Abstract
To study the evolutionary effects of polyploidy on plant gene functions, we analyzed functional genomics data for a large number of duplicated gene pairs formed by ancient polyploidy events in Arabidopsis thaliana. Genes retained in duplicate are not distributed evenly among Gene Ontology or Munich Information Center for Protein Sequences functional categories, which indicates a nonrandom process of gene loss. Genes involved in signal transduction and transcription have been preferentially retained, and those involved in DNA repair have been preferentially lost. Although the two members of each gene pair must originally have had identical transcription profiles, less than half of the pairs formed by the most recent polyploidy event still retain significantly correlated profiles. We identified several cases where groups of duplicated gene pairs have diverged in concert, forming two parallel networks, each containing one member of each gene pair. In these cases, the expression of each gene is strongly correlated with the other nonhomologous genes in its network but poorly correlated with its paralog in the other network. We also find that the rate of protein sequence evolution has been significantly asymmetric in >20% of duplicate pairs. Together, these results suggest that functional diversification of the surviving duplicated genes is a major feature of the long-term evolution of polyploids.
INTRODUCTION
Polyploidy (i.e., genome duplication) is recognized as a common phenomenon in the evolution of plants (Wendel, 2000) and some animal clades (Ohno, 1970). It is estimated that 50 to 80% of angiosperms are polyploids, including crop plants such as alfalfa (Medicago sativa), potato (Solanum tuberosum), wheat (Triticum aestivum), oat (Avena sativa), cotton (Gossypium hirsutum), and coffee (Coffea arabica) (Wendel, 2000). Moreover, comparative mapping and/or large-scale sequencing have provided convincing evidence that some genetically diploid species are in fact ancient polyploids—examples of such paleopolyploid species include yeast (Saccharomyces cerevisiae) (Wolfe and Shields, 1997), vertebrates (Gu et al., 2002a; McLysaght et al., 2002), maize (Zea mays) (Gaut and Doebley, 1997), soybean (Glycine max) (Shoemaker et al., 1996), cabbage (Brassica oleracea) (Lagercrantz and Lydiate, 1996), and Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000). It is therefore understandable that much effort has been made to understand the biological significance and evolution of polyploidy. Studies focusing on the early evolution of synthetic or recently formed allopolyploid plants have shown that the merger of two distinct genomes can be followed by genomic changes (e.g., sequence elimination, sequence homogenization, and repeat invasion) and epigenetic changes (resulting in gene silencing, novel expression, and mobile element derepression) (Wendel, 2000; Liu and Wendel, 2002). Because the Arabidopsis genome underwent several ancient rounds of polyploidy (Vision et al., 2000; Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003) and has been completely sequenced (Arabidopsis Genome Initiative, 2000), this species represents a new and valuable model for studying the long-term evolution of a paleopolyploid.
The remnants of these polyploidy events in the current Arabidopsis genome form a large set of duplicated chromosomal segments, which have been identified and ordered in different age classes by several groups using slightly different approaches (Arabidopsis Genome Initiative, 2000; Blanc et al., 2000, 2003; Paterson et al., 2000; Vision et al., 2000; Simillion et al., 2002; Bowers et al., 2003). The duplicated chromosomal segments corresponding to the most recent polyploidy are the best defined and cover 70 to 89% of the genome (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003). They were formed by a single event that occurred sometime between 20 and 60 million years ago, after the split of the Arabidopsis and cotton lineages, but before the split of the Arabidopsis and Brassica lineages (Lynch and Conery, 2000; Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003; Ermolaeva et al., 2003). Older duplicated chromosomal segments have been found and attributed to two older rounds of polyploidy that occurred before the separation of the Arabidopsis and cotton lineages more than 100 million years ago (Simillion et al., 2002; Bowers et al., 2003).
The duplication of all genes in a genome is the most obvious consequence of polyploidy. Therefore, studying the subsequent fate of these gene pairs is particularly important for understanding the evolution of polyploids. Because duplicated genes should have redundant functions immediately after they are formed, one of the copies can accumulate deleterious mutations and eventually be lost without effect on the fitness of an individual. Classical models predict that gene loss is the most likely outcome (Walsh, 1995), and this has been confirmed by empirical analyses (Lynch and Conery, 2003). The process of loss of duplicated genes is an important aspect of the long-term evolution of polyploids because <27% of the genes in Arabidopsis (Blanc et al., 2003) and 16% of the genes in S. cerevisiae (Wong et al., 2002) are still duplicated between sister genomic regions originating from polyploidy. A less likely outcome is that both copies remain in the genome and evolve under purifying selection after a possible period of relaxed constraint (Lynch and Conery, 2003). This can happen either when the duplicated genes are completely functionally redundant but the dosage effect presents a selective advantage (Osborn et al., 2003) or when their function diverges. Functional divergence can occur by neofunctionalization (a gene copy acquires a new function) or by subfunctionalization (the copies retain different subsets of the functionality of the ancestral gene; Force et al., 1999). However, the incidence of functional divergence among duplicated genes is difficult to quantify because genes exert their biological roles in many different ways. Some gene products are part of subcellular structures, others engage in protein–protein interactions, interactions with DNA or RNA, or catalyze the transformation of small molecules. Moreover, genes with the same biochemical functions may be expressed at different times or in different places. Because the integration of the multiple aspects of gene functionality is very complex, it is impossible to summarize them with a single measure.
Nevertheless, for Arabidopsis genes, more and more functional data of various types are becoming available in public databases. For instance, microarray data consist of expression intensity measures for several thousands of genes under different environmental conditions and tissues. This type of information provides raw material for the large-scale analysis of the expression patterns of genes, which is an important aspect of their function. In addition, efforts have been made to classify Arabidopsis genes into functional categories using the controlled vocabularies developed by the Gene Ontology project (Ashburner et al., 2000; Rhee et al., 2003) and the Munich Information Center for Protein Sequences (MIPS) database (Schoof et al., 2002). Here, we exploited this information to investigate the evolution of Arabidopsis polyploidy-derived duplicated genes in a functional framework. Because acceleration of protein evolution may indicate functional divergence, we also analyzed the rates of amino acid evolution between pairs of duplicated genes. The results of this analysis show that nonrandom loss and functional diversification of the duplicated genes are important features of the long-term evolution of polyploids.
RESULTS
Underrepresentation and Overrepresentation of Arabidopsis Duplicates in Functional Categories
We tested if the loss or retention of duplicated genes is influenced by the function of the proteins they encode. Polyploidy-derived duplicated gene pairs were identified in our previous study (referred to as the Blanc dataset thereafter; Blanc et al., 2003). To examine functional categories, we focused only on the pairs of duplicates formed by the most recent polyploidy. The corresponding duplicated regions cover 80% of the genome, so most of the gene pairs formed by this event have been identified. Although it would have been interesting to study gene pairs formed by the old polyploidy events as well, we did not do this for two reasons. First, the old duplicate pairs result probably from two rounds of polyploidy (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003), and their attribution to either one of the polyploidy events was not well resolved in our previous analysis. Second, all analyses to date have found that the old duplicated chromosomal blocks cover only a minor fraction (26 to 52%) of the genome (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003). This suggests that, if these ancient events were polyploidies, many of the duplicate pairs resulting from them have been missed. Because our statistical method requires that the number of genes in the preduplication genome be estimated, the old gene pairs could not be analyzed.
To study the relationship between gene functions and their loss or retention after the most recent polyploidy, we first utilized the Gene Ontology (GO) classification scheme. The GO consortium provides a standardized and hierarchical vocabulary (GO terms) to describe the function of gene products. In addition, GO classifies genes into functional categories, which fall into three general sections: molecular function, biological process, and cellular component (Ashburner et al., 2000). For 2185 recent duplicate pairs, at least one of the duplicates was associated with GO terms. On average, 78% of the GO terms associated with one of the copies were also associated with the other. Because many of the genes were annotated using computer predictions without supporting laboratory evidence, some GO term associations may be incorrect or missing. Consequently, it is impossible to determine with confidence which cases of differences in the GO terms associated with duplicated gene pairs reflect real divergence between the functions of the two genes.
