Much of the life on earth depends on photosynthesis, a process in which electromagnetic solar energy is used to produce oxygen and carbohydrates from atmospheric CO2 and water. In plants and algae, the photosynthetic reactions are catalyzed by a system that includes several protein-pigment complexes embedded in the thylakoid membranes. These membranes consist of flattened vesicles that exist either as appressed or single nonappressed membranes. In eukaryotic organisms, the biogenesis and activity of the photosynthetic complexes depends on the coordinate action of the nuclear and chloroplast genetic systems. A remarkable feature of the photosynthetic system is its ability to adapt rapidly to changes in environmental cues, such as light. This essay provides a historical outline of some of the key findings obtained through genetic approaches that had a strong impact on the current understanding of the biogenesis and dynamics of the photosynthetic machinery. It is not possible to cover all the important studies in this area within this short article, and the citations chosen reflect my personal bias. Most of the article bears on the system that catalyzes the primary light reactions and electron transfer in the photosynthetic membranes.
GENETICS OF PHOTOSYNTHESIS
Levine was amongst the first to use a genetic approach for studying the photosynthetic apparatus by isolating a large number of mutants deficient in photosynthetic activity (Levine, 1968). Although this approach was met with considerable skepticism at the beginning, it proved highly successful. Levine chose the photosynthetic unicellular organism Chlamydomonas reinhardtii for several reasons. First, it is amenable to genetic analysis because of a well-defined sexual cycle, and nuclear and chloroplast mutations follow distinct segregation patterns. In the progeny of crosses of cells of opposite mating type, nuclear mutations segregate 2:2, whereas chloroplast mutations are transmitted uniparentally from the mating type + parent to the offspring. Second, the photosynthetic function is dispensable when the algae are grown in the presence of a carbon source, such as acetate. Mutants were screened for their inability to grow on medium lacking a carbon source and further tested for their inability to fix CO2 or by using a fluorescence assay based on the fact that photosynthetic mutants display altered fluorescence patterns. These screens produced a large collection of photosynthetic mutants. Some were affected in the different steps of the photosynthetic electron transport chain. In this way, new components of this chain were discovered (e.g., the Rieske protein of the cytochrome b6f complex was first identified genetically through this approach) (Levine, 1968). Moreover, some of the mutants obtained were crucial for determining the correct order of the electron transfer components of the photosynthetic electron transport chain; in this way, the Z-scheme, which proposes that photosystem II (PSII) is functionally linked through the electron transport chain to photosystem I (PSI), was fully confirmed. These wide screens also led to the isolation of mutants deficient in PSII, PSI, ATP synthase, or in some of the enzymes of the Calvin cycle.
Another genetic-biochemical approach was taken by Wildman, who took advantage of the fact that different species of plants can be crossed to form an F1 generation (Chan and Wildman, 1972). He used the differences in the primary structure of proteins between these species to determine the source of genetic coding information for these differences. In this way, he was able to show that the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, called fraction I protein at that time, is inherited maternally; thus, he concluded that this protein is most likely encoded by the chloroplast DNA.
While the biochemical and genetic identification of the photosynthetic components progressed at a rapid pace, studies were initiated on the biosynthesis of the photosynthetic system in the early 1960s. Ris and Plaut (1962) firmly demonstrated the presence of DNA in chloroplasts by the detection of Feulgen-positive regions in the cytoplasm of C. reinhardtii that were DNase sensitive. The discovery of chloroplast DNA was followed by the characterization of an autonomous plastid protein–synthesizing system comprising DNA-dependent RNA polymerase and prokaryote-like 70S ribosomes. Moreover, antibiotics that specifically inhibit the cytosolic 80S or plastid 70S ribosomes were used to determine to what extent photosynthetic reactions are dependent on cytosolic or chloroplast protein synthesis and more generally to establish which of the chloroplast proteins are synthesized in the chloroplast or cytosolic compartments (Chua and Gillham, 1977). These early studies revealed that most photosynthetic activities examined were dependent on both the chloroplast and nucleocytosolic genetic systems.
Combining biochemical and genetic approaches proved extremely powerful for identifying specific photosynthetic components. Comparative high-resolution protein gel electrophoresis from extracts of wild-type and mutant cells deficient in a photosynthetic complex was used to identify subunits belonging to the corresponding complex (Chua and Bennoun, 1975). These assignments were later confirmed by the analysis of the components of the purified complexes solubilized from the membranes by appropriate detergent treatment. The picture that emerged from this work was that the four major photosynthetic complexes, PSII, PSI, the cytochrome b6f complex, and plastid ATP synthase, consist of both nucleus- and chloroplast-encoded subunits.
Thereafter, restriction enzymes became available and were used to establish the first physical maps of chloroplast genomes in the 1970s that revealed a circular structure with a large inverted repeat containing the rRNA genes (Bedbrook and Bogorad, 1976). Soon afterwards, the first chloroplast genes were cloned, and the first plastid protein gene to be sequenced was that of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Mc Intosh et al., 1980). The 1980s were characterized by an intensive study of chloroplast DNA that culminated in 1986 with the first complete sequences of the chloroplast genomes from tobacco (Nicotiana tabacum) and liverwort (Marchantia polymorpha) (Shinozaki and Sugiura, 1986). These sequences revealed that land plant chloroplast genomes contain close to 120 genes involved mostly in chloroplast protein synthesis and photosynthesis. Moreover, additional open reading frames of unknown function were identified. More than a dozen chloroplast genomes were sequenced subsequently. These studies revealed that, with few exceptions, the distribution of photosynthetic genes between chloroplast and nuclear genomes of vascular plants and green algae is conserved, although nongreen algae contain a larger set of chloroplast genes. Another important result arising from the sequencing of multiple plastid genomes was the evidence for a single primary endosymbiosis that gave rise to all eukaryotic algae and plants (Martin et al., 1998).
