Abstract
Although gut host-pathogen interactions are glycan-mediated processes, few details are known about the participating structures. Here we employ high-resolution mass spectrometric profiling to comprehensively identify and quantitatively measure the exact modifications of native intestinal epithelial cell surface N-glycans induced by S. typhimurium infection. Sixty minutes postinfection, select sialylated structures showed decreases in terms of total number and abundances. To assess the effect of cell surface mannosylation, we selectively rerouted glycan expression on the host using the alpha-mannosidase inhibitor, kifunensine, toward overexpression of high mannose. Under these conditions, internalization of S. typhimurium significantly increased, demonstrating that bacteria show preference for particular structures. Finally, we developed a novel assay to measure membrane glycoprotein turnover rates, which revealed that glycan modifications occur by bacterial enzyme activity rather than by host-derived restructuring strategies. This study is the first to provide precise structural information on how host N-glycans are altered to support S. typhimurium invasion.
Comprising the outermost layer of all eukaryotic cells, glycans are optimally positioned as the primary molecular contacts engaged during cellular encounters with viruses, bacteria, antibodies, hormones, toxins, and other host cells (1–3). The cell-specific surface glycome also regulates cellular processes either by modifying the structure and function of membrane proteins or by contributing to the structure of the plasma membrane lipid scaffold. The importance of these interactions is highlighted by the fact that alterations in the cell surface glycome correlate with the onset and progression of a variety of different diseases (4, 5). There is also emerging evidence that glycans play a central role in a variety of immunological processes (6–8).
Within the mammalian gut the luminal surface of the intestine is continuously exposed to foreign substances. Highly glycosylated epithelial cells are the primary boundary separating embedded host tissues from intestinal pathogens. Microbes adapt to this glycan rich environment by employing endogenous glycan-binding proteins and glycan-degrading enzymes (9–11), which participate in microbial invasion and colonization (12). In parallel, carbohydrate-active enzymes in resident symbiotic microbes can liberate host glycans and thereby creating a competitive environment for available resources with pathogenic bacteria (13–15). Given the number and broad heterogeneity of intestinal microorganisms (16), regulation of these interactions is essential for host health.
A common cause of gastrointestinal disturbances in mammals is Salmonella enterica subsp. enterica serovar typhimurium (S. typhimurium) infection. Although significant progress has been made in understanding S. typhimurium invasion strategies (17–25), it remains unclear which types of host cell surface glycans are necessary or inhibitory for its colonization. Previously, it was demonstrated that liberated host sugars facilitate pathogen expansion when the gut microbiota is disrupted (26). Such model studies provide precedents for the importance of host glycans in shaping the gut microbiota. In addition, complete genome sequencing has determined that S. typhimurium is capable of exploiting sugar motifs present on host cell surfaces with certain specificity (27). We therefore sought to precisely define the pathogen-sensitive structural components in the host cell surface glycome as this will provide novel insight to the pathophysiology of S. typhimurium.
To date, there have been technological limitations preventing a full understanding of the glycan structures that mediate intestinal epithelial cell encounter with pathogenic bacteria. Herein, we first examine the glycan products remaining on the host cell surface after brief contact with bacteria using high resolution nano-LC/MS-based analytical tools capable of isomeric-level differentiation. We then introduce kifunensine, an alpha-mannosidase I inhibitor, to construct live cells with nearly exclusive expression of high mannose type glycans, enabling reliable structure-phenotype correlative experiments. Our study constitutes the first comprehensive and quantitative profiling of the S. typhimurium-induced structural changes of oligosaccharides associated with human intestinal epithelial cells. Monitoring for specific structural changes during the course of infection affords identification of glycan-derived targets for the development of pathogen-specific therapeutics. Furthermore, inducing a specific glycan expression pattern with kifunensine provides novel insight into the physiologic consequences of the observed changes in glycosylation.
EXPERIMENTAL PROCEDURES
Cell Culture
The human colorectal epithelial cell line, Caco-2, was obtained from American Type Culture Collection (ATCC, Manassas, VA) and grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 16.6% fetal bovine serum (FBS), nonessential amino acids, 10 mm MOPS, 10 mm TES, 15 mm HEPES, and 2 mm NaH2PO4. For kifunensine studies, cells were grown in Eagle's Minimum Essential Medium (EMEM) supplemented with 10% FBS, nonessential amino acids, 2 mm L-glutamine, 1 mm sodium pyruvate, and 1.5 g/L sodium bicarbonate. Cells were seeded at 80% confluency and incubated at 37 °C with 5% CO2. Prior to infection, Caco-2 cells were allowed to differentiate in culture 14 days postconfluency (28). Uninfected and infected cell samples were maintained and analyzed in triplicates.
Salmonella enterica subsp. enterica serovar typhimurium LT2 (ATCC 700720) were grown at 37 °C in Luria-Bertani (LB) liquid medium (Difco, Detroit, MI) with continuous shaking for 14–16 h. Bacterial cells were washed with PBS and resuspended in the epithelial cell growth medium to the appropriate optical density measured at 600 nm.
Bacterial Infection
To measure bacterial association with host cells, S. typhimurium were added to Caco-2 cells grown in 96-well plate at a multiplicity of infection (MOI) of 1000 for 60 min. Adhered bacteria were measured after the cell culture medium was removed after washing the cells with PBS. Cells were lysed prior to quantitative PCR to determine the absolute amount of host and invaded bacteria.
For glycomic analyses, bacteria were added to Caco-2 cells grown in 75 cm2 flasks containing an MOI of 1000. After co-incubation for specified times, bacteria were removed by washing three times with cold PBS and Caco-2 cells were harvested by scraping. In all infection experiments, flasks or plates were centrifuged at 500 rpm for 1 min prior to incubation to allow close contact of bacteria with host cells.
Bacterial Gene Deletion
The mutant strain was constructed as described by Datsenko and Wanner (29). Plasmid pKD46 containing ampicillin resistance and λ Red recombinase genes were isolated from E. coli BW25141 (CGSC 7634) and electroporated into the wildtype Salmonella strain. The parent strain was then grown in LB broth with 100 μg/ml ampicillin and 100 mm L-arabinose to induce λ Red recombinase production. Plasmid pKD3 carrying chloramphenicol resistance gene from E. coli BW25141 (CGSC 7631) was used as a template to generate PCR products for deletion of the gene of interest. The purified PCR products were used for transformation by electroporation (Bio-Rad Gene Pulser, Bio-Rad Laboratories, Hercules, CA) of electrocompetent Salmonella WT with induced λ Red recombinase. The transformants were selected on LB agar with 10 μg/ml chloramphenicol. The gene deletion/interruption was confirmed using PCR for each junction site created by the insertion.
