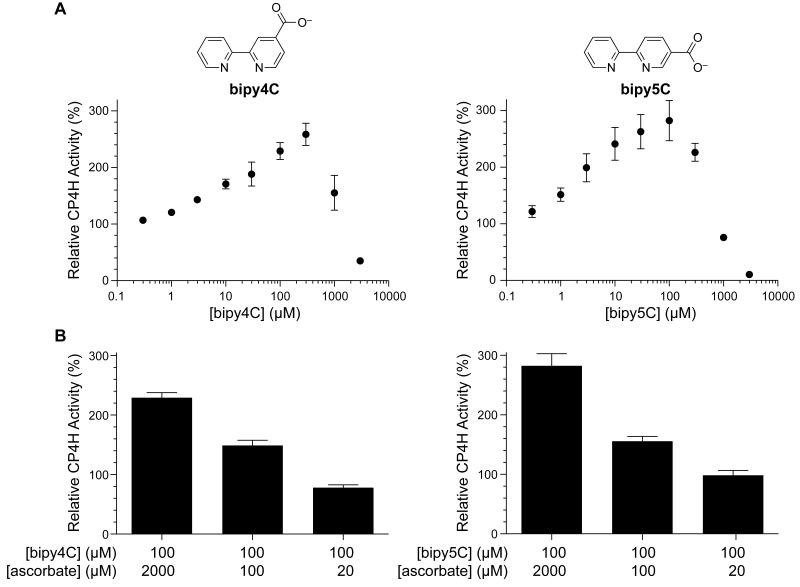
In previous investigations of bipyridinedicarboxylates as inhibitors of human CP4H1, we also investigated the monocarboxylates bipy4C and bipy5C (Fig. 1A), expecting that they would serve as modest inhibitors of the enzyme. In these investigations, we used a standard liquid chromatography-based assay in which we determined the hydroxylation of the model tetrapeptide substrate: dansyl-Gly-Pro-Pro-Gly-OEt.20,23 To our surprise, we found that standard reactions in which bipy4C or bipy5C were added at a concentration of 10 μM produced significantly more hydroxylated product compared to reactions to which vehicle was added. In subsequent investigations, we found that this activation was dose-dependent (Fig. 1A) and not outcompeted by α-ketoglutarate, which is a cosubstrate. At high concentrations (millimolar), bipy4C and bipy5C did inhibit catalysis. Neither bipy4C nor bipy5C could replace other assay components, such as α-ketoglutarate or VitCred. Whereas the levels of many assay components did not perturb activation by bipy4C and bipy5C, activation did require VitCred cofactor in a dose-dependent manner (Fig. 1B).
Figure 1.
Activation of CP4H1 by bipy4C and bipy5C is dose-dependent and requires the Vitamin C cofactor. (A) Dose–response curves for activation (and inhibition) of CP4H1 by bipymonoCs. CP4H assays were performed in the presence and absence of activator (0.3–3000 μM) and ascorbate (2 mM), and in the presence of a saturating concentration of α-ketoglutarate (1 mM). Relative activity values are the mean (±SE) of three independent experiments) and represent the ratio of enzymatic activity in the presence and absence of activator. (B) Dependence of the activation observed by bipy4C and bipy5C on ascorbate concentration. CP4H assays were performed in the presence and absence of activator (100 μM) and ascorbate (20 μM–2 mM), and in the presence of 1 mM α-ketoglutarate. Relative activity values are the mean (±SE) of three independent experiments) and represent the ratio of CP4H activity observed in the presence and absence of activator.
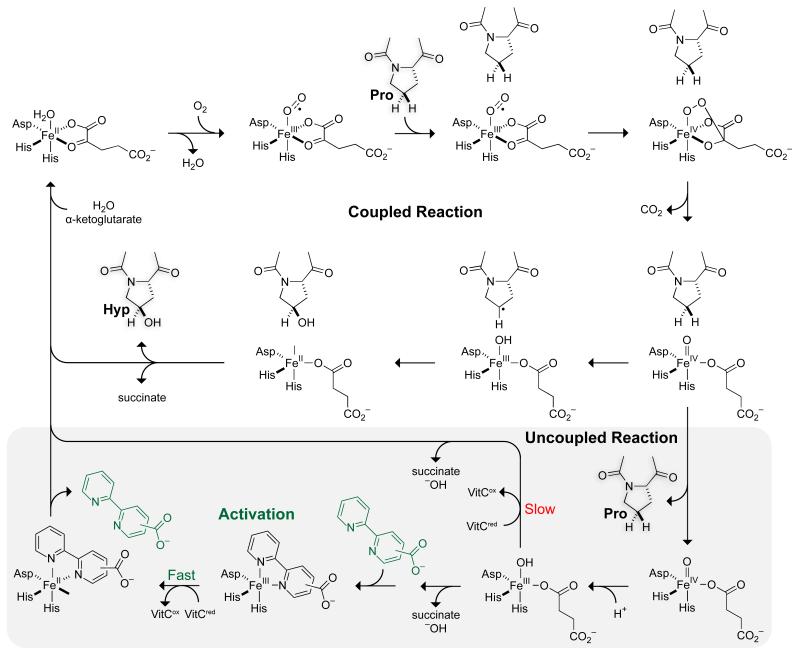
The discovery that activation of CP4H1 by bipy4C and bipy5C required VitCred was interesting to us, as such a phenomenon has not been reported previously for an enzyme of the FAKGD family. To seek an explanation for the activation, we considered the kinetic mechanism of catalysis by CP4Hs (Fig. 2). CP4Hs display an ordered Ter Ter mechanism in which α-ketoglutarate first binds to a CP4H•Fe(II) complex, after which O2 and the peptide substrate bind in an ordered manner.28 Subsequently, the oxidative decarboxylation of α-ketoglutarate facilitates the formation of a highly reactive Fe(IV)=O species (ferryl ion) that hydroxylates the peptide substrate via a radical rebound process.29-31 Whereas all CP4Hs appear to require VitCred for activity, this cofactor is not required