Once genes involved in photosynthesis had been identified, an important question was to elucidate their role. A spectacular technical feat was achieved when Boynton et al. (1988) established a robust chloroplast transformation system in Chlamydomonas. They used a particle gun for bombarding cells with transforming DNA bound to tungsten particles. Because of the highly active homologous recombination system in the chloroplast, specific gene disruptions and precise chloroplast gene surgery could be achieved. The chloroplast transformation system was later also developed for tobacco plants (Svab et al., 1990). This opened the door for chloroplast reverse genetics, for a structure-function analysis of photosynthetic proteins, and for extensive studies of chloroplast gene expression. This system was especially useful for elucidating the role of several conserved open reading frames of unknown function in the chloroplast genome (Rochaix, 2003). As an example, two open reading frames, ycf3 and ycf4, were shown to encode factors specifically required for the assembly of PSI. This analysis also led to the identification of novel subunits of the cytochrome b6f complex that had escaped biochemical detection and to the identification of a novel factor involved in cytochrome f maturation.
Besides plants and algae, cyanobacteria also provided important insights into the photosynthetic machinery. These prokaryotic organisms are capable of performing oxygenic photosynthesis, and their photosynthetic apparatus is very similar to that of plants and algae. The strain Synechocystis spp PC 6803 became the workhorse of cyanobacterial photosynthesis research after it was shown that it was transformable and that homologous recombination allows for easy gene replacements (Williams, 1988). Moreover, the determination of the complete sequence of the genome of this cyanobacterium provided invaluable help for establishing a catalog of the entire set of genes involved in oxygenic photosynthesis (Kaneko et al., 1996).
Whereas inactivation of specific chloroplast genes could be achieved readily, this proved more difficult with nuclear genes. Attempts to inactivate nuclear genes through homologous recombination met with little success both in Chlamydomonas and land plants. However, new tools for nuclear reverse genetics were developed. An RNA antisense approach was used successfully in Arabidopsis thaliana to study the role of several PSI subunits (Scheller et al., 2001). Later, RNA interference based on the use of double-stranded RNA was used to inactivate additional PSI genes. The sequencing of entire plant genomes also provided a new source of information on genes involved in photosynthesis. Moreover, a large effort was invested in generating large collections of T-DNA insertion lines of Arabidopsis (Alonso et al., 2003), which provided powerful tools for the identification and functional characterization of novel genes involved in photosynthesis.
BIOGENESIS OF THE PHOTOSYNTHETIC APPARATUS
The primary reactions of photosynthesis occur on the thylakoid membrane. The origin of this internal plastid membrane system has intrigued researchers for many years. Although its lipid composition is similar to that of the inner plastid envelope membrane, no convincing fusion between these two membranes could be detected. Vesicle budding from the inner envelope and accumulation of vesicles in the chloroplast stroma could be observed under special conditions, such as cold treatment of leaves or after mild heat shock of yellow mutants of Chlamydomonas that are unable to synthesize chlorophyll in the dark (Vothknecht et al., 2001). The role of these vesicles in thylakoid formation became clearer with the isolation of a mutant of Arabidopsis affected in a gene encoding vesicle inducing plastid protein (VIPP), in which loss of thylakoid formation correlated with the loss of vesicle formation. The VIPP1 gene is related to the bacterial phage shock gene that is responsible for membrane repair upon phage infection. This gene was apparently duplicated in cyanobacteria that still possess both the phage shock and the VIPP1 gene. Inactivation of VIPP1 in cyanobacteria also led to thylakoid deficiency. Vesicles could be induced by treatment of chloroplasts with microcystin, a known inhibitor of protein phosphatase I that is involved in vesicle fusion (Westphal et al., 2001). Although these studies have not yet revealed how VIPP1 functions and whether the cargo of these vesicles includes lipids, proteins, or both, they provide a new important entry site into thylakoid biogenesis.
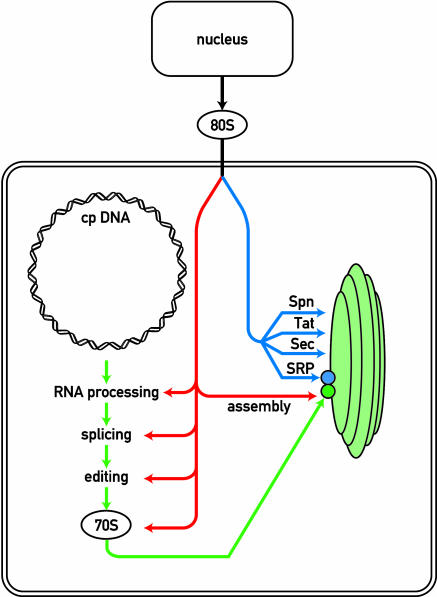
Photosynthetic complexes of land plants and algae consist of subunits encoded by chloroplast genes that are synthesized by plastid ribosomes and of subunits encoded by nuclear genes, translated on cytosolic ribosomes and specifically imported into the chloroplast. These proteins are synthesized as precursors containing an N-terminal transit peptide that is required for the entry into the chloroplasts and that is cleaved after import. Nucleus-encoded thylakoid polypeptides contain a bipartite transit peptide with the N-terminal part required for import into the chloroplast and the C-terminal part required for integration in or transport across the thylakoid membrane. Biochemical and genetic approaches revealed surprisingly that there are four different pathways for this process: spontaneous protein insertion, a pathway related to the bacterial Sec pathway that requires ATP and an electrochemical gradient, the signal recognition particle pathway, and the pH gradient–dependent pathway (see Robinson et al., 2001) (Figure 1).
Figure 1.
Biosynthesis of the Photosynthetic Apparatus by the Nucleocytosolic and Chloroplast Genetic Systems.
The red line refers to nucleus-encoded factors that are required for the different chloroplast posttranscriptional steps. The blue and green lines refer to nucleus and chloroplast-encoded proteins, respectively, that are part of the photosynthetic machinery. Thylakoids are indicated as green vesicles. The four pathways for protein insertion/translocation in the thylakoid membranes are indicated (SRP, Sec, Tat [ΔpH-dependent pathway], and Spn [spontaneous insertions]). 70S, chloroplast ribosomes; 80S, cytosolic ribosomes.