Cell Membrane Extraction
Details of the isolation of the cell membrane fraction have been described previously (30, 31). In brief, harvested cells were resuspended in 20 mm homogenization buffer containing 0.25 m sucrose, 20 mm HEPES-KOH (pH 7.4), and 1:100 protease inhibitor mixture (EMD Millipore, Billerica, MA). Cells were lysed on ice using a probe sonicator (Qsonica, Newtown, CT) and lysates were centrifuged at 2000 × g for 10 min to remove the nuclear fraction and debris and ultracentrifuged in a series at 200,000 × g for 45 min at 4 °C to remove the cytoplasmic fraction. The resulting membrane pellet was isolated and stored at −20 °C until further processing.
N-Glycan Release and Enrichment
Membrane pellets were suspended with 100 μl of 100 mm NH4HCO3 in 5 mm dithiothreitol and heated for 10 s at 100 °C to thermally denature the proteins. To release the glycans, 2 μl of peptide N-glycosidase F (PNGase F) (New England Biolabs, Ipswich, MA) were added to the samples and incubated at 60 °C in a microwave reactor (CEM Corporation, Matthews, NC) for 10 min at 20 watts. After addition of 400 μl of ice-cold ethanol, samples were frozen for 1 h at −80 °C and centrifuged for 20 min at 21,000 × g to precipitate deglycosylated proteins.
Released N-glycans were purified by solid phase extraction (SPE)1 using porous graphitized carbon (PGC) packed cartridges (Grace, Chicago, IL). Cartridges were first equilibrated with alternating washes of nanopure water and a solution of 80% (v/v) acetonitrile and 0.05% (v/v) trifluoroacetic acid in water. Samples were loaded onto the cartridges and washed with nanopure water at a flow rate of 1 ml/min to remove salts and buffer. N-Glycans were eluted with a solution of 40% (v/v) acetonitrile and 0.05% (v/v) trifluoroacetic acid in water and dried.
Exoglycosidase Digestion
Sequencing of glycosidic linkages using exoglycosidases was performed as described by Song et al (38). Briefly, released N-glycans were chemically reduced with 1 m NaBH4 in a 65 °C water bath for 1.5 h and desalted by automated SPE-PGC. Reduced glycan compounds were fractionated from the total released glycans using HPLC and digested with highly specific exoglycosidase mixtures, including α-2,3-neuraminidase, α-1,2/3-mannosidase, β-N-acetylglucosaminidase (GlcNAcase), β-1,4-galactosidase, and α-1,3/4-fucosidase (New England Biolabs). Incubation conditions were optimized based on the concentration of the collected fractions and specific enzyme activity. For cell surface glycan analysis, isolated sialylated compounds were digested with α-2,3-neuraminidase. Glycans that did not lose a sialic acid residue after the 1.5 h digestion time were annotated as having an α-2,6-linked sialic acid. To locate the position of the sialic acid, the same isolated fractions were digested with a mixture of exoglycosidases to trim the glycan to the chitobiose core. Depending on the intact monosaccharides and their sequence, the sialic acid was positioned on the terminal end of the α-1,3- or the α-1,6-mannosyl antennae.
Kifunensine Treatment
Caco-2 cells were grown 14 days postconfluency with media renewal twice per week. Kifunensine (Santa Cruz Biotechnology, Dallas, TX) was dissolved into the cell media at specified concentrations and incubated for 72 h. For time point experiments, cells were grown and treated with kifunensine at time 0 and harvested in replicates at consecutive times following treatment (12 h, 24 h, 36 h, 48 h, 60 h, and 72 h).
Real Time PCR Analysis
Untreated and kifunensine-treated Caco-2 cells (2 × 106 cells per preparation) were harvested, washed twice with PBS, and resuspended in RNAlater (Life Technologies, Carlsbad, CA). Total RNA were extracted using RNeasy plus mini kit (Qiagen, Valencia, CA) and the quantity and quality of RNA were determined by using a Qubit Fluorometer (Life Technologies) and TapeStation 2200 (Agilent Technologies, Santa Clara, CA) following manufacturer's protocol. Total RNA were reverse transcribed to cDNA using iScript Reverse Transcription Supermix (Bio-Rad Laboratories) following manufacturer's instructions. The predesigned human glycosylation PrimePCR plates (Bio-Rad Laboratories) were used for real-time PCR using the CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories) and the analysis was performed using the CFX Manager 3.1 software (Bio-Rad Laboratories). Each 96-well plate has 88 target genes relevant to the glycosylation pathway and 3 reference genes (GAPDH, TBP, and HPRT1). The gene expression was normalized to the reference genes and presented as fold changes.
Chromatographic Separation and Mass Spectrometric Analysis
Glycan samples were reconstituted in nanopure water and analyzed using a nano-HPLC-chip/TOF-MS system (Agilent Technologies). Samples were introduced to the MS with a microfluidic chip, which consists of PGC packed enrichment and analytical columns and a nanoelectrospray tip. A binary gradient was applied to separate and elute glycans at a flow rate of 0.4 μl/min: (A) 3% (v/v) acetonitrile and 0.1% (v/v) formic acid in water and (B) 90% (v/v) acetonitrile in 1% (v/v) formic acid in water. MS spectra were acquired at 1.5 s per spectrum over a mass range of m/z 600–2000 in positive ionization mode. Mass inaccuracies were corrected with reference masses m/z 622.029, 922.010, 1221.991, and 1521.971.
Collision-induced dissociation (CID) was performed with nitrogen gas using a series of collision energies (Vcollision) dependent on the m/z values of the N-glycans, based on the equation:
| (Eq. 1) |
where the slope and offset were set at (1.8/100 Da) V and −2.4 V, respectively.
Data Analysis
N-Glycan compounds were identified with an in-house retrosynthetic library of all possible glycan compositions according to accurate mass (human library, 369 entries). Subtypes including high mannose, complex and hybrid were grouped accordingly by knowledge of the mammalian N-glycan biosynthetic pathway. Quantitative reproducibility and tandem MS confirmation of library matches were previously validated, enabling rapid and accurate assignment of glycan compounds (32–34). Signals above a signal-to-noise ratio of 5.0 were filtered and deconvoluted using MassHunter Qualitative Analysis version B.03.01 (Agilent Technologies). Deconvoluted masses were compared with theoretical masses using a mass tolerance of 20 ppm and a false discovery rate of 0.6%. Area under the peak was used to represent the data as follows. Absolute abundances refer to the integrated ion counts divided by the total number of cells to account for intersample variations. Relative abundances were determined by integrating ion counts of all assigned charge states for observed glycan masses and normalizing to the summed ion counts of all glycans detected. Statistical evaluation of significant glycan abundance changes after bacterial infection was performed using an unpaired, two-tailed Student's t test.
