Abstract
Hmt1 is the major type I arginine methyltransferase in the yeast Saccharomyces cerevisiae and facilitates the nucleocytoplasmic transport of mRNA-binding proteins through their methylation. Here we demonstrate that Hmt1 is recruited during the beginning of the transcriptional elongation process. Hmt1 methylates Yra1 and Hrp1, two mRNA-binding proteins important for mRNA processing and export. Moreover, loss of Hmt1 affects interactions between mRNA-binding proteins and Tho2, a component of the TREX (transcription/export) complex that is important for transcriptional elongation and recruitment of mRNA export factors. Furthermore, RNA in situ hybridization analysis demonstrates that loss of Hmt1 results in slowed release of HSP104 mRNA from the sites of transcription. Genome-wide location analysis shows that Hmt1 is bound to specific functional gene classes, many of which are also bound by Tho2 and other mRNA-processing factors. These data suggest a model whereby Hmt1 affects transcriptional elongation and, as a result, influences recruitment of RNA-processing factors.
Keywords: Hmt1, arginine methylation, genome-wide localization
In eukaryotic cells, pre-messenger RNAs (pre-mRNAs) must be fully processed and packaged into mature messenger ribonucleoparticles (mRNPs) before export to the cytoplasm as fully translatable mRNAs. Intranuclear RNA processing steps, such as 5′-capping, splicing, 3′-end cleavage, and polyadenylation, are accomplished through the association of numerous RNA-binding proteins (RBPs) such as serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) with the pre-mRNA (for review, see Dreyfuss et al. 2002; Lei and Silver 2002b; Reed and Hurt 2002). Many RBPs that participate in RNA processing and export contain a variety of posttranslational modifications such as phosphorylation and methylation. The dynamic interactions between RBPs and pre-mRNAs suggest that their binding to and dissociation from RNAs and other proteins may be regulated by these posttranslational modifications.
One type of posttranslational modification commonly found in RNA-binding proteins is the methylation of arginine residues, usually in the context of arginine- and glycine-rich motifs (for review, see Gary and Clarke 1998). The enzymes that catalyze this process are called protein arginine methyltransferase, or PRMTs. hnRNPs are a major substrate of PRMT1 in yeast and mammalian cells. Methylated hnRNPs contain at least one N-terminal RRM-type (RNA recognition motif) RNA-binding motif in conjunction with RGG-rich (arginine-glycine-glycine) repeats, the sites of arginine methylation, in the C-terminal domains (Liu and Dreyfuss 1995).
Recent studies have shown that arginine methylation is important for modulating protein-protein interactions. For example, loss of arginine methylation on the STAT1 protein inhibits association with its inhibitor PIAS, resulting in decreased interferon responses mediated by STAT1 (Mowen et al. 2001). Methylation of the Src kinase substrate Sam68 has been shown to change its affinity for SH3-containing proteins, resulting in alteration of its function (Bedford et al. 2000). In addition, arginine methylation of the transcriptional elongation factor Spt5 regulates its interaction with RNA polymerase (Pol) II, thereby globally affecting transcription (Kwak et al. 2003). However, the precise role of methylation of the many RBPs remains unclear.
In Saccharomyces cerevisiae, Hmt1 (also termed RMT1) has been previously identified as the major type I arginine methyltransferase and is functionally equivalent to the mammalian PRMT1 (Gary et al. 1996; Henry and Silver 1996; Gary and Clarke 1998). Hmt1 has been shown to be important for nuclear transport of the hnRNPs Nab2, Hrp1, and Npl3 (Shen et al. 1998; McBride et al. 2000; Green et al. 2002). Nab2, found in a complex with Hrp1, controls mRNA poly(A) tail length and its in vivo methylation state is dependent on the presence of Hmt1 (Gavin et al. 2002; Green et al. 2002; Hector et al. 2002; Ho et al. 2002). Hrp1, also an in vitro substrate of Hmt1 (Shen et al. 1998), is a component of the cleavage factor I (CFI) that is involved in 3′-end premRNA processing (Kessler et al. 1997; Gross and Moore 2001). Npl3, a major SR-like RNA-binding protein important for mRNA export (Bossie and Silver 1992; Kadowaki et al. 1994), is methylated on its RGG repeats by Hmt1 and also phosphorylated by the SR protein kinase Sky1 (Henry and Silver 1996; Siebel and Guthrie 1996; Gilbert et al. 2001). The observation that Sky1 localizes to the cytoplasm whereas Hmt1 is predominately nuclear suggests that the regulation of import and export of substrate proteins may be determined by the “opposing effects” of methylation and phosphorylation (Siebel et al. 1999; Yun and Fu 2000).
Given that several pre-mRNA processing factors and components of the transcriptional machinery are substrates of arginine methyltransferases, arginine methylation may play a role in coordinating the cotranscriptional assembly of the mRNP. Several lines of evidence for the coupling of splicing and transcription include the removal of the spliced 5′-end intron on the nascent RNAs in Chironomus tentans (Wieslander et al. 1996), as well as the cotranscriptional recruitment of U1 sn-RNP (Kotovic et al. 2003) and the splicing factor Sub2 (Lei and Silver 2002a) in S. cerevisiae. In S. cerevisiae, the 3′-end processing factors are recruited by RNA Pol II near the promoter, suggesting that they are recruited during the early stages of transcription (Licatalosi et al. 2002; Ryan et al. 2002). Interestingly, mutation of RNA15, another component of CFI, affects the recruitment of the mRNA export factor Yra1 to nascent RNAs (Lei and Silver 2002a). Further underscoring the extensive coupling between transcription and mRNA export is the cotranscriptional recruitment of Npl3, Yra1, and components of the TREX complex (transcription/export; Lei et al. 2001; Strasser et al. 2002). A yeast homolog of Aly, Yra1 belongs to a family of evolutionarily conserved hnRNP-like proteins (Stutz et al. 2000). Unlike Npl3, both RNA Pol II transcription and other pre-mRNA processing events such as 3′-end formation must occur for proper Yra1 recruitment to take place (Lei and Silver 2002a). Together, these lines of evidence support the idea that early recruitment of 3′-end processing factors and mRNA export factors to transcriptionally active genes facilitate the formation of proper mRNPs.
In this study, we demonstrate that Hmt1 is recruited to genes during transcription and that it functions to modulate protein-protein interactions within an mRNP in an RNA-dependent manner. We have used genome-wide location analysis to identify the functional gene classes with which Hmt1 and its substrates associate. Additionally, we have determined the effect of Hmt1 on the chromatin-binding specificity of its substrates. Together, these results demonstrate how an arginine methyltransferase affects mRNP dynamics.
Results
Hmt1 is recruited to genes cotranscriptionally
We used chromatin immunoprecipitation (ChIP) of Hmt1 at the galactose-inducible GAL10 gene to demonstrate that, like many of its substrates, Hmt1 can be cotranscriptionally recruited to genes. For ChIP experiments, yeast cells expressing a Myc epitope-tagged version of Hmt1 were generated (see Materials and Methods). This Myc-tagged Hmt1 was determined to be functional by its ability to methylate one of its substrates, Npl3 (data not shown).
