Abstract
It is often anticipated that many of today's diploid plant species are in fact paleopolyploids. Given that an ancient large-scale duplication will result in an excess of relatively old duplicated genes with similar ages, we analyzed the timing of duplication of pairs of paralogous genes in 14 model plant species. Using EST contigs (unigenes), we identified pairs of paralogous genes in each species and used the level of synonymous nucleotide substitution to estimate the relative ages of gene duplication. For nine of the investigated species (wheat [Triticum aestivum], maize [Zea mays], tetraploid cotton [Gossypium hirsutum], diploid cotton [G. arboretum], tomato [Lycopersicon esculentum], potato [Solanum tuberosum], soybean [Glycine max], barrel medic [Medicago truncatula], and Arabidopsis thaliana), the age distributions of duplicated genes contain peaks corresponding to short evolutionary periods during which large numbers of duplicated genes were accumulated. Large-scale duplications (polyploidy or aneuploidy) are strongly suspected to be the cause of these temporal peaks of gene duplication. However, the unusual age profile of tandem gene duplications in Arabidopsis indicates that other scenarios, such as variation in the rate at which duplicated genes are deleted, must also be considered.
INTRODUCTION
Genome duplication (polyploidy) is common in flowering plants (Wendel, 2000). Estimates for the incidence of polyploidy in angiosperms vary from 30 to 80%, and 2 to 4% of speciation events can be attributed to genome duplications (Otto and Whitton, 2000). It is therefore likely that many if not all plant species have had at least one polyploid ancestor at some point during their evolution. However, an ancient polyploidy is difficult to detect because time erases the traces of duplication. The long-term evolution of polyploids is generally associated with extensive genome reorganization both at the gene and chromosome level (Wolfe, 2001). After genome duplication, an antagonistic process of gene loss eliminates a large fraction of the duplicate genes (Lynch and Conery, 2000, 2003), counteracting this expansion of genetic information. For example, after several tens of millions of years of evolution since their respective polyploidizations, the Arabidopsis thaliana and Saccharomyces cerevisiae genomes show that ∼70 to 90% of duplicated genes formed by genome duplication have returned to a single copy state (Seoighe and Wolfe, 1999; Arabidopsis Genome Initiative, 2000; Wong et al., 2002; Blanc et al., 2003). Furthermore, genomic rearrangements split up and relocate pieces of duplicated chromosomes around the genome, which further scrambles the intragenomic synteny. This therefore restricts the timeframe in which polyploidy events can be inferred from cytological or mapping approaches (Otto and Whitton, 2000).
Up to now, only sequence analyses performed on a genome scale (in yeast and Arabidopsis) have provided relatively strong evidence for ancient polyploidies (Wolfe and Shields, 1997; Arabidopsis Genome Initiative, 2000; Blanc et al., 2000, 2003; Paterson et al., 2000; Vision et al., 2000; Simillion et al., 2002; Wong et al., 2002; Bowers et al., 2003). However, because of the relatively large sizes of most plant genomes, Arabidopsis and rice (Oryza sativa) are the only (nearly) fully sequenced plant species (Arabidopsis Genome Initiative, 2000; Buell, 2002). This has prevented systematic investigations of when and which plant lineages sustained ancient polyploidy events. Partial sequencing of randomly chosen cDNA clones (ESTs) has emerged as an alternative to explore the gene content of complex genomes (Adams et al., 1991). Although ESTs are relatively short sequences that are error prone and often highly redundant, they can reveal a substantial portion of the expressed genes of a genome. Moreover, several bioinformatic algorithms have been developed to assemble overlapping EST into contigs (see Parkinson et al., 2002 and references therein) and derive consensus sequences for each transcript. This data condensation step outputs a set of longer and nonredundant transcribed sequences (referred to as unigenes) with higher overall quality, which provides a preliminary base for the identification of gene families in a species (Van der Hoeven et al., 2002) and the analysis of gene duplications. Here, we used intraspecies comparisons of unigenes to study the timing of gene duplications in species whose genomes have not been sequenced.
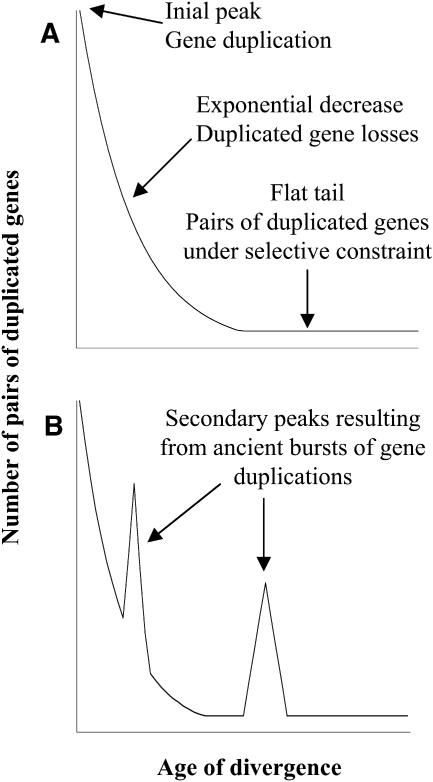
The level of divergence between homologous nucleotide sequences is widely used as a molecular clock to estimate the relative age of their separation. Nucleotide substitutions in protein-coding sequences can either result in amino acid change (nonsynonymous substitutions) or not (synonymous substitutions). Because natural selection acts mainly on protein sequences, synonymous codon positions are probably largely free from selection and so accumulate changes in a neutral manner, at a rate similar to the mutation rate. Thus, it is generally assumed that the level of synonymous substitutions (Ks) between two homologous sequences increases approximately linearly with time, at least for relatively low levels of sequence divergence before saturation with multiple substitutions becomes an issue (Li, 1997). Pairs of duplicated (paralogous) sequences found in a genome can be sorted in order of their relative ages of duplication by estimating Ks for each pair (Lynch and Conery, 2000, 2003). This type of representation gives an overview of the pattern of accumulation of duplicates during the evolution of a lineage, which is mainly influenced by gene duplication and loss processes. If we assume that gene duplications and gene deletions are random and have relatively steady rates during the course of evolution, such a distribution is expected to show an L shape (Figure 1A). Because gene duplications occur in the present, an initial peak of density of duplicates must be contained within the youngest age classes. The elimination of duplicated sequences results in an exponential decrease of density along with increasing age (Lynch and Conery, 2000, 2003). Finally, a long and nearly flat tail is expected to account for those pairs of older duplicates that escaped this tragic fate, with both copies evolving under selective constraints (Prince and Pickett, 2002).
Figure 1.
Theoretical Age Distributions of Pairs of Duplicated Genes in a Genome.