RESULTS
Variations in Host Glycan Abundances during Salmonella Infection
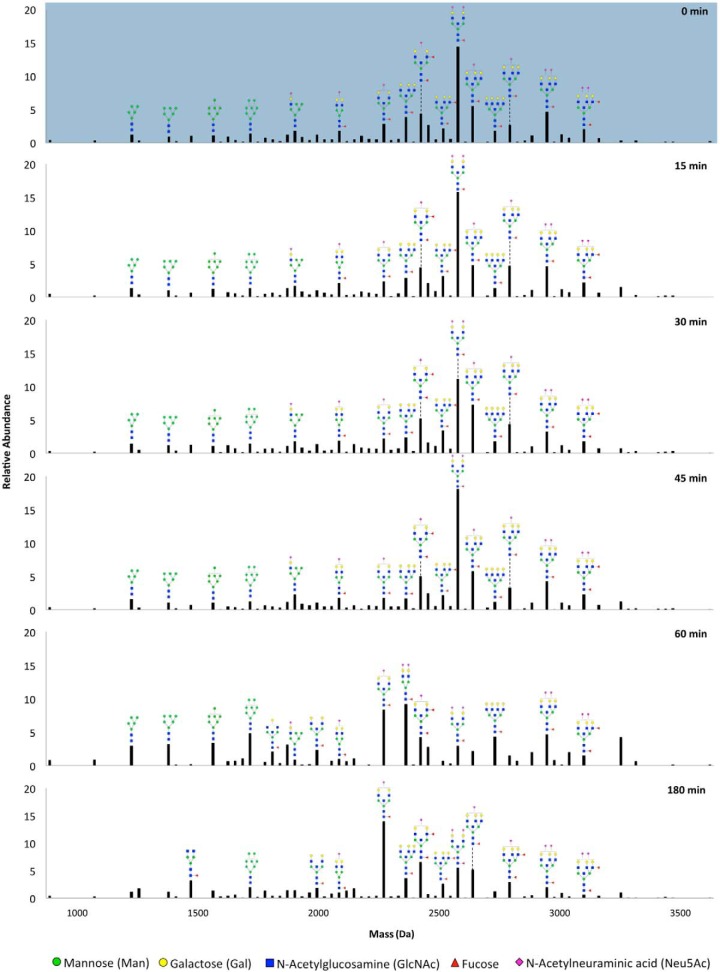
The effect of microbe interactions on host glycosylation can be readily characterized by global profiling of liberated host glycans. To first determine the time point following bacterial encounter when alterations of host cell surface glycans occur, Caco-2 cells were coincubated with S. typhimurium for varying time lengths that would correspond to early phases of infection (0, 15, 30, 45, 60 min) and a later time point of 180 min. The cell surface N-glycan profiles of Caco-2 at the specified infection times are shown in Fig. 1, where the relative abundances of every glycan composition are represented from lowest to highest mass. Between 0 to 45 min, glycan distributions remain unchanged. However, beginning at 60 min, large restructuring occurs, where mainly, glycans with lower masses (high mannose glycans) become higher in abundance while a distinguishing composition (Hex5HexNAc5Fuc1NeuAc2, mass 2571.92) decreases dramatically. At time points later than 60 min, S. typhimurium interactions induced further changes with ultimately another composition (Hex5HexNAc5Fuc1NeuAc1, mass 2281.85) becoming the most abundant at 180 min post-infection.
Fig. 1.
Relative abundances of glycan compositions found in the glycocalyx of Caco-2 after interactions with S. typhimurium for varying time durations. Values were averaged for replicate experiments. Putative structures are drawn for abundant peaks. Symbol nomenclature is used for representing glycan structures (52).
Following this time course study, we selected the initial point of glycan modification for further in-depth analysis. Thus, in the subsequent discussions below, uninfected and infected states refer to 0 and 60 min post-infection, respectively. To validate the reproducibility of the described analytical workflow, i.e. the sample preparations and the instrument response, total compound chromatograms (TCCs) of released glycans from a set of bio-replicates are shown in supplemental Fig. S1. From statistical assessment of individual compositions, 76 showed significant differences (p < 0.05) in abundances after infection, corresponding to ∼40% of all (nonredundant) compositions identified in uninfected and infected cells, as listed in supplemental Table S1.
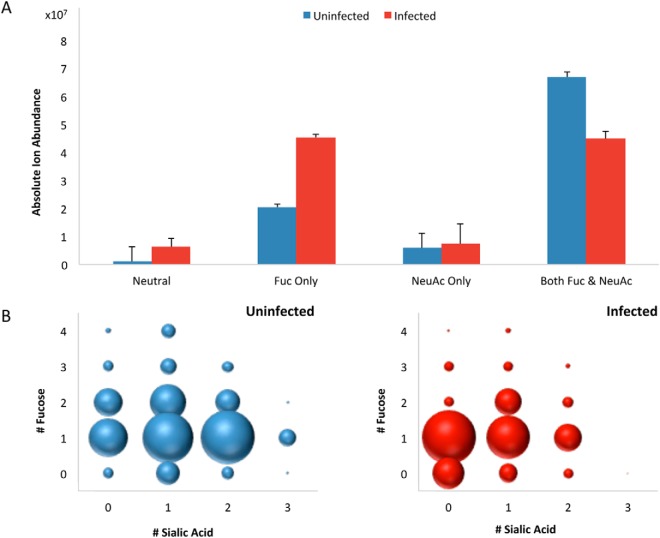
Based on the sum of all glycan signals, compositional profiles were generated for uninfected and infected cells to examine general trends according to glycan types (Fig. 2). For this analysis, complex and hybrid type glycans were combined to distinguish them from high mannose type. To enable quantitative comparisons between infection states, absolute abundances were normalized to the total cell counts per sample. Fig. 2A shows the summed absolute abundances of all complex and hybrid type structures grouped by the type of decoration (i.e. extension of the oligosaccharide chain by fucose, sialic acid, both, or neither). Among these, species that were both fucosylated and sialylated were reduced by 33% in infected cells. This decrease in abundance is reflected in the number of fucosylated and sialylated compositions, which decreased from 66 to 30 after infection. In parallel, the number of asialylated species bearing only fucose increased by 122%. Changes in sialylated species are particularly noteworthy given the expression of sialidases by the bacteria (27). The concurrent decrease of sialylated structures and increase of asialylated glycans is consistent with enzymatic degradation. We further note that desialylation appears to occur primarily on species that are both fucosylated and sialylated, given that afucosylated glycans bearing only sialic acid showed no significant changes in absolute abundances and in the number of compositions post-infection. To further account for outside variables, relative abundances were calculated based on the sum of all peak areas, which are detailed in supplemental Fig. S2. To examine the decreases in abundances of fucosylated and sialylated glycans in more detail, the degrees of fucosylation and sialylation in uninfected and infected cells are compared in Fig. 2B. Mono- and difucosylated as well as mono- and disialylated structures represent the majority of decorated glycans on the host cell surface prior to infection. After infection, decreases were observed such that asialylated structures became the most abundant. Generally, disialylated and trisialylated glycans showed the greatest decrease in abundances of 4-fold and 221-fold, respectively.