A major breakthrough occurred when the maize (Zea mays) mutant hcf106 was found to be deficient in the ΔpH pathway (Voelker and Barkan, 1995). This mutant lacks Hcf106, a protein that is conserved in prokaryotes and whose function was unknown at that time. The bacterial homolog, named TatB, is a product of the Tat operon that consists of the TatA, TatB (Hcf106), and TatC genes. Tat stands for twin Arg translocation because of a conserved twin Arg motif in the transit peptides, a hallmark of all proteins that use this pathway. Another maize mutant was found to be deficient in the TatA homolog. Further studies revealed that TatC is also involved in this pathway and that homologs of this protein exist in plants. Thus, a genetic approach in maize coupled with genomic analysis led to the rapid identification of the principal components of this new pathway. The ΔpH/Tat pathway appears to be an ancient protein translocation process that was formed before the endosymbiotic origin of plastids. A remarkable feature of this pathway is that it does not require ATP, that it relies only on the trans-membrane pH gradient, and that it is able to transport folded proteins while maintaining membrane impermeability (see Robinson et al., 2001).
After synthesis and proper targeting to the thylakoid membrane, the subunits of the photosynthetic complexes need to be coordinately inserted into the membrane and assembled into functional units together with numerous pigments and redox cofactors. How this cross talk between nuclear and chloroplast genomes is coordinated and how the different processes are controlled has remained a challenging problem. Genetic analysis of numerous mutants of Chlamydomonas, maize, and Arabidopsis revealed the existence of a surprisingly large set of nucleus-encoded factors that are involved in several posttranscriptional steps of chloroplast gene expression (Barkan and Goldschmidt-Clermont, 2000). Thus, mutants deficient in plastid RNA stability, in RNA processing, in RNA editing, splicing, translation initiation and elongation, in cofactor integration, and in the assembly of the photosynthetic complexes were isolated and characterized. A rather surprising observation made first in Chlamydomonas was that most of these nucleus-encoded factors act in a gene-specific way.
One of the first mutants of C. reinhardtii to be characterized at the molecular level, nac2, was found to be specifically deficient in the accumulation of the psbD mRNA (Boudreau et al., 2000). All other plastid mRNAs accumulate normally in this mutant. At least one target site of the mutant factor was localized to the 5′ untranslated region (UTR) of the psbD mRNA. The Nac2 gene was shown to encode a protein containing 10 tetratricopeptide repeat-like domains that are usually involved in protein–protein interactions. However, in this case, they appear to play a role in the recognition of the psbD 5′UTR (Boudreau et al., 2000). Interestingly, tetratricopeptide repeat domains resemble pentatricopeptide repeats (PPRs) that are found amongst members of a large family of nucleus-encoded proteins of Arabidopsis (Small and Peeters, 2000). That PPR domains are important for plastid RNA metabolism was shown by the isolation of a maize mutant deficient in petA translation and petD RNA processing. The gene deficient in this mutant encodes a PPR protein (Fisk et al., 1999). Recent results indicate that PPR domains play a role in RNA binding (Nakamura et al., 2003). Other studies revealed that some of the factors involved in plastid posttranscriptional steps are highly conserved in algae, land plants, and cyanobacteria and most likely in all oxygenic photosynthetic organisms. One example is the Tab2 protein of Chlamydomonas that binds specifically to the psaB 5′UTR and is involved in psaB mRNA translation (Dauvillee et al., 2003).
Molecular genetic approaches were especially rewarding for studying the splicing of chloroplast group II introns. Although these introns are catalytic RNAs, in most cases proteins are required for efficient splicing. Analysis of chloroplast splicing mutants of maize led to the identification of two nucleus-encoded factors, Crs1 and Crs2, that are involved in the splicing of chloroplast group II introns (Jenkins and Barkan, 2001). Crs2 is related to peptidyl-tRNA hydrolases and is part of distinct ribonucleoprotein complexes that include either the Caf1 or Caf2 protein (Ostheimer et al., 2003). Each of these complexes has distinct intron specificities. Moreover, it was found that Caf1 and Caf2 are members of a protein family that includes Crs1. They share repeated domains that were derived by amplification and diversification from an ancient RNA binding module found in prokaryotes. Analysis of mutants deficient in splicing of the two group II introns of the psaA gene of Chlamydomonas led to the identification of three splicing factors that are also part of large molecular weight complexes but unrelated to the maize chloroplast splicing factors. The psaA gene of Chlamydomonas was found to consist of three exons that are widely separated on the chloroplast genome and that are transcribed independently (Kück et al., 1987). Assembly of the mature psaA mRNA requires two trans-splicing reactions that splice together exons 1 and 2 and exons 2 and 3. At least 14 nucleus-encoded factors are required for these reactions. Moreover, the fragmentation is not limited to the exons but also concerns the first psaA intron that consists of at least three parts that are transcribed independently (Goldschmidt-Clermont et al., 1991). Thus, new insights into splicing, a basic biological process, resulted from the genetic analysis of photosynthesis in both land plants and Chlamydomonas.
Another rather surprising finding was RNA editing in land plant chloroplasts, a process in which some plastid transcripts are posttranscriptionally modified by C-to-U conversion (Tsudzuki et al., 2001). As many as 18 editing sites were identified in tobacco. The existence of RNA editing added a new twist in the identification of chloroplast open reading frames from DNA sequences because both initiation and termination codons can be created from sense codons by C-to-U editing.
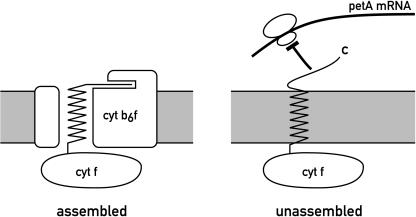
Given the complexity of photosynthetic complexes, the ordered assembly of their subunits into functional units in the thylakoid membrane raised many challenging questions, in particular on the regulatory mechanisms of synthesis of these subunits that ensures their stoichiometric accumulation. In this area, too, genetic approaches provided important insights. Analysis of numerous mutants deficient in the synthesis of a single core subunit of a complex showed that the remaining subunits are rapidly degraded. However, the study of the biosynthesis of the cytochrome b6f complex in mutants of Chlamydomonas deficient in one of the core subunits of this complex revealed more subtle regulatory mechanisms and gave rise to the control by epistasy of synthesis (CES) model (Choquet et al., 1998). In the absence of either cytochrome b6 or the PetD subunit (called dominant subunit), synthesis of cytochrome f (CES subunit) is strongly reduced, but the accumulated cytochrome f is stable. It was shown that the rate of translation of cytochrome f depends on the assembly state of the complex. In the unassembled state, the C-terminal end of cytochrome f inhibits the initiation of translation of its own mRNA most likely indirectly by interacting with a trans-acting factor (Figure 2). In the assembled state, this C-terminal end is shielded by the other subunits of the complex and, thus, unavailable for blocking translation. Further studies showed that the CES model may also be valid for the assembly of PSI (with PsaB and PsaA acting as the dominant and CES subunits, respectively) and PSII, where these roles are taken by D2 and D1, respectively.