for the typical Ter Ter mechanism, but has been shown to participate in the “uncoupled reaction” in which CP4H decarboxylates α-ketoglutarate in the absence of the peptide substrate.9 EPR spectroscopy indicates that the ferryl ion decays via an unknown mechanism to an inactive Fe(III) state,10 and VitCred is required to reduce the iron center back to the Fe(II) state that is necessary for catalysis.9,10 The finding that VitCred is required for activation by bipy4C and bipy5C suggests that these activators operate during the uncoupled reaction to assist VitCred in the rescue of CP4H molecules that have suffered oxidative inactivation.
Figure 2.
Putative mechanism of catalysis by CP4H. O2 and the peptide substrate bind to a CP4H•Fe(II)•α-ketoglutarate complex in an ordered fashion.28 Oxidative decarboxylation of α-ketoglutarate leads to the formation of a highly reactive Fe(IV)=O species (ferryl ion) that hydroxylates the peptide substrate via a radical rebound process.29-31 For prolonged activity, CP4H requires VitCred during an “uncoupled reaction” in which CP4H decarboxylates α-ketoglutarate in the absence of the peptide substrate.9 The ferryl ion decays to an inactive Fe(III) state,10 and VitCred reduces the iron center back to the requisite Fe(II) state.10,9 We propose that bipy4C and bipy5C activate CP4H1 during this uncoupled reaction by serving as a ligand that enhances the rate of the VitCred-dependent reduction of the iron center.
Diimine ligands, such as 2,2′-bipyridine (bipy) and 1,10-phenanthroline, are known to catalyze the VitCred-dependent reduction of Fe(III) to Fe(II) in solution.32,33 Whereas bipy itself does not serve as an activator of CP4H,20,22 we reasoned that bipy4C and bipy5C (which resemble α-ketoglutarate) could serve as iron ligands that enhance the rate of the VitCred-dependent reduction. Still, as such a mechanism had never been reported for an FAKGD enzyme, we sought to identify aspects of bipy4C and bipy5C that are required for activation.
Determinants for the Activation of CP4H
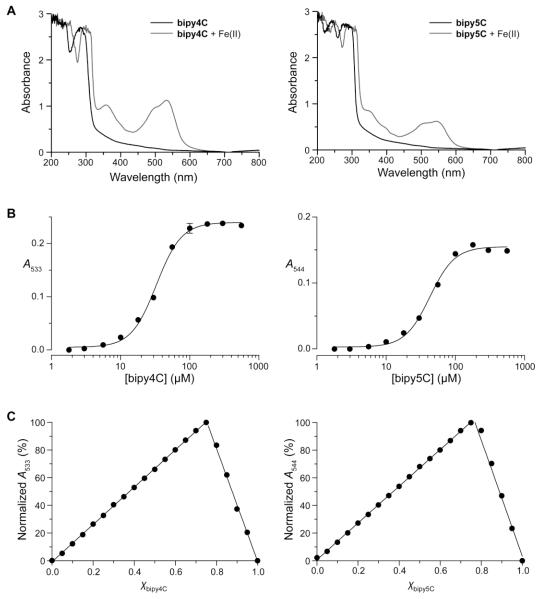
We began by investigating the iron-binding properties of bipy4C and bipy5C compared to that of the parent bipy. We performed titration experiments to determine the value of Fe20-EC50, which is the half-maximal concentration required to form a complex with 20 μM Fe(II) (Fe20-EC50) at pH 7.0. These experiments took, advantage of the strong absorption typically observed for complexes of bipy and related analogues with Fe(II). Together with an estimation of stoichiometry obtained with Job’s method, the Fe20-EC50 value provides a comparative metric for iron affinity.20 Bipy itself forms an Fe(bipy)32+ complex with an Fe20-EC50 of (43 ± 2) μM.20 We found that, like bipy, bipy4C and bipy5C served as ligands for Fe(II), forming distinct red Fe(ligand)3 type complexes (Fig. 3) with Fe20-EC50 values of (33 ± 0.8) μM and (43 ± 2) μM, respectively.