Fig. 2.
Global compositional profiling of complex/hybrid (C/H) signals in uninfected (blue) versus infected (red) cells. A, Absolute ion abundances summed together for each type of decorated and nondecorated C/H structures. Error bars represent S.D. (n = 3). B, Distribution of C/H type glycans containing fucose and/or sialic acid. The size of the dots represents the relative abundances of the indicated groups of glycans.
Determination of Sialyl Linkages and Connectivity
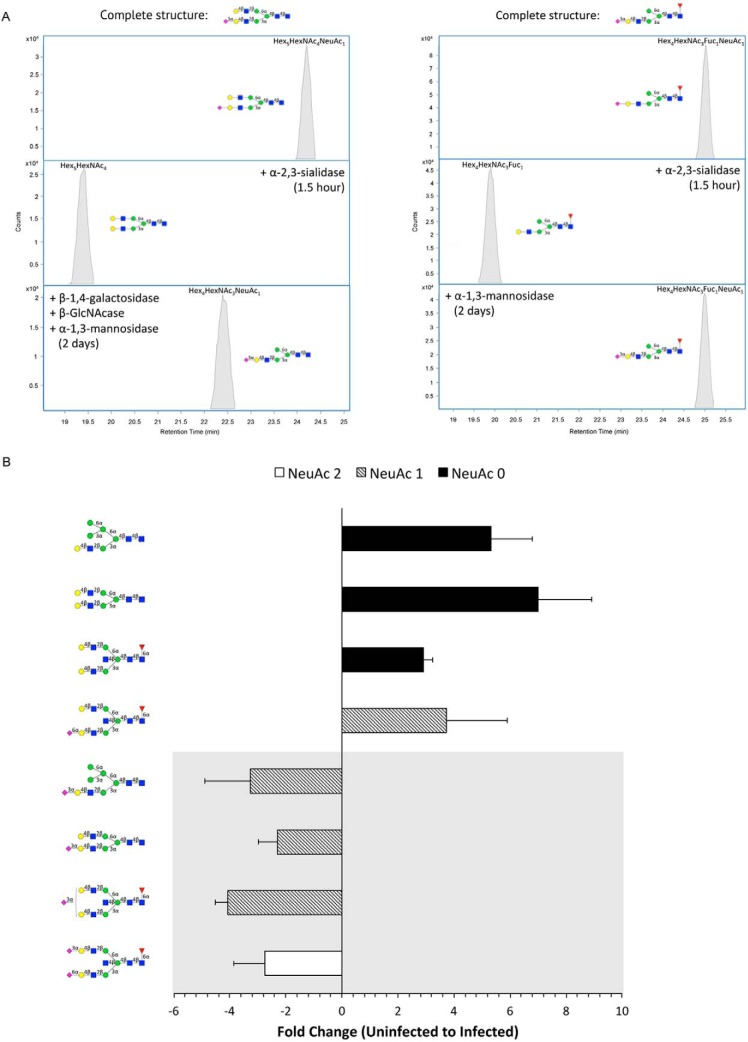
In addition to searching for S. typhimurium-induced glycan alterations by grouping glycan structures together by class (Fig. 2), compound-by-compound analyses were also conducted to obtain a better understanding of the activity of bacterial glycosidases. Porous graphitized carbon (PGC) nano-LC provides superior isomeric-level separation of glycan structures that can be resolved and quantified accordingly (35–37) (supplemental Fig. S3). Since a given glycan composition often has multiple isomers, nano-LC/MS analysis dramatically increases the number of structures that can be monitored. To annotate individual structures, we utilized a rapid characterization technique that combines HPLC fractionation with exoglycosidase digestion and MS analysis (38). The details of the enzymatic treatments and the elucidated compounds are given in supplemental Table S2. Furthermore, glycan structures were verified by MS/MS (supplemental Fig. S4). As an example, the extracted compound chromatograms (ECC) of an isolated fraction containing a single isomer of a complex type glycan, Hex5HexNAc4NeuAc1, before and after enzymatic digestion are shown in Fig. 3A (left). From the observed loss of sialic acid upon digestion with α-2,3-sialidase (Fig. 3A left, middle panel), we could determine the linkage of the sialic acid moiety. To locate its position, the same fraction was incubated with a mixture of α-1,4-galactosidase, α-GlcNAcase, and α-1,3-mannosidase for 2 days. After digestion, the structure Hex4HexNAc3NeuAc1 was detected, following a loss of hexose and GlcNAc (Fig. 3A right, bottom panel). Because one hexose residue still retained, we concluded that the sialic acid must be positioned on the terminal end of the α-1,3-mannosyl antennae. Similarly, the digestions used to determine the structure of a hybrid type glycan isomer are depicted in Fig. 3A (right).
Fig. 3.
Exoglycosidase digestion of sialyl glycans. A, Extracted compound chromatograms (ECCs) of HPLC fractionated glycan compounds before and after digestion with a mixture of exoglycosidases. Location and linkage determination of sialic acid is shown for Hex5HexNAc4NeuAc1 (m/z 1931.69) (left) and Hex4HexNAc3Fuc1NeuAc1 (m/z 1712.61) (right). Elucidated structures are displayed at the top of each panel. B, Fold changes of representative isomeric-specific forms of sialylated and desialylated structures of different subtypes (bisecting complex, biantennary complex, and monosialylated hybrid type glycans). Negative values indicate decreases in abundance and positive values indicate increases from uninfected to infected cell samples. Sequentially lower orders of sialylation are distinguished by color. Glycans with an α-2,3-sialyl feature are displayed in the gray panel. Symbol nomenclature is used for representing glycan structures (52).
Among the annotated structures, compounds from various sialylated glycan subclasses (i.e. bisecting complex, biantennary complex, and hybrid type) that showed high-fold abundance changes are highlighted in Fig. 3B. Interestingly, glycans containing at least one α-2,3-linked sialic acid decreased significantly after infection. Simultaneously, the corresponding desialylated species each showed more than 3-fold increases. For example, the composition Hex5HexNAc5Fuc1NeuAc1 yielded three isomers, one isomer with α-2,6-linked sialic acid and two isomers with α-2,3-sialic acid (one on each antennae). While the former decreased, the latter two isomers experienced relative increases in abundances following S. typhimurium infection. Our results precisely identify the α-2,3-sialyl-containing glycan structures that are enzymatically cleaved by the bacterial sialidase.