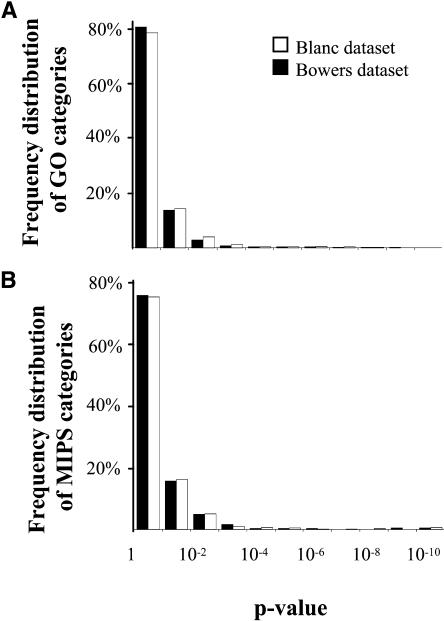
The genes retained in duplicate are not distributed evenly in several GO functional categories (Table 1, Blanc dataset columns), which indicates types of genes that have been preferentially kept or lost during the subsequent evolution of a polyploid. Although, as one would expect, most (93%) of the functional categories tested were not significantly overduplicated or underduplicated (at α = 0.01; Figure 1A); these results indicate that the loss of duplicated genes is not entirely random and is partly determined by the role of the gene in the cell. Other laboratories have identified polyploidy-derived duplicated genes in Arabidopsis using different methods that resulted in lists of duplicate pairs that are similar but not identical to the ones we previously reported. To test how robust the finding of overduplicated and underduplicated functional categories in Table 1 is to the input set of duplicated genes, we repeated the analysis using the duplicated gene pairs identified by Bowers et al. (2003) (referred to as the Bowers dataset hereafter). For this dataset, we analyzed 2434 pairs attributed to the most recent polyploidy and for which at least one of the genes was annotated by GO. As for the Blanc dataset, most (94%) of the functional categories tested with the Bowers dataset were not significantly overduplicated or underduplicated (α = 0.01; Figure 1A). For each GO category, the degree of departure from the model of random loss of duplicated genes was measured for the two datasets using the formula S = ɛ × log(P), where P is the P value calculated using the binomial distribution to test the hypothesis that duplicated genes are lost randomly. Epsilon (ɛ) is equal to −1 or 1 if the observed frequency of retained duplicated genes is lower or higher than the expected frequency, respectively. The correlation coefficient of the S values calculated for all GO categories between the Blanc and Bowers datasets is r = 0.96 (P = 0), indicating that the two datasets give essentially the same results. Among the 43 GO categories found significantly underduplicated or overduplicated (α = 0.01) in at least one of two datasets (Table 1), 28 (65%) have a significant P value for both datasets, and many others have a significant P value for one dataset and are borderline significant P value for the other dataset.
Table 1.
GO Functional Categories for Which Arabidopsis Genes Remained in Duplicate after the Most Recent Polyploidy Are Found Significantly Overrepresented or Underrepresented (α = 0.01) for Either the Blanc or Bowers Datasets
| Blanc Dataset
|
Bowers Dataset
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GO Term ID | GO Term Description | GO Subdivisiona | Estimated Number of Genes in Preduplication Genome | Percentage of Pairs in Category (Expected 14.4%) | P Valueb | Estimated Number of Genes in Preduplication Genome | Percentage of Pairs in Category (Expected 16.3%) | P Value b | |
| Over duplicated Functional Categories | GO:0006534 | Cys metabolism | BP | 9 | 55.6% | 0.005 | 10 | 40.0% | 0.065 |
| GO:0009225 | Nucleotide-sugar metabolism | BP | 23 | 47.8% | 1.3E-04 | 23 | 47.8% | 4.3E-04 | |
| GO:0019642 | Anaerobic glycolysis | BP | 27 | 37.0% | 0.003 | 27 | 37.0% | 0.008 | |
| GO:0019650 | Butanediol fermentation | BP | 31 | 32.3% | 0.009 | 31 | 32.3% | 0.022 | |
| GO:0006334 | Nucleosome assembly | BP | 46 | 30.4% | 0.004 | 44 | 36.4% | 0.001 | |
| GO:0030003 | Cation homeostasis | BP | 54 | 27.8% | 0.008 | 52 | 32.7% | 0.003 | |
| GO:0007264 | Small GTPase mediated signal transduction | BP | 69 | 26.1% | 0.007 | 69 | 26.1% | 0.026 | |
| GO:0006511 | Ubiquitin-dependent protein catabolism | BP | 155 | 23.2% | 0.002 | 152 | 25.7% | 0.002 | |
| GO:0006355 | Regulation of transcription, DNA-dependent | BP | 838 | 21.7% | 6.4E-09 | 816 | 25.0% | 1.6E-10 | |
| GO:0006468 | Protein amino acid phosphorylation | BP | 726 | 20.4% | 6.3E-06 | 711 | 22.9% | 3.2E-06 | |
| GO:0009654 | Oxygen evolving complex | CC | 6 | 50.0% | 0.042 | 5 | 80.0% | 0.003 | |
| GO:0005837 | 26S proteasome | CC | 33 | 39.4% | 3.7E-04 | 33 | 39.4% | 0.001 | |
| GO:0005718 | Nucleosome | CC | 40 | 32.5% | 0.003 | 39 | 35.9% | 0.002 | |
| GO:0008287 | Protein Se/Thr phosphatase complex | CC | 85 | 25.9% | 0.004 | 83 | 28.9% | 0.003 | |
| GO:0005886 | Plasma membrane | CC | 81 | 24.7% | 0.010 | 82 | 23.2% | 0.068 | |
| GO:0005840c | Ribosome | CC | 270 | 22.6% | 1.9E-04 | 266 | 24.4% | 4.3E-04 | |
| GO:0005634 | Nucleus | CC | 1134 | 20.7% | 4.0E-09 | 1100 | 24.5% | 3.1E-12 | |
| GO:0004680 | Casein kinase activity | MF | 10 | 50.0% | 0.008 | 10 | 50.0% | 0.014 | |
| GO:0008514 | Organic anion transporter activity | MF | 22 | 36.4% | 0.009 | 22 | 36.4% | 0.019 | |
| GO:0005187 | Storage protein | MF | 17 | 35.3% | 0.026 | 16 | 43.8% | 0.009 | |
| GO:0004840 | Ubiquitin conjugating enzyme activity | MF | 33 | 33.3% | 0.005 | 33 | 33.3% | 0.013 | |
| GO:0015385 | Sodium:hydrogen antiporter activity | MF | 21 | 33.3% | 0.023 | 20 | 40.0% | 0.010 | |
| GO:0005216 | Ion channel activity | MF | 54 | 27.8% | 0.008 | 54 | 27.8% | 0.023 | |
| GO:0015268 | α-Type channel activity | MF | 80 | 27.5% | 0.002 | 78 | 30.8% | 0.001 | |
| GO:0003928 | RAB small monomeric GTPase activity | MF | 67 | 26.9% | 0.005 | 67 | 26.9% | 0.019 | |
| GO:0005509 | Calcium ion binding activity | MF | 204 | 23.0% | 0.001 | 200 | 25.5% | 0.001 | |
| GO:0003700 | Transcription factor activity | MF | 565 | 22.7% | 8.8E-08 | 552 | 25.5% | 2.2E-08 | |
| GO:0003735c | Structural constituent of ribosome | MF | 277 | 22.4% | 2.2E-04 | 272 | 24.6% | 2.8E-04 | |
| GO:0004713 | Protein tyrosine kinase activity | MF | 627 | 22.2% | 1.0E-07 | 613 | 25.0% | 3.0E-08 | |
| GO:0004674 | Protein serine/threonine kinase activity | MF | 652 | 22.1% | 7.7E-08 | 638 | 24.8% | 3.1E-08 | |
| GO:0003677 | DNA binding activity | MF | 1462 | 19.4% | 5.0E-08 | 1431 | 21.9% | 8.3E-09 | |
| GO:0005515 | Protein binding activity | MF | 448 | 18.5% | 0.009 | 437 | 21.5% | 0.003 | |
| Under duplicated Functional Categories | GO:0006915 | Apoptosis | BP | 83 | 4.8% | 0.005 | 82 | 6.1% | 0.005 |
| GO:0006418 | Amino acid activation | BP | 113 | 4.4% | 0.001 | 111 | 6.3% | 0.001 | |
| GO:0006281 | DNA repair | BP | 89 | 1.1% | 1.6E-05 | 86 | 4.7% | 8.9E-04 | |
| GO:0005739 | Mitochondrion | CC | 2176 | 12.0% | 0.002 | 2145 | 13.7% | 0.001 | |
| GO:0009507 | Chloroplast | CC | 2985 | 11.5% | 7.9E-06 | 2937 | 13.3% | 1.1E-05 | |
| GO:0019825 | Oxygen binding activity | MF | 135 | 9.6% | 0.069 | 136 | 8.8% | 0.009 | |
| GO:0003793 | Defense/immunity protein activity | MF | 154 | 6.5% | 0.002 | 153 | 7.2% | 0.001 | |
| GO:0004888 | Transmembrane receptor activity | MF | 103 | 5.8% | 0.006 | 100 | 9.0% | 0.026 | |
| GO:0008246 | Electron transfer flavoprotein | MF | 108 | 5.6% | 0.003 | 107 | 6.5% | 0.002 | |
| GO:0004812 | tRNA ligase activity | MF | 100 | 4.0% | 0.001 | 98 | 6.1% | 0.002 | |
| GO:0003685 | DNA repair protein | MF | 51 | 2.0% | 0.004 | 49 | 6.1% | 0.031 | |
GO subdivisions: BP, biological process; CC, cellular component; MF, molecular function.