Figure 2.
CES Model for Assembly of the Cytochrome b6f Complex.
The N-terminal lumenal domain (bottom part), the trans-membrane region, and the stromal C-terminal domain of cytochrome f (top part) are indicated. In the fully assembled state, the C-terminal domain of cytochrome f is shielded by the other subunits of the complex. In the unassembled state, this C-terminal end represses the initiation of translation of the petA mRNA of cytochrome f most likely indirectly through a trans-acting factor.
The complexity in the molecular design of the PSII antenna is quite remarkable. The light-harvesting antenna of PSII consists of the inner antenna of CP43/47, which forms the core complex together with the D1/D2 reaction center polypeptides, surrounded by a peripheral antenna that contains the Lhcb proteins as well as additional Lhcb-related proteins (Jansson, 1999). The six Lhcb proteins (Lhcb1-6) are arranged in two groups. Lhcb1-3 form the major Lhcb antenna complex consisting of trimers and binding 70% of the PSII chlorophyll, whereas Lhcb4-6 form the minor antenna complexes CP29, CP26, and CP24, which are monomeric and contain 10% of the chlorophyll in PSII. Each core dimer binds two copies of the minor antenna complexes and two to four light-harvesting complex II (LHCII) trimers to form a PSII supercomplex in the grana membranes of the chloroplast. During acclimation to changes in light quantity and quality, these supercomplexes undergo conformational changes and are reorganized.
ACCLIMATION OF THE PHOTOSYNTHETIC APPARATUS TO CHANGES IN LIGHT CONDITIONS: NONPHOTOCHEMICAL QUENCHING AND STATE TRANSITIONS
Approaches based on molecular biology, biochemistry, and crystallography provided a well-defined picture of the photosynthetic complexes. A major breakthrough was the determination of the atomic structure of the photosynthetic reaction center of Rhodopseudomonas viridis, which paved the way for the structural and functional analysis of the other complexes involved in photosynthesis (Deisenhofer et al., 1984). However, the picture that emerged from these studies was mostly static and did not directly address the remarkable dynamic properties of the photosynthetic apparatus.
A particularly interesting feature of the photosynthetic membrane is its ability to adapt to changes in light quantity and quality. Although light is essential for photosynthesis, too much light can cause serious damage to the reaction centers and other cellular constituents through overreduction of the electron carriers and the production of reactive oxygen species. Upon light absorption by the antennae, the excitation energy can be dissipated in several ways (Krause and Weis, 1992). It can be reemitted as chlorophyll fluorescence or it can be transferred to the photochemical reaction center or dissipated as heat in the antennae complexes. Because fluorescence is decreased in both of the latter processes, they are referred to as photochemical quenching and nonphotochemical quenching (NPQ), respectively. NPQ is especially important because it allows plants and algae to dissipate excess excitation energy when the capacity of the photosynthetic system is saturated (Demmig-Adams and Adams, 1992). In this way, damage to the photosynthetic apparatus is prevented. In the absence of NPQ, excess excitation energy in the antennae can cause the production of triplet state chlorophyll molecules that interact with molecular oxygen to produce highly reactive singlet oxygen.
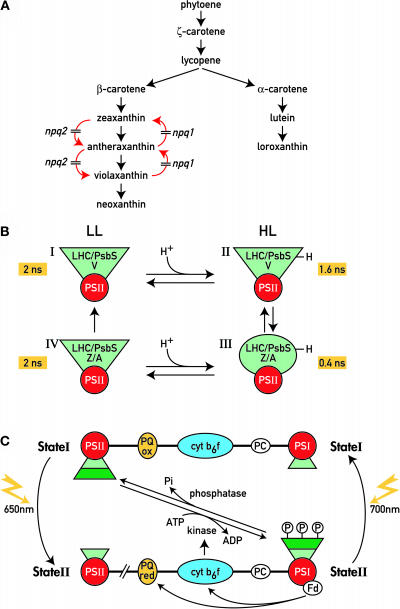
NPQ has at least three different components based on their relaxation kinetics in darkness after a period of illumination. The major component, qE, the energy-dependent component, requires a proton gradient across the thylakoid membrane and relaxes within minutes to seconds. The second component, qT, relaxes within minutes and is a result of state transition (see below). The third component, qI, is a result of photoinhibition and relaxes very slowly. NPQ (qE) occurs upon acidification of the thylakoid lumen as a result of excessive illumination. The elevated proton concentration in the thylakoid lumen leads to the activation of the violaxanthin deepoxidase that converts violaxanthin first to antheraxanthin and then to zeaxanthin. These xanthophylls are bound to LHC polypeptides and participate in a cycle that involves their reversible deepoxidation and epoxidation (Demmig-Adams and Adams, 1992) (Figure 3A). The increased pH gradient also results in protonation of acidic amino acid side chains of specific polypeptides associated with PSII. These processes appear to cause a conformational change in the thylakoid membrane that can be monitored by absorbance changes at 535 nm (Horton et al., 2000) and that lead to the dissipation of excess energy through heat. These conformational changes could also be deduced from measurements of chlorophyll a fluorescence lifetime distributions (Gilmore, 1997). A lifetime shift from 2 to 1.6 ns was induced by the trans-thylakoid pH gradient alone. Upon subsequent binding of zeaxanthin, the 1.6-ns component was replaced by a fluorescence lifetime component of 0.4 ns. The amount of this component is directly related to qE.
Figure 3.
NPQ and State Transition.
(A) Xanthophyll biosynthetic pathway. The reactions of the xanthophyll cycle are marked with red arrows. The reactions that are affected in the npq1 and npq2 mutants are indicated.
(B) Model for NPQ (adapted from Muller et al., 2001). I to IV correspond to the different PSII fluorescence lifetime states. I. In low light (LL), no quenching occurs. II. In high light (HL), protonation of PsbS and other LHC proteins occurs accompanied by a change in the fluorescence lifetime to 1.6 ns. III. Binding of zeaxanthin and protons leads to the formation of a quenching complex with altered conformation detectable as an absorbance change at 535 nm and with a faster 0.4-ns fluorescence lifetime component. IV. Upon a shift from high to low light, deprotonation occurs faster than epoxydation of zeaxanthin to violaxanthin. Fluorescence lifetimes are highlighted. V, violaxanthin; A, antheraxanthin; Z, zeaxanthin.