Glycan Remodeling Precedes Invasion
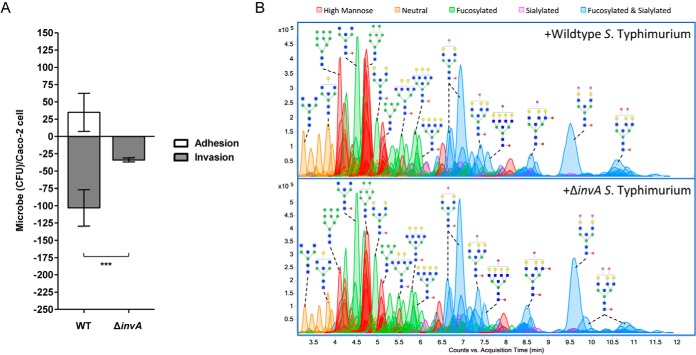
The display of intestinal epithelial cell surface glycans can theoretically be altered either by external factors (i.e. pathogen-encoded glycosidases) or by internal factors (i.e. host-derived genes of glycosylation). To differentiate between these two possibilities, we sought to assess host glycan changes when bacteria are not internalized. For this experiment, we constructed ΔinvA (STM2896) S. typhimurium isolates, which are unable to assemble the needle-like complex of the type III secretion system and are thus considered noninvasive (39). After 60 min of infection, we characterized the extent to which the mutant S. typhimurium associates with Caco-2. Bacterial adhesion and invasion of Caco-2 cells were confirmed by a Q-RT-PCR-based gentamicin protection assay (40–43). When co-incubated with invA-deficient S. typhimurium, the number of bacteria that invade Caco-2 was significantly reduced compared with that of the wildtype strain (Fig. 4A). Mutant strains are furthermore compromised in their binding to host cells, promoting discussion on the adhesive activities of the InvA protein (39, 44, 45).
Fig. 4.
Infection of Caco-2 with ΔinvA S. typhimurium. A, Comparison of the CFU of wildtype and mutant S. typhimurium that adhered (white bars) and invaded (gray bars) per Caco-2 cell. Error bars show S.E. between 3 biological replicates. Astericks indicate statistical significance, where ***, p < 0.001. B, Chromatograms of identified glycan compounds on Caco-2 after co-incubation with WT and ΔinvA S. typhimurium. Abundant peaks are annotated with putative structures. Symbol nomenclature is used for representing glycan structures (52).
When glycomic profiling was conducted following ΔinvA S. typhimurium infection, cell surface glycans were similarly distributed as when Caco-2 cells were infected with wildtype bacteria. Representative annotated chromatograms from triplicate analyses are shown in Fig. 4B. On average, glycans showed equal or less than 1.2-fold change when comparing infections with WT and mutant strains. Information related to identified glycans is listed in supplemental Table S3. These results strongly support the S. typhimurium-induced glycan alterations originating from extracellular host-pathogen interactions, which occur prior to bacterial entry.
Overexpression of High Mannose Type Glycans on Host Cells Increases Bacterial Invasion
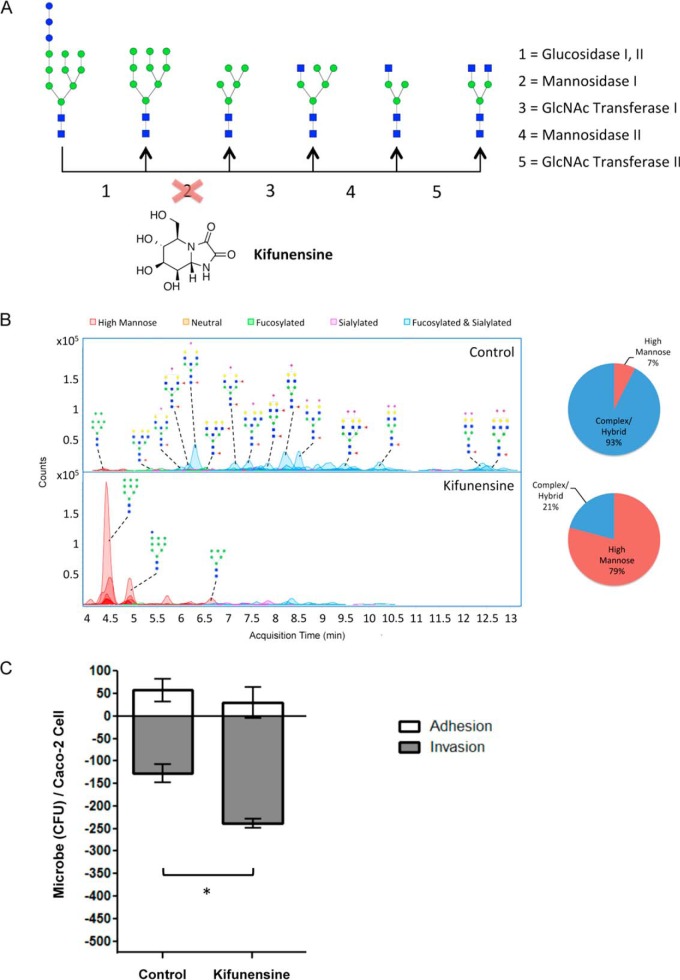
It is known that two steps are involved in S. typhimurium infection: The first is desialylation, as evidenced above, and the second is bacterial binding to mannose units. To determine the effect of the presence of mannosylated substrates on the cell surface, we augmented the expression of high mannose from the native amount (7%, relative abundance) to differing degrees. For this experiment, Caco-2 treatment with kifunensine, a cell permeable α-mannosidase I inhibitor, was effective in interrupting the glycan biosynthetic pathway prematurely, preventing the formation of hybrid and complex type glycans (Fig. 5A). A key feature of our profiling strategy is the ability to determine the structures that are present on the transformed cell surface to quantitatively assess the effectiveness and extent of the treatment. To do this, we performed mass spectrometric analysis on cell membrane glycans extracted following varying additions of kifunensine. With each increase in concentration, the percent of high mannose type glycans increased on the Caco-2 cell surface (supplemental Fig. S5). The greatest change was observed after addition of 100 μg/ml of kifunensine, at which point the relative abundances of high mannose glycans summed to 79% of all glycans (Fig. 5B). Of the high mannose glycan structures, Man 9 was the most abundant. This effect was dominant regardless of the amount of kifunensine added. As expected, hybrid and complex type glycans collectively decreased in abundances after addition of kifunensine, showing that existing membrane proteins are replaced with newly synthesized proteins bearing unprocessed high mannose glycans.
Fig. 5.