Figure 3.
Iron-binding properties of bipy4C and bipy5C. (A) Absorption spectra of activators (300 μM) in the presence and absence of Fe(II)SO4 (100 μM). Complexes of bipy4C and bipy5C with Fe(II) showed local absorption maxima (λmax) of 533 nm and 544 nm, respectively. (B) Titrations of Fe(II) with bipy4C and bipy5C. Values of Fe20-EC50 were (33 ± 0.8) μM and (43 ± 2) μM, respectively. (C) Job’s plots for the complexes of Fe(II) with bipy4C and bipy5C. Values of χ at the point intersection were 0.75 ± 0.02 and 0.76 ± 0.06, respectively, consistent with the formation of Fe(ligand)3 complexes.
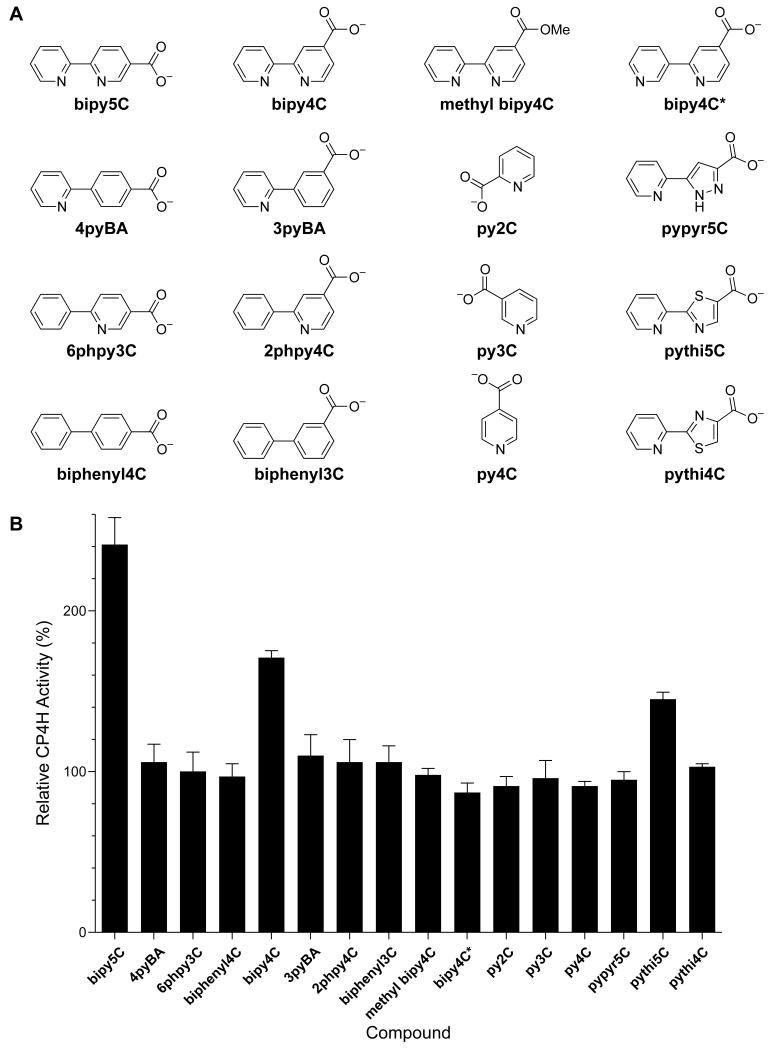
Next, we explored structure–activity relationships (SARs) within a library of chemical analogues (Fig. 4). We found that replacement of one or both of the pyridine rings with a phenyl ring (as in 4pyBA, 6Phpy3C, biphenyl4C, 3pyBA, 2Phpy4C, and biphenyl3C) abolished activation. Likewise, repositioning of the pyridyl nitrogen in bipy4C to a non-chelating geometry (as in bipy4C*) abolished activation, as did esterification of bipy4C (as in methyl bipy4C). To distinguish between the importance of a carboxylate in the appropriate geometry and the importance of a net negative charge, we also tested a series of pyridine carboxylates (as is py2C, py3C, and py4C) and found that none of these compounds served as activators for CP4H1. Collectively, these SARs confirm the importance of both the bipyridyl chelate and a pendant carboxylate group with appropriate regiochemistry for the activation of CP4H1.
Figure 4.
Structure–activity relationships of bipymonoCs and related chemical analogues for the activation of human CP4H1. (A) Chemical structures and abbreviations. (B) Screen (10 μM) of the compounds depicted in panel A against human CP4H1. CP4H1 assays were performed in the presence of 100 μM α-ketoglutarate. Relative activity values are the mean (±SE) of three independent experiments and represent the ratio of CP4H1 activity observed in the presence and absence of the activator or chemical analogue.
Activation by Free Ligand or Fe(Ligand)3 Complex?