(A) Age distribution of pairs of paralogs expected under constant rates of gene duplication and duplicated gene deletion. Three main phases can be outlined. An initial peak accounts for the most recently duplicated genes. The distribution then drops off after an exponential decrease because of the deletion of duplicated genes that are not under selective constraints. A long tail corresponds to older pairs of duplicates, where both genes evolve under selective constraints.
(B) Age distribution of pairs of paralogs expected for a species that sustained two ancient large-scale duplication events. The overrepresentation of duplicated genes at periods corresponding to the large-scale duplication events gives rise to two secondary peaks.
However, large-scale duplication events, such as segmental duplication, aneuploidy, or polyploidy, lead to a punctuated, dramatic increase in the number of duplicated genes. The resulting excess of pairs of paralogs of a particular age is expected to give rise to a secondary peak in the distribution (Figure 1B). Lynch and Conery (2000, 2003) analyzed the frequency distributions of pairs of paralogs in several eukaryotic genomes as a function of the time of duplication estimated by their level of synonymous substitutions. For Arabidopsis, the distribution displayed a secondary peak of high density for relatively old pairs of paralogs, which was proposed to reflect an ancient polyploidy event. This was confirmed later by analyses of the genomic sequences (Simillion et al., 2002; Zhang et al., 2002; Blanc et al., 2003; Bowers et al., 2003).
A large body of data generated by EST and genome sequencing projects is now available for various species of angiosperms. We took this opportunity to identify gene families and investigate the timing of the underlying gene duplication events in 14 model plant species. Nine of them show clear temporary increases in the accumulation of duplicated genes during a period of their evolution. The possible interpretations are discussed.
RESULTS
Identification of Pairs of Paralogous Transcribed Sequences
We downloaded sets of unigene sequences available for 10 model plants (barrel medic [Medicago truncatula], soybean [Glycine max], tomato [Lycopersicon esculentum], potato [Solanum tuberosum], sunflower [Helianthus annuus], lettuce [Lactuca sativa], ice plant [Mesembryanthemum crystallinum], wheat [Triticum aestivum], maize [Zea mays], and barley [Hordeum vulgare]) from The Institute for Genomic Research (TIGR) gene indices database (Quackenbush et al., 2000). Each dataset consists of consensus sequences from transcript (EST and full-length cDNA) clusters as well as singleton sequences. The predicted gene coding sequences for Arabidopsis (Arabidopsis Genome Initiative, 2000) and rice (Yuan et al., 2003) were also retrieved from the GenBank and TIGR databases, respectively (Table 1). We noticed that the TIGR cotton unigene dataset is a mixture of sequences from several Gossypium species, so we did not use it in our analysis. Instead, we independently constructed unigenes for tetraploid cotton (Gossypium hirsutum) and diploid cotton (G. arboreum) from EST sequences following the same protocol as in the TIGR database. Finally, Arabidopsis and rice unigenes were also constructed for control analyses. After cleaning each dataset for repeated elements and sequence redundancy, the number of sequences ranged from 6920 for ice plant to 55,762 for soybean (Table 1).
Table 1.
Numbers of Sequences and Paralogs Found for Each of the 14 Model Plants Investigated
| Species | Sequences in Initial Dataseta | Sequences in Cleaned Datasetb | Paralogsc | Percentage of Paralogsd | Gene Familiese | Gene Family Sizef | Duplication Event with Median Ks < 2g |
|---|---|---|---|---|---|---|---|
| M. crystallinum (ice plant) | 6,975 | 6,920 | 1,334 | 19% | 519 | 2.57 | 380 |
| H. annuus (sunflower) | 15,248 | 15,196 | 2,713 | 18% | 1,007 | 2.69 | 625 |
| H. vulgare (barley) | 39,667 | 39,108 | 6,388 | 16% | 2,062 | 3.10 | 1,523 |
| L. sativa (lettuce) | 21,960 | 21,803 | 5,160 | 24% | 1,905 | 2.71 | 1,634 |
| Z. mays (maize) | 32,362 | 32,272 | 10,346 | 32% | 3,767 | 2.75 | 2,015 |
| L. esculentum (tomato) | 32,317 | 30,838 | 7,963 | 26% | 2,876 | 2.77 | 2,222 |
| S. tuberosum (potato) | 23,561 | 23,418 | 6,597 | 28% | 2,452 | 2.69 | 2,462 |
| G. hirsutum (tetraploid cotton) | 8,660 | 8,646 | 2,212 | 26% | 797 | 2.78 | 799 |
| G. arboreum (diploid cotton) | 18,962 | 18,791 | 8,721 | 46% | 2,686 | 3.25 | 2,600 |
| M. trunculata (barrel medic) | 33,765 | 33,380 | 7,961 | 24% | 2,813 | 2.83 | 2,653 |
| T. aestivum (wheat) | 52,352 | 52,197 | 19,128 | 37% | 5,719 | 3.34 | 4,362 |
| G. max (soybean) | 55,990 | 55,762 | 17,663 | 32% | 6,067 | 2.91 | 5,076 |
| O. sativa (rice) gene models | 56,056 | 18,562 | 9,149 | 49% | 2,334 | 3.92 | 4,977 |
| O. sativa (rice) unigenes | 30,087 | 29,857 | 7,006 | 23% | 2,652 | 2.64 | 1,250 |
| Arabidopsis gene models | 26,157 | 25,557 | 11,937 | 47% | 3,978 | 3.00 | 6,801 |
| Arabidopsis unigenes | 23,458 | 19,554 | 3,708 | 19% | 1,483 | 2.50 | 1,562 |
Number of sequences in dataset after download.
Number of sequences in dataset after removing redundant entries of the same gene and transposable element sequences. For rice, hypothetical protein gene models were also removed in the cleaning process.
Number of paralogous sequences found in the cleaned dataset using nucleotide alignment search.
Percentage of paralogous sequences found in the cleaned dataset.
Number of gene families constructed with paralogous sequences from column 3 using single linkage clustering.
Average gene family size (number of genes per family).
Number of duplication events used in the distributions in Figure 2 and for which median Ks values are < 2.
Pairs of paralogous sequences were searched for using nucleotide alignments. The fractions of duplicates found in each dataset (after cleaning) range from 16% for barley to 49% for rice. Nevertheless, these figures must not be regarded as an exact representation of the extent of duplications in each species because we applied relatively stringent cutoffs for duplicate identification, which probably excluded pairs of paralogous sequences with higher levels of divergence. In certain instances, multiple unigenes can represent a single gene transcript (e.g., as a result of nonoverlapping EST sequences), and these could potentially be placed incorrectly into separate pairs with the same paralogous transcript. Furthermore, some overlapping unigene sequences originating from the same gene may not have been filtered out at the cleaning step and would increase the number of duplicates in the dataset. After organizing paralogs in gene families (using single linkage clustering), the average number of paralogous sequences per gene family ranges from 2.50 to 3.92 (Table 1). The percentages of paralogs for rice and Arabidopsis are consistently smaller in the unigene datasets (23 and 19%, respectively) than in the gene model datasets (49 and 47%, respectively). The main reason for this is a smaller sampling of gene family members in the unigene dataset.