(C) State transitions. In state I, the plastoquinone pool is oxidized, and the mobile part of LHCII (dark green) is associated with PSII. After excitation with 650-nm light that is preferentially absorbed by LHCII, a transition to state II occurs: the plastoquinone pool (PQ) is reduced and activation of the LHCII kinase occurs through the cytochrome b6f complex leading to the phosphorylation of LHCII. The phosphorylated mobile part of LHCII is displaced from PSII and reconnected to PSI. Excitation with 700-nm light preferentially absorbed by LHCI oxidizes the plastoquinone pool, triggering a transition from state II to state I. The LHCII kinase is inactivated, and a phosphatase dephosphorylates LHCII, which moves back to PSII. In Chlamydomonas, a switch from linear to cyclic electron flow occurs upon transition from state I to state II.
A genetic approach using a digital video imaging system led to the isolation and characterization of several npq mutants of Chlamydomonas and Arabidopsis (Niyogi et al., 1997a, 1997b). Some of these mutants, npq1 and npq2, were found to be deficient in the xanthophyll cycle, thus confirming the involvement of this pathway in NPQ (Figure 3A). However, closer analysis of these mutants revealed that the xanthophyll cycle does not account for all ΔpH-dependent NPQ. Moreover, mutants deficient in lutein and loroxanthin synthesis (α-branch) were also found to be partly deficient in NPQ, whereas double mutants deficient in the synthesis of antheraxanthin and zeaxanthin (β-branch) and in the α-branch were found to be nearly completely deficient in NPQ and highly sensitive to light. The most interesting mutants were those not visibly affected in photosynthesis or in the xanthophyll cycle (i.e., those that were normal in high light-induced deepoxidation of violaxanthin). The gene affected in one of these Arabidopsis mutants, PsbS, encodes a protein associated with PSII (Li et al., 2000). PsbS belongs to the superfamiliy of LHC proteins, although it contains four rather than three trans-membrane domains, and the protein has a symmetrical topology. Parts of the protein exposed to the thylakoid lumen are acidic and could be involved in proton binding. Indeed, mutagenesis of two lumen-exposed glutamate residues of PsbS abolished the rapidly inducible NPQ and the characteristic leaf absorbance change at 535 nm, and the chlorophyll fluorescence lifetimes were altered, in particular the 0.4-ns component was absent (Li et al., 2004). Moreover, binding of dicyclohexylcarbodiimide to the wild type occurs, but not to the mutant PsbS, indicating that PsbS is the target of this qE inhibitor that reacts with proton active residues. It was also shown that zeaxanthin binds to PsbS.
Based on these data, a model was proposed in which PsbS acts as a sensor of excess light (Muller et al., 2001) (Figure 3B). Under low light conditions the excitation energy is transferred from the PSII antenna to the reaction center, and qE is not induced. Upon exposure to high light, proteins in the antenna, in particular PsbS, become protonated, causing a change in the fluorescence halftime form 2 to 1.6 ns. Protonation of the two glutamate residues of PsbS is followed by binding of two zeaxanthin molecules. This leads to a conformational change within the PSII antenna and to the formation of a quenching complex with a further decrease in the fluorescence lifetime to 0.4 ns. A shift to low light is expected to lead to rapid deprotonation of the antenna proteins and to the disassembly of the quenching complex, which is associated with a return of the fluorescence lifetime to 2 ns. A role for zeaxanthin as direct quencher of excess excitation energy was provided recently through ultrafast spectroscopy and by taking advantage of various npq mutants (Ma et al., 2003).
Analysis of another Chlamydomonas mutant, npq5, revealed that it was deficient in Lhcbm1, a representative of the family of the LHCII polypeptides that is present in the trimers of the PSII antenna (Elrad et al., 2002). The analysis of this mutant suggests that the peripherally associated trimeric LHC polypeptides play an important role in NPQ. These results are particularly important because they seriously question the prevailing view that all LHCs perform similar functions. Rather, they point to the fact that although the major LHCIIs have nearly identical sequences, they can perform distinct functions. Whether this occurs through processing or covalent or noncovalent modifications remains to be determined and constitutes a challenging problem for future studies. These examples illustrate the power of a forward genetic approach for the dissection of photosynthetic processes. It would have been extremely difficult to identify these proteins as important players in NPQ by purely biochemical means or through an approach based uniquely on reverse genetics. It is interesting to compare these results with those obtained with Arabidopsis antisense plants in which Lhcb1 and Lhcb2, the most abundant LHCII proteins, are absent (Andersson et al., 2003). Although NPQ is reduced to some degree in these plants, the relative reduction is not as pronounced as in the Chlamydomonas Lhcbm1 mutant. These results point to differences in NPQ between Chlamydomonas and plants.
In oxygenic photosynthesis, the two reaction centers of PSII and PSI are connected to their antennae and are linked in series through the electron transport chain. Because these antennae have a different pigment composition and, therefore, different light absorption properties, excitation of the two photosystems can be unbalanced depending on the light conditions. However, the balance can be restored through a process called state transition (Allen, 1992). This leads to a reorganization of the antennae so that the two photosystems are excited to the same extent, thereby increasing the photosynthetic yield especially under low light (Figure 3C). This process was discovered independently by Bonaventura and Myers (1969) and by Murata (1969). They showed that upon illumination of photosynthetic unicellular organisms with 650-nm light that is absorbed preferentially by PSII, PSI becomes sensitized within a few minutes so that it receives more light excitation energy, whereas illumination with 700-nm light that is absorbed preferentially by PSI sensitizes PSII (Figure 3C). The meaning of this sensitization became clearer when it was found that the two photosystems are distributed unequally on the thylakoid membrane with PSII localized in the appressed grana regions and PSI in the unappressed regions (Andersson and Anderson, 1980). This meant that the light excitation energy had to be redistributed over several hundreds of nanometers on the thylakoid membrane. The next step forward was made by Bennett and Allen in the early 1980s. They showed that reduction of the plastoquinone pool through overexcitation of PSII relative to PSI leads to the activation of a protein kinase that specifically phosphorylates the light-harvesting proteins that are subsequently laterally displaced in the thylakoid membrane and connected to PSI (Allen et al., 1981). This provided the framework of the current model of state transition and triggered a biochemical hunt for the kinase.