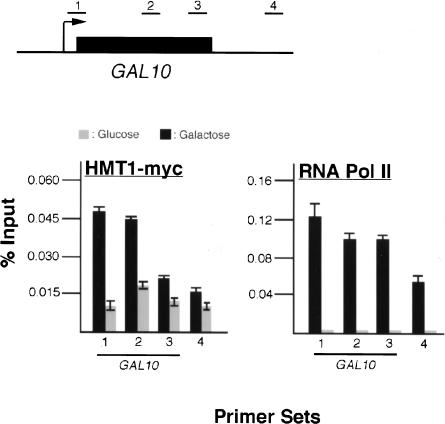
To determine whether and where Hmt1 associates with the transcribed GAL10 gene, quantitative PCR was performed on immunoprecipitated DNA fragments using primer sets spanning the GAL10 coding region as well as the downstream intergenic region (Fig. 1, top). Upon galactose induction, α-Myc antibodies immunoprecipitated a significant level of the 5′-end of GAL10 and central part of its coding sequence in comparison with cells grown in glucose (Fig. 1, bottom left, cf. black and the gray bars). The amount of GAL10 from the central part of the ORF immunoprecipitated by α-Myc antibodies is slightly less than the 5′-end of the coding sequence under galactose induction, but still considerably more than cells grown in glucose (Fig. 1, bottom left). No significant level of Hmt1 occupancy was detected at the 3′-end of GAL10 or the intergenic region in either growth condition (Fig. 1, bottom left). As a control to show that Hmt1 recruitment is not a function of polymerase occupancy, we performed the same experiment using α-Rpb3, a monoclonal antibody directed against a subunit of yeast RNA Pol II. Rpb3 was found to occupy the entire GAL10 gene with approximately equal levels. A high level of Rpb3 was also found to be present at the GAL10/7 intergenic region, consistent with previously published results (Greger and Proudfoot 1998). These data indicate that Hmt1 is cotranscriptionally recruited to genes at the 5′-end and up to the middle of the coding region. The decreased Hmt1 occupancy at the 3′-coding sequence implies that Hmt1 is recruited during transcriptional initiation or early stages of transcriptional elongation.
Figure 1.
Hmt1 is cotranscriptionally recruited to the GAL10 gene. Hmt1-Myc is cross-linked to the coding sequence of GAL10 in a transcription-dependent manner. ChIP was performed using either α-Myc or α-RPB3 on a strain containing Hmt1-Myc under galactose-inducible conditions. Both α-Myc and α-RPB3 immunoprecipitated chromatin fragments were subjected to quantitative PCR. (Top) The schematic representation of the GAL10 gene with primer sets used in quantitative PCR is shown. (Bottom) The quantitative PCR results from cells grown in both glucose (gray bar) and galactose (black bar) are shown in the bar graphs. The numbers under each graph correspond to the amplified sequence from the GAL10 gene by the specific primer sets indicated above.
Hmt1 methylates the mRNA export factor Yra1 and the 3′-processing factor Hrp1 in vivo
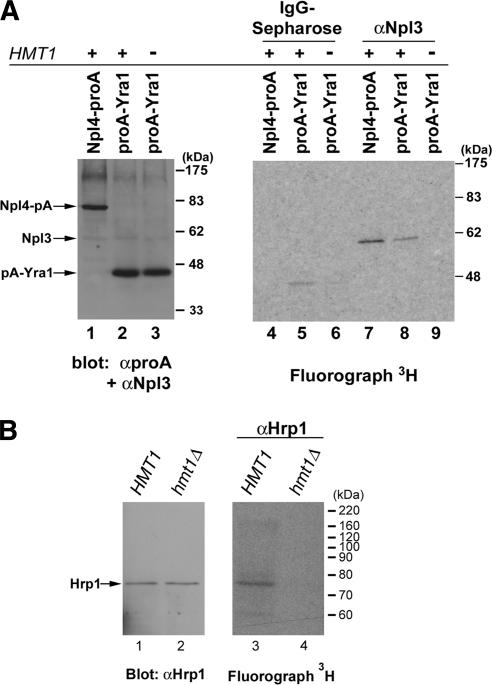
Comparison of the amino acid sequences of the RNA-binding protein Yra1 (Aly/REF in metazoans) across several species reveals a cluster of arginine and glycine residues in the N terminus region (data not shown). To examine whether Yra1 is arginine-methylated in yeast, we used the in vivo methylation assay (see Materials and Methods). In a strain that contains protein-A-tagged Yra1, immunoprecipitates from IgG-coupled sepharose beads included a single, major band that corresponds to methylated protein-A-tagged Yra1, as confirmed by immunoblotting with α-protein A antibody (Fig. 2A, lanes 2,5). Methylation of Yra1 is dependent on Hmt1 because protein-A-tagged Yra1 is no longer methylated in a strain that lacks Hmt1 (Fig. 2A, lanes 3,6). To determine if methylation of Yra1 is caused by the presence of the protein A tag, a resident ER protein Npl4 tagged with protein A was also tested. As expected, protein-A-tagged Npl4 is not methylated (Fig. 2A, lane 4). As a positive control for the methylation assay, methylated Npl3 can be immunoprecipitated from extracts of both protein-A-tagged Npl4 and Yra1 strains (Fig. 2A, lanes 7,8), but not from extracts in a strain that lacks Hmt1 (Fig. 2A, lane 9).
Figure 2.
Hmt1 methylates RNA-processing factors Yra1 and Hrp1. All immunoprecipitates were either resolved by 7% or 10% SDS-PAGE and immunoblotted with antibody as described (left panel) or visualized by fluorography (right panel). The migration of molecular mass markers is shown at right. (A) Yra1 is arginine-methylated by Hmt1 in vivo. A CEN-plasmid carrying protein-A-tagged Yra1 (proA-Yra1) in both HMT1 and hmt1Δ cells was subjected to the in vivo methylation assay in the presence of [methyl-3H]-SAM. Protein-A-tagged Yra1 was immunoprecipitated from an equivalent amount of lysate by incubating with IgG sepharose or α-Npl3-conjugated beads. To determine the efficiency of Yra1 immunoprecipitations, α-PrA antibody was used in immunoblotting (lanes 2,3). A band corresponding to [methyl-3H]-labeled protA-Yra1 is observed only in the presence of Hmt1 (lane 5) but not in the absence of Hmt1 (lane 6). Protein-A-tagged Npl4 (Npl4-proA) was used as a negative control (lanes 1,4). As a positive control for methylation, [methyl-3H]-labeled Npl3 was visualized with approximately equal intensity in both Npl4-proA and ProA-Yra1 strains (lanes 7,8), whereas in the absence of Hmt1, no methylated Npl3 was visualized (lane 9). (B) Hrp1 is an in vivo substrate of Hmt1. After an in vivo methylation reaction in the presence of [methyl-3H]SAM, immunoprecipitation was performed from equivalent amount of lysate in either HMT1 or hmt1Δ by incubating with α-Hrp1-conjugated beads. In the fluorograph, the band corresponding to [methyl-3H]-labeled Hrp1 is observed (lane 3), whereas no [methyl-3H]-labeled Hrp1 is observed in the absence of Hmt1 (lane 4). (Lanes 1,2) Equal loading and immunoprecipitation efficiency from both HMT1 and hmt1Δ was determined by immunoblotting with α-Hrp1 antibody.
As it was previously shown to be a substrate of purified Hmt1 in vitro (Shen et al. 1998), we examined whether Hrp1, a factor that participates in the 3′-end formation and polyadenylation (Kessler et al. 1997), can be arginine-methylated by Hmt1 in vivo. Using a rabbit polyclonal antibody generated against Hrp1, we performed in vivo methylation assays in both wild-type and hmt1Δ mutants. Immunoblotting of the immunoprecipitates using α-Hrp1 antibody showed approximately equal amounts of Hrp1 being immunoprecipitated from both HMT1 and hmt1Δ strains (Fig. 2B, cf. lanes 1 and 2). Visualization of the corresponding fluorograph reveals [methyl-3H]-labeled Hrp1 only in the HMT1 cells (Fig. 2B, lane 3) but not in the hmt1Δ cells (Fig. 2B, lane 4). Taken together, the above results demonstrate that mRNP components Yra1 and Hrp1 are arginine-methylated in vivo.