Age Distributions of Gene Duplication Events
Assuming that the number of silent substitutions per site increases approximately linearly with time, we assessed the relative chronology of duplications of pairs of sequences. The level of synonymous substitutions (Ks) was estimated between the coding sequences of each paralogous pair. The number of possible pairs of paralogs within a gene family with more than two members is greater than the number of duplication events. Therefore, when multiple Ks values could be estimated for a single duplication event, we attributed it the median Ks value. We only retained Ks values < 2.0 because higher Ks values are associated with a large degree of uncertainty because of saturation of substitutions (Li, 1997).
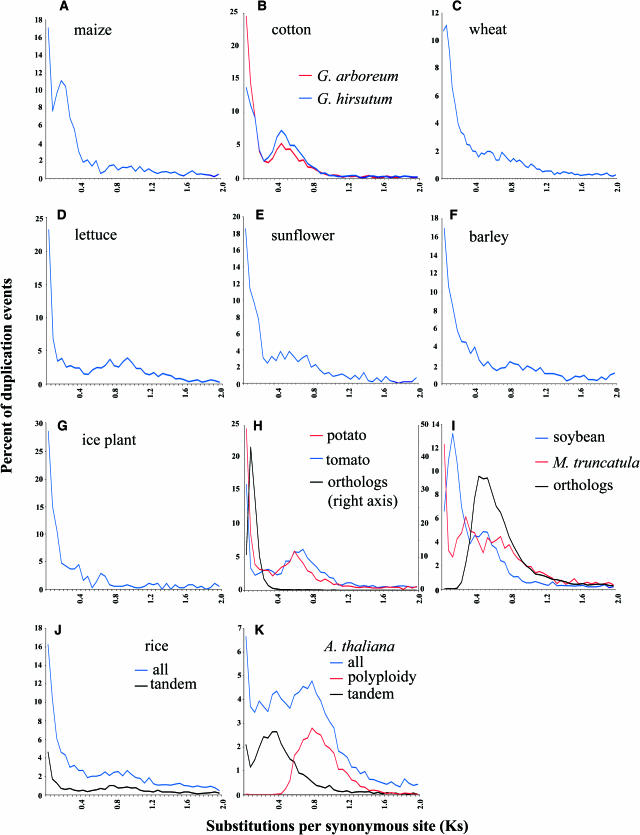
For each organism, we analyzed the shape of the distribution of the gene duplication events as a function of Ks. All distributions show a relatively high density of duplicates in the smallest Ks classes (Figure 2), as observed for other eukaryotes (Lynch and Conery, 2000, 2003), indicating that gene duplication is an ongoing process in all species studied. However, the shapes of the distributions vary from one species to another, which reflect different evolutionary patterns of gene duplication.
Figure 2.
Distributions of the Fraction of Duplication Events as a Function of Their Levels of Synonymous Substitution for 14 Model Plant Species.
Data was grouped into bins of 0.05 Ks units for graphing, except for wheat, where bins represent intervals of 0.03 Ks units (C). For G. hirsutum and G. arboreum (B), the Ks distributions of duplication events are shown in blue and red, respectively. For potato and tomato (H) and for soybean and M. truncatula (I), the Ks distribution of orthologs compared between the species is plotted (green line) as well as the paralog distributions within each species (blue and red lines). For rice and Arabidopsis ([J] and [K]), the distributions of all pairs of duplicated genes and pairs of tandem duplicates are shown in blue and green, respectively. The distribution of Arabidopsis duplicate genes resulting from the most recent polyploidy event (Blanc et al., 2003) is shown in red in (K).
Maize, Cotton, and Wheat
For maize, the distributions present a clear secondary peak (Figure 2A). Although maize behaves genetically as a diploid, mapping data indicates that its genome results from an ancestral tetraploidy event (Helentjaris et al., 1988; Moore et al., 1995). Gaut and Doebley previously reported that the levels of synonymous substitution in duplicated gene pairs formed by this event range from Ks = 0.10 to 0.30 (Gaut and Doebley, 1997). Therefore, the secondary peak with a mode at Ks = 0.15 to 0.20 in the distribution for maize (Figure 2A) certainly represents the traces of this large-scale duplication event.
G. hirsutum is an allotetraploid cotton resulting from the hybridization of two parental diploid species (genomes A and D; Wendel and Cronn, 2003). An average Ks value of 0.042 has been reported for 42 pairs of paralogous genes resulting from this genome duplication event (Senchina et al., 2003). However, there is a large discrepancy between this Ks average and the position of the secondary peak we observe in the distribution for cotton (mode Ks = 0.40 to 0.45; Figure 2B). The most likely explanation is that this secondary peak represents a much older large-scale duplication event as already suspected from mapping data (Rong et al., 2004). Because of the relatively low level of divergence between paralogous sequences resulting from the recent allotetraploidy event in G. hirsutum, the expected secondary peak is probably obscured within the initial peak of duplicate genes with low Ks values, which makes it invisible in this analysis. A complementary analysis of duplicated unigenes from the diploid G. arboreum, a close relative of the A genome progenitor (Wendel and Cronn, 2003), yielded a similarly shaped distribution of Ks values with a clear secondary peak at Ks = 0.40 − 0.45 (Figure 2B).
The fact that the unigene data from G. hirsutum and G. arboreum yield coincident peaks indicates that our analytical method is robust. Nevertheless, we observe nearly twice as many duplication events in the youngest age category for G. arboreum than for G. hirsutum, whereas the opposite would be expected because of recent allopolyploidy in the latter. Although this observation might reflect a high rate of gene duplications in G. arboreum, another alternative is that there is a higher fraction of redundant sequences for G. arboreum. In particular, we flagged unigene sequences that overlap as originating from the same gene only when the level of synonymous substitutions between them was Ks = 0 (see Methods). However, because ESTs, and consequently unigenes, may contain sequencing errors, some pairs of overlapping unigenes may have Ks > 0. Inclusion of these unigene pairs would result in an overestimation of the fraction of recent duplication events. This emphasizes some methodological issues that may be encountered when working with EST-type data. Although the general shape of the distributions provides accurate information on the occurrence and timing of large-scale duplication events, errors in unigene sequences and sequence redundancy preclude comparative analysis of the distributions in quantitative terms.