Although several kinase activities associated with the thylakoid membrane were found, the LHCII kinase remained elusive for many years. Kohorn developed an elegant screen searching for proteins that interact with the N-terminal end of the light-harvesting proteins known to contain the target Thr that is phosphorylated during a transition from state I to state II (Snyders and Kohorn, 1999). This screen identified a kinase of Arabidopsis that was associated with the cell wall rather than with the thylakoid membrane. Antiserum raised against this kinase was then used to isolate another kinase associated with the thylakoid membrane called thylakoid-associated kinase (TAK), which is part of a small family of kinases that bear some resemblance to the human TGF β1 receptor. TAK activity was shown to be enhanced by reductant and to be associated with PSII and the cytochrome b6f complex. Antisense TAK lines were found to be deficient in state transition and light sensitive (Snyders and Kohorn, 2001). However, no homolog of TAK could be found amongst the ESTs and in the nuclear genome sequence of Chlamydomonas. Screens for mutants deficient in state transition were performed in this alga (Fleischmann et al., 1999; Kruse et al., 1999). Because transition from state I to state II is accompanied by a large fluorescence decrease, mutants can simply be screened for the loss of this fluorescence change using a fluorescence video imaging system. Amongst several mutants isolated independently, two were found to lack a novel Ser-Thr protein kinase, called Stt7, that is localized in the thylakoid membrane (Depège et al., 2002). This kinase is nucleus encoded and produced as a precursor protein with a characteristic transit peptide. It is firmly associated with the thylakoid membrane and contains a putative trans-membrane domain. Interestingly this kinase is conserved in land plants. Arabidopsis mutants deficient in this kinase are also deficient in state transition and unable to phosphorylate the light-harvesting system (S. Bellafiore and J.D. Rochaix, unpublished results). These data raise the question on the relationship between the TAK and Stt7 kinases and whether a kinase cascade is involved in the phosphorylation of LHCII.
A further important discovery was that in the absence of the cytochrome b6f complex, algae and plants are blocked in state I, and the kinase is no longer activated even under conditions in which the plastoquinone pool is fully reduced (Wollman, 2001). Moreover, it was shown that it is not the redox state of the plastoquinone pool per se that is critical for the activation of the kinase, but the occupancy of the QO site by plastoquinol on the luminal side of the cytochrome b6f complex (Zito et al., 1999). Further insights into how the cytochrome b6f complex activates the kinase came from the spectacular progress of the three-dimensional crystal structure of the mitochondrial bc1 (Zhang et al., 1998) and more recently of the closely related chloroplast cytochrome b6f complex (Kurisu et al., 2003; Stroebel et al., 2003). A highly interesting finding was that the position of the Rieske protein, a key subunit of the complex, changes depending on whether the Qo site is occupied by stigmatellin, an inhibitor of the cytochrome bc1 complex. Further analysis of two-dimensional crystals of the cytochrome b6f complex confirmed the changing position of the Rieske protein and showed that it involves a reorganization of the trans-membrane portion of the complex. These dynamics of the cytochrome b6f complex could play an important role in the activation of the kinase (Wollman, 2001).
During transition from state I to state II, the mobile phosphorylated fraction of LHCII is displaced from PSII in the appressed region to PSI in the nonappressed region. How this is achieved is still unclear, although several explanations have been proposed, such as electrostatic repulsion between the phosphorylated LHCII and the negative charges on the stromal grana surface or the light-induced conformational changes of LHCII (Allen, 1992). Moreover, the reversible phosphorylation of several PSII subunits could also play a role. Two important observations led to a model in which the association of phosphorylated LHCII with PSI and PSII is a competitive process. First, LHCII is still phosphorylated under state II conditions in PSI-deficient mutants, and it stays connected with PSII (Delosme et al., 1996). Second, in mutants of Arabidopsis deficient in the PSI subunit PsaH, LHCII is phosphorylated under state II conditions but remains associated with PSII, suggesting that PsaH is part of a high affinity binding site for phosphorylated LHCII (Lunde et al., 2000). It is interesting that the recently established plant PSI-LHCI crystal structure reveals that PsaH is on the side of PSI that lacks LHCI and that would thus be available for docking of LHCII during a state I to state II transition (Ben-Shem et al., 2003).
Although state transition was originally defined as a process that balances the size of the LHCII and LHCI antennae in response to changes in the spectral quality of light so as to optimize the photosynthetic yield, in Chlamydomonas, it is also involved in the cellular response to reduced levels of ATP. Chlamydomonas cells deficient in ATP undergo a state I to state II transition in darkness (Bulte et al., 1990). In this case, reducing power arising from glycolysis is shuttled through the chlororespiratory chain into the plastoquinone pool and thereby activates the LHCII kinase and promotes transition to state II. It is interesting to link this finding with the observation that state transition involves a switch from linear electron flow under state I conditions to cyclic electron flow under state II conditions. This was shown by Finazzi et al. (2002), who observed that under state II conditions, the PSII-specific inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea no longer prevents the rereduction of cytochrome f. To rule out the possibility that this effect is mediated through increased influx of reducing power from the chlororespiratory chain, the stt7 kinase-deficient mutant that is blocked in state I was examined. Under state II conditions, 3-(3,4-dichlorophenyl)-1,1-dimethylurea still blocks the rereduction of cytochrome f in this mutant, indicating that remodeling of the electron transfer chain to a cyclic mode has occurred in state II in wild-type cells (Finazzi et al., 2002). In this respect, it is interesting that a change in distribution between appressed and nonappressed regions also occurs for the cytochrome b6f complex, although to a lesser extent than for LHCII. Moreover, this displacement no longer occurs in mutants blocked in state I.