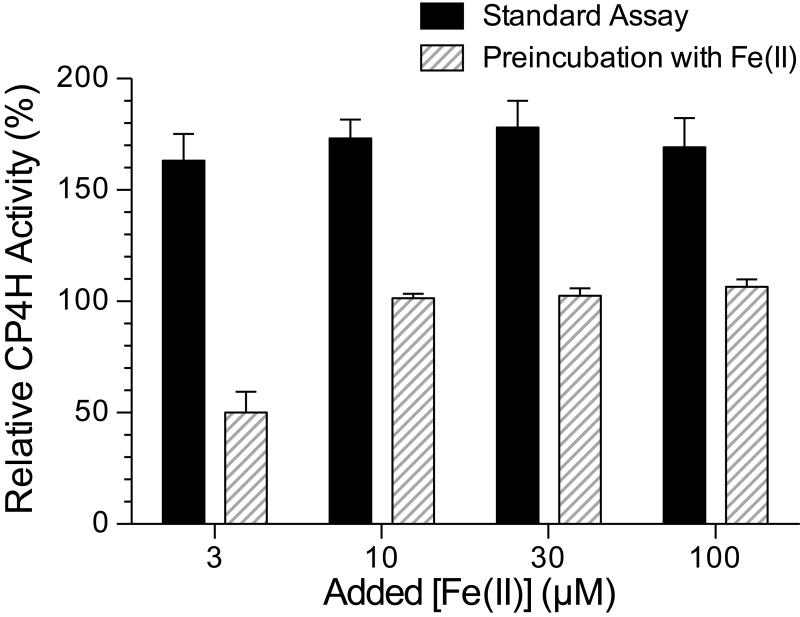
Given that bipy4C and bipy5C form strong complexes with iron, we next determined whether free ligand or an Fe(ligand)3 type complex32,33 was involved in the activation. To do so, we performed experiments in which bipy4C (10 μM) was preincubated with Fe(II) prior to addition to the assay mixture, and compared the activation to that when bipy4C was added last. We found that regardless of the amount of free Fe(II) added to the assay, preincubation of bipy4C with the Fe(II) abolished activation (Fig. 5). Moreover, at the lowest iron concentration tested (3 μM), we found that bipy4C appeared to inhibit the enzyme, suggesting that the preformed Fe(ligand)3 type complex could sequester iron from CP4H1 over the time-course of our assay. These findings suggested that the free ligand activates CP4H1.
Figure 5.
Preincubation of bipy4C with iron abolishes activation of CP4H1. CP4H1 assays were performed in the presence of 100 μM α-ketoglutarate and 10 μM bipy4C that was either added last (“Standard Assay”) or preincubated with Fe(II)SO4 prior to addition to the assay. Relative activity values are the mean (±SE) of three independent experiments and represent the ratio of CP4H activity observed in the presence and absence of activator.
Next, we determined whether bipy4C and bipy5C bind in the CP4H1 active site. Bipy-4,5′-dicarboxylate (bipy45′DC) and bipy-5,5′-dicarboxylate (bipy55′DC) are potent inhibitors of human CP4H1 and thought to inhibit the enzyme by competing with α-ketoglutarate.20 Moreover, the bipy core in these molecules can be replaced with chelating biheteroaryls.21 For example, a thiazole analog (pythi) is a potent competitive inhibitor of CP4H1, whereas a pyrazole analog (pypyr) is not.
Given the known SARs for biheteroaryl dicarboxylates, we reasoned that replacement of the bipy core of bipy4C and bipy5C with either pythi or pypyr should also show similar SARs for the activation of CP4H1 if these activators are interacting with the CP4H1 iron center. Accordingly, we tested the monocarboxylates pythi5C and pypyr5C, as well as the regioisomer pythi4C, as activators of CP4H1 under standard assay conditions. We found that pythi5C served as an activator of CP4H1, whereas the regioisomer pythi4C and pypyr5C did not (Fig. 4).
Chemical Mechanism for Activation of CP4H
Based upon the cumulative data presented above, we put forth the mechanism depicted in Fig. 2 for the activation of CP4H1 by bipymonoCs. During the uncoupled decarboxylation of α-ketoglutarate, the CP4H1 iron center becomes trapped in an unproductive Fe(III) state,10 which normally requires reduction by the VitCred cofactor to return to the catalytically active Fe(II) state.9,10 We propose that bipy4C and bipy5C bind to the unproductive Fe(III) center, displacing any occupying ligands such as water, hydroxide, or the succinate byproduct. Once bound, these ligands serve to enhance the rate of the VitCred-dependent reduction of the iron center back to a catalytically active state, after which they dissociate to begin another catalytic cycle.
Our data suggest that the VitCred-dependent reduction of the CP4H1 iron center is the rate-limiting step in the overall reaction catalyzed by CP4H1, at least under our assay conditions. CP4H can catalyze its reaction at a high initial rate in the absence of VitCred.10 Yet, that rate decreases gradually within 30 s due to the oxidation of the enzyme during the sporadic uncoupled decarboxylation of α-ketoglutarate.10 Subsequent addition of the VitCred cofactor allows catalysis to continue at a rate that is ~35% of that observed initially,10 suggesting that a step in the pathway for the uncoupled reaction becomes rate-limiting for overall catalysis. Our finding that bipy4C and bipy5C activate CP4H1 by ~3-fold under standard conditions supports our mechanism and the notion that the VitCred-dependent reduction of the CP4H1 iron center can limit the rate of catalysis by CP4H1.