The distributions for wheat displays a small secondary peak for Ks = 0.03 to 0.06 (Figure 2C). Wheat (T. aestivum) is an allohexaploid (2n = 6x) and contains three sets of homeologous chromosomes (A, B, and D) that diverged from each other an estimated 2.5 to 3.1 million years ago (Huang et al., 2002). For genes that have been cloned from the A, B, and D genomes, such as Acc-1, Pgk-1, and Sut-1 (Aoki et al., 2002; Huang et al., 2002), the levels of synonymous or intron divergence are <0.08. Hence, the small secondary peak observed in the distribution for wheat is likely the mark, rather subtle, left by these genome duplications. As for the tetraploid cotton (G. hirsutum; Figure 2B), the two secondary peaks expected from polyploidy are mostly hidden in the most recent duplicate classes because of their very short time of divergence (Figure 2C).
Lettuce, Sunflower, Barley, and Ice Plant
The distributions for lettuce, sunflower, barley, and ice plant do not present any clear evidence of increased accumulation of duplicates (Figures 2D to 2G, respectively). However, the variation in chromosome numbers between species of the Helianthineae subtribe suggested that a polyploidy event also occurred in the sunflower lineage (Sossey-Alaoui et al., 1998), but this supposed event is not clearly evidenced by the distribution for H. annuus (Figure 2E). Nevertheless, it is possible that the signal of this event, provided it is relatively recent, is not dissociable from the initial peak.
Tomato-Potato and M. truncatula-Soybean
Tomato and potato are two closely related plants from the nightshade family and both present a secondary peak at approximately Ks = 0.60 in their respective distributions (Figure 2H). To assess the relative age of their separation, we also estimated the level of synonymous substitutions for 6838 pairs of orthologs (sequences separated by a speciation event) identified between the two species. The Ks values for ortholog pairs are distributed in a narrow peak (mode Ks = 0.05 to 0.10; Figure 2H), which delimits on its right, the period of evolution corresponding to their common ancestor, and on its left, the time frame elapsed since the separation of the two species. Therefore, the two nearly superimposed secondary peaks observed for tomato and potato probably represent a single ancient episode of increased accumulation of duplicates that occurred in their common ancestor.
It is likely that this episode during Solanaceae evolution was a large-scale duplication event because synteny comparisons between Arabidopsis and potato revealed multiple highly degenerate and probably old duplicated chromosomal segments in the potato genome (Gebhardt et al., 2003). Moreover, although potato is a young autotetraploid, no signal for a recent genome duplication event is apparent in the distribution. Contrary to allopolyploids, an autotetraploid has four virtually identical sets of chromosomes that form tetravalents during meiosis. Because the resulting duplicated genes segregate as alleles and do not diverge independently, these pairs of sequences must be contained in the youngest age class of the distribution.
We performed the same type of comparison for M. truncatula and soybean, two closely related plants from the legume family (Figure 2I). The distribution for soybean displays an obvious first secondary peak (mode Ks = 0.10 to 0.15) followed by a smaller bulge (mode Ks = 0.45 to 0.50; Figure 2I). The distribution of 7930 putative ortholog pairs between soybean and M. truncatula (mode Ks = 0.40 to 0.45) clearly indicates that the first secondary peak for soybean, which lies to the left of the ortholog peak, represents a burst of gene duplications that occurred in soybean after the separation between the two lineages (Figure 2I). This observation is consistent with conclusions drawn from mapping data that the soybean ancestor experienced a genome duplication (Shoemaker et al., 1996). The smaller older bulge in soybean (Ks = 0.45 to 0.50) probably represents the signature of an older burst of gene duplications. Recent analysis of intragenomic synteny in soybean supports this hypothesis in showing that several chromosomal regions exist in more than two copies (Lee et al., 2001; Yan et al., 2003). This level of segmental duplications cannot be accounted for by just one tetraploidy event and implies other large-scale duplication events.
Evidence of ancient segmental duplications has also been found for the M. truncatula genome (Yan et al., 2003; Zhu et al., 2003), suggesting that its ancestor may have sustained large-scale duplication event(s), too. This hypothesis is corroborated by the presence of two overlapping secondary peaks in the M. truncatula distribution (Figure 2I). The younger one (mode Ks = 0.25 to 0.30) is located to the left of the ortholog peak and so corresponds probably to a burst of gene duplications that occurred in the M. truncatula lineage after its separation from that of soybean. The second secondary peak (Ks = 0.65 to 0.70) is more difficult to interpret because it lies to the right of the ortholog peak (mode Ks = 0.40 to 0.45). This evolutionary event may therefore have occurred before the separation between M. truncatula and soybean. However, there is no clear equivalent in the soybean distribution. Beside uncertainties in the peak position resulting from sampling variance and Ks estimation, other alternatives could explain this inconsistency. One hypothesis is that the ancestor of Medicago may have been an allotetraploid resulting from the merger of two diverged genomes (i.e., A and B). In this case, the date of divergence between these two subgenomes—corresponding to the secondary peak at Ks = 0.65 to 0.70 in the M. truncatula distribution (Figure 2I)—is older than the actual polyploidy event. To account for the absence of equivalent secondary peak in the soybean distribution at Ks = 0.65 to 0.70, this scenario requires that the soybean lineage split from one of the diploid genome lineages (i.e., A) before their hybridization leading to the Medicago lineage. A similar evolutionary scenario has been proposed for the separation between maize and sorghum (Gaut and Doebley, 1997; White and Doebley, 1998). If this scenario is correct, one would also expect the distribution of ortholog (reciprocal best BLAST hit) Ks values between soybean and Medicago to be bimodal, with a major peak corresponding to the soybean versus A distance and a smaller peak corresponding to the larger soybean versus B distance as a result of the loss of some genes from the Medicago A subgenome. If the A and B distances are not too dissimilar, their peaks could blur together, which might explain why the distribution of ortholog Ks values in Figure 2I is quite broad and overlaps with the older secondary peak apparent in Medicago. It is apparent that a complex set of events occurred in the legume lineage at around the time of the soybean/Medicago divergence, and more genomic sequence data may be required before they can be properly resolved.
Arabidopsis and Rice
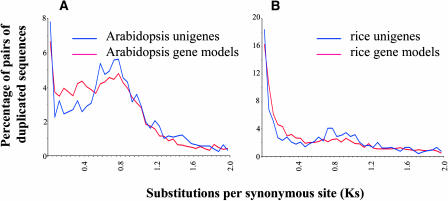
For Arabidopsis and rice, we used complete genome sequence data (Arabidopsis Genome Initiative, 2000; Yuan et al., 2003) in preference to unigenes because this allowed us to distinguish between tandemly duplicated genes on chromosomes and other types of duplicate genes. However, we first verified that the distributions of Ks values for Arabidopsis and rice as calculated from unigenes were essentially identical to those calculated from complete genome sequence data (Figure 3), which lends support to the validity of the method used for other species in Figure 2. For both Arabidopsis and rice, we consistently observe more gene duplications in the youngest age category for the unigene dataset than for gene model dataset. As for G. arboreum, this discrepancy probably results from the inclusion of pairs of unigenes corresponding to the same gene but having Ks > 0.