Because correction for the significance of repeated statistical tests could not be applied (see Methods), the significance of each individual statistical test must be taken with caution. Rather, the P values must be interpreted as a measure of the importance of the bias in the representation of duplicated genes.
The two GO categories “ribosome” and “structural constituent of ribosome” (GO:0005840 and GO:0003735, respectively) comprise nearly the same set of genes but appear in two different subdivisions of GO.
Figure 1.
Frequency Distributions of Functional Categories in Function of the P Value Attached to the Hypothesis That Polyploidy-Derived Duplicated Genes Are Lost Randomly.
(A) Frequency distributions for GO functional categories.
(B) Distributions for MIPS functional categories. Analyses with the Blanc and Bowers datasets are shown in white and black, respectively.
Table 1 indicates that genes coding for basic cellular machinery, such as ribosomal proteins, Cys metabolism, the proteasome, anaerobic glycolysis butanediol fermentation, photosystem (oxygen evolving complex), and nucleotide-sugar metabolism, have survived in duplicate more often than expected by chance. Furthermore, genes in categories with signal transduction and regulatory functions, such as transcription factors, protein kinases, protein phosphatases, and calcium binding proteins, are also overduplicated. Also, overduplicated are genes involved in ion transport and storage ubiquitin-dependant catabolism. On the other hand, most of the genes involved in DNA repair, apoptosis, defense, tRNA ligation, flavoproteins, and transmembrane receptors have returned to a single copy state. Globally, it appears that duplicated genes encoding proteins targeted to the nucleus and the plasma membrane have been preferentially kept, whereas those encoding proteins targeted to organelles have been preferentially lost (Table 1).
To test how robust these results are relative to gene annotation, we analyzed the same gene pairs using the MIPS functional category definitions (Table 2; Schoof et al., 2002). Again, most (91% for both the Blanc and Bowers datasets) of the MIPS categories are not significantly (α = 0.01) underduplicated or overduplicated (Figure 1B). The Blanc and Bowers datasets were highly congruent between each other: the correlation coefficient of the S values calculated for the MIPS categories with the Blanc and Bowers datasets was r = 0.93 (P = 0). The MIPS classification differs substantially from the GO scheme in terms of function definition and category size. Nevertheless, many common trends can be observed between the two types of classifications, which reinforces our findings. For example, categories of genes significantly (P ≤ 0.01) overduplicated in both the GO and MIPS classifications include transcription control, ribosome, proteasome, protein modification (mainly kinases and phosphatases), calcium binding proteins, ion transport, proteins targeted to the nucleus, and defense proteins. The GO and MIPS schemes both show DNA repair, aminoacyl-tRNA synthetase, and defense genes to be significantly underduplicated. However, we note that the MIPS apoptosis and transmembrane signal transduction categories are found significantly overduplicated, whereas the corresponding GO categories (apoptosis and transmembrane receptor activity, respectively) are found significantly underduplicated. In fact, although these GO categories and their MIPS counterparts refer to the same biological processes, they are totally different subsets of the proteome. Only three genes are shared between the MIPS and GO apoptosis categories (out of 376 and 87, respectively), and 31 genes are shared between the MIPS and GO transmembrane signal transduction protein categories (out of 1561 and 109, respectively). In addition, the MIPS analysis reveals some other categories of genes that may have been preferentially kept in duplicate, including phosphate metabolism, mitotic cell cycle, and nuclear division, and identifies metabolism of vitamins, cofactors, and prosthetic groups as significantly underduplicated (Table 2).
Table 2.
MIPS Functional Categories for Which Arabidopsis Genes Remained in Duplicate after the Most Recent Polyploidy Are Found Significantly Overrepresented or Underrepresented (α = 0.01) for Either the Blanc or Bowers Datasets
| Blanc Dataset
|
Blowers Dataset
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Category ID | Category Description | Estimated Number of Genes in Preduplication Genome | Percentage retained in Duplicate (Expected 14.4%) | P Valuea | Estimated Number of Genes in Preduplication Genome | Percentage Retained in Duplicate (Expected 16.4%) | P Valuea | |
| Over duplicated Functional Categories | 01.04 | Phosphate metabolism | 747 | 20.7% | 1.6E-06 | 731 | 23.4% | 7.5E-07 |
| 03.03.01.01.09 | G2/M transition of mitotic cell cycle | 43 | 32.6% | 0.002 | 43 | 32.6% | 0.007 | |
| 03.03.04.07 | Nuclear division | 177 | 24.9% | 1.7E-04 | 177 | 24.9% | 0.003 | |
| 04.05.01.04 | Transcriptional control | 2083 | 20.2% | 6.3E-14 | 2042 | 22.6% | 5.9E-14 | |
| 05.01.01 | Ribosomal proteins | 272 | 22.1% | 4.4E-04 | 268 | 23.9% | 0.001 | |
| 06.07 | Protein modification | 1421 | 18.3% | 2.9E-05 | 1391 | 20.8% | 9.0E-06 | |
| 06.10 | Assembly of protein complexes | 507 | 19.1% | 0.002 | 503 | 20.1% | 0.017 | |
| 06.13.01.01 | Proteasomal degradation | 81 | 24.7% | 0.010 | 80 | 26.3% | 0.017 | |
| 08.07 | Vesicular transport | 374 | 20.1% | 0.002 | 369 | 21.7% | 0.005 | |
| 08.19 | Cellular import | 131 | 23.7% | 0.003 | 129 | 25.6% | 0.005 | |
| 10.01.01 | Unspecified signal transduction | 1314 | 20.4% | 2.2E-09 | 1283 | 23.3% | 1.3E-10 | |
| 10.01.09 | Second messenger mediated signal transduction | 153 | 24.2% | 0.001 | 150 | 26.7% | 0.001 | |
| 10.05 | Transmembrane signal transduction | 1334 | 17.0% | 0.004 | 1305 | 19.6% | 0.001 | |
| 13.07 | Cell adhesion | 155 | 23.2% | 0.002 | 151 | 26.5% | 0.001 | |
| 13.11 | Cellular sensing and response | 853 | 17.6% | 0.005 | 839 | 19.5% | 0.009 | |
| 14.04 | Cell differentiation | 1706 | 16.7% | 0.007 | 1677 | 18.7% | 0.011 | |
| 14.10.02 | Apoptosis | 315 | 19.4% | 0.009 | 303 | 24.1% | 3.8E-04 | |
| 20 | Systemic regulation/interaction with environment | 1383 | 17.4% | 0.001 | 1360 | 19.3% | 0.002 | |
| 25 | Development | 2557 | 17.1% | 8.1E-05 | 2512 | 19.2% | 1.4E-04 | |
| 30.01 | Cell wall | 580 | 18.6% | 0.003 | 576 | 19.4% | 0.030 | |
| 30.04 | Cytoskeleton | 578 | 18.9% | 0.002 | 564 | 21.8% | 0.001 | |
| 35 | Tissue differentiation | 455 | 18.9% | 0.005 | 446 | 21.3% | 0.004 | |
| 40.03 | Cytoplasm | 2724 | 17.5% | 4.6E-06 | 2685 | 19.2% | 1.1E-04 | |
| 40.07 | Endoplasmic reticulum | 599 | 18.2% | 0.006 | 592 | 19.6% | 0.023 | |