Hmt1 can modulate protein-protein interactions
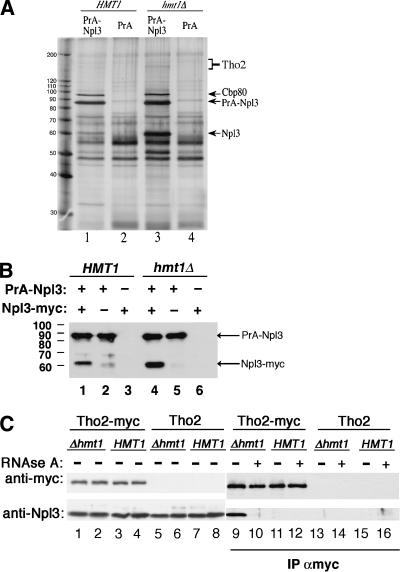
The presence of Hmt1 at the promoter and the central region of a gene during transcription suggests that protein methylation could be involved in modulating protein complexes early in mRNP formation. To identify proteins that interact with Npl3 in a methylation-state-dependent manner, a protein A Npl3 fusion protein (PrANpl3) was expressed in a strain that either contained a functional Hmt1 (HMT1) or lacked Hmt1 (hmt1Δ). Silver staining of proteins that copurified with PrA-Npl3 but not PrA alone was compared in both HMT1 and hmt1Δ cells (Fig. 3A). Although several bands with different degrees of intensity copurified with PrA-Npl3, the only reproducible differences were the bands that migrated at -60 kDa and 150-180 kDa in hmt1Δ. These interactions were reduced in HMT1 cells (Fig. 3A, lanes 1,3). In addition, a 97-kDa band was observed in both HMT1 and hmt1Δ cells (Fig. 3A, lanes 1,3). Subsequent identification of the 97-kDa band by mass spectrometry yielded Cbp80, the largest subunit of mRNA cap-binding protein. Cbp80 and Npl3 have previously been shown to interact (Shen et al. 1998).
Figure 3.
Hmt1 modulates protein-protein interactions. (A) Silver stain of immunoprecipitates from protein A-Npl3 (PrANpl3) in HMT1 and hmt1Δ cells. Increased PrA-Npl3 association was observed with untagged Npl3, Cbp80, and the TREX component Tho2 (lane 3) in hmt1Δ compared with HMT1 cells (lane 1). (Lanes 2,4) Negative controls in both HMT1 and hmt1Δ cells were performed with PrA-only beads. (B) The immunoprecipitates from PrA-Npl3 and protein A alone (PrA) expressed either in HMT1 or hmt1Δ cells were analyzed in the context of Npl3-Myc. Total lysate (see Materials and Methods) was resolved by 10% SDS-PAGE prior to immunoblotting using polyclonal anti-Myc antiserum. Both PrA-Npl3 and Npl3-Myc fusion proteins were recognized by the rabbit antiserum. The migration of the molecular mass marker is shown at left. (C) The interaction between Npl3 and Tho2 is increased in the absence of Hmt1. The genomic copy of THO2 in HMT1 and hmt1Δ strains was tagged with a Myc sequence. Parental THO2 strains (PSY 867, PSY 865) and Tho2-Myc strains (PSY 3210, PSY 3211) were grown in YPD and lysed, and Tho2-Myc was immunoprecipitated from equivalent amounts of lysate by incubation with α-Myc-conjugated beads. Samples were subsequently divided and treated (+) or not treated (-) with RNase A for 20 min at 25°C. The proteins isolated were loaded in equal amounts and resolved by 7% SDS-PAGE. Tho2-Myc and Npl3 were detected by probing the top of the blot with α-Myc antibody and the bottom of the blot with α-Npl3, respectively. The inputs (lanes 1,3,5,7) and supernatants post IP (lanes 2,4,6,8) are shown.
Interestingly, the prominent 60-kDa band was identified as endogenous, untagged Npl3. It is possible that the increase in Npl3 observed in the hmt1Δ strain was caused by an increased interaction of PrA-Npl3 with untagged, endogenous Npl3. Alternatively, there could be selective proteolytic clipping of the PrA-Npl3 in the hmt1Δ strain. To address these two possibilities, PrANpl3 and Myc-tagged Npl3 were expressed in cells lacking endogenous Npl3. Proteins that copurified with PrANpl3 were analyzed by immunoblotting with polyclonal α-Myc antibody, which binds to the protein A domain of the fusion protein as well as to the Myc epitope (Fig. 3B). The Npl3-Myc band migrating above 60 kDa was observed specifically in lanes containing both PrA-Npl3 and Npl3-Myc, demonstrating that these two proteins interact (Fig. 3B, lane 1). As with intact Npl3, the intensity of the Npl3-Myc band was also increased in the hmt1Δ strain (Fig. 3B, cf. lanes 1 and 4). This interaction is RNA-independent as RNAse A treatment did not abolish the association between PrA-Npl3 and Npl3-Myc (data not shown). The minor bands migrating slightly lower than that of Npl3-Myc in lanes 2 and 5 in Figure 3B are degradation products of PrA-Npl3. Together, these data suggest that methylation of Npl3 modulates its self-association.
In addition to the prominent bands described above, one of the minor copurifying bands in the 150-180-kDa range was subsequently identified by mass spectrometry as Tho2, a component of the TREX complex. To confirm the role of Hmt1 in the biochemical association of Npl3 and Tho2, a nine-copy Myc tag was integrated at the 3′-end of the THO2 gene in a wild-type strain and then genetically crossed with an hmt1-null strain to create a Tho2-Myc hmt1Δ strain. Immunoprecipitation using α-Myc antibodies on the extracts of untagged and Myc-tagged Tho2 strains was followed by immunoblotting with α-Myc and α-Npl3 antibodies (Fig. 3C). In the absence of Hmt1, the interaction between Npl3 and Tho2 increased (Fig. 3C, cf. lanes 9 and 11). This interaction is RNA-dependent because treatment with RNAse A abolishes the increased association (Fig. 3C, cf. lanes 10 and 12). Because the amino acid sequence of Tho2 contains possible sites of arginine methylation, we performed an in vivo methylation assay and found that Tho2 can also be methylated (data not shown). However, we were not able to determine whether this methylation is dependent on Hmt1 because the signal for methylated Tho2 is faint.