The picture that emerges from these studies is that the electron transfer chain is a highly dynamic system that operates either in a linear mode generating ATP and NADPH or in a cyclic mode as ATP generator (Wollman, 2001). The cytochrome b6f complex plays a central role in this process because it is involved in the activation of the LHCII kinase. The recently determined crystal structure of this complex at 3 Å resolution identified a novel haem that had escaped biochemical detection (Stroebel et al., 2003). This haem is localized near the stromal face in the Qi site where plastoquinone is reduced through the low potential bL and bH chain. Because this haem is accessible from the stromal side, it has been postulated that it could participate in cyclic electron flow. This would lend further support to the proposal that electrons are reinjected directly through the cytochrome b6f complex. Genetic studies in Arabidopsis identified a novel stromal potein, Pgr 5, that is essential for cyclic electron flow (Munekage et al., 2002). Thus, several pieces of the cyclic electron flow puzzle, one of the last mysteries of photosynthesis, are gradually falling into place.
CONCLUSIONS AND PERSPECTIVES
A rather remarkable feature of photosynthetic processes is that because of the unique composition of the photosynthetic complexes, they can be studied over a wide range of time domains. These range from femtosecond during exciton transfer processes in the antennae, from picosecond to microsecond during the various electron transfers in the reaction centers, from milliseconds to seconds during enzymatic turnovers, from minutes during short term adaptations such as NPQ and state transition, from hours to days for thylakoid differentiation, and from months to years for plant development to millennia for the evolution of the photosynthetic apparatus. Combining spectroscopic, biochemical, molecular genetic, crystallographic, and physiological approaches has proven to be highly successful for elucidating the photosynthetic processes over these vast time domains. The built-in pigments and radicals formed during the photosynthetic reactions provide specific probes for following these reactions and can be studied even in vivo in the nanosecond range through newly developed and sensitive spectroscopic methods (Joliot and Joliot, 1999).
More than two hundred years of photosynthetic research have led to a comprehensive picture of the four major complexes involved in photosynthesis: PSII, PSI and their associated antennae, the cytochrome b6f complex, and the ATP synthase. Each of these complexes has been characterized in great depth biochemically and genetically; more recently, impressive advances in the crystallization of membrane complexes have yielded high-resolution structures of these complexes and provided new insights into their function.
Molecular genetic and biochemical studies have revealed that the biogenesis of the photosynthetic apparatus is a complicated process involving the participation of two distinct genetic systems that cooperate closely with each other in the coordinated assembly of the photosynthetic complexes. Although these studies have provided a wealth of information on the components of the biosynthetic machinery and on how some of these factors operate, we still know little about how a functional complex is assembled in the thylakoid membrane together with its numerous pigment and redox cofactors. The photosynthetic system is a dynamic machinery that can adapt to a wide range of conditions by controlling the synthesis and turnover of some of its components and by remodeling the electron transport chain. This flexibility is particularly important for plants that are sessile organisms. Now, at the start of the 21st century, research on photosynthesis is ready to tackle the novel challenging task of understanding the molecular mechanisms that underlie the multiple dynamic responses of the photosynthetic system to changes in environmental conditions.
Acknowledgments
I thank M. Goldschmidt-Clermont for helpful comments and N. Roggli for preparing the figures. Work in the author's laboratory was supported by grants from the Swiss National Foundation.
References
- Allen, J.F. (1992). Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta 1098, 275–335. [DOI] [PubMed] [Google Scholar]
- Allen, J.F., Bennett, J., Steinback, K.E., and Arntzen, C.J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291, 25–29. [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Andersson, B., and Anderson, J.M. (1980). Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta 593, 427–440. [DOI] [PubMed] [Google Scholar]
- Andersson, J., Wentworth, M., Walters, R.G., Howard, C.A., Ruban, A.V., Horton, P., and Jansson, S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II: Effects on photosynthesis, grana stacking and fitness. Plant J. 35, 350–361. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bedbrook, J.R., and Bogorad, L. (1976). Endonuclease restriction sites mapped on Zea mays chloroplast DNA. Proc. Natl. Acad. Sci. USA 73, 4309–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem, A., Frolow, F., and Nelson, N. (2003). Crystal structure of plant photosystem I. Nature 426, 630–635. [DOI] [PubMed] [Google Scholar]
- Bonaventura, C., and Myers, J. (1969). Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta 189, 366–383. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.L., Ossenbühl, F., and Rochaix, J.-D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton, J.E., Gillham, N.W., Harris, E.H., Hosler, J.P., Johnson, A.M., Jones, A.R., Randolph-Anderson, B.L., Robertson, D., Klein, T.M., Shark, K.B., and Sanford, J. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240, 1534–1538. [DOI] [PubMed] [Google Scholar]
- Bulte, L., Gans, P., Rebeille, F., and Wollman, F.A. (1990). ATP control on state transitions in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1020, 72–80. [Google Scholar]
- Chan, P.H., and Wildman, S.G. (1972). Chloroplast DNA codes for the primary structure of the large subunit of fraction I protein. Biochim. Biophys. Acta 277, 677–680. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., Stern, D.B., Wostrikoff, K., Kuras, R., Girard-Bascou, J., and Wollman, F.A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95, 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, N.H., and Bennoun, P. (1975). Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc. Natl. Acad. Sci. USA 72, 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, N.H., and Gillham, N.W. (1977). The sites of synthesis of the principal thylakoid membrane polypeptides in Chlamydomonas reinhardtii. J. Cell Biol. 74, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillee, D., Stampacchia, O., Girard-Bascou, J., and Rochaix, J.D. (2003). Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 22, 6378–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer, J., Epp, O., Miki, K., Huber, R., and Michel, H. (1984). X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J. Mol. Biol. 180, 385–398. [DOI] [PubMed] [Google Scholar]
- Delosme, R., Olive, J., and Wollman, F.A. (1996). Changes in light energy distribution upon state transitions: An in vivo photoacoustic study of the wild type and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1273, 150–158. [Google Scholar]
- Demmig-Adams, B., and Adams, I.W.W. (1992). Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 599–626. [Google Scholar]