| 40.10 | Nucleus | 3187 | 18.5% | 4.9E-11 | 3133 | 20.5% | 5.6E-10 | |
| 62.02 | Target of regulation | 623 | 18.0% | 0.008 | 611 | 20.3% | 0.007 | |
| 63.01 | Protein binding | 1843 | 17.0% | 0.002 | 1811 | 19.1% | 0.003 | |
| 63.03 | Nucleic acid binding | 2400 | 16.5% | 0.003 | 2353 | 18.8% | 0.002 | |
| 63.17.01 | Calcium binding | 249 | 20.1% | 0.009 | 245 | 22.0% | 0.013 | |
| 67.01.01 | Ion channels | 75 | 25.3% | 0.009 | 74 | 27.0% | 0.014 | |
| 67.04.01 | Cation transporters | 216 | 22.2% | 0.001 | 213 | 23.9% | 0.003 | |
| 67.04.01.01 | Heavy metal ion transporters | 81 | 24.7% | 0.010 | 80 | 26.3% | 0.017 | |
| Under duplicated Functional Categories | 01.07 | Metabolism of vitamins, cofactors, and prosthetic groups | 452 | 11.3% | 0.032 | 448 | 12.3% | 0.009 |
| 03.01.03 | DNA synthesis and replication | 329 | 10.0% | 0.012 | 328 | 10.4% | 0.001 | |
| 03.01.05.01 | DNA repair | 267 | 9.4% | 0.009 | 262 | 11.5% | 0.015 | |
| 05.10 | Aminoacyl-tRNA-synthetases | 62 | 4.8% | 0.016 | 62 | 4.8% | 0.006 | |
| 11.05.03 | Defense related proteins | 332 | 10.8% | 0.035 | 334 | 10.2% | 0.001 | |
Because correction for the significance of repeated statistical tests could not be applied (see Methods), the significance of each individual statistical test must be taken with caution. Rather, the P values must be interpreted as a measure of the importance of the bias in the representation of duplicated genes.
Divergence of Transcription Profiles of Polyploidy-Derived Duplicated Genes
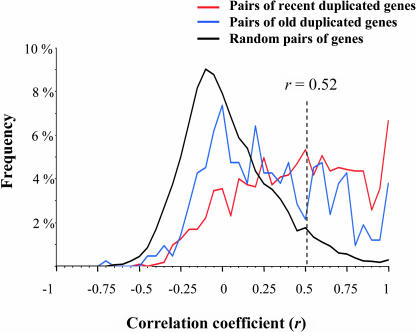
We obtained expression data for most Arabidopsis genes from 62 Affymetrix microarray analyses representing various environmental conditions and tissues. Genes weakly expressed or subject to cross-hybridization were excluded. The duplicated pairs formed by the recent polyploidy event and the old polyploidy events were analyzed separately. We only present results from the Blanc dataset because the same analysis with the Bowers dataset gave identical results (data not shown). For 1137 and 420 pairs of young and old duplicates, respectively, expression profiles were available for both genes. The degree of similarity between the expression profiles for each pair of duplicated genes across all experiments was measured using the Pearson correlation coefficient (r; Figure 2). The expression profiles of two duplicated genes should be virtually identical just after duplication, so their correlation coefficient is initially 1 (Gu et al., 2002b). Hence, if two duplicated genes do not show evidence of coregulation (i.e., have a low r value), they must have acquired divergent expression patterns. To determine a cutoff r value below which duplicated gene pairs can be considered divergent, we calculated r between the expression profiles of 10,000 pairs of randomly chosen genes. Because most of the genes in random pairs are not functionally related and not coregulated, the associated distribution of r can be used to test the null hypothesis that the duplicated genes are not coregulated. Ninety five percent of the r values obtained from random gene pairs have r < 0.52 (Figure 2). Any gene pairs with r ≥ 0.52 can be considered to be significantly coregulated at α = 0.05, so we used r < 0.52 as a criterion for determining that two duplicated genes have diverged in expression. Note that with r ≤ 0.52, r2 is lower than 0.27, so that knowing the pattern of expression of one gene provides little information for predicting the expression pattern of the other gene (Gu et al., 2002b). Our criterion is close to that of Gu et al. (2002b) who used r < 0.5 to indicate expression divergence in a similar analysis of S. cerevisiae duplicates. Based on this condition (r < 0.52), we find that 57% (653) of the pairs of young duplicates and 73% (306) of the pairs of old duplicates, have diverged in expression (Figure 2). For 15% (174) of the young pairs and 29% (129) of the old pairs, the correlation coefficient is negative (r < 0).
Figure 2.
Correlations of Expression Profiles between Pairs of Arabidopsis Genes.
Frequency distributions of the correlation coefficient (r) values obtained from the expression profiles of pairs of duplicated genes formed by the most recent polyploidy event (red), pairs of duplicated genes formed by the old duplication events (blue), and randomly chosen genes (black). Ninety-five percent of the r values obtained from random pairs of genes are smaller than r = 0.52 and appear at the left of the vertical dotted line.
One potential concern with microarray experiments is that cross-hybridization between duplicated genes will cause an artifactual correlation of their expression profiles. We tried to minimize this by using only Affymetrix oligonucleotide array data, which should be less susceptible to cross-hybridization than cDNA arrays, and by excluding probes flagged by Affymetrix as potentially cross-hybridizing. We do observe a weak but significant correlation between the level of sequence similarity of duplicated genes and their expression profile correlation (r = 0.10, P = 1.6E-4; 1137 data points; Blanc dataset for young pairs). Although this could result from cross-hybridization between duplicates with high similarity level, it could also reflect a genuine biological feature (i.e., highly similar duplicates tend to have redundant functions). Finally, it must be noted that cross-hybridization will only make duplicated genes appear to have artificially high correlations, so our estimates that 57 to 73% of pairs have diverged in expression is an underestimate if cross-hybridization is occurring at a high rate.
Concerted Divergence of Expression in Groups of Polyploidy-Derived Paralogs
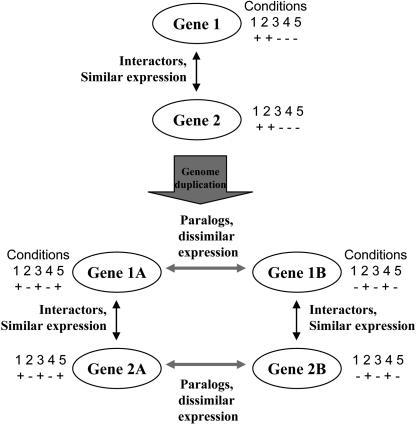
Because many genes exert their function through interactions with other genes, a change in the expression pattern of one gene could drag along changes in the expression patterns of the genes it interacts with to maintain the integrity of the interaction network. After a polyploidy, it is theoretically possible that single members of each duplicated gene pair in an interaction network could diverge in expression in a correlated way, resulting in two parallel versions of the network that are expressed in two different cell types, developmental stages, or environmental conditions. We refer to this process (diagrammed in Figure 3) as concerted divergence of gene expression.
Figure 3.
Illustration of the Concept of Concerted Divergence of Pairs of Paralogs.
In an ancestor, genes 1 and 2 interacted and were coexpressed. After genome duplication, the two pairs of paralogs (1A/1B and 2A/2B) form two separate interacting networks, each with its own expression profile.