Loss of Hmt1 affects the kinetics of HSP104 mRNA production
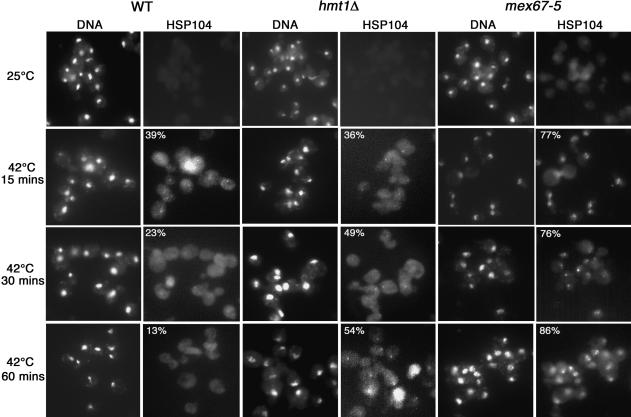
It is possible that biochemical association between Npl3 and Tho2 in the absence of Hmt1 affects the subsequent recruitment of mRNA-processing factors and thereby results in the improper release of transcripts from the site of transcription or release of the proteins from the transcripts. To address this, we examined the fate of a heat-shock protein transcript, Hsp104. It was previously shown that a block of mRNA nuclear export leads to the retention of transcripts at the site of transcription and that this accumulation can be visualized as a “nuclear dot” using mRNA fluorescent in situ hybridization (FISH) analysis with a transcript-specific probe (Jensen et al. 2001). We therefore used this assay to examine the kinetics of HSP104 mRNA production in hmt1Δ cells. Using a Cy3-conjugated oligonucleotide probe against the 5′-end of HSP104 mRNA, the appearance of a “nuclear dot” is detectable in both wild-type and hmt1Δ cells 5 min after a temperature shift from 25°C to 42°C (data not shown). In wild-type cells, the percentage of cells with a “nuclear dot” peaks at 15 min (Fig. 4, HSP104 panel of wild-type cells, 15 min at 42°C). The population with a “nuclear dot” gradually decreased with longer incubation at 42°C (Fig. 4, HSP104 panel of wild-type cells, 30 and 60 min at 42°C).
Figure 4.
Cells harboring hmt1Δ retain HSP104 transcript localization with time. Localization of HSP104 mRNAs at intranuclear foci after temperature shift is visualized as a “nuclear dot” in a cell. FISH analysis using Cy3-labeled oligonucleotides (THJ361) against the 5′-end of the HSP104 transcript was performed on cultures grown at 25°C or temperature-shifted to 42°C for times indicated. mex67-5 was used as a positive control for this experiment. The percentage of cells observed with a “nuclear dot” is denoted in the upper corner of the HSP104 panel.
The percentage of hmt1Δ cells with a “nuclear dot” after a 15-min temperature shift is similar to wild-type cells (Fig. 4, HSP104 panel of hmt1Δ cells, 15 min at 42°C). However, in contrast to wild-type cells, the percentage of hmt1Δ cells with a “nuclear dot” did not decrease with longer incubation at 42°C (Fig. 4, HSP104 panel of hmt1Δ cells, 30 and 60 min at 42°C). As a positive control, the same experiment was performed in mex67-5 cells; these cells exhibited a strong nuclear retention phenotype, as described previously (Fig. 4, mex67-5 panels; Jensen et al. 2001). Thus, the mRNA-FISH analysis demonstrates that Hmt1 influences the kinetics of HSP104 transcript production.
Hmt1 is recruited to highly transcribed genes
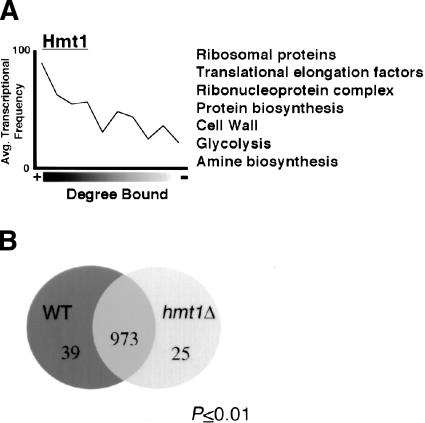
Because Hmt1 can be cotranscriptionally recruited to genes, we sought to determine which genes are associated with Hmt1 to better understand its role in gene regulation. To this end, we used genome-wide location analysis to determine the occupancy profile for Hmt1. This approach reflects the association of a specific protein with genes through the combination of ChIP and full-length cDNA microarray analysis (Ren et al. 2000; Iyer et al. 2001). With this method, the genome-wide occupancy is surmised for each array spot as a ratio expressed as the fluorophore intensity from the chromatin fragments enriched by immunoprecipitation (IP) over the fluorophore intensity from unenriched chromatin fragments (Input). The final confidence value (P-value) for each spot from a combined set of three arrays was calculated using an error model provided by the Rosetta Resolver (see Materials and Methods). Spots that had both a P-value threshold of 0.02 and a ratio intensity >1.0 were included in the final data set (see Supplemental Material) and used to identify statistically significant gene classes as listed in the Yeast Proteome Database and the Gene Ontology Consortium (Ashburner et al. 2000) for S. cerevisiae (Berriz et al. 2003).
As shown in Figure 5A, genomic occupancy of Hmt1 correlates with transcriptional frequency (Holstege et al. 1998), suggesting that Hmt1 is recruited to actively transcribed genes. When the data were examined for enrichment of particular gene classes, we found that Hmt1 is bound to genes involved in a variety of functions, including general protein synthesis, metabolic and catabolic pathways, creation and maintenance of cell wall, and glycolysis (Fig. 5A). Because many of the glycolysis and ribosomal protein gene classes constitute some of the most highly transcribed genes in yeast, this supports the observed transcriptional bias seen in Figure 5A.
Figure 5.
Enrichment of gene functional groups bound by Hmt1 and the effect of Hmt1 on gene expression profile. (A) Correlation between degree of Hmt1-binding and their average transcriptional frequency. The average transcription frequency of genes bound by the studied proteins versus the degree of binding is plotted as a solid line. A partial list of enriched functional gene classes is listed next to the plot. (B) Gene expression changes due to the loss of Hmt1. The Venn diagram indicates the number of genes that are increased (hmt1Δ circle) or decreased (wild-type [WT] circle) at least twofold in Hmt1-null mutants. The number of genes not affected is indicated at the intersection of both circles. Only genes with P-values ≤0.01 are subjected to analysis.
Effect of Hmt1 on genome-wide binding of its substrates
To understand the interplay between Hmt1 and components of the mRNP, we extended the genome-wide location analysis approach to determine how Hmt1 affects mRNP formation. We therefore compared the genome-wide location profiles of mRNP components in wild-type (HMT1) versus Hmt1-null (hmt1Δ) cells. To ensure that any genome-wide occupancy changes observed are not caused by the difference in the gene expression profile resulting from the loss of Hmt1, we compared the gene expression profiles from both wild-type and hmt1Δ cells (Fig. 5B). We found that the absence of Hmt1 did not greatly impact gene expression levels as only 64 genes had levels that changed twofold or more (Fig. 5B). Thus, any changes we observed in the genome-wide binding studies are independent of changes in gene expression when HMT1 is deleted.
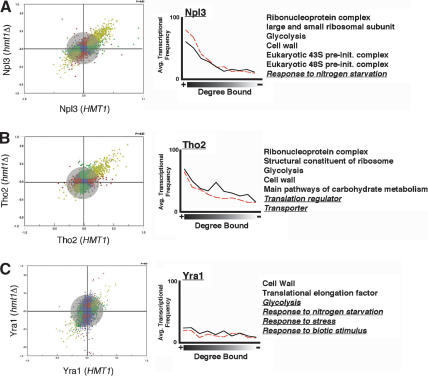
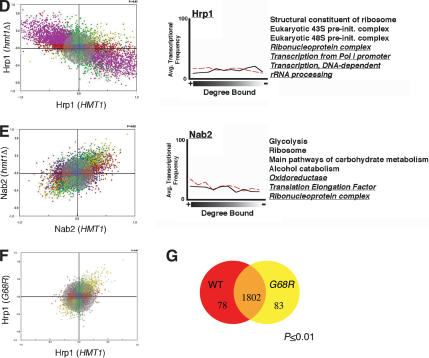
For each comparison of protein occupancy, we generated a scatterplot representing all protein-bound genes and their degree of occupancies in both backgrounds. In both Npl3 and Tho2, only a slight change in the occupancy is observed as evidenced by the clustering of the majority of spots within the gray circle (Fig. 6A,B). Absence of Hmt1 causes minor changes in the genes bound by Yra1 and Nab2, as demonstrated by a greater degree of scattered datapoints away from the center (Fig. 6C,E). The absence of HMT1 had the most profound effect on Hrp1 genomic occupancy (Fig. 6D); many Hrp1-bound genes are shifted from a low degree of binding in the HMT1 background to a high degree of binding in the hmt1Δ cells and vice versa (Fig. 6D, purple spots).