Figure 3.
Frequency Distributions of Ks Values Obtained from Pairs of Duplicated Genes Identified in Unigene Data (Blue Line) and Complete Genome Sequence Data (Red Line) Are Essentially Identical.
(A) Distributions of Ks values for Arabidopsis.
(B) Distributions of Ks values for rice.
The age distribution of all gene duplications in Arabidopsis presents a secondary peak (mode Ks = 0.75 to 0.80; Figure 2K) as previously reported (Lynch and Conery, 2000, 2003), whereas the distribution for rice (Figure 2J) is similar to that expected under relatively steady rates of gene duplications and deletions of duplicates (Figure 1A). We then used genomic data to investigate these distributions in more detail. For both Arabidopsis and rice, we calculated the Ks distributions for pairs of duplicates created by tandem duplication (<20 genes apart in the genome). For Arabidopsis, we also plotted the Ks values of duplicate gene pairs formed by its most recent polyploidy event (Blanc et al., 2003). This analysis shows that the secondary peak in the overall distribution for Arabidopsis is mainly caused by the genome duplication event (Figure 2K). However, there is a striking peak in the Ks distribution of tandemly duplicated genes in Arabidopsis, centered on Ks = 0.30 to 0.35 and somewhat to the left of the polyploidy peak (Figure 2K). These two secondary peaks—one caused by polyploidy and the other by tandem duplicates—become blurred into one another in the Ks distribution for Arabidopsis when all paralogs are considered. In rice, there is no equivalent peak in the Ks distribution of tandem duplicates (Figure 2J).
The origin of this pattern of accumulation of tandemly duplicated genes in Arabidopsis is intriguing and puzzling. We would have expected the age distribution of tandem duplicates to follow an L-shaped distribution as in Figure 1A and suggest three hypotheses. (1) One possible explanation is that a transient increase in the rate of tandem duplication took place during the evolution of the Arabidopsis lineage. However, such a hypothesis implies that the rate of tandem duplication can be modulated, which remains to be shown. (2) The size of a tandem array evolves by means of unequal crossing-over between two repeated units. This process simultaneously expands the array on one chromosome, shrinks it on the other chromosome, and generates two reciprocal recombinant genes (Jelesko et al., 1999). A newly formed recombinant unit is a chimera between the recombining parental sequences and therefore is only partially identical to either of its parents. Hence, new pairs of tandemly duplicated genes can be created with Ks > 0, and this process might be responsible for the excess observed between Ks ∼0.20 and 0.40. To assess the possible effect of chimerism on the age distribution of Arabidopsis tandem duplicates, we reestimated Ks considering only one end of each gene. We used only the first 25% of codons from the 5′ end of each pairwise sequence alignment (or alternatively, only the last 25% from the 3′ end). Assuming that recombination breakpoints are distributed randomly, this would reduce by 75% the number of Ks estimates affected by chimeric sequences. The distributions for the 5′ end, the 3′ end, and the full-length alignments are nearly identical (data not shown), which indicates that chimeric duplicates do not contribute significantly to the observed pattern. (3) The secondary peak observed in the distribution of tandem paralogs could be an indirect consequence of a deficit of young duplicates. This could happen if, during the relatively recent past, tandem arrays of reduced size were preferentially fixed after recombination. Because the level of sequence identity between the recombining sequences is a key factor, young pairs of tandem duplicates would be more prone to recombine and disappear. We favor this third explanation (see Discussion).
DISCUSSION
Interpreting the histograms in Figure 2 to decide which distributions deviate from the null model of steady rates of duplication and deletion involves a certain degree of subjectivity. We were unable to find a statistical test that could estimate the significance of a potential secondary peak. However, for nine of the investigated species, a secondary peak emerged clearly above the background level, so we are confident that they reflect real periods of increased accumulation of duplicate genes. For maize, soybean, and M. truncatula, they probably result from large-scale duplication events like polyploidy or aneuploidy. It is very possible that large-scale duplications are also at the origin of the secondary peaks observed in the distributions for cotton, potato, and tomato. However, because the pattern of accumulation of paralogs is dependent on multiple factors, mainly the rate of gene duplication and the rate of gene deletion, other scenarios must be considered.
An illustration is given by the secondary peak in the distribution of Arabidopsis tandem duplicates (Figure 2K), which obviously cannot be explained by a single event engendering tandem duplications at numerous loci at the same time. For now, the only known sources of rapid and massive genome expansion via DNA duplication mechanisms are occasional large-scale duplication events and induced transpositions of repeated elements (SanMiguel et al., 1996, 1998; Wendel, 2000). Alternatively, the rate of sequence deletion can play an important role in determining the pattern of accumulation of duplicates (Devos et al., 2002). Arabidopsis has a significantly smaller genome than many of its relatives in the family Brassicaceae, and its genome may have shrunk significantly during the past ∼50 million years. Interestingly, Zhang and Gaut (2003) made the observation that most (87%) of Arabidopsis tandem gene arrays contains only two or three members. They found that tandem arrays tend to be located in regions of the genome with higher local recombination rates. They suggested that stabilizing selection could control the size of arrays via selection against individuals with overly large (or short) arrays, but whether Arabidopsis is substantially different from other species in terms of array size is currently unknown. Genome shrinkage in Arabidopsis could have occurred through an increased rate of DNA loss, but whether it was driven by natural selection is a matter of debate; there is an increasing body of data indicating that genome size is correlated with a wide range of important phenotypic characters (Bennett, 1998; Petrov, 2001), which gives it a potential adaptive role. Regardless of whether it was an adaptive or a neutral phenomenon, an accelerated rate of DNA deletion in Arabidopsis could have led to a tendency to lose genes from tandem arrays. Tandem pairs with high sequence similarity would be the best candidates for such deletions because they are more likely to recombine and less likely to have a severe phenotype when one gene is deleted. We therefore suggest that the difference between the age distributions of tandemly duplicated genes in Arabidopsis and rice is attributable to a recent increase in the rate of DNA deletion in Arabidopsis.