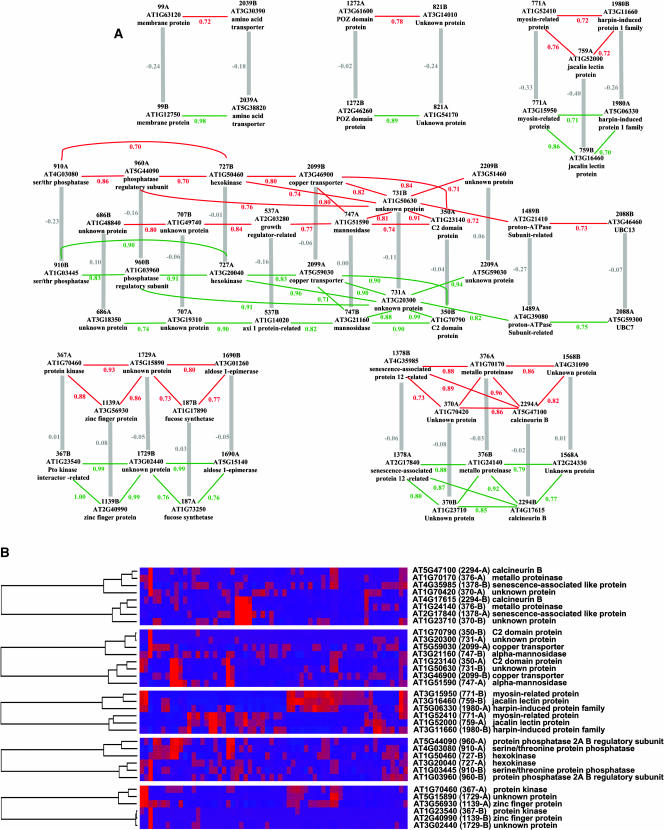
To search for examples of concerted divergence, we restricted our analysis to 248 recent duplicate pairs from the Blanc dataset, where the expression profiles of the two paralogs were highly divergent, and looked for cases where two pairs of paralogs had diverged in parallel directions. Specifically, we searched for associations between pairs of paralogs, such that for two pairs (pair 1 consisting of genes 1A and 1B, and pair 2 consisting of genes 2A and 2B) the interpair correlations were both high (r1A-2A ≥ 0.7 and r1B-2B ≥ 0.7) even though the intrapair correlations were both low (r1A-1B < 0.1 and r2A-2B < 0.1). Because genes with highly similar expression patterns (r ≥ 0.7) are probably coregulated, they are likely to be involved in the same biological pathway (Wu et al., 2002).
We found 37 concerted divergence associations of this type, involving 30 distinct pairs of duplicates (12% of the studied pairs). These can be organized into six networks (Figure 4A) using single-linkage clustering (i.e., a pair of genes is included in the network when it is associated with at least one other pair from the network). A more conservative clustering of these putatively coevolving genes can be obtained by only grouping pairs where all the interpair correlation coefficients are consistently ≥0.70 (complete linkage clustering), which defines five clusters with three to four gene pairs each (Figure 4B). Although very little experimental information is available for most of the genes in these coevolving clusters, careful analysis of the putative gene functions and the literature reveals possible relevant associations. For example, we found an association between a pair of Ser/Thr phosphatase genes and a pair of phosphatase regulatory subunit genes (pairs 910 and 960, respectively; Figure 4B). These two pairs are themselves closely associated with a pair of hexokinase genes (pair 727). It has been shown that phosphatase proteins modulate the activity of hexokinases in yeast (Randez-Gil et al., 1998). Also, two duplicated genes coding for proteins similar to a senescence-associated protein in daylily (Hemerocallis hybrid) (pair 1378; Figure 4B) are strongly correlated with a pair of metalloproteinase genes (pair 376), one of which is known to be involved in senescence in Arabidopsis (Golldack et al., 2002). Finally, we found a pair of membrane amino acid transporter genes (pair 2039) undergoing concerted divergence with two other membrane protein genes (pair 99) (Figure 4A).
Figure 4.
Groups of Duplicated Genes Showing Evidence of Concerted Divergence in Their Expression Profiles.
(A) Schematic representation of clusters of codiverging duplicate pairs. Thick gray lines link paralogous gene pairs (formed by genome duplication) with r ≤ 0.1. Red and green lines link nonhomologous genes with r ≥ 0.7. The r values are indicated beside each line.
(B) Clusters of codiverging duplicate pairs for which all interpair r values are ≥ 0.7. Gene names are indicated for each cluster at the right. Paralogous gene pairs are identified by numbers in parentheses with suffixes A and B. Normalized expression intensities for each gene in 62 public microarray experiments are represented by colored squares in the middle grid. Bright red and bright blue correspond to relative high and low expression levels, respectively. The complete hierarchical clustering of the genes is shown to the left of each cluster.
Asymmetric Rates of Protein Sequence Evolution in Pairs of Duplicated Genes
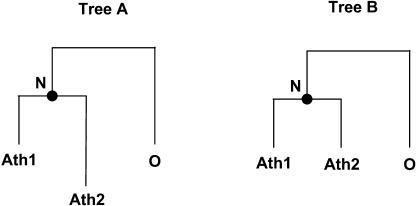
To further characterize the evolution of duplicated gene pairs formed by polyploidy, we examined their rates of protein sequence divergence. To detect significant rate differences between the two members of each pair, we constructed triplets of sequences composed of the two duplicated proteins and an outgroup sequence. We focused our study on the duplicated gene pairs formed by the most recent polyploidy event (Blanc dataset) because the phylogenetic context of this genome duplication is well established (i.e., after the Arabidopsis/cotton split and before the Arabidopsis/Brassica split; Blanc et al., 2003; Bowers et al., 2003), which allows the identification of reliable outgroups. We then compared the log likelihood (lnL) of the aligned triplets under two competing models of evolution that are represented schematically by the trees in Figure 5: one model where all branch lengths are unconstrained (i.e., all sequences are free to evolve at their own rate; Tree A) and a second model where the lengths of the branches leading from node N to the duplicated proteins are forced to be equal (i.e., the duplicated proteins evolve at clock-like rates; Tree B). Twice the difference of the log likelihood of the aligned triplets under the two models [2ΔlnL, where ΔlnL = lnL(no constraint) – lnL(clock)] follows a χ2 distribution with one degree of freedom (Goldman and Yang, 1994). Hence, a probability can be attached to the null hypothesis that the two duplicated protein sequences evolve at the same rate (this is the basis of the Likelihood Ratio Test of Goldman and Yang, 1994). For 173 of the 833 triplets analyzed (21%), we found a significant difference in the rates of divergence of the duplicated protein sequences (P < 0.05). However, with a α = 0.05 significance cutoff in repeated statistical tests, we would expect 5% of the tested triplets to falsely reject the hypothesis of symmetrical evolution. Nevertheless, the actual number of asymmetric triplets is significantly higher than the expected number of false positives (P = 0, binomial distribution with parameter P = 5%; Conant and Wagner, 2003). Therefore, this indicates that a substantial fraction of the pairs of duplicated genes have evolved at unequal rates.
Figure 5.
Schematic Representation of the Two Competing Trees Used to Detect Differences in the Rates of Sequence Divergence among Duplicated Proteins.
Ath1, Ath2, and O stand for two paralogous Arabidopsis proteins formed by genome duplication and an outgroup protein. In Tree A, the branch lengths are unconstrained, whereas in Tree B, the branches leading from node N to Ath1 and Ath2 are forced to be equal. A significantly better maximum likelihood of the triplet of sequences under the model of Tree A compared with the model of Tree B (tested with the Likelihood Ratio Test) indicates that the two paralogous proteins evolved at different rates.
If we combine the results from the two indicators of functional divergence (i.e., expression divergence and asymmetrical sequence divergence), 62% of the recent duplicate pairs present evidence for functional diversification (316 pairs with r < 0.52 and/or significant asymmetric sequence divergence at P < 0.05, out of 511 for which both pieces of information are available). Thus, our analysis suggests that a large majority of the Arabidopsis polyploidy-derived duplicates have acquired divergent functions. The median correlation coefficient of expression profiles for gene pairs with asymmetrical rates of sequence evolution (median r = 0.34) is smaller than that for pairs evolving under a molecular clock (median r = 0.48). This suggests that pairs of asymmetrically evolving genes tend to have more divergent expression, though the temporal order between the two processes cannot be established. Nevertheless, this result must be taken with caution because the difference in median correlation coefficients is only of borderline statistical significance (Wilcoxon two-sample test, P = 0.015). We also tested whether some functional categories have a higher or lower number of functionally divergent pairs of duplicates than expected by chance. Only ribosomal protein genes clearly emerged from these tests with only 13% of the duplicated pairs showing asymmetrical sequence and/or expression divergence (4 pairs out of 31), which represents a significant underrepresentation of functional divergence (P = 2 × 10−08; binomial distribution with parameter P = 0.62). This indicates that this family of genes tends to retain redundant function, which is unsurprising given their very specific role in the cell.