Figure 6.
Effects of Hmt1 on genome-wide localization profiles of its substrates. Genome-wide location analysis is used to determine whether recruitment of Hmt1 substrates is influenced by the absence of Hmt1 expression. The scatterplot represents the degree of occupancy by Npl3 (A), Tho2 (B), Yra1 (C), Hrp1 (D), and Nab2 (E) in the hmt1Δ strain (vertical axis) versus the occupancy in the HMT1 strain (horizontal axis). (F) The degree of occupancy by Hrp1 in the hmt1G68R mutant versus the HMT1 strain. The scale on each axis represents the degree of protein binding in a specific genetic background, wherein the scale of 0.0 to 1.0 represents the degree bound and 0.0 to -1.0 represents the degree unbound for any given gene. Blue spots represent the protein-bound genes unchanged in the degree of occupancy in both strains independent of that of the defined P-value. The red spots represent genes bound by the protein in the hmt1Δ or hmt1G68R cells, whereas the green spots are ones bound in the HMT1 cells whose calculated P-value is <0.02. The purple spots represent protein-bound genes with anticorrelated properties (i.e., bound in one background but not bound in the other background). The olive spots represent protein-bound genes in both backgrounds whose calculated P-value is <0.02. Lastly, the distance a gene spot deviates away from the center reflects the degree of change in the occupancy. Gene spots within the gray circle represent insignificant changes of occupancy in both backgrounds. In order for a gene spot to be considered as having a significant positive change (i.e., high degree of occupancy by a protein in the absence of Hmt1), it must possess a P-value of <0.02 in both HMT1 and hmt1Δ/hmt1G68R. In addition, the ratio of IP/Input must have a positive change of greater than twofold in the hmt1Δ/hmt1G68R cells as well as a negative change of greater than twofold in the HMT1 cells. The effect of Hmt1 on the distribution of its substrates across the genome is plotted next to the scatterplot. The average transcription frequency of genes bound by the studied protein versus the degree of binding is plotted as a solid (in wild-type cells) or dotted (in hmt1Δ cells) line. A partial list of enriched functional gene classes is displayed next to the plot. Functional gene classes that are changed between wild-type and hmt1Δ cells are underlined and italicized.
We next determined the enrichment of gene functional groups bound by these mRNP components. In each case, we found that the top bound populations are enriched for the same gene classes as Hmt1 (Figs. 5A, 6). Npl3 and Tho2, for example, are recruited to genes involved in protein synthesis such as genes involved in ribosome biogenesis (Fig. 6A,B). There are minor changes in Yra1-bound gene classes as a result of Hmt1 deletion (Fig. 6C). Nevertheless, genes involved in cell wall biosynthesis, a highly enriched gene class bound by Yra1, is also bound by Hmt1 (Figs. 5A, 6C). In hmt1Δ cells, Nab2 is recruited to several new gene classes in addition to the ones observed in the wild-type cells such as genes encoding oxidoreductases (Fig. 6E, underlined gene groups). Hrp1 no longer binds to genes involved in the ribonucleoprotein complex and rRNA-processing pathway in the hmt1Δ cells (Fig. 6D, underlined gene classes). When we compared the average transcriptional frequency of a gene with the degree of binding, we found that the loss of Hmt1 did not significantly alter the degree of correlation between occupancy and transcriptional frequency for all substrates examined (Fig. 6, cf. solid and dotted lines in all right panels).
To determine whether the changes in genomic occupancy are a result of methylation by Hmt1, we examined the genome-wide binding of Hrp1 in a previously characterized catalytic mutant of HMT1, hmt1G68R (McBride et al. 2000). Interestingly, the scatterplot indicated that the degree of change in Hrp1 binding as a result of hmt1G68R is less dramatic than in hmt1Δ cells (Fig. 6F). Nevertheless, some of the gene functional groups bound by Hrp1 in the wild type are lost in the hmt1G68R, such as genes involved in oxidoreductase and gluconeogenesis. The presence of Hmt1 catalytic point mutation thus appears to influence the genomic interaction of Hrp1, albeit to a less extent than the null mutation. This is consistent with hmt1G68R mutants not having a complete loss of function (see Discussion).
Similar to hmt1Δ cells, the hmt1G68R mutation did not significantly alter genome-wide transcription levels (Fig. 6G) and thus does not contribute to the degree of changes observed in the Hrp1-bound population. Together, these data support a role for Hmt1 in cotranscriptional recruitment of mRNP components to ensure proper association with their genomic targets.
Validation of genome-wide binding analysis
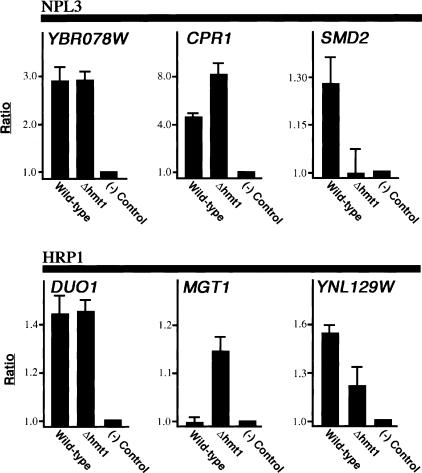
To validate the results of our genomic-binding studies, we performed directed ChIP on genes bound either by Hrp1 or Npl3 in both wild-type and hmt1Δ cells. In each case, we chose genes from the genome-wide data that show either different or equal binding, but with equal expression level, between wild-type and hmt1Δ cells. For Npl3, YBR078W displayed almost equal occupancy by Npl3 (Fig. 7, YBR078W panel), whereas binding to CPR1 is decreased in the wild-type cells when compared with hmt1Δ cells (Fig. 7, CPR1 panel). In contrast, Npl3 occupancy of SMD2 is higher in hmt1Δ cells (Fig. 7, SMD2 panel). All three results concur with the genome-wide analysis data.
Figure 7.
Validation of genome-wide localization studies. ChIPs were performed on either wild-type or Hmt1-null cells using α-Npl3 or α-Hrp1. The choice of genes tested is based on genome-wide localization data. The bar graph depicts the normalized ratio of experimental signal to the intergenic region.
Hrp1 showed unchanged binding to DUO1, increased binding to MGT1, and decreased binding to YNL129W in hmt1Δ cells (Fig. 7, bottom panels). This is also in agreement with the genomic-binding data. Thus, we have demonstrated that the degree of binding determined using our genome-wide approach is recapitulated in directed studies.