Effects of kifunensine treatment on cell surface glycosylation and bacterial infection. A, Kifunensine inhibits the mannosidase I enzyme that participates in N-glycan biosynthesis. B, Chromatograms display each compound at its column retention time. Pie charts show the summed abundances of high mannose and complex/hybrid type glycans before and after treatment. C, S. typhimurium adhesion (white bars) and invasion (gray bars) plot of untreated versus kifunensine-treated Caco-2 cells. S.E. is represented by error bars (n = 3). Astericks indicate statistical significance, where *, p < 0.05. Symbol nomenclature is used for representing glycan structures (52).
S. typhimurium interactions with high mannose-rich Caco-2 cells were then studied to observe whether these extensive changes in the host cell surface glycan landscape affect the nature of bacterial association. To this end, S. typhimurium were introduced into the medium of untreated and kifunensine-treated Caco-2 cells to assess any alterations in adhesion and invasion rates. As shown in Fig. 5C, infection was accompanied by a significant increase in the number of internalized bacteria in kifunensine-treated compared with untreated cells (p < 0.05). Thus, increasing the amount of expression of high mannose type glycans, particularly of larger structures containing nine mannose residues, resulted in enhanced bacterial invasion. Cell surface glycan remodeling coupled with detailed characterization provides the means to examine the involvement of specified glycan structures in cellular properties and functions. Using this strategy, we demonstrate that the epithelial cell glycocalyx layer profoundly influences the nature of host-bacterial interactions.
Membrane Protein Turnover Rates Inferred from Glycan Expression
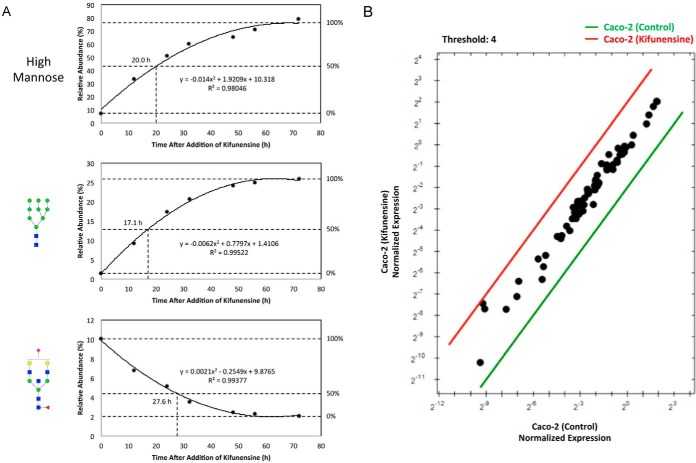
As an extension of the cell glycan remodeling strategy, we additionally developed an assay to determine if the restructuring of the host glycome observed after S. typhimurium infection is part of a host response to the pathogen, i.e. whether S. typhimurium induce dysregulation of the host's glycan biosynthetic pathway, resulting in newly synthesized proteins to be decorated with altered glycans. To investigate this possibility, we measured the rate at which cell membrane proteins are turned over. Because Man 9 was consistently the compound with the highest abundance when kifunensine was introduced into cells, we selectively monitored its levels at various time points following initiation of kifunensine treatment. Concurrently, an existing high abundant complex type glycan compound on the cell surface, Hex5HexNAc5Fuc1NeuAc1, was monitored to examine its decrease as a function of time following the initial addition of kifunensine. The abundances of these two compounds were tracked over time until a plateau was observed around 72 h, at which point the measured abundance was designated as 100% expression (or 0%). Furthermore, all high mannose type glycans were grouped together and assessed as a function of time. Graphical representations of the observed rates are shown in Fig. 6A. The amount of time to replace 50% of cell surface proteins with those presenting Man 9 was ∼17 h, as indicated by the interpolated line. Collectively, the sum of all high mannose structures over time revealed nearly the same time trend (20 h). Importantly, this rate is an indication of the average time it takes for cell membrane proteins to be inserted into the membrane upon their synthesis and processing in the ER. In the presence of kifunensine, all newly synthesized proteins bear only high mannose structures (mainly Man 9) before being expressed on the surface. Therefore, we can estimate the changes in Man 9 expression to be within the time frame of the turnover of membrane glycoproteins.
Fig. 6.
Changes in glycan expression over time after addition of kifunensine. A, Rate lines are interpolated based on the average abundances of high mannose, Man 9, and Hex5HexNAc5Fuc1NeuAc1 following kifunensine treatment. B, Expression level comparison of total RNA extracted from untreated and kifunensine-treated cells. Data are shown as fold changes relative to reference genes. Symbol nomenclature is used for representing glycan structures (52).
Q-RT-PCR was conducted in parallel to confirm that kifunensine treatment itself did not alter the expression of host glycosylation genes. Using a Q-RT-PCR array, human glycosylation gene expression profiles were compared in the presence and absence of kifunensine. Following the addition of kifunensine, meaningful changes in glycosylation genes were not observed (Fig. 6B). These results provide further support that the S. typhimurium-induced alterations in host cell glycosylation are the result of bacterial-expressed sialidases rather than changes in protein expression. Furthermore, this technique offers a unique and effective means for determining membrane protein turnover rates.
DISCUSSION
The comprehensive glycan profiling method employed in this study reveals mechanistic features of the early phases of S. typhimurium infection. Namely, bacteria recognize and trim sialic acid-containing glycan structures as they near the host surface, altering the extracellular glycan heterogeneity to favor nonsialylated glycans. While it is known that S. typhimurium encodes endogenous glycosyl hydrolases, some annotations remain putative. Our study is the first to demonstrate that S. typhimurium-derived sialidases act on specific glycan structures exposed on the host surface that affect the nature of bacterial invasion. As opposed to the standard use of synthesized substrates (46), structural detail and linkage specificity was observed on cell membrane sialo-glycoconjugates extracted from live human intestinal cells. Moreover, the nature of the sialic acid cleavage provides evidence that the enzyme is secreted during an infection, which has been merely speculated by its crystal structure (47). Given the nature of the cleavage, the sialidase encoded by STM0928 is likely responsible for the majority of host desialylation by S. typhimurium. In addition, the putative sialidase encoded by STM1252 may act as a general sialidase. Structural elucidation before and after infection revealed that sialic acid is diminished from high abundant and therefore more accessible cell surface N-glycans. It is likely that sialic acid residues, which make up the bulk of glycan structures on intestinal epithelial cell surfaces and reside on the terminal ends of glycoconjugates, confer a physical decoy for S. typhimurium before they dock onto the host cell's exterior. Once the enzyme enables sialic acid release, a sialic acid permease (nanT) enables uptake (27). We demonstrate that enzymatic activity occurs chiefly on the cell surface. Alternatively, restructuring of cell surface glycans as part of a host defense or stress response would not be evident within 60 min, as we demonstrated that the protein turnover in the cell membrane requires a significantly longer time. We further note that S. typhimurium encodes other glycosidases that may be able to degrade existing N-glycans into smaller structures, which were not pursued in this study.
