Abstract
Cadmium (Cd) is a widespread pollutant that is toxic to plant growth. However, only a few genes that contribute to Cd resistance in plants have been identified. To identify additional Cd(II) resistance genes, we screened an Arabidopsis cDNA library using a yeast (Saccharomyces cerevisiae) expression system employing the Cd(II)-sensitive yeast mutant ycf1. This screening process yielded a small Cys-rich membrane protein (Arabidopsis plant cadmium resistance, AtPcrs). Database searches revealed that there are nine close homologs in Arabidopsis. Homologs were also found in other plants. Four of the five homologs that were tested also increased resistance to Cd(II) when expressed in ycf1. AtPcr1 localizes at the plasma membrane in both yeast and Arabidopsis. Arabidopsis plants overexpressing AtPcr1 exhibited increased Cd(II) resistance, whereas antisense plants that showed reduced AtPcr1 expression were more sensitive to Cd(II). AtPcr1 overexpression reduced Cd uptake by yeast cells and also reduced the Cd contents of both yeast and Arabidopsis protoplasts treated with Cd. Thus, it appears that the Pcr family members may play an important role in the Cd resistance of plants.
Cadmium (Cd) is a widespread soil pollutant, including in agricultural areas. The sources of Cd pollution are industry, sewage sludge disposal, pesticides, fungicides, and phosphate fertilizers. Cd is very toxic to all organisms, including plants, as it causes cellular damage (1) by inactivating or denaturing proteins by binding to free sulfhydryl residues, (2) by displacing cofactors from a variety of proteins, including transcription factors and enzymes, and (3) by generating reactive oxygen species (Liao et al., 2002). In plants, these damaging effects of Cd inhibit root growth, reduce stomatal conductance, and induce chlorosis.
Several Cd detoxification mechanisms have been elucidated in plants and other organisms. In animals, small Cys-rich proteins named metallothioneins (MTs) are the main chelators of heavy metals (Clemens, 2001). In plants, MTs are thought to play a role mainly in heavy metal homeostasis (Zhou and Goldsbrough, 1994). In addition to MTs, plants synthesize the glutathione-derived phytochelatins (PCs), which bind Cd and thereby reduce the interaction of the free ionic form of Cd with important cytosolic proteins (Clemens et al., 1999; Ha et al., 1999). Cd is also eliminated from metabolically active compartments into compartments with low metabolic activity by being transported into the vacuole either as PC-Cd, bis-glutathione-Cd, or by a Cd2+/H+ exchanger. This activity can therefore be regarded as a detoxification step. In Schizosaccharomyces pombe, a one-half-size ATP-binding cassette transporter has been shown to transport PC-Cd complexes (Ortiz et al., 1995). In Saccharomyces cerevisae, which does not synthesize phytochelatins, a multidrug resistance-associated protein-like ATP-binding cassette transporter transports bis-glutathione-Cd into the vacuole (Szczypka et al., 1994; Li et al., 1997). In plants, the corresponding transporters have not been identified, but CAX, a Ca2+/H+ exchanger, has been shown to exchange Cd(II) with protons (CAX2; Hirschi et al., 2000). A putative vacuolar metal ion transporter in Thlaspi goesingense was also found to improve tolerance to Cd when expressed in yeast (Saccharomyces cerevisiae; Persans et al., 2001). In addition, heavy metals can be pumped out of the cell by heavy metal transporting P-type ATPases (CPx-type ATPase) at the plasma membrane, such as ZntA in Escherichia coli (Rensing et al., 1998) and CAD2 in S. cerevisiae (Shiraishi et al., 2000).
Plants show varying levels of Cd resistance, but the factors responsible for this variability have not been identified. It is likely that apart from the well-described heavy metal tolerance mechanisms that have already been described, plants have other proteins that participate in novel Cd tolerance mechanisms. In this paper, we report a novel gene family that confers a strong Cd resistance at the plasma membrane level. We show that the product of one of these genes reduces the Cd levels in Cd-treated yeast and plant cells, and we identified the domains that are important in the Cd resistance conferred by this gene family.
RESULTS
Screening for Arabidopsis Genes That Confer Cd Resistance to the Cd-Sensitive ycf1 Yeast Strain
To screen for Cd-resistance genes, an Arabidopsis cDNA library was introduced into the YCF1-null (ycf1) yeast mutant strain DTY167, which is sensitive to Cd(II). Approximately 1 × 106 primary transformants were plated onto Cd(II)-containing one-half strength (1/2) synthetic dextrose (SD) agar medium and grown for 4 d. Plasmids were rescued from the surviving 400 colonies and again transformed into ycf1 cells and subjected to a second round of screening. Transformed cells that grew better than empty vector-transformed ycf1 cells in Cd(II)-containing medium (Fig. 1A) were identified, and their plasmids were rescued and sequenced. Of the 21 plasmids sequenced, 10 encoded MT 1a, 8 MT 2a, 1 MT 3, 1 phytochelatin synthase, and 1 encoded an unknown gene. This new gene encodes a 16-kD membrane protein that we named AtPcr1 (Arabidopsis plant cadmium resistance 1).
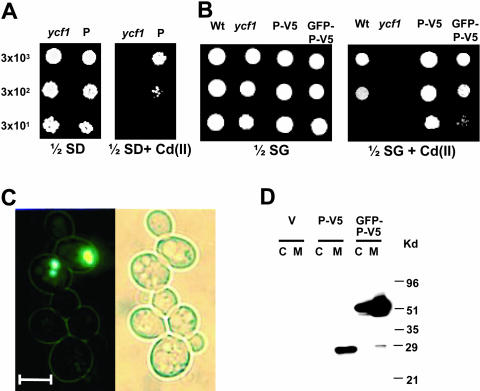
Figure 1.
AtPcr1 overexpression confers Cd(II) resistance in yeast and AtPcr1 is localized in the plasma membrane. A and B, Growth in the presence or absence of Cd(II) of ycf1 cells carrying the empty pFL61 or pYES2/NTC vector (ycf1), the pFL61-AtPcr1 (P), the pYES2/NTC-AtPcr1-V5 (P-V5), or the pYES2/NTC-GFP-AtPcr1-V5 (GFP-P-V5) construct. The yeast strains were grown at 30°C for 4 d on 1/2 SD or 1/2 SG plates with or without 50 μm Cd(II). As a control, an isogenic wild-type (wt) strain was also examined. C, Localization of AtPcr1 in cells overexpressing GFP-AtPcr1-V5. Left, a fluorescent image of GFP-AtPcr1-V5-expressing yeast cells. Right, a bright field image of the same cells shown in the left panel. Bar represents 5 μm. D, Presence of AtPcr1 in the membrane fraction of AtPcr1-V5 (P-V5) and in both the membrane and cytosolic fractions of GFP-AtPcr1-V5 (GFP-P-V5)-expressing ycf1 cells grown on SG medium. The microsomal and cytosolic fractions were isolated and their proteins (50 μg each) were electrophoresed on SDS-PAGE and detected by the V5 tag antibody. The predicted AtPcr1-V5 and GFP-AtPcr1-V5 sizes are 25 and 50 kD, respectively.
To determine the cellular function and localization of AtPcr1 in yeast, ycf1 yeast cells were transformed with the green fluorescence protein (GFP)- and V5-tagged AtPcr1 constructs GFP-AtPcr1-V5 and AtPcr1-V5. The AtPcr1-V5-transformed yeast strain was significantly more resistant to Cd(II) than ycf1 transformed with the empty pYES2/NTC vector or even the isogenic wild-type strain DTY165 with the empty vector (Fig. 1B). However, the expression of AtPcr1 did not change the resistance of the yeast cells to 1 to 3 mm Pb(II), 0.5 to 1.0 mm Ni(II), 1 to 4 mm Fe(II), 20 to 80 μm Cr(VI), 2 to 6 mm Zn(II), or 0.5 to 4 mm Cu(II) (data not shown). The GFP-AtPcr1-expressing yeast strain also showed enhanced Cd resistance (Fig. 1B), although its effect was not as strong as that of AtPcr1-V5. The green fluorescence of the GFP-AtPcr1 protein in these yeasts was evenly spread throughout the plasma membrane and, in some cells, as bright spots in the cytoplasm (Fig. 1C). The bright spots in the cytoplasm did not coincide with either nuclei or vacuoles and most likely were aggregates of proteins not properly targeted or partially digested.