Discussion
Although protein arginine methylation was first discovered more than 30 years ago, much remains unknown with respect to the substrates as well as the mechanism by which these enzymes influence their substrates' functions. In this study, we present evidence regarding the role of the arginine methyltransferase Hmt1 in defining the architecture of mRNP complexes by modulating the interactions among mRNA processing factors and with the RNA substrate. Using a ChIP assay, we have shown that Hmt1 is cotranscriptionally recruited to genes with a preference toward the 5′-coding region. By characterizing the composition of Npl3 immunoprecipitates from both HMT1 and hmt1Δ cells, we have identified an RNA-dependent association between Npl3 and the TREX component Tho2 that is affected by Hmt1. Further supporting the hypothesis that Hmt1 affects proper mRNP formation, we also show aberrant association of Npl3 with itself in the absence of Hmt1. This supports the observation that loss of Hmt1 affects the kinetics of heat shock message production. Using a genome-wide localization approach, we determined the functional gene classes bound by Hmt1 as well as its substrates. Interestingly, genome-wide binding profiles are changed for Hrp1 and Nab2 in the absence of Hmt1.
Arginine methylation is generally thought to affect the function of its substrates by modulating interactions with other proteins. PRMT1, the mammalian homolog of Hmt1, has been shown to regulate transcriptional elongation by interacting with Spt5, a transcriptional elongation factor (Kwak et al. 2003). Association between Spt5 and RNA Pol II is altered in nonmethylated Spt5 mutants (Kwak et al. 2003). Recently, recruitment of histone H4-specific methyltransferase activity has been attributed to an interaction between PRMT1 and transcription factor Ying Yang 1 (YY1; Rezai-Zadeh et al. 2003). The degree of arginine methylation is proportional to the extent of the interaction between YY1 and PRMT1 (Rezai-Zadeh et al. 2003). These reports place arginine methyltransferases at sites of transcription. We find that Hmt1 may operate on its substrates during transcription as we show Hmt1 is recruited to the transcriptionally active GAL10 gene. Using a more detailed ChIP analysis, we further resolve Hmt1 recruitment to the promoter and the central region rather than the 3′-end of the gene.
Hmt1 associates with genes at the beginning of transcriptional elongation. However, Hmt1 most likely does not affect transcriptional initiation, because similar numbers of wild-type and hmt1Δ cells with HSP104 nuclear signal are present 15 min after heat shock. The kinetics of wild-type HSP104 message production is reflected by a gradual decrease in the number of cells with HSP104 nuclear signal between 15 min and 30 min after heat shock. Indeed, the HSP104 message level peaks at 30 min postshift (Halladay and Craig 1995). Because the heat-shock response is robust and the polymerase must be cleared from the gene prior to additional rounds of transcription, the increased number of hmt1Δ cells with HSP104 nuclear signal 30 min after temperature shift demonstrates a change in the kinetics of HSP104 message production in the hmt1Δ cells. Null mutants of Hmt1 have no obvious defect in mRNA processing and export (Shen et al. 1998). Thus, the change in the kinetics of HSP104 message biosynthesis observed in hmt1Δ cells could be caused by a perturbation of transcriptional elongation brought about by a failure of mRNA export and transcriptional elongation factors to dissociate.
Many of the Hmt1 substrates are recruited to the same functional gene classes as Hmt1. Both Hrp1 and Nab2 exhibited significant changes in their genomic occupancy in the absence of Hmt1. One possible explanation for this effect is the increase in the level of Hrp1 and Nab2 in the nucleus, because Hmt1 has been previously shown to affect the nuclear export of Hrp1 and Nab2 (Shen et al. 1998; Green et al. 2002). However, the absence of Hmt1 had much less of an impact on the genomic binding of Npl3 and nuclear export of Npl3 is also inhibited in hmt1Δ cells. Because arginine methylation has previously been shown to modulate the biochemical properties of RNA-binding proteins (Denman 2002), it is also possible that change in the genome-wide recruitment of Nab2 and Hrp1 in Hmt1-null cells is caused by altered RNA-binding affinity. This is unlikely in the case of Hrp1, however, because we have previously shown that methylation does not affect the ability of Hrp1 to bind RNA (Valentini et al. 1999).
To determine whether methylation is responsible for the change in the Hrp1-binding profile, we repeated the analysis using a catalytic mutant of Hmt1, hmt1G68R. The results revealed only nominal changes in the Hrp1 binding in the presence of the catalytic mutation. One explanation for the difference observed in the null versus the point-mutant is that hmt1G68R is not completely catalytically inactive and that trace amounts of methyltransferase activity are sufficient for proper Hrp1 recruitment. This view is supported by the observation that hmt1G68R on a high-copy plasmid can suppress the synthetic lethality between hmt1Δ and npl3-1 (A.E. McBride and P.A. Silver, unpubl.). Alternatively, Hmt1 may have a role in mediating protein-protein interaction within an mRNP independent of its catalytic functions.
Because Hrp1 and Nab2 occupancies display a similar correlation to transcriptional frequency as Yra1, this suggests that RNA Pol II transcription events may not be sufficient for the recruitment of Hrp1 and Nab2. Consistent with this notion is the previous report that RNA Pol II transcription is not sufficient for the cotranscriptional recruitment of Yra1 to mRNA (Lei and Silver 2002a). Nevertheless, other studies have implicated stable mRNP formation as a reason for the recruitment of Yra1 with transcribed genes (Libri et al. 2002; Zenklusen et al. 2002).
One of the underlying benefits of our genome-wide approach is the ability to formulate additional hypotheses regarding the physiological significance of Hmt1. In the case of Npl3 and Yra1, loss of Hmt1 results in the binding of these proteins to genes involved in the general stress response such as nitrogen starvation. This points to the potential role for Hmt1 in the regulation of stress response. Additionally, loss of Hrp1 binding to genes involved in rRNA-processing pathways in the absence of Hmt1 suggests that Hmt1 plays a role in these processes as well.
Taken together, our data support a model in which Hmt1 influences the binding of its hnRNP substrates. Hmt1 associates with a transcribed gene at the beginning of transcription initiation/elongation, and this association is important for the subsequent mRNP formation as loss of Hmt1 disrupts proper dissociation between the mRNA export factor Npl3 and the transcriptional elongation factor Tho2. This failed dissociation may be the reason for the altered kinetics observed in HSP104 message production. Additionally, the failed dissociation between Npl3 and Tho2 may create an unstable mRNP that influences the recruitment of 3′-end RNA-processing factors Hrp1, Nab2, and Yra1. Thus, a protein methyltransferase affects the dynamics of mRNP formation by modulating protein-protein interactions and influencing the genomic recruitment of certain substrates.
Materials and methods
Yeast strains and genetic manipulations
Standard yeast methods and media were used (Guthrie and Fink 1991). Strains used in this study include PSY 2156 (MATa, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3), PSY 2881 (MATa, hmt1-9xMyc::TRP, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3), PSY 865 (MATα, hmt1:: HIS3, ade2, his3, leu2, lys1, ura3), PSY 867 (MATa, ade2, his3, leu2, lys1, ura3), PSY 3210 (MATa, tho2-9xMyc::TRP, his3-11,15, leu2-3,112, lys1, trp1-1, ura3), PSY 3211 (MATα, hmt1::HIS3, tho2-9xMyc::TRP, lys1, ura3), PSY 2673 (MATa, sub2-3HA::KANMX6, leu2Δ1, trp1Δ63, ura3-52), PSY 3212 (MATa, hmt1::HIS3, yra1-3HA::KANMX6, ade2, his3-11,15, leu2-3,112, lys1, ura3), PSY 3213 (MATα, yra1-3HA::KANMX6, ade2, leu2, lys1, ura3). PSY 3272 (MATα, ura3, ade2-1, hmt1G68R::HIS3:LEU2), PSY 3273 (MATa, ura3, ade2-1, his3-11,15, trp1-1, leu2-3,112). C-terminal tagging with either nine copies of the Myc tag or three copies of the HA tag was described in either Longtine et al. (1998; PSY 3212 and PSY 3213) or Knop et al. (1999; PSY 2881, PSY 3210, PSY 3211).