In addition to host glycan degradation, S. typhimurium exhibits mannose-binding activity, which facilitates infection. Among strains of enterobacteria, S. typhimurium preferentially binds oligomannose chains using a type 1 fimbrial adhesin (48–50). Our results indicate that bacteria catabolize complex and hybrid type compounds while reserving high mannose structures strictly for anchoring to the cell surface. Large high mannose glycans on the host cell surface not only enables bacterial adherence but also promotes more rapid and effective invasion. Enhancing high mannose production with kifunensine treatment supports this model. This experiment further reveals that S. typhimurium are able to infect cells that are nearly devoid of sialylated structures when high mannose is abundant, a key aspect of secondary infection (51).
Herein, we have developed and demonstrated a method to identify sensitive changes in host cell surface N-glycans during the early stages of bacterial infection. This method provides detailed structural characterization of human glycan alterations following encounter with pathogenic S. typhimurium, an important step to link known bacterial gene expression to the pathophysiology of host cell invasion (27). Similar methods can be applied to investigate the effects of bacterial interactions on other glycoconjugates, including O-glycans and glycolipids.
CONCLUSION
At the bacterial-eukaryotic interface, a dense layer of glycosylated structures protrudes from the host cell surface and act as the first molecules of contact for bacteria, which express specific glycan-recognizing enzymes. Importantly, glycosylation is species-specific and different host cells are decorated by a different array of glycans. This work provides new insight into the role of host glycosylation during pathogenesis. We have determined that the abundance of a certain type of substrates (high mannose) can even enhance bacterial invasion. There are no reports to date regarding glycan changes in host cells during bacterial infection. Structural characterization of the glycans that participate in infection may provide targets for development of glycan-derived therapeutics to regulate infections. Our results indicate that bacteria show preference for the types of cells they infect depending on the host glycome. Therefore, it will be crucial to establish the animal model glycosylation in performing infection studies.
Supplementary Material
Footnotes
Author contributions: D.P., N.A., C.C.W., B.C.W., and C.B.L. designed the research. D.P. and C.C.W. performed the cell membrane enrichment and glycomics experiments. N.A. conducted the bacterial infection and knockout experiments. T.S. determined linkages with exoglycosidases. D.P. analyzed the LC/MS data. A.M. performed the real time PCR experiments. D.P., E.M., and C.B.L. wrote the paper.
* This work was supported by the National Institutes of Health under award number GM049077 (to CBL), 1R01HD065122-01A1 (to BCW), and DP2OD008752 (to EM). EM is also supported by the Burroughs Wellcome Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
Raw data files are available online at ftp://massive.ucsd.edu/ MSV000080199 (MassIVE ID: MSV000080199).
1 The abbreviations used are:
- SPE
- Solid Phase Extraction
- HM
- High Mannose
- C
- Complex
- H
- Hybrid
- Hex
- Hexose
- HexNAc
- N-Acetylhexosamine
- Man
- Mannose
- GlcNAc
- N-Acetylglucosamine
- Fuc
- Fucose
- NeuAc
- N-Acetylneuraminic Acid
- PGC
- Porous Graphitized Carbon
- TCC
- Total Compound Chromatogram
- ECC
- Extracted Compound Chromatogram.
REFERENCES
- 1. Holgersson J., Gustafsson A., and Breimer M. E. (2005) Characteristics of protein-carbohydrate interactions as a basis for developing novel carbohydrate-based antirejection therapies. Immunol. Cell Biol. 83, 694–708 [DOI] [PubMed] [Google Scholar]
- 2. Marcobal A., Southwick A. M., Earle K. A., and Sonnenburg J. L. (2013) A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23, 1038–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karlsson K. A. (2001) Pathogen-host protein-carbohydrate interactions as the basis of important infections. Adv. Exp. Med. Biol. 491, 431–443 [DOI] [PubMed] [Google Scholar]
- 4. Ohtsubo K., and Marth J. D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 5. Dube D. H., and Bertozzi C. R. (2005) Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 4, 477–488 [DOI] [PubMed] [Google Scholar]
- 6. Marth J. D., and Grewal P. K. (2008) Mammalian glycosylation in immunity. Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Kooyk Y., and Rabinovich G. A. (2008) Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 [DOI] [PubMed] [Google Scholar]
- 8. Maverakis E., Kim K., Shimoda M., Gershwin M. E., Patel F., Wilken R., Raychaudhuri S., Ruhaak L. R., and Lebrilla C. B. (2015) Glycans in the immune system and The Altered Glycan Theory of Autoimmunity: a critical review. J. Autoimmun. 57, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paulson J. C., Blixt O., and Collins B. E. (2006) Sweet spots in functional glycomics. Nat. Chem. Biol. 2, 238–248 [DOI] [PubMed] [Google Scholar]
- 10. Karlsson K. A. (1998) Meaning and therapeutic potential of microbial recognition of host glycoconjugates. Mol. Microbiol. 29, 1–11 [DOI] [PubMed] [Google Scholar]
- 11. Karlsson K. A. (1999) Bacterium-host protein-carbohydrate interactions and pathogenicity. Biochem. Soc. Trans. 27, 471–474 [DOI] [PubMed] [Google Scholar]
- 12. Baumler A. J., Tsolis R. M., and Heffron F. (1996) Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64, 1862–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., and Gordon J. I. (2005) Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 14. Pickard J. M., Maurice C. F., Kinnebrew M. A., Abt M. C., Schenten D., Golovkina T. V., Bogatyrev S. R., Ismagilov R. F., Pamer E. G., Turnbaugh P. J., and Chervonsky A. V. (2014) Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El Kaoutari A., Armougom F., Gordon J. I., Raoult D., and Henrissat B. (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504 [DOI] [PubMed] [Google Scholar]
- 16. Savage D. C. (1977) Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31, 107–133 [DOI] [PubMed] [Google Scholar]
- 17. Finlay B. B., and Cossart P. (1997) Exploitation of mammalian host cell functions by bacterial pathogens. Science 276, 718–725 [DOI] [PubMed] [Google Scholar]
- 18. Sansonetti P. (2002) Host-pathogen interactions: the seduction of molecular cross talk. Gut 50, III2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haraga A., Ohlson M. B., and Miller S. I. (2008) Salmonellae interplay with host cells. Nat. Rev. Microbiol 6, 53–66 [DOI] [PubMed] [Google Scholar]
- 20. Galan J. E., and Curtiss R. 3rd. (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 86, 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ginocchio C., Pace J., and Galan J. E. (1992) Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 89, 5976–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sherry A. E., Inglis N. F., Stevenson A., Fraser-Pitt D., Everest P., Smith D. G., and Roberts M. (2011) Characterisation of proteins extracted from the surface of Salmonella Typhimurium grown under SPI-2-inducing conditions by LC-ESI/MS/MS sequencing. Proteomics 11, 361–370 [DOI] [PubMed] [Google Scholar]