To further investigate where in the cell AtPcr1 acts, the GFP-AtPcr1-V5- and AtPcr1-V5-expressing yeast strains were homogenized, separated into membrane and cytoplasmic fractions, and immunoblotted using the V5 tag antibody. Only the membrane fraction and not the cytosolic fraction prepared from the AtPcr1-V5 yeast, which show higher Cd(II) resistance than the GFP-AtPcr1-V5-expressing yeast (Fig. 1B), contained a band (28 kD) that was recognized by the V5 antibody (Fig. 1D). In contrast, both the cytosolic and membrane fractions of the GFP-AtPcr1-V5 yeast contained a band (51 kD) that was recognized by the V5 tag antibody (Fig. 1D). These observations are consistent with the microscopy data (Fig. 1C). In both cases the proteins were slightly larger than predicted. This is often observed for membrane proteins. Thus, it appears that AtPcr1 present in the plasma membrane of yeast can detoxify Cd(II).
Many AtPcr Family Members Are Found in the Plant Kingdom
Our search of the Arabidopsis genome database (www.arabidopsis.org) identified 9 genes that encode proteins homologous to AtPcr1 (Fig. 2A). AtPcr2, which has the highest homology to AtPcr1, was found next to AtPcr1 in chromosome 1, while AtPcr4, 5, and 7 are tandemly positioned in chromosome 3. Amino acid sequence similarity searching identified other possible members of this family in other plants, including tomato (Lycopersicon esculentum; AAF74287), petunia (Petunia hybrida; AAD02554), and rice (Oryza sativa; AAK52582). We named the rice AtPcr homolog OsPcr1.
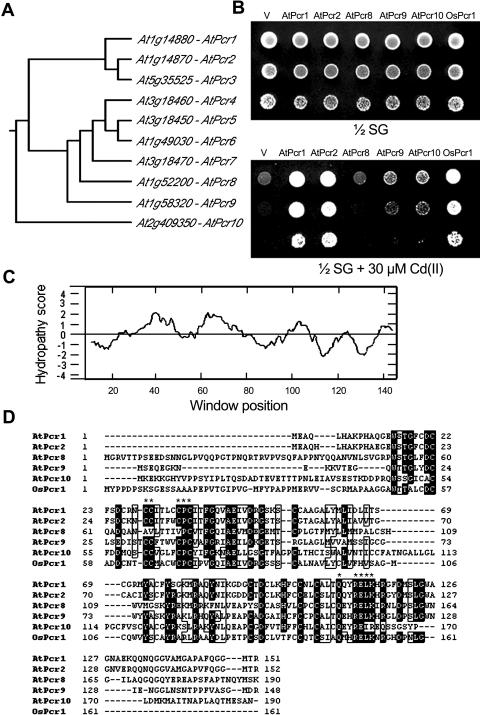
Figure 2.
Other members of the Pcr gene family confer Cd(II) resistance in yeast. A, Dendrogram of the AtPcr family in Arabidopsis. The tree was generated based on amino acid sequence homologies using CLUSTALW (origin2). B, Growth in the presence or absence of Cd(II) of ycf1 yeast cells expressing the empty pYES2/NTC vector (V), AtPcr1, AtPcr2, AtPcr8, AtPcr9, AtPcr10, or OsPcr1. The cells were grown on 1/2 SG medium with or without 30 μm Cd(II). C, Hydropathy profile of AtPcr1. Hydrophobicity was determined by the method of Kyte and Doolittle, using a moving window of 11 amino acid residues. D, The amino acid sequences of the Pcr protein in Arabidopsis (AtPcrs 1, 2, 8, 9, and 10) and rice (OsPcr1). Identical amino acid residues are shown by black boxes. CLUSTALW program was used for this alignment. An asterisk marks regions that are highly conserved among the Pcr family members.
To determine whether these AtPcr1 homologs also confer Cd(II) resistance, their cDNAs were amplified by reverse transcription (RT)-PCR and cloned into the yeast vector pYES2/NTC. We obtained clones encoding AtPcrs 1, 2, 8, 9, and 10 and OsPcr1 from rice, but could not obtain AtPcrs 3, 4, 5, 6, and 7, probably because of their low expression. Comparison of the predicted amino acid sequences of the cloned cDNAs revealed that AtPcr1 is 80%, 35%, 40%, 29%, and 36% identical to its AtPcr2, AtPcr8, AtPcr9, AtPcr10, and OsPcr1 homologs, respectively. Yeast cells expressing AtPcr1, 2, 9, 10, or OsPcr1 grew better on Cd(II)-containing plates than the empty vector-expressing strain but the AtPcr8-expressing yeast strain did not (Fig. 2B). Thus, many members of the Pcr family confer Cd(II) resistance.
Structural Analysis of AtPcr1
Membrane topology analysis using the TopPred 2 and Kyte and Doolittle programs showed that the predicted protein of Arabidopsis, AtPcr1, has two hydrophobic segments, AA28-48 and AA51-71, which form helix structures that may function as transmembrane domains (Fig. 2C). A protein domain search identified a DUF614 Cys-rich region common to Pcr family members (AA15-116 in AtPcr1). DUF614 is a Cys-rich domain of unknown function found in a number of eukaryotes. None of the Pcr family members or DUF614-containing proteins have been characterized in detail.
The Putative Transmembrane Domains of the Pcr Family Are Important for Cd(II) Resistance
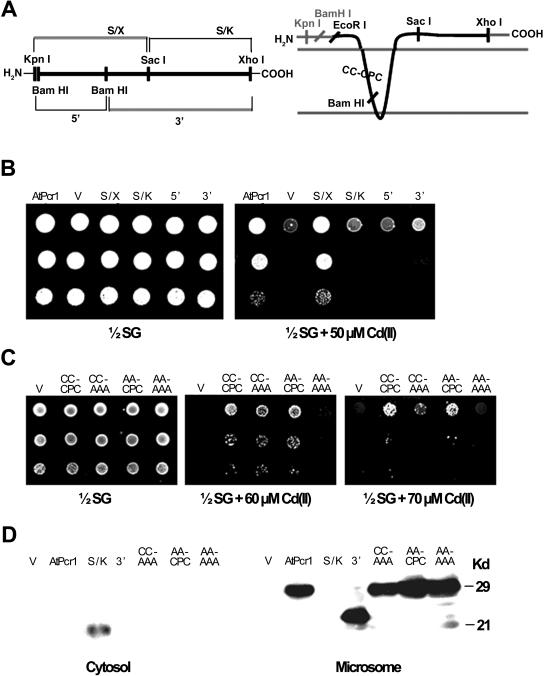
To identify the domains that are important for Cd(II) resistance, we aligned the amino acid sequences of AtPcr1 and its homologs (Fig. 2D). High homology was found in the CCXXXXCPC domain in the N-terminal first transmembrane region and in the QXXRELK domain of the C-terminal cytosolic region. These motifs are labeled with an asterisk in Figure 2D. To determine the importance of these domains in Cd(II) resistance, we tested the effect of their deletion in AtPcr1 on Cd(II) resistance. The ycf1 yeast cells overexpressing the AtPcr1 mutant that lacks the C-terminal 66 amino acids (S/X; AA1-84) showed the same Cd(II) resistance as the yeast expressing the full-length AtPcr1 (Fig. 3B). However, ycf1 yeast cells overexpressing AtPcr1 that lacks its N-terminal 84 amino acids (S/K; AA85-151) displayed Cd(II) resistance that was similar to that of the empty vector-transformed ycf1 control. Yeast cells expressing the AtPcr1 fragments containing amino acids 1 to 50 (5′ BamHI-BamHI fragment; AA1-50) or amino acids 51 to 151 (3′ BamHI-BamHI deletion; AA51-151) were more similar to the empty vector-transformed ycf1 control than the full-length AtPcr1-expressing strain in their sensitivity to Cd(II). From the western blot shown in Figure 3D, the S/K and 3′ segments are clearly expressed in yeasts transformed with the corresponding genes. The S/K segment was found in the cytosol instead of the microsome (Fig. 3D), as can be predicted from the hydropathy plot (Fig. 2C). Thus, the Cd(II) resistance-conferring domain of AtPcr1 is situated in the N-terminal region of the protein that includes the two putative transmembrane domains. This conclusion is consistent with the result presented in Figure 1, B and D that AtPcr1 localized at the membrane confers resistance to Cd(II).