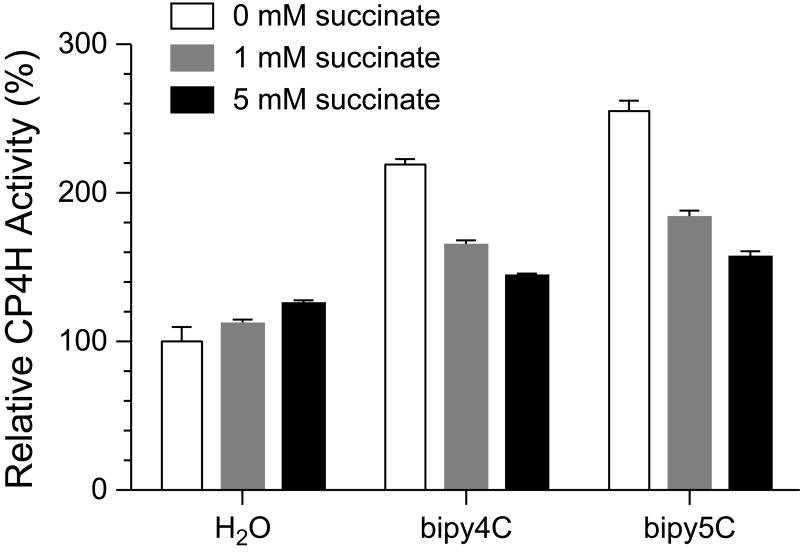
In an additional experiment, we explored the effect of added succinate product on activation by bipy4C and bipy5C. As succinate is a known inhibitor that competes with α-ketoglutarate, we performed these reactions at high and saturating concentrations of α-ketoglutarate (≥1 mM) so as to prevent the added succinate from inhibiting the enzyme. Moreover, comparisons were always made to control reactions lacking the activator so as to dissect out any effects associated with adding succinate itself. We found that succinate (1 or 5 mM) was indeed able to outcompete the activating effect of the bipymonoCs (100 μM) in a dose-dependent manner (Fig. 6), providing further support for our proposed mechanism (Fig. 2).
Figure 6.
Activation of CP4H1 by bipymonoCs is competitive with succinate. CP4H1 assays were performed in the presence of 100 μM activator and 1 mM α-ketoglutarate. Relative activity values are the mean (±SE) of three independent experiments and represent the ratio of CP4H activity observed in the presence and absence of the activator.
Reconciling Activation and Inhibition of CP4H
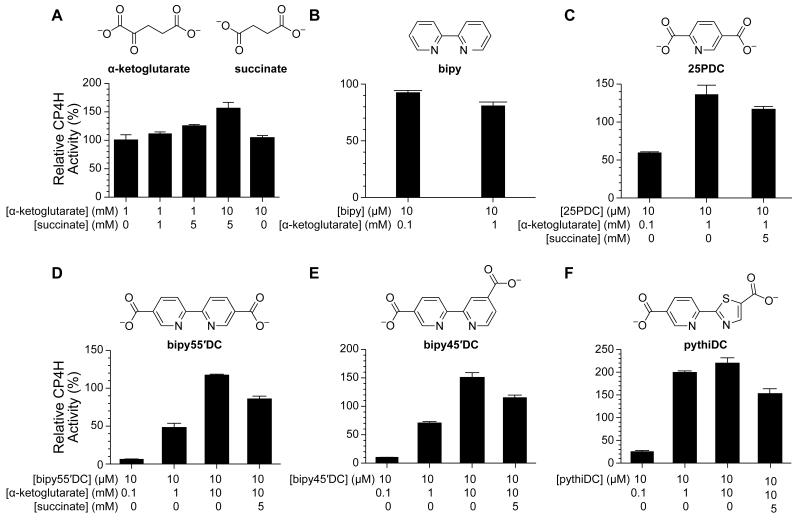
At high concentrations of α-ketoglutarate, succinate itself appeared to activate the enzyme modestly (Fig. 7A), a phenomenon that, to our knowledge, had not been reported previously. This finding suggested to us that this mode of activation might be available to other inhibitors of CP4H1, so long as inhibition was prevented by high levels of α-ketoglutarate. We probed a variety of compounds at 10 μM, including the modest inhibitor 2,5-pyridinedicarboxylate (25PDC) and the potent inhibitors bipy45′DC, bipy55′DC, and pythiDC. Whereas the effect of bipy itself (Fig. 7B) was not dependent on the amount of α-ketoglutarate, 25PDC (Fig. 7C) displayed modest inhibition at lower concentrations of α-ketoglutarate and activation in the presence of millimolar α-ketoglutarate. Moreover, succinate was able to disrupt the activation effect (Fig. 7C), consistent with the proposed mechanism (Fig. 2). Inhibition by bipy55′DC (Fig. 7D), bipy45′DC (Fig. 7E), and pythiDC (Fig. 7F), also depended on the amount of α-ketoglutarate, serving as potent inhibitors at low α-ketoglutarate concentrations (100 μM), but apparent activators at high α-ketoglutarate concentrations (up to 10 mM). Again, succinate was able to disrupt the activation effects, consistent with the proposed mechanism (Fig. 2). Lastly, high concentrations of α-ketoglutarate did not appear to provide any additional activating effect (Fig. 7A), suggesting that α-ketoglutarate itself does not bind to the Fe(III) species. This finding explains why the activating effect observed for biheteroarylcarboxylate ligands is not outcompeted by α-ketoglutarate, despite their common enzymic binding site.