Although we did not observe any obvious secondary peak in the rice distribution, a small flat peak can be discerned in our data for Ks = 0.6 to 1.0 (Figure 2J). Vandepoele et al. made the same observation in a similar analysis of rice duplicated genes (Vandepoele et al., 2003). They were able to attribute this to a possible aneuploidy based on the detection of duplicated chromosomal segments in rice, for which nearly half of the pairs of duplicated genes have Ks values falling in this range. This shows how analysis of complete genome sequences has the power to detect large-scale duplications that are older or encompass fewer genes (Wolfe and Shields, 1997; Arabidopsis Genome Initiative, 2000; Goff et al., 2002; Vandepoele et al., 2003) than the events that can be detected by the unigene method. However, it is not currently possible to apply whole-genome methods to plant species other than Arabidopsis and rice. Estimation of the level of synonymous substitutions is statistically reliable only when Ks < 1 (Li, 1997), which suggests that the parts of the distributions where Ks > 1 in Figure 2 could be merely uninformative. Furthermore, levels of synonymous substitution among genes duplicated at the same time show unexpectedly high variation (Zhang et al., 2002). The impact on the shape of a distribution is that the older a large-scale duplication is, the wider and smaller the associated secondary peak will be. This could obscure the signal from an old polyploidy to the extent that it cannot be clearly observed. Generally speaking, all these factors limit the timeframe in which large-scale duplications can be efficiently detected by the method used here.
Although wheat (Huang et al., 2002), G. hirsutum (Senchina et al., 2003), and possibly sunflower (Sossey-Alaoui et al., 1998) are recent polyploids, we did not find clear evidence of these events in their distributions (Figures 2B, 2C, and 2E, respectively), probably because the associated secondary peaks are hidden in the initial peak of young duplicates. This pinpoints another limitation of our approach, which requires that the secondary peak corresponding to a large-scale duplication event must be sufficiently dissociated from (i.e., older than) the initial peak to be observable in the distributions. Nevertheless, we can note that for G. hirsutum and sunflower, the rate of decay of gene duplications in the youngest age categories is less pronounced than for other species that are not young polyploids (Figure 2). This suggests that these two species have a higher fraction of moderately young duplicates than expected under steady rates of duplicate birth and loss, which can be explained by recent large-scale duplication events.
To estimate absolute dates for the large-scale gene duplications, we estimated the ages of the modes of the secondary peaks of gene duplication (Table 2), assuming clock-like rates of synonymous substitution of 6.5 × 10−9 substitutions/synonymous site/year for cereals (Gaut et al., 1996) and 1.5 × 10−8 substitutions/synonymous site/year for dicots (Koch et al., 2000). All of these duplication episodes are estimated to have occurred in the last 30 million years (Table 2). However, it is important to mention that the purpose of these estimates is to give an idea of the time scale involved because they are certainly highly approximate. The rate of synonymous substitution varies substantially among genes (Zhang et al., 2002), and generation time is known to affect the overall mutational rate (Gaut, 1998). These phenomena are likely to substantially affect the calibration of the molecular clock for each plant. Furthermore, most of our analyses relied on EST sequences, which are error-prone and may slightly overestimate Ks. Finally, estimating the age of large-scale duplications using the mode of secondary peaks is a rather crude method and may be biased by sampling issues or the proximity of other secondary peaks in the distribution. Thus, although different modal Ks values were found for the two older soybean and M. truncatula secondary peaks and the distribution of orthologs between the two species (Table 2), we cannot exclude the possibility that a single large-scale duplication event occurred just before the separation of the two lineages. It is even possible that a large-scale duplication was involved in the speciation event that gave rise to these two lineages because duplications followed by deletions and chromosomal rearrangements lead to the reassignment of gene locations, which is a powerful mechanism for creating reproductive isolation between species (Lynch, 2002). These two phenomena have been observed in several resynthesized polyploid species (Feldman et al., 1997; Liu et al., 1998; Comai et al., 2000; Ozkan et al., 2001; Shaked et al., 2001), so polyploidy is certainly a prominent mechanism of speciation in plants (Wendel, 2000).
Table 2.
Estimated Ages of the Observed Secondary Peaks and the Potato-Tomato and Soybean-M. truncatula Speciation Events
| Species | Mode Peak of Paralog Ks | Estimated Age in Myra |
|---|---|---|
| Maize | 0.15 to 0.20 | 11.5 to 15.4 |
| G. hirsutum and G. arboreum | 0.40 to 0.45 | 13.3 to 15.0 |
| Potato and tomato | 0.55 to 0.70 | 18.3 to 23.3 |
| Soybean young peak | 0.10 to 0.15 | 3.3 to 5.0 |
| Soybean old peak | 0.45 to 0.50 | 15.5 to 16.7 |
| M. truncatula young peak | 0.25 to 0.30 | 8.3 to 10.0 |
| M. truncatula old peak | 0.65 to 0.70 | 21.6 to 23.3 |
| Arabidopsis peak of duplicates formed by polyploidy | 0.75 to 0.80 | 25.0 to 26.7 |
| Arabidopsis peak of tandem duplicates | 0.30 to 0.35 | 10.0 to 11.7 |
| Lineage Separation Events | Mode Peak of Ortholog Ks | Estimated Age in Myr |
| Potato-tomato | 0.05 to 0.10 | 1.6 to 3.3 |
| Soybean-M. truncatula | 0.40 to 0.45 | 13.3 to 15.0 |
Myr, millions of years.
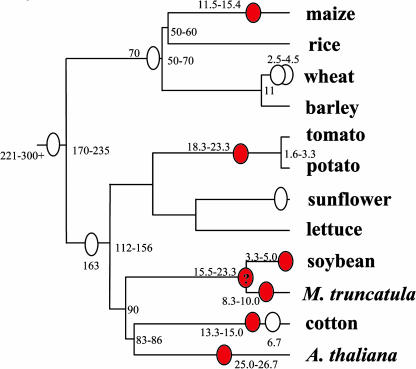
In conclusion, nine of the 14 species studied here have age distributions of paralogous genes that are incompatible with a null model of gradual gene duplication and loss but similar to what is expected from large-scale duplications such as polyploidy or aneuploidy. The events proposed here are summarized in Figure 4. We estimate that all these large-scale duplications occurred within the past 30 million years, and we note that our method underestimates the true level of polyploidy in the plant kingdom because it is unable to detect either very recent polyploidies (as have occurred in cotton, wheat, and possibly sunflower) or very old polyploidies (such as the older events detected by analysis of the Arabidopsis genome; Vision et al., 2000; Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003). This illustrates the prevalence of polyploidy in angiosperms, reaffirms the general consensus that many of the flowering plants are in fact paleopolyploids, and has ramifications for the definition of orthology across species and, hence, for the use of nuclear genes in plant molecular systematics.
Figure 4.
Suspected Large-Scale Duplication Events Presented in Phylogenetic Context.