Figure 3.
The transmembrane domains of AtCdr1 are important for Cd(II) resistance in yeast. A, The map of the truncated AtPcr1 constructs used (left) and the topology of AtPcr1 as predicted by the SOSUI program (right). On the left, thin lines indicate vector portions, and the thick line is the AtPcr1 gene. On the right, the epitope tag regions at both the amino and carboxy termini are shaded gray, and the AtPcr1 protein is in black. B, Growth in the presence or absence of Cd(II) of the yeast strains expressing the full-length and truncated AtPcr1 constructs. The yeast cells were grown at 30°C for 4 d on 1/2 SG plates with or without 50 μm Cd(II). C, Growth in the presence or absence of Cd(II) of the yeast strains expressing the wild-type AtPcr1 and AtPcr1 mutated at the CC-CPC sequence in the first putative transmembrane region. The yeast cells were grown at 30°C for 4 d on 1/2 SG plates with or without 60 or 70 μm Cd(II). D, Western blot of the wild-type AtPcr1 and mutated AtPcr1 proteins. The microsomal and cytosolic fractions were isolated and their proteins (50 μg each) were electrophoresed on SDS-PAGE and detected by the V5 tag antibody.
Cys residues are often important for Cd(II) resistance. In addition, the CPX domain is important in CPX-type metal pumps (Solioz and Vulpe, 1996; Rensing et al., 2000). With the exception of AtPcr8, the CPC motif in the N-terminal transmembrane domain is highly conserved among the AtPcr family members (Fig. 2D). To evaluate the importance of the CPC motif to the Cd(II) resistance function of AtPcr1, Cys→Ala and Pro→Ala replacements were introduced by oligonucleotide-directed site-specific mutagenesis, and yeast cells expressing the corresponding constructs were tested for Cd(II) resistance. The CPA, CAC, and APC single amino acid mutations did not change the Cd(II) resistance function of AtPcr1 (data not shown), but the AAA mutation slightly decreased it (Fig. 3C, CC-AAA).
Other Cys residues that may be important in the Cd(II) resistance function of AtPcr1 were identified by comparing the amino acid sequences of AtPcrs 1, 2, 9, and 10 and OsPcr1 (which all confer Cd(II) resistance to transformed ycf1 yeast) to that of AtPcr8 [which does not confer Cd(II) resistance]. The Cd(II) resistance-conferring proteins have two Cys residues four amino acids upstream of the CPC motif, while AtPcr8 has Ala and Val in those positions, respectively (Fig. 2B). However, when we substituted the two Cys residues with Ala, the Cd(II) resistance induced by AtPcr1 expression was not altered (Fig. 3C, AA-CPC).
To further test the importance of the CC and CPC residues to Cd(II) resistance, CC and CPC were simultaneously mutated to AA and AAA, respectively. This mutant could not confer Cd(II) resistance in medium containing 60 μm Cd (Fig. 3C), although Cd(II) resistance was observed at lower Cd(II) concentrations (data not shown). Moreover, the AA-AAA mutant was much less resistant to Cd than the CC-AAA mutant described above at 60 and 70 μm Cd(II) (Fig. 3C). These differences are not due to changes in the expression level or localization of the proteins, since CC-AAA, AA-CPC, and AA-AAA mutant proteins were expressed at similar levels as the wild-type AtPcr1, and their membrane localization was the same as the wild-type AtPcr1 (Fig. 3D). Thus, the complete Cys-rich region (but not a single Cys) in the first putative transmembrane domain of AtPcr1 is important for Cd(II) resistance.
AtPcr1 and AtPcr2 Are Expressed in Arabidopsis
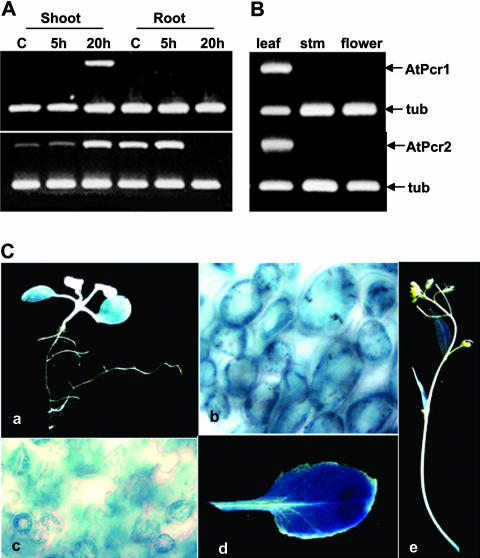
To determine the expression patterns of AtPcr1 and AtPcr2 in Arabidopsis, we performed northern-blot analysis using RNA from 3- or 8-week-old plants. However, we were not able to detect any message, even after Cd(II) treatment (data not shown). Thus, RT-PCR was performed with total RNA extracted from 3-week-old Arabidopsis plants treated with or without 100 μm CdCl2 for 20 h and AtPcr gene-specific primers plus tubulin primers for the internal standard. The roots and shoots of untreated plants did not express AtPcr1, but AtPcr2 was constitutively expressed in both tissues (Fig. 4A). CdCl2 treatment for 20 h caused AtPcr1 expression in the shoots and up-regulated the AtPcr2 transcript levels in the shoots (Fig. 4A). Thus, AtPcr1 and AtPcr2 expression is up-regulated in the shoots of 3-week-old Arabidopsis plants by Cd(II) treatment. However, Cd(II) did not up-regulate AtPcr1 in roots and after 20 h there was no AtPcr2 expression in roots. In the leaves of 8-week-old plants that had not been treated with Cd(II), both AtPcr1 and AtPcr2 were expressed (Fig. 4B). However, transcripts were not detected in the stems and flowers (Fig. 4B).
Figure 4.
AtPcr1 and AtPcr2 expression in Arabidopsis is Cd-inducible and tissue-specific. A, AtPcr1 and AtPcr2 transcript levels in 3-week-old Arabidopsis seedlings treated with 100 μm CdCl2 for 20 h. B, AtPcr1 and AtPcr2 transcript levels in 8-week-old Arabidopsis plants not treated with Cd(II). RT-PCRs were performed with total RNA as a template and gene-specific primers described in “Materials and Methods,” and the products were loaded on agarose gels. C, Histochemical assay of GUS activity in 10-d-old Arabidopsis seedlings treated with 100 μm CdCl2 for 2 d (a–c) and in mature 8-week-old untreated Arabidopsis plants (d and e).
AtPcr1 expression was further investigated using Arabidopsis plants transformed with an AtPcr1 promoter::GUS construct. Ten-day-old seedlings treated with 100 μm CdCl2 for 2 d and 8-week-old untreated plants were assessed for GUS activity. The untreated seedlings exhibited low GUS activity, but Cd(II) treatment resulted in high GUS activity in the leaves but not in the roots (Fig. 4C, a). In the leaves of Cd-treated seedlings, the guard and mesophyll cells showed GUS activity, while the epidermal cells were only slightly colored (Fig. 4C, b and c). In the untreated 8-week-old plants, the rosette leaves had high GUS activity, while the cauline leaves, flower-bearing stems, and pedicels showed low but detectable GUS activity (Fig. 4C, d and e).