DISCUSSION
Use It or Lose It
Most of the genetic redundancy originating from polyploidy events in Arabidopsis has been erased by massive loss of the duplicated genes. However, duplicated genes are not all equal regarding their loss or retention in the genome. We found several functional categories for which duplicated genes have been preferentially kept or lost after the most recent polyploidy. A previous analysis of polyploidy-derived genes retained in the yeast (S. cerevisiae) genome reached similar conclusions (Seoighe and Wolfe, 1999). Moreover, some functional categories are found overduplicated in both organisms (ribosomal proteins, phosphatases, and kinases). These observations raise the question of the evolutionary significance of the preferential loss or retention of duplicated genes. A polyploidy event results in a doubling of the nuclear DNA content as well as the number of nuclear genes. A simple and intuitive expectation is that the throughput of all pathways involved in protein and nucleic acid metabolism would need to be raised accordingly, and an increase in energy production is needed to meet the increased demand. Either the corresponding genes return to a single copy state but are able to raise their level of expression, or the presence of the two duplicated genes is required to do so. Thus, the preferential retention of duplicated ribosomal protein genes, proteasome subunit genes, and genes involved in Cys metabolism, nucleotide sugar metabolism, and glycolysis (but not DNA repair) might result from this second type of selection. Additionally, empirical data indicates that regulatory genes involved in signal transduction or transcription tend to be dosage dependant in multicellular eukaryotes (Birchler et al., 2001). The duplication of a regulatory gene is likely to influence phenotypic traits. Consequently, the fate of these duplicate pairs is more likely to be under the control of natural selection (nonrandom loss) than for genes that are not dosage dependant. Hence, selection may have promoted the retention of these duplicates for increased dosage.
Another factor that may govern the propensity of duplicated genes to be retained is that alteration of a gene that codes for a subunit of a protein complex may lead to nonfunctional complexes with dominant negative phenotypes (Gibson and Spring, 1998; Veitia, 2003). In this case, it is expected that selection against deleterious mutations will result in the retention of the duplicated genes in a totally undifferentiated state. The only way for the genome to get rid of this category of genes would be to knock them out without any intermediary state (e.g., significant sequence divergence or gene truncation), which reduces the probability of loss. A theoretical study has pinpointed the detrimental effects of changes in the stoichiometry of complex subunits (Veitia, 2003). Hence, selective constraints may exist to maintain stoichiometric concentrations of all members of a protein complex so that duplicated genes encoding protein subunits must either all be kept or all be lost at the same time. This phenomenon is observed in yeast, where protein subunits are not free to evolve independently of their partners (Papp et al., 2003) and where genes coding for subunits of the same complex tend to be physically clustered in the genome (Teichmann and Veitia, 2004). It is notable that many of the overduplicated functional categories largely comprise proteins that are parts of complexes or that interact with other proteins (e.g., ribosomal proteins, kinases, phosphatases, and nucleosomal and proteasomal proteins).
The fact that several categories of regulatory functions (transcription factors, kinases, phosphatases, and calcium binding proteins) and transporter activities are found overduplicated suggests that polyploidy could be an important means of increasing the complexity of regulatory networks and adaptability to changing environmental conditions, as first suggested by Ohno (1970). These duplicated regulatory genes may evolve to change their targets or their ligands by sequence divergence. Alternatively, the two duplicated copies may diverge in expression to coordinately achieve more complex control of the same genetic network. On the other hand, defense genes are significantly underduplicated. This is perhaps not surprising because this class of genes is thought to evolve under a birth and death process, which proceeds by frequent duplications and deletions of genes (Nei et al., 1997; Michelmore and Meyers, 1998). The fact that other categories of genes—like DNA repair and tRNA synthetase genes—are found significantly underduplicated suggests that the presence of more than one copy of these genes tend to be deleterious for the plant.
Functional Divergence of Duplicated Genes
Assuming that r < 0.52 between expression profiles is an indication of expression divergence (Figure 2), we found that a majority of the duplicate pairs (57 and 73% for the recent and old duplicates, respectively) have acquired divergent expression patterns. This observation is in agreement with previous analyses in S. cerevisiae and human, which showed a rapid divergence in expression between duplicated genes (Gu et al., 2002b; Makova and Li, 2003). It is also consistent with the observation of Adams et al. (2003) of divergent expression patterns in 10 of 40 paralog pairs studied in allotetraploid cotton. Moreover, our results indicate that the fraction of transcriptionally divergent pairs is greater in the set of ancient paralogs than in the set of young paralogs (Figure 2). This suggests a progressive, rather than saltatory, divergence of transcription patterns. Our estimates of transcriptionally divergent pairs may even underestimate the true proportion of divergent pairs in Arabidopsis because of several factors. For example, duplicated genes having largely similar profiles but expressed differentially in a small subset of conditions could still have a significant positive correlation coefficient, which would mask a genuine divergence of their expression patterns. Similarly, the expression datasets analyzed here are necessarily only a sampling of all the possible environmental conditions or tissues where the duplicated genes may be expressed. Obviously, the more conditions that are examined, the more likely it is that two genes will be seen to have different expression patterns. Finally, cross-hybridization between the duplicated genes may increase the correlation of the expression profiles, though this artifact should be minimal with Affymetrix chips. Acting in the opposite direction is the possibility that noise in the microarray data will tend to reduce the true correlation between the expression patterns of duplicated genes and thus lead to an overestimation of the proportion of divergent gene pairs.
Most genes do not work alone. Rather, gene products engage in complex interactions with other proteins or are part of a metabolic chain. Duplication of one of these genes on its own followed by a change in the transcription profile of one copy is probably a dead end in most cases. Although some degree of functional clustering of genes is observed in the Arabidopsis genome (Elo et al., 2003; Lee and Sonnhammer, 2003), genes involved in the same pathway are usually scattered, so small-scale DNA duplications cannot simultaneously duplicate all the components of a pathway. Thus, large-scale duplications like polyploidy or aneuploidy are unique events in terms of their potential for whole pathway evolution. Indeed, the simultaneous duplication of all the genes in a pathway offers a unique opportunity for interacting duplicated genes to diversify their expression patterns concertedly. Biochemical pathways or protein complexes could then be expressed at different times or in different places with two different but specialized sets of genes (Caffrey et al., 1999; Pastor-Satorras et al., 2003; Ihmels et al., 2004). In support of this idea, we found six groups of paralogs formed by the most recent polyploidy event that present evidence for concerted divergence of their transcription profiles (Figure 4). Note that we applied very stringent criteria to identify these groups, so the actual number and size of codiverging clusters may be much greater than those identified here. Thus, our results suggest that the impact of large-scale duplication events on the biology of an organism may be more than the cumulative effect of the duplication of individual genes and that the simultaneousness of the duplications confers an added value with regard to the potential for evolution of interaction networks.
We also found that for 21% of the recent duplicate pairs, the two protein sequences evolve at significantly different rates. Similar proportions of asymmetrically evolving duplicates have been observed for yeasts (21 to 27%), Drosophila melanogaster (30%), C. elegans (28%) (Conant and Wagner, 2003), and in a smaller sample for Arabidopsis (13 to 23%) (Zhang et al., 2002). A fundamental tenet in molecular evolution is that functional divergence after gene duplication is highly correlated with a change of evolutionary rate (Gu, 1999; Dermitzakis and Clark, 2001). Although further experimental work is obviously required to determine whether functional diversification has occurred or not in each of the Arabidopsis duplicate pairs, asymmetric evolution of the protein sequences may be an indicator of functional divergence. For example, the duplicated genes At3g44310 and At5g22300 encode the nitrilase proteins NIT1 and NIT4. We found that NIT1 diverged 4.4 times more rapidly than NIT4. NIT1 is responsible for the metabolism of nitriles originating from the breakdown of glucosinolate and converts indole-3-acetonitrile to the plant hormone indole-3-acetic acid in vivo, whereas NIT4 encodes a β-cyano-l-alanine-hydratase/nitrilase involved in cyanide detoxification. The function of the ancestral preduplicated gene was probably similar to that of NIT4 because the orthologous gene in tobacco (Nicotiana tabacum) performs the same reaction (Kutz et al., 2002). Thus, for this specific case, the acceleration of sequence evolution is clearly correlated with the creation of a new function.