The phylogenetic tree represents the currently accepted phylogeny of the plant species analyzed here (Soltis et al., 1999). Branch lengths are not to scale. The red ovals represent suspected large-scale duplication events (polyploidy or aneuploidy) recovered in this study. White ovals correspond to suspected polyploidy or aneuploidy events inferred in previous publications (see text for details) but not evidenced in our analysis. Estimated dates of duplication and speciation events are given in million years when available (from present or previous publications; see text). The question mark for the oldest large-scale duplication event of M. truncatula and soybean indicates that the phylogenetic position and the number of independent large-scale duplication events are still unclear.
METHODS
Identification of Paralogs and Orthologs
For each species, all-against-all nucleotide sequence similarity searches were done among the transcribed sequences using the program BLASTN (Altschul et al., 1997). Sequences aligned over >300 bp and showing at least 40% identity were defined as pairs of paralogs. To identify putative orthologs between two species (A and B), each sequence from species A was searched against all sequences from species B using BLASTN and, conversely, each sequence from B was searched against all sequences from A. Two sequences were defined as orthologs if each of them was the best hit of the other and if the sequences were aligned over >300 bp.
Contruction of Unigenes
We downloaded EST sequences for Gossypium arboreum, Gossypium hirsutum, Arabidopsis thaliana, and Oryza sativa (japonica cultivar group) from the EST database. For each species, unigenes were constructed following the same protocol as for the construction of the gene indices by TIGR (Quackenbush et al., 2000). Vector contamination and low quality sequences in the ESTs were removed with the program seqclean. EST sequences corresponding to the same transcript were then assembled into unigenes with the program tgicl using default parameters. Both programs can be found at http://www.tigr.org/tdb/tgi/software/.
Estimation of the Level of Synonymous Substitution between Two Sequences
Because unigenes are derived from EST sequences and have no annotated open reading frames and may contain frameshift sequencing errors, the following approach was taken. Each member of a pair of sequences was searched using BLASTX (Altschul et al., 1997) against all plant protein sequences available in GenBank. The best match was considered significant if the alignment length was >100 amino acids and the expect value (E) was <1e-15. If no significant best match was found, the pair of sequences was discarded. The nucleotide sequence was then translated using the Genewise program (which can infer frameshift sites; Birney et al., 1996) with the corresponding best match protein as a guide. For each pair of paralogs, the two translation products were then aligned using the Smith-Waterman algorithm (Smith and Waterman, 1981), and the resulting alignment was used as a guide to align the nucleotide sequences. After removing gaps and N-containing codons, the level of synonymous substitution was estimated using the maximum likelihood method implemented in codeml (Yang, 1999) under the F3x4 model (Goldman and Yang, 1994).
Dataset Cleaning
We first removed and stored separately all sequences annotated as transposable elements from the Arabidopsis and rice predicted genes. The number of pairs of paralogous sequences found in the full rice gene model dataset was too high for subsequent analyses (131,865 pairs). We therefore discarded 33,411 rice gene models (out of 56,056) annotated as “hypothetical protein” to keep the number of pairs reasonable and focus on the most reliable genes. We then compiled a sequence dataset consisting of the Arabidopsis and rice genes annotated as transposable elements and plant repeated sequences downloaded from RepBase (Jurka, 2000). All unigenes were searched against this database using BLASTN (Altschul et al., 1997). Any unigene matching a repeated sequence over >150 bp and with E <1e-15 was removed from the dataset. In all unigene datasets inspected, only a few sequences were removed by this step, which indicates that EST data is largely free from expressed transposable element genes. Searches at the amino acid level using TBLASTX with virtually equivalent parameters led to essentially the same results.
A drawback associated with the analysis of paralogous sequences derived from ESTs is that multiple entries for the same gene can be present in the dataset, leading to redundant Ks measures. First, because EST sequences are partial reads of cDNAs, two ESTs can be derived from nonoverlapping regions of the coding sequence of the same gene but potentially be placed into distinct pairs with paralogous sequences. The longer a coding sequence is, the more likely this outcome is. However, we can reasonably assume that genes of large size did not duplicate more frequently at a particular evolutionary period, so it is likely that redundant Ks measures are randomly distributed among all the Ks values. Second, multiple entries can arise from sequences corresponding to different splicing variants of the same gene, which have not been assembled into the same contig. However, redundant entries of this type can be detected because their aligned regions have identical sequences. Thus, for all datasets downloaded from the TIGR gene index database, we systematically discarded one sequence from a pair of paralogs showing no synonymous substitutions (Ks = 0) as well as all Ks values involving this sequence. Nevertheless, it must be noted that most of the sequences studied here are derived from ESTs, which contain sequencing errors. Hence, redundant entries may not all be filtered out at this step because some pairs of unigene sequences originating from the same gene may have Ks slightly higher than 0. Applying a more stringent cut (i.e., considering any two sequences with a similarity level higher than 95% as redundant entries) resulted in too small a sampling of duplicated unigene pairs for most species and the disappearance of the initial peaks. Because the inclusion of a limited number of redundant entries did not influence the occurrence of secondary peaks, we preferred using the first low-stringency cutoff (i.e., Ks = 0) for sequence redundancy detection.
Discarding Redundant Ks Values in Gene Families
A gene family of n members results from n − 1 gene duplication events. However, the number of possible pairwise comparisons within a gene family (n × (n − 1)/2) can be substantially larger than the number of gene duplications, which results in multiple estimates of the ages of some duplications. To eliminate the redundant Ks values, pairs of duplicated sequences were grouped into gene families using a single linkage clustering method. For example, if (A, B) is one pair of paralogous sequences and (B, C) is another pair, then A, B, and C were defined as members of the same family, even if the pair (A, C) was absent in the whole set of pairs. A hierarchical clustering method was used to reconstruct a tentative phylogeny of each gene family. (1) Initially, all sequences in the family were treated as a separate clusters. (2) Then, the Ks values for all possible pairs of clusters were compared. (3) The pair of clusters having the smallest Ks value was replaced by a single new cluster containing all their sequences. (4) The median Ks value was chosen to represent the duplication event that gave rise to the two merged clusters. (5) Steps 2 to 4 were repeated until all sequences were contained in a single cluster. When two clusters A and B contained more than one sequence, their associated Ks value in step 2 corresponded to the median Ks obtained for all possible pairs of any sequence from A and any sequence from B. This heuristic method was used instead of complete phylogenetic analysis to reduce computation requirements and because phylogenetic trees could not be drawn for many gene families because their sequences are incomplete and do not all overlap in multiple alignments.
Definition of Tandemly Duplicated Genes
For Arabidopsis and rice, two paralogous genes were considered as tandem duplicates when the number of genes separating them in the genomic sequence was <20.