AtPcr1 Is Localized at the Plasma Membrane of Arabidopsis
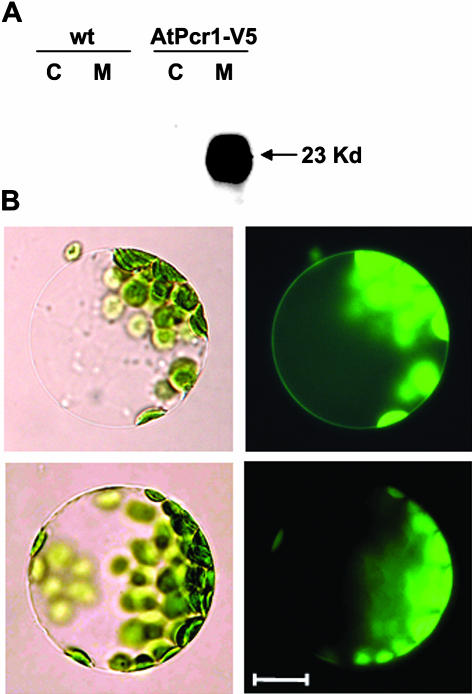
To investigate the subcellular localization of AtPcr1 in planta, transgenic Arabidopsis lines overexpressing the 35S::GFP-AtPcr1-V5 or the 35S::AtPcr1-V5 construct were generated. The soluble and microsomal proteins of the AtPcr1-V5-transgenic plants were separated by SDS-PAGE and western blotting was performed using the V5 tag antibody. Only the microsomal fraction showed a 23-kD band, which is slightly larger than the expected size of 20 kD (Fig. 5A). Moreover, examination of Arabidopsis protoplasts expressing the GFP-AtPcr1-V5 construct revealed fluorescence at the plasma membrane (Fig. 5B). This matches in general the fluoresence observations made with the transformed yeast (Fig. 1C) and Arabidopsis protoplasts (Lee et al., 2003) transformed with a plasma membrane localization control, AtAHA2-RFP. Thus, it appears that AtPcr1 is a plasma membrane protein.
Figure 5.
AtPcr1 is localized at the plasma membrane of Arabidopsis cells. A, AtPcr1-V5 transgenic plants were used to isolate cytosolic and microsomal fractions and the proteins in these fractions were separated on SDS-PAGE and subjected to western blotting using the V5 tag antibody. B, Fluorescence at the plasma membrane of a protoplast isolated from a GFP-AtPcr1-transgenic Arabidopsis plant. Top, a mesophyll cell protoplast from a AtPcr1-V5 transgenic plant; bottom, a mesophyll cell protoplast from a wild-type plant; left, a bright field image; right, a fluorescent image of the same cells. Chlorophyll autofluorescence is observed due to overexposure both from the transformed and wild-type protoplasts. Bar represents 10 μm.
Arabidopsis Plants Overexpressing AtPcr1 Exhibit Increased Cd(II) Resistance
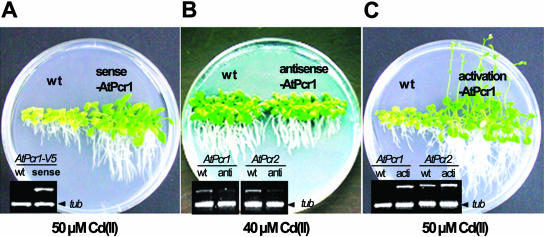
To test whether AtPcr1 is involved in Cd(II) resistance in planta, Arabidopsis plants expressing a sense or antisense 35S::AtPcr1 construct were generated. T2 lines were grown on 40 and 50 μm CdCl2-containing 1/2 Murashige and Skoog plates for 4 weeks and their growth was analyzed. Of the 45 sense lines, 37 were more resistant to Cd(II) compared to the wild-type plants, while 3 of the 25 antisense lines were more sensitive to Cd(II) compared to wild-type plants (data not shown). The T3 lines exhibited the same phenotype as the T2 lines (data not shown). The AtPcr1 mRNA levels in the sense and antisense lines were then assessed by semiquantitative RT-PCT. The sense lines showed elevated AtPcr1 mRNA levels (Fig. 6A, inset), while in the antisense lines, AtPcr1 mRNA levels were decreased to 40% to 70% of those observed in wild-type plants (Fig. 6B, inset). Moreover, examination of the AtPcr2 mRNA levels in the antisense lines revealed a similar decrease (Fig. 6B, inset). Thus, the AtPcr1 antisense construct suppresses the transcription of both AtPcr1 and AtPcr2. That AtPcr2 transcription is also reduced by the expression of an AtPcr1 antisense construct is not surprising as the AtPcr1 antisense construct sequence is 90% identical to the sequence of the same region of AtPcr2 gene. In contrast, the AtPcr1 antisense construct sequence is only about 30% identical to the sequences of the same regions in other Arabidopsis Pcr family members.
Figure 6.
AtPcr1 enhances Cd(II) resistance in Arabidopsis plants. AtPcr1-sense (A) and AtPcr1-antisense (B) lines on the Col-0 background were germinated and grown on 40 or 50 μm Cd(II)-containing 1/2 MS agar plates for 4 weeks. Wild-type (wt) Col-0 was grown as a control (A). The AtPcr1 activation-tagged line and its wild-type control on the WS background (C) were also grown in identical conditions. To determine the AtPcr1 message levels, total RNA was extracted from whole plants grown and RT-PCR was performed using the Pcr1-R1 and V5 tag-primers (A), AtPcr1- or AtPcr2-specific primers (B and C) and tubulin primers for the internal standard (A–C).
To further analyze AtPcr1- and AtPcr2-dependent phenotypic changes in Arabidopsis, an AtPcr1 activation-tagged line was selected from the new Basta resistant lines transformed with an activation-tag vector (Weigel et al., 2000; see “Materials and Methods”). Sequence analysis revealed that the insertion site of the vector was between AtPcr1 and AtPcr2 in chromosome 1 and 60 bases upstream of the AtPcr1 start codon, and RT-PCR analysis confirmed that the mutant produced higher mRNA levels of both AtPcr1 and AtPcr2 compared to the corresponding wild-type plants (Fig. 6C, inset). As with the AtPcr1-overexpressing sense lines (Fig. 6A), the activation-tagged line grew better in the presence of 50 μm Cd(II) than the wild-type plant (Fig. 6C).
Wild type, AtPcr1-overexpressing, AtPcr1-antisense-tagged, and AtPcr1-activation-tagged plants were also checked for growth differences in the presence of other metals at concentrations inhibitory for the growth of wild-type plants. In 1/2 Murashige and Skoog plates containing 30 mm Ca(II), 0.5 mm Zn(II), 10 um Ag(I), 0.3 mm Al(III), 0.1 mm Cr(VI), 30 mm Na+, 30 um Cu(II), 2 mm Mn(II), and 20 um Ni(II), growth did not differ dramatically (data not shown). This result suggests that AtPcr1 has a specific role to play in Cd(II) resistance.