Conclusion
The early phenotypic changes associated with polyploidy are often subtle, although small differences in developmental rates, metabolism, gene regulation, and physiological tolerance can be enormously important for the evolutionary success of newly formed polyploid lineages (Otto and Whitton, 2000). Thus, the evolutionary importance of polyploidy has been questioned (Stebbins, 1950). The development of high-throughput technologies that provide functional genomics information can now shed new light on the impact of paleopolyploidy in plants like Arabidopsis. First, even tens of millions of years after they happened, the polyploidy events still make a significant contribution to the Arabidopsis proteome. Notably, many of the genes retained in duplicate are involved in regulatory functions (Table 1), which seems particularly understandable in terms of the creation of biological novelties or adaptation to ecological environments. Second, our results suggest that, in general, genes that remain duplicated do not tend to retain redundant functions; their divergent expression patterns or rates of sequence evolution indicate that they are becoming specialized. Finally, as exemplified in this analysis, the simultaneous duplication of all genes gives the evolutionary process the opportunity to play with complete duplicated pathways rather than individual genes and consequently to follow more complex paths.
One of the important challenges facing plant biologists is to redirect the progress made in unraveling all aspects of Arabidopsis biology into improving plants of agronomic importance. It is often anticipated that homologous genes and more particularly orthologs are likely to perform similar or identical functions in different organisms, which would considerably facilitate the identification of gene functions in nonmodel species. The observation that even distantly related plants may share some degree of conserved colinearity with segments of Arabidopsis chromosomes makes the idea rather appealing (Acarkan et al., 2000; Grant et al., 2000; Ku et al., 2000; Gebhardt et al., 2003; Zhu et al., 2003). On the other hand, more and more data indicate that polyploidy and aneuploidy occur frequently in plants (Shoemaker et al., 1996; Gaut and Doebley, 1997; Arabidopsis Genome Initiative, 2000; Otto and Whitton, 2000; Wendel, 2000; Goff et al., 2002; Vandepoele et al., 2003). Even the diploid Brassica species, which are the closest crop relatives of Arabidopsis, underwent three probable additional events of genome duplication since their divergence from Arabidopsis (Lagercrantz and Lydiate, 1996). Thus, simple orthologous genes in a 1:1 relationship that have not been duplicated since the separation of Arabidopsis and a species of interest might be relatively rare. Because our results suggest that functional divergence of duplicated genes is the rule rather than the exception, functional equivalence between orthologous genes may be limited, which might complicate the transfer of knowledge from Arabidopsis to crop plants.
METHODS
Sequence Data
The sequences of all Arabidopsis thaliana predicted genes were downloaded from the Institute for Genomic Research (TIGR) database (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/). Transposable element genes and tandem duplicates were removed according to Blanc et al. (2003), leaving 21,999 protein genes for further analysis. The lists of polyploidy-derived duplicated gene pairs used in this study (Blanc and Bowers datasets) and their age classes are available at http://wolfe.gen.tcd.ie/blanc/supp/functional.html. The β and γ age classes from the Bowers dataset (Bowers et al., 2003) were pooled in a single old class. For the recent age class, the Blanc and Bowers datasets contained 2584 and 3802 duplicate pairs, respectively (2474 pairs in common). For the old age class, the datasets contained 914 and 1949 pairs, respectively (497 pairs in common).
GO and MIPS Gene Annotations
We downloaded two sets of GO annotations for Arabidopsis proteins from the Arabidopsis Information Resource (Rhee et al., 2003) and TIGR (Wortman et al., 2003) databases. We also scanned all protein sequences from the Arabidopsis proteome against the InterPro protein signature database with the InterProScan program (Zdobnov and Apweiler, 2001). Using the available mapping of InterPro signatures to GO terms, we constructed a third set of gene annotations and finally combined the three datasets into a single larger one. Among the 21,999 proteins, 19,706 were annotated (14,018, 14,726, and 10,349 in the molecular function, cellular component, and biological process subdivisions, respectively) with 2993 different GO terms. The FunCat gene annotations were downloaded from the MIPS database (Schoof et al., 2002; ftp://ftpmips.gsf.de/catalog/) and edited to remove functional categories irrelevant for plants (categories referring to processes specific to animals, fungi, or prokaryotes). The resulting annotation data contained 18,935 of the studied genes organized in 1006 functional categories. On average, a gene was associated with 11.6 GO terms and 10.7 MIPS categories.
For each functional category, we estimated the number of genes in the preduplication Arabidopsis genome by adding 1 for a gene without polyploidy-derived copy and 0.5 for a gene with a polyploidy-derived duplicate. Among the functional categories with at least five genes in the preduplication genome, we tested if the number of genes retained in duplicate today is significantly higher or lower than expected by chance using the binomial distribution. For the Blanc dataset, the numbers of functional categories tested in the GO annotation scheme were 732 in the biological process subdivision, 129 in the cellular component subdivision, and 558 in the molecular function subdivision. The corresponding numbers for the Bowers dataset were 727, 129, and 555, respectively. In the MIPS annotation scheme, 597 categories were tested with the Blanc dataset and 596 with the Bowers dataset. The proportion of retained genes expected under random loss for a given functional category was set to the overall proportion of genes retained in the corresponding annotation dataset. Because of the hierarchical structure of the GO and MIPS annotation systems (parent–children relationships), the result of the statistical test for any functional category is likely to be correlated with the results for its child categories. This could lead to an over-counting of the number of significantly overduplicated or underduplicated categories. To avoid this, we tested each category after subtracting the genes assigned to all child categories that had significant P values (P ≤ 0.01). Moreover, it must be noted that repeated statistical tests may lead to the inclusion of false positives in the lists presented in Tables 1 and 2. Because of the nonindependence between functional categories, the Bonferroni correction for multiple tests could not be applied to avoid this problem. Therefore, the P value attached to each functional category must be taken with caution.
Expression Data
Expression data obtained with the ATH1 Affymetrix Arabidopsis microarray was retrieved from the NASCArray database (http://ssbdjc2.nottingham.ac.uk/narrays/experimentbrowse.pl). These datasets correspond to expression intensities under various experimental conditions and tissues. For each microarray experiment, the overall intensity mean was calculated excluding the top 2% and bottom 2% of signal intensities. The original signal values were scaled such that the mean was made equal to 100. Expression intensities were averaged among replicates, and the final expression profiles for each gene were composed of 62 expression values representing different experimental conditions and/or tissues. We removed 128 genes with the potential for cross-hybridization (marked with the “x” suffix on their probe ID). We also discarded all expression profiles without any expression signal above 150. To normalize the remaining expression profiles, each intensity value was subtracted by the mean expression intensity of the profile and then divided by the standard deviation.
Detection of Asymmetric Sequence Evolution
We restricted this analysis to pairs of duplicates formed by the recent polyploidy event and for which the structure of both genes is fully supported by full-length cDNA. To identify outgroup sequences, Arabidopsis duplicated gene products were searched against plant protein sequences using the BLASTP program (Altschul et al., 1997). Because the most recent polyploidy event of Arabidopsis has probably occurred in the Brassicaceae lineage (Blanc et al., 2003), only matches with non-Brassicaceae sequences were considered. Moreover, we only kept sequences that aligned with at least 80% of the Arabidopsis protein length for further analysis. We identified an outgroup sequence for 833 pairs of duplicated genes. For each triplet of sequences, multiple alignments were constructed using the T-Coffee program (Notredame et al., 2000) with parameters gapopen = 10 and gapext = 2. Sites containing gaps were subsequently removed from the alignments. We use the codeml program from the PAML package (Yang, 1999) to compute the maximum likelihood estimates of the two competing hypotheses (unconstrained rate of evolution versus clock-like rate of evolution) with the Jones substitution matrix and the γ correction to accommodate variability in substitution rates. For 27 triplets, the distance between the paralogous proteins was larger than the distance between one of the Arabidopsis proteins and the outgroup sequence. Because this potentially indicates an incorrect choice of outgroup, we further examined these triplets. We estimated the levels of synonymous nucleotide substitution (Ks) among the three sequences using the modified Nei and Gojobori method (Nei and Gojobori, 1986). Because synonymous codon positions are largely free from selection, they accumulate substitutions at a roughly constant rate. Hence, Ks can be considered as a much more reliable molecular clock than amino acid distances. For 3 of the 27 triplets, the Ks values between the paralogs were higher than the Ks values between the paralogs and their outgroup. These three triplets were discarded from further analyses. The list of the sequence accessions for each triplet and their log likelihoods are available upon request.