Figure 7.
Inhibition or activation of CP4H1 by biheteroarylcarboxylates and related analogues is dependent upon α-ketoglutarate and succinate. CP4H1 assays were performed in the presence of 2 mM ascorbate. Relative activity values are the mean (±SE) of three independent experiments and represent the ratio of CP4H activity observed in the presence and absence of a modulator. (A) Effect of added succinate and α-ketoglutarate on CP4H1 activity. (B) Effect of bipy on CP4H1 activity. (C) Effect of 25PDC on CP4H1 activity and its dependence upon α-ketoglutarate and succinate. (D, E, F) Effect of bipy55′DC (D), bipy45′DC (E), and pythiDC (F) on CP4H1 activity and their dependence upon α-ketoglutarate and succinate.
These data cause us to revise our thoughts about the mode of action of biheteroaryl carboxylate ligands and related analogues. These compounds can modulate CP4H1 activity at multiple points in the catalytic mechanism. They can serve as competitive inhibitors of α-ketoglutarate to form dead-end complexes with Fe(II), or serve as ligands for the Fe(III) species that enhance the rate of the VitCred-dependent reduction of the iron center back to a catalytically active Fe(II) state. Thus, the observed effect of treatment of CP4H1 with a biheteroaryl carboxylate ligand is the net effect of both activating and inhibiting processes, where the concentrations of the ligand, α-ketoglutarate, the succinate byproduct, and the VitCred cofactor all influence that net effect.
Other P4H Enzymes
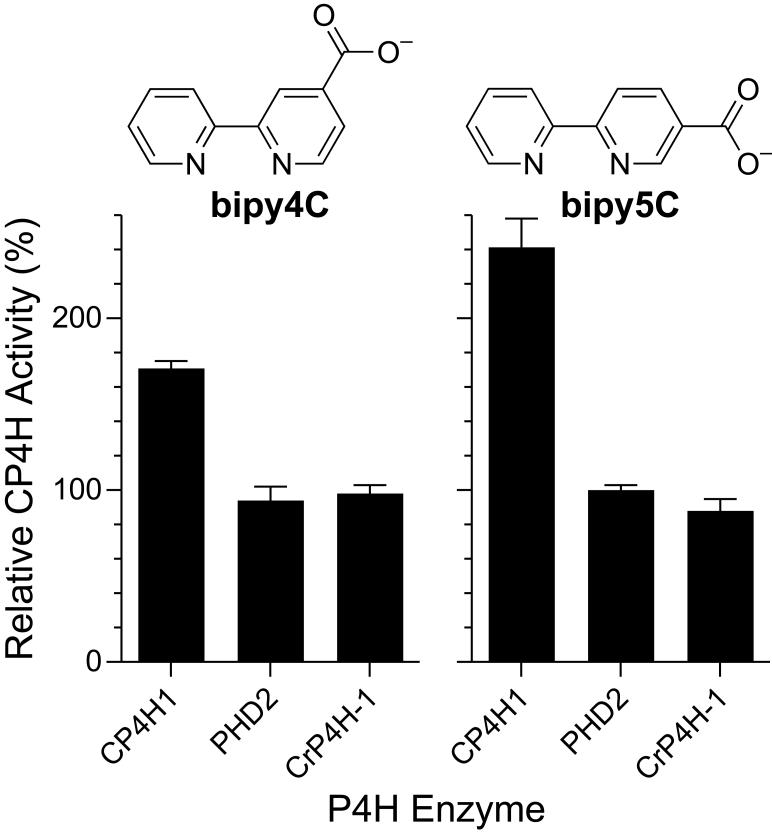
Lastly, we were curious as to whether this mechanism of activation was available to other P4H enzymes. There are a variety of other P4Hs of biological importance in animals, plants, and lower organisms.2,34 The most well characterized P4Hs are those involved in hypoxia signaling (PHDs) and in cell-wall biosynthesis in the algae C. reinhardtii (CrP4Hs). We chose to investigate these two alternative types of P4H enzymes, using the well-characterized PHD isoform 2 (PHD2) and CrP4H isoform 1 (CrP4H-1) as model enzymes. After screening these enzymes under standard assay conditions comparable to those used to assay CP4H1, we found that neither was activated by bipy4C or bipy5C (Fig. 8). Based upon our postulated mechanism for the activation of CP4H1 by these bipymonoCs, this finding suggests that these activators do not bind appreciably in the active site of the other P4Hs, or that the rate-limiting step in the overall kinetic mechanisms of the other P4Hs is not the VitCred-dependent reduction of their enzymic iron center. Considering crystal structures of CrP4H and human PHD2 along with SARs for known active-site inhibitors,20,21 we suspect that CrP4H should be able to accommodate all of the activators reported herein, and that human PHD2 should be able to accommodate bipy4C and pythi5C. Thus, a likely explanation for the selective activation of only human CP4H1 is that the VitCred-dependent reduction of the enzymic iron center does not limit the overall rate of CrP4H and PHD2.