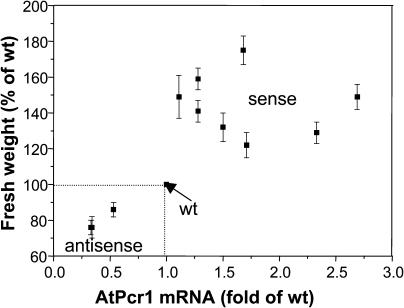
To gain more insights into the relationship between Cd(II) resistance and the transcript levels of AtPcr1, we compared the fresh weights of the wild-type, sense, and antisense plants grown in the presence of 40 or 50 μm Cd(II) with their corresponding AtPcr1 mRNA levels. When grown on 40 or 50 μm Cd(II)-containing 1/2 Murashige and Skoog plates for 4 weeks, the fresh weights of the AtPcr1-V5-overexpressing lines were 1.2- to 1.8-fold higher compared to wild-type plants. In contrast, the fresh weight of the antisense lines was decreased to 0.8-fold of that observed in wild-type plants (Fig. 7). To measure AtPcr1 mRNA levels, total RNA from 7 AtPcr1-V5 sense lines, 3 antisense lines, and wild-type plants grown in the presence of 30 μm Cd(II) was extracted and subjected to RT-PCR. In order to obtain sufficient amounts of mRNA, the Cd(II) concentration was decreased to improve plant growth. Relative transcript levels were determined by using AtPcr1-specific primers, which detects both the endogenous and introduced AtPcr1 gene expression, followed by hybridization of the PCR product with 32P-labeled AtPcr1. Western blots using V5 tag antibodies showed that in Arabidopsis expressing AtPcr1-V5, the amount of AtPcr1 protein is highly correlated to the transcript levels (data not shown). Plotting the transcript levels and fresh plant weights shows clearly that there is a strong correlation (r = 0.7, P < 0.01) between the level of AtPcr1 transcription and the fresh weight of Arabidopsis plants grown in Cd(II)-containing medium (Fig. 7). Thus, expression of AtPcr1 reduces the negative effects of Cd(II) exposure on Arabidopsis growth.
Figure 7.
Plants with higher AtPcr1 mRNA levels grow larger on Cd(II)-containing medium. Wild-type, antisense, and sense lines were grown on Cd(II)-containing agar plates for 4 weeks. The AtPcr1 mRNA levels were analyzed by RT-PCR and compared to the fresh plant weights. RT-PCR was performed with the total RNAs using the Pcr1-R1 and Pcr1-X1 primers and tubulin primers for an internal standard. RT-PCR products were separated on agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled AtPcr1 and tubulin probes. The relative AtPcr1 mRNA levels were quantified on a phosphorimager and normalized by the tubulin gene expression levels.
AtPcr1 Reduces Cd Uptake
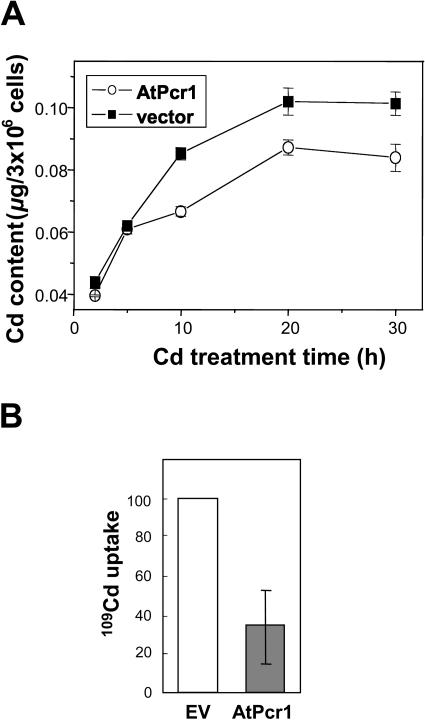
To gain insights into the mechanism by which AtPcr1 mediates Cd(II) resistance, the Cd contents in Cd-treated AtPcr1-expressing and control yeast cells were measured. The total Cd level in yeast cells grown for 10 to 30 h in 10 μm Cd(II)-containing medium was 17% to 29% lower than that in vector control yeast cells (Fig. 8A). Interestingly, a significant difference in Cd levels could only be observed after the yeast cells had been cultivated for a few hours in Cd(II)-containing medium (P < 0.05 at h = 10, 20, and 30).
Figure 8.
AtPcr1-expressing yeast cells reduce their Cd content by decreasing Cd(II) uptake. A, Cd content in Cd-treated yeast cells expressing the empty vector or AtPcr1. The cells were grown in 15 μm Cd(II)-containing SG media for indicated times, harvested, briefly rinsed twice with ice-cold water, and digested with 65% HNO3 at 200°C for 6 h. Their Cd contents were measured using stomic absorption spectroscopy. Shown are averages ± se (n = 3). B, Uptake of 109Cd(II) by yeast cells expressing empty vector (EV) or AtPcr1 that had been preincubated for 6 h with 20 μm CdCl2. See “Materials and Methods” for a description of the experimental details.
To determine whether the reduced Cd content in the cells is due to a reduction in Cd uptake or to an increase in Cd efflux, we compared the uptake of 109Cd by wild-type and AtPcr1-expressing yeast cells that had been preincubated with 20 μm CdCl2 for 6 h. Uptake of 109Cd was reduced to about 35% in the AtPcr1-expressing yeast cells as compared to the empty vector-transformed cells (Fig. 8B). Moreover, the uptake of 109Cd linearly increased with time without any decline (data not shown), supporting that AtPcr1 reduces Cd(II) uptake rather than activating efflux. Ca2+ channels have been shown to be permeable to Cd2+ (Perfus-Barbeoch et al., 2002). Therefore, we tested the possibility that AtPcr1 reduces Cd(II) content via modulating Ca2+ transport by comparing Ca2+ flux in the AtPcr1-expressing yeast cells to that in the empty vector-transformed cells. Ca2+ uptake did not differ between the empty vector control and AtPcr1 expressing yeast (data not shown). This result indicates that AtPcr1 is not closely coupled to a calcium transporter.
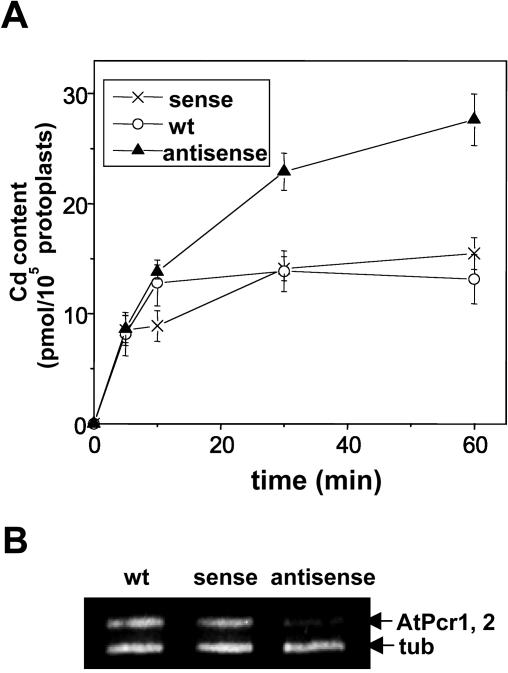
To examine whether the observations in yeast are also true for Arabidopsis, we performed the Cd flux assays with protoplasts isolated from the AtPcr1 antisense plants, the AtPcr1-overexpressing sense plants, and wild-type plants. To obtain enough protoplasts for this assay, the leaves of 8-week-old plants were used. This experiment showed that after incubation for 30 or 60 min in Cd(II)-containing medium, the AtPcr1 antisense protoplasts had higher Cd levels compared to wild-type protoplasts (Fig. 9A, P < 0.001 at h = 30 and 60 min).
Figure 9.
Antisense Arabidopsis protoplasts show elevated Cd contents. A, Cd uptake by protoplasts from wild-type, sense, and antisense Arabidopsis plants was assessed by following the protocol described in “Materials and Methods.” The protoplasts were isolated, purified, and concentrated by a percoll gradient. Shown are averages ± se (n = 6). B, AtPcr1 and AtPcr2 transcript levels in the Arabidopsis plants used in A. To amplify both the AtPcr1 and AtPcr2 messages, the Pcr1-R1 and Pcr1-X1 primers were used for the PCR step of RT-PCR.