- 23. Niemann G. S., Brown R. N., Gustin J. K., Stufkens A., Shaikh-Kidwai A. S., Li J., McDermott J. E., Brewer H. M., Schepmoes A., Smith R. D., Adkins J. N., and Heffron F. (2011) Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antunes L. C., Arena E. T., Menendez A., Han J., Ferreira R. B., Buckner M. M., Lolic P., Madilao L. L., Bohlmann J., Borchers C. H., and Finlay B. B. (2011) Impact of salmonella infection on host hormone metabolism revealed by metabolomics. Infect. Immun. 79, 1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buckner M. M., Croxen M. A., Arena E. T., and Finlay B. B. (2011) A comprehensive study of the contribution of Salmonella enterica serovar Typhimurium SPI2 effectors to bacterial colonization, survival, and replication in typhoid fever, macrophage, and epithelial cell infection models. Virulence 2, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng K. M., Ferreyra J. A., Higginbottom S. K., Lynch J. B., Kashyap P. C., Gopinath S., Naidu N., Choudhury B., Weimer B. C., Monack D. M., and Sonnenburg J. L. (2013) Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., Layman D., Leonard S., Nguyen C., Scott K., Holmes A., Grewal N., Mulvaney E., Ryan E., Sun H., Florea L., Miller W., Stoneking T., Nhan M., Waterston R., and Wilson R. K. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 [DOI] [PubMed] [Google Scholar]
- 28. Pinto M., Robineleon S., Appay M. D., Kedinger M., Triadou N., Dussaulx E., Lacroix B., Simonassmann P., Haffen K., Fogh J., and Zweibaum A. (1983) Enterocyte-like differentiation and polarization of the human-colon carcinoma cell-line Caco-2 in culture. Biol. Cell 47, 323–330 [Google Scholar]
- 29. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. An H. J., Gip P., Kim J., Wu S., Park K. W., McVaugh C. T., Schaffer D. V., Bertozzi C. R., and Lebrilla C. B. (2012) Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteomics 11, M111 010660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park D., Brune K. A., Mitra A., Marusina A. I., Maverakis E., and Lebrilla C. B. (2015) Characteristic Changes in Cell Surface Glycosylation Accompany Intestinal Epithelial Cell (IEC) Differentiation: High Mannose Structures Dominate the Cell Surface Glycome of Undifferentiated Enterocytes. Mol. Cell. Proteomics 14, 2910–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruhaak L. R., Miyamoto S., Kelly K., and Lebrilla C. B. (2012) N-Glycan profiling of dried blood spots. Anal. Chem. 84, 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hua S., An H. J., Ozcan S., Ro G. S., Soares S., DeVere-White R., and Lebrilla C. B. (2011) Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 136, 3663–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hua S., Saunders M., Dimapasoc L. M., Jeong S. H., Kim B. J., Kim S., So M., Lee K. S., Kim J. H., Lam K. S., Lebrilla C. B., and An H. J. (2014) Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J. Proteome Res. 13, 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hua S., Williams C. C., Dimapasoc L. M., Ro G. S., Ozcan S., Miyamoto S., Lebrilla C. B., An H. J., and Leiserowitz G. S. (2013) Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. J. Chromatogr. A 1279, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zaia J. (2008) Mass spectrometry and the emerging field of glycomics. Chem. Biol. 15, 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruhaak L. R., Deelder A. M., and Wuhrer M. (2009) Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 394, 163–174 [DOI] [PubMed] [Google Scholar]
- 38. Song T., Ozcan S., Becker A., and Lebrilla C. B. (2014) In-depth method for the characterization of glycosylation in manufactured recombinant monoclonal antibody drugs. Anal. Chem. 86, 5661–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith A. C., Heo W. D., Braun V., Jiang X., Macrae C., Casanova J. E., Scidmore M. A., Grinstein S., Meyer T., and Brumell J. H. (2007) A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J. Cell Biol. 176, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elsinghorst E. A. (1994) Measurement of invasion by gentamicin resistance. Methods Enzymol. 236, 405–420 [DOI] [PubMed] [Google Scholar]
- 41. Desai P. T., Walsh M. K., and Weimer B. C. (2008) Solid-phase capture of pathogenic bacteria by using gangliosides and detection with real-time PCR. Appl. Environ. Microbiol. 74, 2254–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shah J., Desai P. T., Chen D., Stevens J. R., and Weimer B. C. (2013) Preadaptation to cold stress in Salmonella enterica serovar Typhimurium increases survival during subsequent acid stress exposure. Appl. Environ. Microbiol. 79, 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shah J., Desai P. T., and Weimer B. C. (2014) Genetic mechanisms underlying the pathogenicity of cold-stressed Salmonella enterica serovar typhimurium in cultured intestinal epithelial cells. Appl. Environ. Microbiol. 80, 6943–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hamburger Z. A., Brown M. S., Isberg R. R., and Bjorkman P. J. (1999) Crystal structure of invasin: a bacterial integrin-binding protein. Science 286, 291–295 [DOI] [PubMed] [Google Scholar]
- 45. Pizarro-Cerda J., and Cossart P. (2006) Bacterial adhesion and entry into host cells. Cell 124, 715–727 [DOI] [PubMed] [Google Scholar]
- 46. Hoyer L. L., Roggentin P., Schauer R., and Vimr E. R. (1991) Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl alpha 2—3 linkages. J. Biochem. 110, 462–467 [DOI] [PubMed] [Google Scholar]
- 47. Crennell S. J., Garman E. F., Laver W. G., Vimr E. R., and Taylor G. L. (1993) Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc. Natl. Acad. Sci. U.S.A. 90, 9852–9856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kisiela D., Sapeta A., Kuczkowski M., Stefaniak T., Wieliczko A., and Ugorski M. (2005) Characterization of FimH adhesins expressed by Salmonella enterica serovar Gallinarum biovars Gallinarum and Pullorum: reconstitution of mannose-binding properties by single amino acid substitution. Infect. Immun. 73, 6187–6190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Duguid J. P., Anderson E. S., and Campbell I. (1966) Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 92, 107–138 [DOI] [PubMed] [Google Scholar]
- 50. Firon N., Ofek I., and Sharon N. (1984) Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 43, 1088–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thankavel K., Shah A. H., Cohen M. S., Ikeda T., Lorenz R. G., RCurtiss 3rd, and Abraham S. N. (1999) Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274, 5797–5809 [DOI] [PubMed] [Google Scholar]
- 52. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Marth J. D., Bertozzi C. R., Hart G. W., and Etzler M. E. (2009) Symbol nomenclature for glycan representation. Proteomics 9, 5398–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.