Figure 8.
Activation of prolyl 4-hydroxylases by bipymonoCs. Human CP4H1, human PHD2, and C. reinhardtii P4H-1 (CrP4H-1) were assayed in the presence or absence of bipy4C (A) or bipy5C (B) at 10 μM. Relative activity values are the mean (±SE) of three independent experiments and represent the ratio of CP4H activity observed in the presence and absence of the activator in question.
Conclusions
We have discovered that bipy4C and bipy5C are activators of human CP4H1 via a novel mechanism (Fig. 2) wherein they serve as ligands that enhance the rate of the VitCred-dependent rescue of the enzyme from a catalytically inactive oxidation state at its iron center. To our knowledge, this report is the first of a small-molecule activator of a CP4H since the discovery that VitCred is a required cofactor for the enzyme. Our postulated mechanism for these activators also suggests that the rate-limiting step of the overall reaction catalyzed by CP4H1 (including both the coupled and uncoupled reaction pathways) is likely the VitCred-dependent reduction of the inactive Fe(III) species. Lastly, we found that this mechanism of activation is also available to many potent inhibitors of CP4H1 that are competitive with α-ketoglutarate. These inhibitors, including 25PDC, bipy45′DC, bipy55′DC, and pythiDC, can also serve as activators of the enzyme, if their inhibitory mechanism is prevented by the presence of saturating concentrations of α-ketoglutarate. The finding that these bipyridinecarboxylate ligands can activate human CP4H1 provides new information about catalysis by this essential enzyme, chemical tools to aid in probing its kinetic mechanism, and the potential for a new chemotherapeutic modality.
Supplementary Material
Acknowledgments
Funding
J.D.V. was supported by Molecular Biosciences Training Grant T32 GM007215 (NIH) and a fellowship from the Department of Biochemistry at the University of Wisconsin–Madison. This work was supported by Grant R01 AR044276 (NIH), and made use of the National Magnetic Resonance Facility at Madison, which is supported by Grant P41 GM103399 (NIH), and a Micromass LCT® mass spectrometer, which was obtained with support from Grant CHE-9974839 (NSF).
Footnotes
Procedures for the synthesis of chemical compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Shoulders MD, Raines RT. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gorres KL, Raines RT. Prolyl-4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple helix of collagen. Biochem. Biophys. Res. Commun. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- (4).Sakakibara S, Inouye K, Shudo K, Kishida Y, Kobayashi Y, Prockop DJ. Synthesis of (Pro–Hyp–Gly)n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxyproline. Biochim. Biophys. Acta. 1973;303:198–202. doi: 10.1016/0005-2795(73)90164-5. [DOI] [PubMed] [Google Scholar]
- (5).Hutton JJ, Marglin A, Witkop B, Kurtz J, Berger A, Udenfriend S. Synthetic polypeptides as substrates and inhibitors of collagen proline hydroxylase. Arch. Biochem. Biophys. 1968;125:779–785. doi: 10.1016/0003-9861(68)90514-6. [DOI] [PubMed] [Google Scholar]
- (6).Rhoads RE, Udenfriend S. Decarboxylation of α-ketoglutarate coupled to collagen proline hydroxylase. Proc. Natl. Acad. Sci. U.S.A. 1968;60:1473–1478. doi: 10.1073/pnas.60.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Counts DF, Cardinale GJ, Udenfriend S. Prolyl hydroxylase half reaction—peptidyl prolyl-independent decarboxylation of α-ketoglutarate. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2145–2149. doi: 10.1073/pnas.75.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rao NV, Adams E. Partial reaction of prolyl hydroxylase. (Gly–Pro–Ala)n stimulates α-ketoglutarate decarboxylation without prolyl hydroxylation. J. Biol. Chem. 1978;253:6327–6330. [PubMed] [Google Scholar]
- (9).Myllylä R, Majamaa K, Gunzler V, Hanauske-Abel HM, Kivirikko KI. Ascorbate is consumed stoichiometrically in the uncoupled reactions catalyzed by prolyl 4-hydroxylase and lysyl hydroxylase. J. Biol. Chem. 1984;259:5403–5405. [PubMed] [Google Scholar]
- (10).de Jong L, Albracht SPJ, Kemp A. Prolyl 4-hydroxylase activity in relation to the oxidation state of enzyme-bound iron: The role of ascorbate in peptidyl proline hydroxylation. Biochim. Biophys. Acta. 1982;704:326–332. doi: 10.1016/0167-4838(82)90162-5. [DOI] [PubMed] [Google Scholar]