However, the Cd levels in the wild-type and overexpressing lines did not differ significantly. At first glance, this latter result appears to contradict the data presented in Figures 6A and 7, where AtPcr1 overexpression lines are much more resistant to Cd(II) than the wild-type plants. However, this discrepancy is very probably due to the difference in the developmental stage of the samples used in the two experiments. Supporting this notion is that in the absence of Cd treatment, AtPcr1 and AtPcr2 expression is strongly increased in 8-week-old plants compared to young plants (Fig. 4, A and B). Furthermore, the levels of AtPcr1 mRNA expressed by the AtPcr1-overexpressing lines and wild-type plants at 8 weeks of age are very similar (Fig. 9B), unlike the 6-week-old counterparts, which show a clear difference in AtPcr1 levels (Fig. 6A). In addition, the transcript levels in antisense plants are lower at both the earlier and later stages of development than wild-type plants (Figs. 6B and 9B). In any case, that lower AtPcr1 levels in Arabidopsis protoplasts are associated with a greater uptake of Cd(II) corresponds to the pattern seen with the transformed yeast cells (Fig. 8).
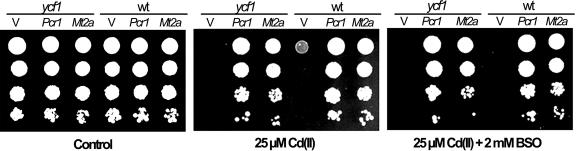
Cd(II) resistance mediated by YCF1 requires the presence of sufficient amounts of glutathione, since the substrate transported by YCF1 has been reported to be bis-glutathione-Cd (Li et al., 1997). In contrast, Cd(II) resistance mediated by MT does not require glutathione (Clemens et al., 1999). To test if AtPcr1 requires glutathione to mediate Cd(II) resistance, we tested the resistance to Cd(II) of AtPcr1-expressing ycf1 yeast cells in the presence of the glutathione biosynthesis inhibitor l-buthionine sulfoxime (BSO). The good Cd(II) resistance of these cells was not affected in the presence of BSO (Fig. 10), indicating that AtPcr1 confers glutathione-independent Cd(II) resistance. This result is similar to that obtained after expression of an Arabidopsis MT gene (AtMT2a) in the ycf1 strain, which also enhanced Cd(II) resistance regardless of the presence of BSO (Fig. 10). In contrast, when BSO was present, the empty vector-bearing wild-type strain became as susceptible to Cd(II) as the empty vector-bearing ycf1 strain (Fig. 10), which confirms that YCF1 requires glutathione to confer Cd(II) resistance to yeast (Li et al., 1997), and that BSO effectively reduced glutathione level in yeasts in this experiment.
Figure 10.
The Cd(II) resistance mechanism of AtPcr1 in yeast is GSH-independent. Empty vector (pYES2/NTC)-bearing wild-type yeast (wt), empty vector-bearing ycf1 yeast (V), AtPcr1-transformed ycf1 yeast (Pcr1), and AtMt2a-transformed ycf1 yeast (Mt2a) strains were grown at 30°C for 4 d on 1/2 SG plates with or without 25 μm Cd(II). The plate on the right also contained 2 mm BSO.
DISCUSSION
Using a functional screening strategy, we identified a novel Arabidopsis gene that mediates Cd(II) resistance in transgenic yeast and Arabidopsis. A search in the database revealed that there are 9 homologs of this gene in Arabidopsis, indicating that this novel Arabidopsis gene family is constituted by 10 members. Expression of 4 of these homologs in the Cd(II)-sensitive yeast strain ycf1 revealed that 3 confer Cd(II) resistance (Fig. 2B), which indicates that Arabidopsis has at least 4 AtPcr genes that mediate Cd resistance. Amino acid sequence similarity searching identified other possible members of this family in other plants and in animals, but not in bacteria, fungi, or other organisms.
The Pcr family members share an N-terminal hydrophobic domain that forms two putative transmembrane α-helices and a hydrophilic C-terminal region. Interestingly, the hydrophobic domain is enriched with Cys residues and contains the CCXXXXCPC motif that is conserved in most of the Pcr family members (located in amino acids 29–37 in the case of AtPcr1). These Cys residues apparently play an important role in the Cd(II) detoxifier function of the Pcr gene products since deletion of this region or mutations of the CC and CPC residues to AA and AAA drastically reduced the capacity of AtPcr1 to confer Cd(II) resistance in transformed yeast (Fig. 3). These findings are supported by the fact that AtPcr8, which does not contain the CCXXXXCPC motif (the corresponding sequence in AtPcr8 is AVXXXXVPC), does not confer Cd resistance to yeast.
That Cd(II) treatment of Arabidopsis seedlings induces the expression of AtPcr1 and AtPcr2 (Fig. 4A) is consistent with the role these genes play in conferring Cd(II) resistance to yeast. AtPcr1 and AtPcr2 may also have additional functions not related to Cd(II) resistance since they are expressed constitutively in mature leaves in the absence of Cd(II) treatment (Fig. 4B and 4C, d and e). Moreover, a putative member of this family in petunia, PGP/D12, which has 45% identity with AtPcr1, is induced during pollen tube germination (Guyon et al., 2000). It is possible that the Pcr proteins may generally be involved in heavy metal homeostasis by sensing the internal concentrations of the heavy metals.
Cd(II)-treated yeast and young Arabidopsis cells expressing AtPcr1 exhibited lower Cd contents than those that do not express the gene (Figs. 8 and 9). However, in older plants, a difference in Cd contents after Cd(II) treatment could not be observed. This was paralleled by transcription levels as Cd(II)-treated 8-week-old wild-type plants express comparable levels of transcripts to those of the Cd-treated 8-week-old AtPcr1-overexpressing lines (Fig. 9B). These observations suggest that genetically engineering plants to overexpress AtPcr1 may help the plants to cope with the heavy metals in polluted soil during their early stages of development.
It is likely that AtPcr1 does not induce Cd(II) resistance by acting as an intracellular chelator, as overexpression of an intracellular chelator would increase the Cd levels in Cd-treated cells rather than reduce them, which is what we observed when AtPcr1 was overexpressed. For example, overexpression of the Cd-chelator phytochelatin caused the transgenic cells to accumulate higher Cd levels after Cd treatment than the control cells (Clemens et al., 1999). As AtPcr1 is localized on the plasma membrane (Fig. 5), we propose that AtPcr1 modulates the transport of Cd(II) at the plasma membrane, and that it probably decreases Cd uptake instead of increasing Cd efflux (Fig. 8B). Although further studies are necessary to elucidate the mechanism used by AtPcr to reduce Cd contents in the cell, AtPcr1 seems to participate in a process that takes time to be set into motion. This is shown by the fact that in both yeast and Arabidopsis cells treated with Cd, the difference in Cd contents between the AtPcr1 mutant and the wild type became apparent only after a lag time (at least 5 h in yeast and 10 min in Arabidopsis; see Figs. 8 and 9). This lag time suggests that AtPcr1 contributes to the change in Cd(II) contents of the transformed cells indirectly, perhaps by regulating Cd transporters. Since AtPcr1 reduces Cd content in both yeast and Arabidopsis, interacting partners of AtPcr1 in these two organisms may be transporters of similar structure and function. We also found that the Cd resistance-inducing mechanism of AtPcr1 does not involve reduced glutathione since the Cd(II) resistance of the AtPcr1-expressing ycf1 yeast did not change in the presence of the glutathione synthesis inhibitor BSO (Fig. 10).
In conclusion, we have isolated a new family of Cd(II) tolerance genes from Arabidopsis. Our data clearly show that AtPcr1 is located at the plasma membrane and reduces the Cd levels in Cd-treated Arabidopsis cells and AtPcr1-transformed yeast. Since this gene does not contain any recognizable domains that are already known to contribute to Cd(II) resistance, the mechanism by which it confers Cd(II) tolerance may also be novel. Thus, further studies on this family of genes may unravel a new mechanism of Cd(II) resistance.
MATERIALS AND METHODS
Plant Materials
Arabidopsis plants (ecotypes Columbia [Col-0] and Wassilewskija [WS]) were grown in a controlled environment with 16 h light/8 h dark cycles at 22°C, except for the experiments testing Cd transport. In this case, the plants were grown under 8 h light/16 h dark cycles to obtain the broad leaves that are ideal for isolating healthy protoplasts. Using a previously established method (Krysan et al., 1999), an AtPcr1 activation-tagged line was selected from the new Basta-resistant lines in ecotype WS transformed with the activation-tag vector pSK1015 (Weigel et al., 2000). These lines were generated in Rick Amasino's laboratory and were provided through the University of Wisconsin Biotechnology Center.