- Depège, N., Bellafiore, S., and Rochaix, J.D. (2002). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and sate transition in Chlamydomonas. Science 299, 1572–1575. [DOI] [PubMed] [Google Scholar]
- Elrad, D., Niyogi, K.K., and Grossman, A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi, G., Rappaport, F., Furia, A., Fleischmann, M., Rochaix, J.D., Zito, F., and Forti, G. (2002). Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep. 3, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk, D.G., Walker, M.B., and Barkan, A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann, M.M., Ravanel, S., Delosme, R., Olive, J., Zito, F., Wollman, F.A., and Rochaix, J.D. (1999). Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J. Biol. Chem. 274, 30987–30994. [DOI] [PubMed] [Google Scholar]
- Gilmore, A.M. (1997). Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leafs. Physiol. Plant 99, 197–209. [Google Scholar]
- Goldschmidt-Clermont, M., Choquet, Y., Girard-Bascou, J., Michel, F., Schirmer-Rahire, M., and Rochaix, J.D. (1991). A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65, 135–143. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Wentworth, M. (2000). Allosteric regulation of the light-harvesting system of photosystem II. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jenkins, B.D., and Barkan, A. (2001). Recruitment of a peptidyl-tRNA hydrolase of group II intron splicing in chloroplasts. EMBO J. 20, 872–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot, P., and Joliot, A. (1999). In vivo analysis of the electron transfer within photosystem I: Are the two phylloquinones involved? Biochemistry 38, 11130–11136. [DOI] [PubMed] [Google Scholar]
- Kaneko, T., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. [DOI] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 313–349. [Google Scholar]
- Kruse, O., Nixon, P.J., Schmid, G.H., and Mullineaux, C.W. (1999). Isolation of stae transition mutants of Chlamydomonas reinhardtii by fluorescence video imaging. Photosyn. Res. 61, 43–51. [Google Scholar]
- Kück, U., Choquet, Y., Schneider, M., Dron, M., and Bennoun, P. (1987). Structural and transcriptional analysis of two homologous genes for the P700 chlorophyll a apoproteins in Chlamydomonas reinhardtii: Evidence for trans-splicing. EMBO J. 6, 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu, G., Zhang, H., Smith, J.L., and Cramer, W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 302, 1009–1014. [DOI] [PubMed] [Google Scholar]
- Levine, R.P. (1968). Genetic dissection of photosynthesis. Science 162, 768–771. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Bjorkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Gilmore, A.M., Caffarri, S., Bassi, R., Golan, T., Kramer, D., and Niyogi, K.K. (2004). Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279, 22866–22874. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., and Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408, 613–615. [DOI] [PubMed] [Google Scholar]
- Ma, Y.Z., Holt, N.E., Li, X.P., Niyogi, K.K., and Fleming, G.R. (2003). Evidence for direct carotenoid involvement in the regulation of photosynthetic light harvesting. Proc. Natl. Acad. Sci. USA 100, 4377–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hapsmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- McIntosh, L., Poulsen, C., and Bogorad, L. (1980). Chloroplast gene sequence for the large subunit of ribulose bisphosphate carboxylase of maize. Nature 288, 556–560. [Google Scholar]
- Muller, P., Li, X.P., and Niyogi, K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M., and Shikanai, T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110, 361–371. [DOI] [PubMed] [Google Scholar]
- Murata, N. (1969). Control of excitation transfer in photosynthesis. I. Light-induced change of chlorophyll a fluorescence in Porphyridium cruentum. Biochim. Biophys. Acta 172, 242–251. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Meierhoff, K., Westhoff, P., and Schuster, G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Bjorkman, O., and Grossman, A.R. (1997. a). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi, K.K., Bjorkman, O., and Grossman, A.R. (1997. b). The roles of specific xanthophylls in photoprotection. Proc. Natl. Acad. Sci. USA 94, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G.J., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., and Barkan, A. (2003). Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 22, 3919–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris, H., and Plaut, W. (1962). Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 13, 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C., Thompson, S.J., and Woolhead, C. (2001). Multiple pathways used for the targeting of thylakoid proteins in chloroplasts. Traffic 2, 245–251. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.D. (2003). Functional analysis of plastid genes through chloroplast reverse genetics in Chlamydomonas. In Photosynthesis in Algae, A.W.D. Larkum, S. Douglas, and J.A. Raven, eds (Dordrecht/Boston/London: Kluwer Academic Publishers), pp. 83–94.
- Scheller, H.V., Jensen, P.E., Haldrup, A., Lunde, C., and Knoetzel, J. (2001). Role of subunits in eukaryotic photosystem I. Biochim. Biophys. Acta 1507, 41–60. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Sugiura, M. (1986). Organization of chloroplast genomes. Adv. Biophys. 21, 57–78. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Snyders, S., and Kohorn, B.D. (1999). TAKs, thylakoid membrane protein kinases associated with energy transduction. J. Biol. Chem. 274, 9137–9140. [DOI] [PubMed] [Google Scholar]
- Snyders, S., and Kohorn, B.D. (2001). Disruption of thylakoid-associated kinase 1 leads to alteration of light harvesting in Arabidopsis. J. Biol. Chem. 276, 32169–32176. [DOI] [PubMed] [Google Scholar]
- Stroebel, D., Choquet, Y., Popot, J.L., and Picot, D. (2003). An atypical haem in the cytochrome b(6)f complex. Nature 426, 413–418. [DOI] [PubMed] [Google Scholar]
- Svab, Z., Hajdukiewicz, P., and Maliga, P. (1990). Stable tranformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 87, 8526–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki, T., Wakasugi, T., and Sugiura, M. (2001). Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 53, 327–332. [DOI] [PubMed] [Google Scholar]
- Voelker, R., and Barkan, A. (1995). Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 14, 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht, U.C., Westhoff, P., Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., and Soll, J. (2001). Biogenesis and origin of thylakoid membranes. Biochim. Biophys. Acta 1541, 91–101. [DOI] [PubMed] [Google Scholar]
- Westphal, S., Soll, J., Vothknecht, U.C., and Heins, L. (2001). A vesicle transport system inside chloroplasts. FEBS Lett. 506, 257–261. [DOI] [PubMed] [Google Scholar]
- Williams, J.G.K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Meth. Enzymol. 167, 766–778. [Google Scholar]
- Wollman, F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20, 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Huang, L., Shulmeister, V.M., Chi, Y.I., Kim, K.K., Hung, L.W., Crofts, A.R., Berry, E.A., and Kim, S.H. (1998). Electron transfer by domain movement in cytochrome bc1. Nature 392, 677–684. [DOI] [PubMed] [Google Scholar]
- Zito, F., Finazzi, G., Delosme, R., Nitschke, W., Picot, D., and Wollman, F.A. (1999). The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 18, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]