- (11).de Jong L, Kemp A. Stoichiometry and kinetics of the prolyl 4-hydroxylase partial reaction. Biochim. Biophys. Acta. 1984;787:105–111. doi: 10.1016/0167-4838(84)90113-4. [DOI] [PubMed] [Google Scholar]
- (12).Lind A. Treatise of the Scurvy in Three Parts. Containing an Inquiry into the Nature, Causes and Cure of that Disease, Together with a Critical and Chronological View of what has been Published on the Subject. A. Millar; London, UK: 1753. [Google Scholar]
- (13).De Vreese L. Casual (mis)understanding and the search for scientific explanations: A case study from the history of medicine. Stud. Hist. Phil. Biol. Biomed. Sci. 2008;39:14–24. doi: 10.1016/j.shpsc.2007.12.016. [DOI] [PubMed] [Google Scholar]
- (14).Carpenter KJ. The History of Scurvy and Vitamin C. Cambridge University Press; New York, NY: 1986. [Google Scholar]
- (15).Guo H, Tong P, Liu Y, Xia L, Wang T, Tian O, Li Y, Hu Y, Zheng Y, Jin X, Li Y, Xiong W, Tang B, Feng Y, Li J, Pan O, Hu Z, Xia Z. Mutations of P4HA2 encoding prolyl 4-hydroxylase 2 are associated with nonsyndromic high myopia. Genet. Med. 2015;17:300–306. doi: 10.1038/gim.2015.28. [DOI] [PubMed] [Google Scholar]
- (16).Torpe N, Pocock R. Regulation of axonal midline guidance by prolyl 4-hydroxylation in Caenorhabditis elegans. J. Neurosci. 2014;34:16348–16357. doi: 10.1523/JNEUROSCI.1322-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol. Cell Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J. Loss of assembly of the main basement membrane collagen, Type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J. Biol. Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- (20).Vasta JD, Raines RT. Selective inhibition of prolyl 4-hydroxylases by bipyridinedicarboxylates. Bioorg. Med. Chem. 2015;23:3081–3090. doi: 10.1016/j.bmc.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Vasta JD, Andersen KA, Deck KM, Nizzi CP, Eisenstein RS, Raines RT. Selective inhibition of collagen prolyl 4-hydroxylase in human cells. ACS Chem. Biol. 2016;11:193–199. doi: 10.1021/acschembio.5b00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hales NJ, Beattie JF. Novel inhibitors of prolyl 4-hydroxylase. 5. The intriguing structure–activity relationships seen with 2,2′-bipyridine and its 5,5′-dicarboxylic acid derivatives. J. Med. Chem. 1993;36:3853–3858. doi: 10.1021/jm00076a014. [DOI] [PubMed] [Google Scholar]
- (23).Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Expr. Purif. 2004;38:279–291. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- (24).Hewitson KS, Schofield CJ, Ratcliffe PJ. Hypoxia-inducible factor prolyl-hydroxylase: Purification and assays of PHD2. Methods Enzymol. 2007;435:25–42. doi: 10.1016/S0076-6879(07)35002-7. [DOI] [PubMed] [Google Scholar]
- (25).Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Job P. Formation and stability of inorganic complexes in solution. Annali di Chimica Applicata. 1928;9:113–203. [Google Scholar]
- (27).Huang CY. Determination of binding stoichiometry by the continuous variation method: The Job plot. Methods Enzymol. 1982;87:509–525. doi: 10.1016/s0076-6879(82)87029-8. [DOI] [PubMed] [Google Scholar]
- (28).Myllylä R, Tuderman L, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 2. Kinetic analysis of the reaction sequence. Eur. J. Biochem. 1977;80:349–357. doi: 10.1111/j.1432-1033.1977.tb11889.x. [DOI] [PubMed] [Google Scholar]
- (29).Hanauske-Abel HM, Günzler V. A stereochemical concept for the catalytic mechanism of prolylhydroxylase: Applicability to classification and design of inhibitors. J. Theor. Biol. 1982;94:421–455. doi: 10.1016/0022-5193(82)90320-4. [DOI] [PubMed] [Google Scholar]
- (30).Hausinger RP. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- (31).Costas M, Mehn MP, Jensen MP, Que LJ. Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- (32).Younathan ES, Frieden E. Catalysis of ascorbate oxidation by 1,10-phenanthroline and related ligands. Biochim. Biophys. Acta. 1961;46:51–58. doi: 10.1016/0006-3002(61)90645-x. [DOI] [PubMed] [Google Scholar]
- (33).Kimura M, Yamamoto M, Yamaba S. Kinetics and mechanism of the oxidation of L-ascorbic acid by tris(oxalato)cobaltate(III) and tris(1,10-phenanthroline)iron(III) complexes in aqueous solution. J. Chem. Soc. Dalton Trans. 1982:423–427. [Google Scholar]
- (34).Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann. Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.