Identification of Cd(II)-Resistance Genes
The Cd-sensitive ycf1 yeast (Saccharomyces cerevisiae) mutant DTY167 (MATa ura3 leu2 his3 trp3 lys2 suc2 ycf::hisG) and its isogenic wild-type DTY165 were kindly provided by Dr. Dennis Thiele. An Arabidopsis leaf cDNA library constructed in pFL61 (Piao et al., 1999) was introduced into DTY167 and transformants were selected on SD plates lacking uracil. The transformants were spread on a 30 to 100 μm CdCl2-containing one-half-strength SD lacking uracil plate and grown for 5 d. The plasmids from the surviving yeasts were rescued in Escherichia coli, extracted, and grouped by their restriction enzyme mapping patterns. Thirty plasmids were selected and sequenced with an automatic sequencing machine (ABI, Columbia, MD).
Synthesis of AtPcr1-V5 and GFP-AtPcr1-V5 Vectors for Transformation of Yeast and Arabidopsis
The open reading frame (ORF) of AtPcr1 was amplified by PCR using a plasmid containing the AtPcr1 gene as the template and the Pcr1-R1 (5′-GAATTCATGGAAGCTCAACTTCATGCCAAG-3′) and Pcr1-X1 (5′-CTCGAGGCGGGTCATGCCGCCTTGGAAGGC-3′) primers. The PCR products were digested by the EcoRI and XhoI restriction enzymes and inserted in frame into the EcoRI and XhoI sites of the pYES2/NTC vector (Invitrogen, Carlsbad, CA), which contains many tags, including V5 at both the N and C termini. The pYES2/NTC vector with the AtPcr1 gene was then introduced into yeast. To express GFP-AtPcr1 in yeast, the AtPcr1 gene was inserted into the GFP-coding region of the 326 GFP vector (Jin et al., 2001), after which the resulting GFP-AtPcr1 gene was inserted into the pYES2 yeast vector.
To overexpress AtPcr1-V5 or GFP-AtPcr1-V5 in Arabidopsis, the AtPcr1-V5 or GFP-AtPcr1-V5 fragments were excised from the relevant AtPcr1-containing pYES2/NTC vectors using the HindIII and PmeI restriction enzymes. The fragments were then ligated into the binary vector PGA1535 (ClaI-blunted/HindIII), which contains the cauliflower mosaic virus 35S promoter. The constructs were introduced into the Agrobacterium LBA4404 strain, which was then used to transform Arabidopsis (col-0) by the dipping method (Clough and Bent, 1988). To detect the expression of the introduced AtPcr1 gene in the transgenic plants apart from the endogenous AtPcr1 gene, the Pcr1-R1 and V5 tag primers were used (Fig. 6A).
Synthesis of Pcr-V5 Constructs and Testing Cd-Resistance Conferred by Pcr Family Members in Yeast
Primers for other Pcr genes were made according to their published ORF sequences. Since the AtPcr2 gene is very similar to AtPcr1, cloning of the AtPcr2 ORF required two-step PCR. Thus, AtPcr2 was first amplified using cDNA made from RNA extracted from Cd-treated-Arabidopsis as the template and the Pcr1-R1 and AtPcr2 (5′-TTTAACACTCGTAACAATTGTGATCCA-3′) primers. This produced the AtPcr2 gene, including its 3′ untranslated region. To obtain only the AtPcr2 ORF, a second PCR was performed using the AtPcr2 PCR product as the template and the Pcr1-R1 and Pcr1-X1 primers. AtPcr8, AtPcr9, and AtPcr10 were amplified using cDNA made from RNA extracted from Cd-treated Arabidopsis plants and the following specific primer pairs: AtPcr8, 5′-AACATATGAATTCATGGGTCGTGTCACTACTCCATC-3′ and 5′-CTAAAATCAAACTCGAGCTTAGACATATATTGATTTG-3′; AtPcr9, 5′-ACCAAAAGAATTCATGTCCGAACAAGAAGGCAAAAA-3′ and 5′-ATTTGTGATGTCTCGAGACGGTCCATGCCTGACGCTA-3′; AtPcr10, 5′-CATCAGAGAATTCATGAAAGAGAAGAAGGGTCATTA-3′ and 5′-ATGAGACAAAGCTCGAGGTTAGCTGATTCCATGGTTT-3′. The OsPcr1 ORF was amplified using cDNA made from RNA extracted from rice roots as the template and the OsPcr1-R1 (5′-GAATTCATGTATCCCCCTGATCCGTCCAAGTCC-3′) and OsPcr1-X1 (5′-CTCGAGACCAAGGTTAGGGTCGTGGCCGCGGTT-3′) primers. The PCR products were digested as described for AtPcr1 above and inserted into the pYES2/NTC vector. The pYES2/NTC vector with or without a Pcr family member gene was introduced into the DTY167 and DTY165 yeast strains. The transformants were spotted onto Cd-containing 1/2 synthetic galactose (SG) plates lacking uracil (SG-ura) and grown for 3 to 5 d. The AtPcr1, AtPcr2, AtPcr8, AtPcr9, AtPcr10, and OsPcr1 ORFs in the transformants were checked by sequencing.
Subcellular Localization Assays in Yeast and Arabidopsis
Yeast cells were transformed with the pYES2-GFP-AtPcr1 vector and GFP-AtPcr1 expression was induced by growing the cells in SG-ura medium. Wild-type Arabidopsis (col-0) plants were transformed with the pGA1535-GFP-AtPcr1 vector by the dipping method and protoplasts were isolated from T2 generation GFP-AtPcr1 transgenic plants. The localization of GFP-AtPcr1 fluorescence in the protoplasts and yeasts was observed using a Zeiss Axioskop2 fluorescence microscope equipped with the filter set 44 (exciter, 455–495 nm; emitter, 505–555) for green fluorescence (Jena, Germany).
AtPcr1 Western Blot-Analysis
Total protein was extracted from AtPcr1-V5 transgenic yeast and Arabidopsis using an extraction buffer (50 mm HEPES-KOH [pH 7.4], 5 mm MgCl2, 1 mm EDTA, 10 mm dithiothreitol, 0.7 μg/mL pepstatin A, 5 μg/mL aprotinin, 20 μg/mL leupeptin, and 0.5 mm phenylmethylsulfonyl fluoride). The total protein was centrifuged at 10,000g for 5 min at 4°C. The resulting supernatant was further centrifuged at 100,000g for 1 h at 4°C to separate the membranes from the soluble fraction. The proteins (10–50 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes by electro-blotting. The membranes were blocked in 1× TBST (0.1% (v/v) Tween 20 in 1× Tris-buffered saline) with 7.5% nonfat dry milk for 1 h at room temperature, washed twice in 1× TBST for 5 min each, and then incubated for 3 h at room temperature with the anti-V5 antibody. After three washes for 15 min each in 1× TBST, the membrane was incubated for 1 h with sheep anti-mouse IgG conjugated to horseradish peroxidase, and washed three times with 1× TBST. Chemiluminescence was detected using the ECL reagent according to the instructions of the manufacturer (Amersham Pharmacia Biotech, Uppsala) and the signals were developed with x-ray film.