Data Availability
Raw and analyzed data can be found at http://wolfe.gen.tcd.ie/blanc/supp/functional.html and on The Plant Cell Web site (www.plantcell.org).
Supplementary Material
Acknowledgments
This study was supported by Science Foundation Ireland.
Online version contains Web-only data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Guillaume Blanc (g_blanc@univ-perp.fr).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.021410.
References
- Acarkan, A., Rossberg, M., Koch, M., and Schmidt, R. (2000). Comparative genome analysis reveals extensive conservation of genome organisation for Arabidopsis thaliana and Capsella rubella. Plant J. 23, 55–62. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Cronn, R., Percifield, R., and Wendel, J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100, 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., et al. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J.A., Bhadra, U., Bhadra, M.P., and Auger, D.L. (2001). Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234, 275–288. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of`chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Caffrey, D.R., O'Neill, L.A., and Shields, D.C. (1999). The evolution of the MAP kinase pathways: Coduplication of interacting proteins leads to new signaling cascades. J. Mol. Evol. 49, 567–582. [DOI] [PubMed] [Google Scholar]
- Conant, G.C., and Wagner, A. (2003). Asymmetric sequence divergence of duplicate genes. Genome Res. 13, 2052–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T., and Clark, A.G. (2001). Differential selection after duplication in mammalian developmental genes. Mol. Biol. Evol. 18, 557–562. [DOI] [PubMed] [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and Mackenzie, S.A. (2003). Nuclear genes that encode mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva, M.D., Wu, M., Eisen, J.A., and Salzberg, S. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 51, 859–866. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, C., Walkemeier, B., Henselewski, H., Barakat, A., Delseny, M., and Stüber, K. (2003). Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J. 34, 529–541. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., and Spring, J. (1998). Genetic redundancy in vertebrates: Polyploidy and persistence of genes encoding multidomains proteins. Trends Genet. 14, 46–49. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Yang, Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736. [DOI] [PubMed] [Google Scholar]
- Golldack, D., Popova, O.V., and Dietz, K.-J. (2002). Mutation of the matrix metalloproteinase At2-MMP inhibits growth and causes late flowering and early senescence in Arabidopsis. J. Biol. Chem. 277, 5541–5547. [DOI] [PubMed] [Google Scholar]
- Grant, D., Cregan, P., and Shoemaker, R.C. (2000). Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. (1999). Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 16, 1664–1674. [DOI] [PubMed] [Google Scholar]
- Gu, X., Wang, Y., and Gu, J. (2002. a). Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat. Genet. 31, 205–209. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Nicolae, D., Lu, H.H., and Li, W.H. (2002. b). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18, 609–613. [DOI] [PubMed] [Google Scholar]
- Ihmels, J., Levy, R., and Barkai, N. (2004). Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 22, 86–92. [DOI] [PubMed] [Google Scholar]
- Ku, H.M., Vision, T., Liu, J., and Tanksley, S.D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz, A., Muller, A., Hennig, P., Kaiser, W.M., Piotrowski, M., and Weiler, E.W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30, 95–106. [DOI] [PubMed] [Google Scholar]
- Lagercrantz, U., and Lydiate, D.J. (1996). Comparative genome mapping in Brassica. Genetics 144, 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.M., and Sonnhammer, E.L. (2003). Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 13, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., and Wendel, J.F. (2002). Non-Mendelian phenomena in allopolyploid genome evolution. Curr. Genomics 3, 489–505. [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2003). The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics 3, 35–44. [PubMed] [Google Scholar]
- Makova, K.D., and Li, W.H. (2003). Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13, 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght, A., Hokamp, K., and Wolfe, K.H. (2002). Extensive genomic duplication during early chordate evolution. Nat. Genet 31, 200–204. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- Nei, M., Gu, X., and Sitnikova, T. (1997). Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., and Heringa, J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag).
- Osborn, T.C., Pires, J.C., Birchler, J.A., Auger, D.L., Chen, Z.J., Lee, H.-S., Comai, L., Madlung, A., Doerge, R.W., Colot, V., and Martienssen, R.A. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. [DOI] [PubMed] [Google Scholar]
- Otto, S.P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. [DOI] [PubMed] [Google Scholar]
- Papp, B., Pal, C., and Hurst, L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. [DOI] [PubMed] [Google Scholar]
- Pastor-Satorras, R., Smith, E., and Sole, R.V. (2003). Evolving protein interaction networks through gene duplication. J. Theor. Biol. 222, 199–210. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.X., Katsar, C.S., Lan, T.H., Lin, Y.R., Ming, R., and Wright, R.J. (2000). Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randez-Gil, F., Sanz, P., Entian, K.-D., and Prieto, J.A. (1998). Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol. Cell. Biol. 18, 2940–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.Y., et al. (2003). The Arabidopsis information resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., Zaccaria, P., Gundlach, H., Lemcke, K., Rudd, S., Kolesov, G., Arnold, R., Mewes, H.W., and Mayer, K.F. (2002). MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 30, 91–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe, C., and Wolfe, K.H. (1999). Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 2, 548–554. [DOI] [PubMed] [Google Scholar]
- Shoemaker, R.C., Polzin, K., Labate, J., Specht, J., Brummer, E.C., Olson, T., Young, N., Concibido, V., Wilcox, J., Tamulonis, J.P., Kochert, G., and Boerma, H.R. (1996). Genome duplication in soybean (Glycine subgenus soja). Genetics 144, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C.E., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99, 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G. (1950). Variation and Evolution in Plants. (New York: Columbia University Press).
- Teichmann, S.A., and Veitia, R.A. (2004). Genes encoding subunits of stable complexes are clustered on the yeast chromosomes. Genetics, in press. [DOI] [PMC free article] [PubMed]
- Vandepoele, K., Simillion, C., and Van de Peer, Y. (2003). Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15, 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R.A. (2003). Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 220, 19–25. [DOI] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Walsh, J.B. (1995). How often do duplicated genes evolve new functions? Genetics 139, 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. [PubMed] [Google Scholar]
- Wolfe, K.H., and Shields, D.C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- Wong, S., Butler, G., and Wolfe, K.H. (2002). Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl. Acad. Sci. USA 99, 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman, J.R., Haas, B.J., Hannick, L.I., Smith, R.K., Jr., Maiti, R., Ronning, C.M., Chan, A.P., Yu, C., Ayele, M., Whitelaw, C.A., White, O.R., and Town, C.D. (2003). Annotation of the Arabidopsis genome. Plant Physiol. 132, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L.F., Hughes, T.R., Davierwala, A.P., Robinson, M.D., Stoughton, R., and Altschuler, S.J. (2002). Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet 31, 255–265. [DOI] [PubMed] [Google Scholar]
- Yang, Z. (1999). Phylogenetic analysis by maximum likelihood (PAML), version 2. (London, UK: University College).
- Zdobnov, E.M., and Apweiler, R. (2001). InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Vision, T.J., and Gaut, B.S. (2002). Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 19, 1464–1473. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Kim, D.-J., Baek, J.-M., Choi, H.-K., Ellis, L.C., Kuester, H., McCombie, W.R., Peng, H.-M., and Cook, D.R. (2003). Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol. 131, 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and analyzed data can be found at http://wolfe.gen.tcd.ie/blanc/supp/functional.html and on The Plant Cell Web site (www.plantcell.org).