ChIPs
ChIPs in biological duplicates were performed as previously described (Lei and Silver 2002a). For the immunoprecipitations, monoclonal α-Myc (9E11; Santa Cruz) or monoclonal α-HA (12CA5; Santa Cruz) antibody was precoupled to protein A sepharose beads for each immunoprecipitation, followed by extensive washing. In the case of RPB3, monoclonal α-RPB3 (catalog #W0012; Neoclone) antibody was precoupled to protein G sepharose beads. Immunoblotting was performed to confirm consistent protein levels and IP efficiency in each experiment. For galactose induction, galactose was added to raffinose cultures to a final concentration of 2%, and cultures were induced for 30 min prior to the harvest of cells.
In vivo methylation assays
Cells (15-20 mL) were grown to a density of 1 × 107 cells per mL at 30°C in minimal media (2% glucose) supplemented with either all amino acids or lacking uracil, if plasmid selection was required. Cyclohexamide and chloramphenicol were then added to a final concentration of 100 μg/mL and 40 μg/mL, respectively, for 15 min at 30°C. Following this incubation, cells were washed twice in minimal media (2% glucose) lacking either methionine or methionine and uracil in the presence of protein synthesis inhibitors. Cells were then resuspended in the same media supplemented with 150 μCi/mL of [3H]-methionine (Amersham) for 90 min at 30°C. Following two washes in phosphate buffer saline (PBS), cells were lysed in cold RIPA buffer with 600 mM NaCl. Protein extracts were then subjected to immunoprecipitation using either a polyclonal α-Myc (A-14; Santa Cruz), an affinity-purified rabbit polyclonal α-Hrp1, an affinity-purified rabbit polyclonal α-Npl3 (Bossie and Silver 1992), or IgGSepharose (Amersham). After extensive washing in lysis buffer, immunoprecipitates were resolved by SDS-PAGE followed by either Coomassie staining and fluorography or immunoblot analysis. α-Hrp1 antibody was generated as described previously (Bossie and Silver 1992).
Npl3 interaction studies
Strains containing either pPS2389 or vector plasmid (Hellmuth et al. 1998) only were grown in synthetic dropout media to mid-log phase and lysed with glass beads as described previously (McBride et al. 2000). For initial identification of Npl3-binding proteins, cells were lysed in PBSM buffer (137 mM NaCl/1.76 mM KH2PO4/5.4 mM Na2HPO4/5.7 mM KCl/2.5 mM MgCl2 at pH 7.2) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 2.5 μg/mL each leupeptin, chymostatin, antipain, pepstatin A, and aprotinin).
Lysates were normalized for total protein and the protein ANpl3 fusion protein was purified by 1-2 h of incubation at 4°C with 25 μL packed volume of IgG sepharose beads that had been prepared as described previously (Siniossoglou et al. 1998). Beads were washed five times with 1 mL of lysis buffer prior to an ammonium acetate (5 mM at pH 5.0) wash and acetic acid (0.5 M at pH 3.4) elution. Following precipitation with 12.5% trichloroacetic acid, protein samples were dried, resuspended in protein sample buffer, and resolved by 10% SDS-PAGE. Proteins were visualized either by silver-staining (Shen et al. 1998) or by immunoblotting with a polyclonal α-Myc antiserum (A-14; Santa Cruz) followed by enhanced chemiluminesence detection (Amersham). For RNAase experiments, lysates were treated with 8.7 μg/mL RNAase A prior to the addition of IgG-sepharose. Mass spectrometry was performed by the Molecular Biology Core Facilities Group at the Dana-Farber Cancer Institute.
Tho2-Myc immunoprecipitation
The Tho2-Myc (PSY 3210, PSY 3211) and parental strains (PSY 867, PSY 865) were grown to mid-log phase, and cells were lysed with glass beads in PBSM buffer supplemented with 0.1% Triton X-100 and protease inhibitors (as above) according to published protocols (McBride et al. 2000). Lysates were normalized to 6 mg/mL and incubated with 35 μL of agarose-conjugated α-Myc antibody (12CA5; Santa Cruz, sc-40) for 2 h at 4°C with rotation. Beads were washed five times with 1 mL of lysis buffer and divided in half, and aliquots were incubated in PBSM with or without 25 μg/mL RNAse A for 20 min at 25°C. After washing once with lysis buffer and once with PBSM, bound proteins were eluted by boiling in SDS-PAGE sample buffer, resolved by 10% SDS-PAGE, and transferred to nitrocellulose. The upper section of the blot was probed with a polyclonal α-Myc antibody (1:2000), the lower section of the blot was probed with a polyclonal α-Npl3 antiserum (1:5000; Bossie and Silver 1992), and proteins were visualized by enhanced chemiluminescence.
RNA-FISH hybridization analysis
Yeast cells were grown in YPD at 25°C to an O.D.600 of 0.1. Cultures were shifted by diluting with an equal volume of 59°C prewarmed media for a temperature shift to 42°C. Cells were harvested at different time points after the temperature shift and RNA-FISH analysis was carried out essentially as described (Jensen et al. 2001) using Cy3-conjugated oligonucleotide THJ361 (kind gift from T. Jensen, Aarhaus University, Aarhaus, Denmark). At least 100 cells were counted to calculate the percentage of cells with a nuclear dot.
Genome-wide localization studies
Genome-wide localization studies were performed essentially as described (Casolari et al. 2004). All the experiments were performed in triplicates. Immunoprecipitation was performed as described previously (Casolari et al. 2004) using either pan-mouse IgG or pan-rabbit IgG Dynal beads (Dynal Co.). In all of the immunoprecipitations, antibody was preincubated with the beads in 1× PBS/5 mg/mL BSA for a minimal of 2 h. For Myc-tagged strains, 10 μL of 9E11 was used. For the HA-tagged strain, 20 μL of 12CA5 was used. For Npl3, 10 μL of rabbit polyclonal α-Npl3 antibody was used. For Hrp1, 20 μL of rabbit polyclonal α-Hrp1 antibody was used. For Nab2, 3 μL of mouse monoclonal α-Nab2 antibody was used (kind gift from M. Swanson, University of Florida, Gainesville).
Cy3- and Cy5-labeled probes were prepared by linker-mediated PCR of the immunoprecipitated templates and sonicated genomic DNA fragments present in the starting chromatin. Two-color competitive hybridization experiments with S. cerevisiae cDNA microarrays (University Health Networks at Toronto) were performed essentially as described (Ren et al. 2000) and followed the manufacturer's recommendations. An Axon Genepix 4000B scanner and accompanying software were used for scanning microarrays and quantitating the fluorescence intensities before being uploaded into the Rosetta Resolver microarray analysis platform. Of the three arrays used in a single experiment, the fluorophore incorporation for one of the arrays was reversed. For data analysis, only the gene spots with P-value <0.02 and the IP/input ratio >1.0 were selected for comparison.