Data Availability
Raw and analyzed data is available at http://wolfe.gen.tcd.ie/blanc/supp/polyploidy_in_plants.html.
Supplementary Material
Acknowledgments
This work was supported by Science Foundation Ireland. We thank two anonymous reviewers for helpful comments.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Guillaume Blanc (g_blanc@univ-perp.fr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021345.
References
- Adams, M.D., et al. (1991). Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, N., Whitfeld, P., Hoeren, F., Scofield, G., Newell, K., Patrick, J., Offler, C., Clarke, B., Rahman, S., and Furbank, R.T. (2002). Three sucrose transporter genes are expressed in the developing grain of hexaploid wheat. Plant Mol. Biol. 50, 453–462. [DOI] [PubMed] [Google Scholar]
- Bennett, M.D. (1998). Plant genome values: How much do we know? Proc. Natl. Acad. Sci. USA 95, 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Thompson, J.D., and Gibson, T.J. (1996). PairWise and SearchWise: Finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 24, 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Buell, C.R. (2002). Current status of the sequence of the rice genome and prospects for finishing the first monocot genome. Plant Physiol. 130, 1585–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., Tyagi, A.P., Winter, K., Holmes-Davis, R., Reynolds, S.H., Stevens, Y., and Byers, B. (2000). Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K.M., and Bennetzen, J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M., Liu, B., Segal, G., Abbo, S., Levy, A.A., and Vega, J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. (1998). Molecular clocks and nucleotide substitution rates in higher plants. In Evolutionary Biology, M.K. Hecht, ed (New York: Plenum Press), pp. 93–120.
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, C., Walkemeier, B., Henselewski, H., Barakat, A., Delseny, M., and Stüber, K. (2003). Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J. 34, 529–541. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Yang, Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736. [DOI] [PubMed] [Google Scholar]
- Helentjaris, T., Weber, D., and Wright, S. (1988). Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Sirikhachornkit, A., Su, X., Faris, J., Gill, B., Haselkorn, R., and Gornicki, P. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99, 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesko, J.G., Harper, R., Furuya, M., and Gruissem, W. (1999). Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka, J. (2000). Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16, 418–420. [DOI] [PubMed] [Google Scholar]
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- Lee, J.M., Grant, D., Vallejos, C.E., and Shoemaker, R.C. (2001). Genome organization in dicots. II. Arabidopsis as a ‘bridging species’ to resolve genome evolution events among legumes. Theor. Appl. Genet. 103, 765–773. [Google Scholar]
- Li, W.H. (1997). Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Liu, B., Vega, J.M., and Feldman, M. (1998). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41, 535–542. [DOI] [PubMed] [Google Scholar]
- Lynch, M. (2002). Genomics. Gene duplication and evolution. Science 297, 945–947. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2003). The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3, 35–44. [PubMed] [Google Scholar]
- Moore, G., Foote, T., Helentjaris, T., Devos, K., Kurata, N., and Gale, M. (1995). Was there a single ancestral cereal chromosome? Trends Genet. 11, 81–82. [DOI] [PubMed] [Google Scholar]
- Otto, S.P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. [DOI] [PubMed] [Google Scholar]
- Ozkan, H., Levy, A.A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, J., Guiliano, D.B., and Blaxter, M. (2002). Making sense of EST sequences by CLOBBing them. BMC Bioinformatics 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.X., Katsar, C.S., Lan, T.H., Lin, Y.R., Ming, R., and Wright, R.J. (2000). Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D.A. (2001). Evolution of genome size: New approaches to an old problem. Trends Genet. 17, 23–28. [DOI] [PubMed] [Google Scholar]
- Prince, V.E., and Pickett, F.B. (2002). Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3, 827–837. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. (2000). The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, J., et al. (2004). A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166, 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Senchina, D.S., Alvarez, I., Cronn, R.C., Liu, B., Rong, J., Noyes, R.D., Paterson, A.H., Wing, R.A., Wilkins, T.A., and Wendel, J.F. (2003). Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20, 633–643. [DOI] [PubMed] [Google Scholar]
- Seoighe, C., and Wolfe, K.H. (1999). Updated map of duplicated regions in the yeast genome. Gene 238, 253–261. [DOI] [PubMed] [Google Scholar]
- Shaked, H., Kashkush, K., Ozkan, H., Feldman, M., and Levy, A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, R.C., Polzin, K., Labate, J., Specht, J., Brummer, E.C., Olson, T., Young, N., Concibido, V., Wilcox, J., Tamulonis, J.P., Kochert, G., and Boerma, H.R. (1996). Genome duplication in soybean (Glycine subgenus soja). Genetics 144, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C.E., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99, 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S. (1981). Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197. [DOI] [PubMed] [Google Scholar]
- Soltis, P.S., Soltis, D.E., and Chase, M.W. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui, K., Serieys, H., Tersac, M., Lambert, P., Schilling, E., Griveau, Y., Kaan, K., and Bervillé, A. (1998). Evidence for several genomes in Helianthus. Theor. Appl. Genet. 97, 422–430. [Google Scholar]
- Vandepoele, K., Simillion, C., and Van de Peer, Y. (2003). Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15, 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. [PubMed] [Google Scholar]
- Wendel, J.F., and Cronn, R.C. (2003). Polyploidy and the evolutionary history of cotton. Adv. Agron. 78, 139–186. [Google Scholar]
- White, S., and Doebley, J. (1998). Of genes and genomes and the origin of maize. Trends Genet. 14, 327–332. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H. (2001). Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2, 333–341. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., and Shields, D.C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Wong, S., Butler, G., and Wolfe, K.H. (2002). Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. USA 99, 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H.H., Mudge, J., Kim, D.-J., Shoemaker, R.C., Cook, D.R., and Young, N.D. (2003). Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 106, 1256–1265. [DOI] [PubMed] [Google Scholar]
- Yang, Z. (1999). Phylogenetic Analysis by Maximum Likelihood (PAML), Version 2. (London, UK: University College).
- Yuan, Q., Ouyang, S., Liu, J., Suh, B., Cheung, F., Sultana, R., Lee, D., Quackenbush, J., and Buell, C.R. (2003). The TIGR rice genome annotation resource: Annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 31, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., and Gaut, B.S. (2003). Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13, 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Vision, T.J., and Gaut, B.S. (2002). Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 19, 1464–1473. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Kim, D.-J., Baek, J.-M., Choi, H.-K., Ellis, L.C., Kuester, H., McCombie, W.R., Peng, H.-M., and Cook, D.R. (2003). Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol. 131, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and analyzed data is available at http://wolfe.gen.tcd.ie/blanc/supp/polyploidy_in_plants.html.