Assaying AtPcr1 and AtPcr2 Expression Patterns in Arabidopsis
Total RNA was extracted from 3- and 8-week-old plants using RNA extraction buffer (250 mm Tris HCl [pH 9.0], 250 mm NaCl, 50 mm EDTA, 345 mm p-aminosalicylic acid, 27 mm triisopropyl naphthalene sulfonic acid, 250 mm β-mercaptoethanol, 0.024% [v/v] phenol) and phenol/chloroform. cDNA was synthesized using an RT-PCR kit employing the SuperScript First-Strand Synthesis System (Invitrogen). Since the AtPcr1 and AtPcr2 ORFs are 88% identical, we used the 3′ untranslated region sequences of these genes as the reverse primers in the PCR step to specifically detect their messages. Thus, to amplify AtPcr1 cDNA, Pcr1-R1 was used as the forward primer, and the reverse primer was 5′-TTTAACACTCGTAACAATTGTGATCCA-3′. To amplify AtPcr2 cDNA, Pcr1-R1 was again used as the forward primer and the reverse primer was 5′-TTTAATTCTTGTAACCAAATAGTGGAATAT-3′. To amplify both the AtPcr1 and AtPcr2 messages simultaneously, the Pcr1-R1 and Pcr1-X1 primers were used.
Assaying AtPcr1 Promoter Activity in Arabidopsis
To make the AtPcr1 promoter::GUS construct, 3.2 kb of the AtPcr1 promoter region was amplified using genomic DNA from Arabidopsis (col-0) and the primers 5′-CTGTTTGTTTTTGAAAGCTAGCACATGAGT-3′ and 5′-TGAAGGTGTTGAGGATCCAAGAAGAGAG-3′. The PCR product was digested with NheI and BamHI and inserted into the XbaI and BamHI sites of the pBI101 binary vector. This construct was introduced into Arabidopsis (col-0) by the dipping method. Kanamycin-selected T2 transformants were then assayed for GUS activity.
Generation of Arabidopsis with Reduced Levels of ArPcr1 Expression
To reduce endogenous AtPcr1 expression in Arabidopsis, an AtPcr1-antisense vector was made by inserting the AtPcr1 fragment (BamHI/XbaI) from the pYES2/NTC-AtPcr1-V5 vector into the BglII and XbaI sites of the PGA1535 vector. The AtPcr1 construct was introduced into Arabidopsis (col-0) by the dipping method. Note that the antisense lines also show reduced levels of AtPcr2 expression.
Measurement of Cd Contents and Cd Uptake Activity in Yeast
To measure Cd contents in yeast cells, the cells were cultured in SG-ura medium to an OD600 of 3 and then pelleted by centrifugation. The cells were diluted to an OD of 1 with SG-ura medium containing 15 μm Cd(II) and grown for indicated times at 30°C. They were then harvested, washed twice with ice-cold water for 2 min, and digested with 65% HNO3 at 200°C for 6 h, and their Cd contents were measured using an atomic absorption spectrometer (SpectrAA-800, Varian, Palo Alto, CA).
To investigate 109Cd-uptake activity of Δycf1/Δyllo15 (Klein et al., 2002), yeast cells that had been freshly transformed with AtPcr1 or the empty vector were grown overnight in SG-ura medium to an OD of 1 to 2. The cultures were subsequently washed once in water and resuspended at an OD of 5. The yeast were then grown for a further 6 h in the presence of 20 μm Cd(II), after which they were collected and washed twice in SG-ura medium. The OD was adjusted to 5 and the cells were then incubated for up to 30 min in the presence of 5 μm 109Cd(II) (specific activity 0.12 μCi/nmol). The 109Cd uptake was measured using nitrocellulose filters and was linear for at least 30 min.
Cd Flux in Arabidopsis Protoplasts
Wild-type and transgenic protoplasts were isolated, purified, and concentrated using a percoll gradient, and used in uptake assays according to the protocol described previously with slight modifications (Martinoia et al., 1993). Thus, 2 × 107 protoplasts were incubated in 1.8 mL of a bathing solution (0.5 m Gly-betaine, 10 mm CaCl2, 10 mm MES-KOH [pH 5.6], 10 mm CdCl2) containing 2 kBq of 3H2O and 6.7 Bq of 109Cd for 0, 5, 10, 30, or 60 min. After each time interval, 100 μL of the sample was taken and transferred to a centrifuge tube. The healthy protoplasts were collected by centrifugation at 10,000g for 20 s using percoll and silicon oil gradients. Only the healthy protoplasts thus obtained were counted for their 109Cd and 3H2O contents using a gamma-ray counter (1470 Wizard, Wallac, Boston) and a liquid scintillation counter (Tri-CARB2100TR, Packard BioScience, Meriden, CT), respectively. The 3H2O levels were counted at the energy window of 0 to 12 keV and corrected for the contamination from the beta rays of the 109Cd at the same energy window. To obtain the time-dependent uptake into the cells only, the radioactivity at t = 0 min was subtracted from all data values.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers NM_101357 for AtPcr1, NM_101356 for AtPcr2, NM_148032 for AtPcr3, NM_112731 for AtPcr4, NM_112730 for AtPcr5, NM_103796 for AtPcr6, NM_112732 for AtPcr7, NM_104100 for AtPcr8, NM_104612 for AtPcr9, NM_129657 for AtPcr10, and AAK52582 for OsPcr1.
Acknowledgments
We thank Dr. Dennis Thiele for kindly providing us with the Cd-sensitive ycf1 mutant and its isogenic wild-type yeast. We also thank Rick Amasino and his laboratory and the University of Wisconsin Biotechnology Center for distributing and screening the activation-tagged Arabidopsis lines.
This work was supported by grants awarded from POSCO and the National Research Laboratory program of the Ministry of Science and Technology of Korea (to Y.L.), from the Bundesamt fuer Bildung und Wissenschaft (BBW C99060 and BBW00.0413/EU proposal Metallophytes, QLRT–2001–02894 to E.M.), and from the Creative Research Initiative Program of the Ministry of Science and Technology of Korea (M10116000005–02F0000–00310 to I.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037739.
References
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212: 475–486 [DOI] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J 18: 3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1988) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Guyon VN, Astwood JD, Garner EC, Dunker AK, Taylor LP (2000) Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in petunia. Plant Physiol 123: 699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S-B, Smith AP, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ (2000) Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like Protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Mamnun Y, Eggmann T, Schüller C, Wolfger H, Martinoia E, Kuchler K (2002) The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett 520: 63–67 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JK, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bae H, Jeong J, Lee J-Y, Yang Y-Y, Hwang I, Martinoia E, Lee Y (2003) Functional expression of a bacterial heavy metal transporter in Arabidopsis enhances resistance to and decreases uptake of heavy metals. Plant Physiol 133: 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z-S, Lu Y-P, Zhen R-G, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao VH, Dong J, Freedman JH (2002) Molecular characterization of a novel, cadmium-inducible gene from the nematode Caenorhabditis elegans. A new gene that contributes to the resistance to cadmium toxicity. J Biol Chem 277: 42049–42059 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Grill E, Tommasini R, Kreuz K, Amrhein N (1993) ATP-dependent glutathione S-conjugate ‘export’ pump in the vacuolar membrane of plants. Nature 364: 247–249 [Google Scholar]
- Ortiz DF, Kreppel L, Spaser DM (1995) Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J Biol Chem 270: 4721–4727 [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J 32: 539–548 [DOI] [PubMed] [Google Scholar]
- Persans MW, Nieman K, Salt DE (2001) Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA 98: 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Pih KT, Lim JH, Kang SG, Jin JB, Kim SH, Hwang I (1999)) An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol 119: 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Fan B, Sharma R, Mitra B, Rosen BP (2000) CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA 97: 652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Sun Y, Mitra B, Rosen BP (1998) Pb(ll)-translocating P-type ATPases. J Biol Chem 273: 32614–32617 [DOI] [PubMed] [Google Scholar]
- Shiraishi E, Inouhe M, Joho M, Tohoyama H (2000) The cadmium-resistant gene, CAD2, which is a mutated putative copper-transporter gene (PCA1), controls the intracellular cadmium-level in the yeast S. cerevisiae. Curr Genet 37: 79–86 [DOI] [PubMed] [Google Scholar]
- Solioz M, Vulpe C (1996) CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci 21: 237–241 [PubMed] [Google Scholar]
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ (1994) Yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem 269: 22853–22857 [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB (1994) Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]