Genome-wide expression profiling studies
The total cDNA was prepared from PSY 865 and PSY 867 as described previously (Casolari et al. 2004). Equal amounts of total cDNA (1 μg) prepared from each strain were subjected to Cy3/Cy5 incorporation using the BioPrime Kit (Invitrogen). This was followed by a two-color competitive hybridization experiments with S. cerevisiae cDNA microarrays (University Health Networks at Toronto) as described above.
GenePix error model
GenePix .GPR files were processed in the Rosetta Resolver Gene Expression Data Analysis system. Processing in this system consists of error correction and calculation of a P-value of differential expression using the intensity-error estimation from the .GPR file. Error correction consists of a simplified version of the algorithm described previously in which a piecewise linear function replaces smoothing splines to fit and correct intensity nonlinearity (Schadt et al. 2001). Calculation of P-values consists of a statistic that combines additive and multiplicative error components in both channels of a two-colored experiment. The resulting ratio profiles (i.e., two-channel, error processed microarray scans) were combined into ratio experiments in the Resolver system as described (Stoughton and Dai 2002).
Acknowledgments
We are indebted to the Harvard Biopolymer Facility for the use of the microarray scanner and the Bauer Center for Genomic Research at Harvard University for the use of Rosetta Resolver. We thank G. Adalmant, K. Auld, A. Brodsky, E.P. Lei, and W. Yang for critical reading of the manuscript; H. Hieronymus for the use of Excel macros; M. Swanson for the α-Nab2 antibody; E. Shen for making the α-Hrp1 antibody; T. Jensen for providing FISH reagents; and members of the Silver laboratory for discussion. M.C.Y. was supported by an NIH postdoctoral fellowship. F.B. was supported by a fellowship from CIHR and the Human Frontier Science Program. J.M.C. was supported by a training grant from the National Eye Institute. This work was supported by grants from the NIH to P.A.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1223204.
Supplemental material is available at http://www.genesdev.org.
References
- Ashburner M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. The Gene Ontology Consortium. 2000. Gene ontology: Tool for the unification of biology. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford M.T., Frankel, A., Yaffe, M.B., Clarke, S., Leder, P., and Richard, S. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275: 16030-16036. [DOI] [PubMed] [Google Scholar]
- Berriz G., King, O.D., Bryant, B., Sander, C., and Roth, F.P. 2003. Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502-2504. [DOI] [PubMed] [Google Scholar]
- Bossie M.A. and Silver, P.A. 1992. Movement of macromolecules between the cytoplasm and the nucleus in yeast. Curr. Opin. Genet. Dev. 2: 768-774. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown, C.R., Komili, S., West, J., Hieronymus, H., and Silver, P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427-439. [DOI] [PubMed] [Google Scholar]
- Denman R.B. 2002. Methylation of the arginine-glycine-rich region in the fragile X mental retardation protein FMRP differentially affects RNA binding. Cell. Mol. Biol. Lett. 7: 877-883. [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Gary J.D. and Clarke, S. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61: 65-131. [DOI] [PubMed] [Google Scholar]
- Gary J.D., Lin, W.J., Yang, M.C., Herschman, H.R., and Clarke, S. 1996. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 271: 12585-12594. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Siebel, C.W., and Guthrie, C. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7: 302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D.M., Marfatia, K.A., Crafton, E.B., Zhang, X., Cheng, X., and Corbett, A.H. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277: 7752-7760. [DOI] [PubMed] [Google Scholar]
- Greger I.H. and Proudfoot, N.J. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17: 4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S. and Moore, C.L. 2001. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21: 8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink, G.R. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- Halladay J.T. and Craig, E.A. 1995. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol. Cell. Biol. 15: 4890-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K., Lau, D.M., Bischoff, F.R., Kunzler, M., Hurt, E., and Simos, G. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18: 6374-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M.F. and Silver, P.A. 1996. A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol. 16: 3668-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717-728. [DOI] [PubMed] [Google Scholar]
- Iyer V.R., Horak, C.E., Scafe, C.S., Botstein, D., Snyder, M., and Brown, P.O. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533-538. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887-898. [DOI] [PubMed] [Google Scholar]
- Kadowaki T., Chen, S., Hitomi, M., Jacobs, E., Kumagai, C., Liang, S., Schneiter, R., Singleton, D., Wisniewska, J., and Tartakoff, A.M. 1994. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126: 649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M.M., Henry, M.F., Shen, E., Zhao, J., Gross, S., Silver, P.A., and Moore, C.L. 1997. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 11: 2545-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., and Schiebel, E. 1999. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast 15: 963-972. [DOI] [PubMed] [Google Scholar]
- Kotovic K.M., Lockshon, D., Boric, L., and Neugebauer, K.M. 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 23: 5768-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y.T., Guo, J., Prajapati, S., Park, K.J., Surabhi, R.M., Miller, B., Gehrig, P., and Gaynor, R.B. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11: 1055-1066. [DOI] [PubMed] [Google Scholar]
- Lei E.P. and Silver, P.A. 2002a. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes & Dev. 16: 2761-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002b. Protein and RNA export from the nucleus. Dev. Cell 2: 261-272. [DOI] [PubMed] [Google Scholar]
- Lei E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger, G., Minet, M., Schroeder, S., Cilli, K., McNeil, J.B., and Bentley, D.L. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9: 1101-1111. [DOI] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss, G. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15: 2800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCT-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- McBride A.E., Weiss, V.H., Kim, H.K., Hogle, J.M., and Silver, P.A. 2000. Analysis of the yeast arginine methyltransferase Hmt1p/Rmt1p and its in vivo function. Cofactor binding and substrate interactions. J. Biol. Chem. 275: 3128-3136. [DOI] [PubMed] [Google Scholar]
- Mowen K.A., Tang, J., Zhu, W., Schurter, B.T., Shuai, K., Herschman, H.R., and David, M. 2001. Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104: 731-741. [DOI] [PubMed] [Google Scholar]
- Reed R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523-531. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert, F., Wyrick, J.J., Aparicio, O., Jennings, E.G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306-2309. [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N., Zhang, X., Namour, F., Fejer, G., Wen, Y.D., Yao, Y.L., Gyory, I., Wright, K., and Seto, E. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes & Dev. 17: 1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K., Murthy, K.G., Kaneko, S., and Manley, J.L. 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 22: 1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Li, C., Ellis, B., and Wong, W.H. 2001. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. Suppl. Suppl. 37: 120-125. [DOI] [PubMed]
- Shen E.C., Henry, M.F., Weiss, V.H., Valentini, S.R., Silver, P.A., and Lee, M.S. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes & Dev. 12: 679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C.W. and Guthrie, C. 1996. The essential yeast RNA binding protein Np13p is methylated. Proc. Natl. Acad. Sci. 93: 13641-13646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C.W., Feng, L., Guthrie, C., and Fu, X.D. 1999. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. 96: 5440-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa, H., Rappsilber, J., Mann, M., and Hurt, E. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17: 6449-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoughton R.S. and Dai, H. 2002. Statistical combining of cell expression profiles. In US Patent no. 6,351,712.
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304-308. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi, A., Doerks, T., Braun, I.C., Seraphin, B., Wilm, M., Bork, P., and Izaurralde, E. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini S.R., Weiss, V.H., and Silver, P.A. 1999. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA 5: 272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L., Bauren, G., Bernholm, K., Jiang, W.Q., and Wetterberg, I. 1996. Processing of pre-mRNA in polytene nuclei of Chironomus tentans salivary gland cells. Exp. Cell Res. 229: 240-246. [DOI] [PubMed] [Google Scholar]
- Yun C.Y. and Fu, X.D. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150: 707-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22: 8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]