Abstract
Uncombable hair syndrome (UHS), also known as “spun glass hair syndrome,” “pili trianguli et canaliculi,” or “cheveux incoiffables” is a rare anomaly of the hair shaft that occurs in children and improves with age. UHS is characterized by dry, frizzy, spangly, and often fair hair that is resistant to being combed flat. Until now, both simplex and familial UHS-affected case subjects with autosomal-dominant as well as -recessive inheritance have been reported. However, none of these case subjects were linked to a molecular genetic cause. Here, we report the identification of UHS-causative mutations located in the three genes PADI3 (peptidylarginine deiminase 3), TGM3 (transglutaminase 3), and TCHH (trichohyalin) in a total of 11 children. All of these individuals carry homozygous or compound heterozygous mutations in one of these three genes, indicating an autosomal-recessive inheritance pattern in the majority of UHS case subjects. The two enzymes PADI3 and TGM3, responsible for posttranslational protein modifications, and their target structural protein TCHH are all involved in hair shaft formation. Elucidation of the molecular outcomes of the disease-causing mutations by cell culture experiments and tridimensional protein models demonstrated clear differences in the structural organization and activity of mutant and wild-type proteins. Scanning electron microscopy observations revealed morphological alterations in hair coat of Padi3 knockout mice. All together, these findings elucidate the molecular genetic causes of UHS and shed light on its pathophysiology and hair physiology in general.
Introduction
UHS (MIM: 191480), was first described as a distinctive hair shaft defect in 1973.1, 2 However, the phenotype had been recognized far earlier and had obtained notoriety as the famous literary character “Struwwelpeter” (“Shockheaded Peter”) from the children’s story published by the German physician Heinrich Hoffmann in 1845. This was later translated by Mark Twain to English as “Slovenly Peter.” Up to now about 100 UHS cases have been reported.3, 4, 5 Most of the cases are simplex occurrences but autosomal-dominant or -recessive inheritance patterns were also observed. In majority of the cases, UHS is an isolated condition of the hair, but it has occasionally been observed with additional symptoms, such as ectodermal dysplasias, retinopathia pigmentosa, juvenile cataract, and polydactyly. Isolated UHS is characterized by silvery, blond, or straw-colored scalp hair that is dry, frizzy, and wiry, has a characteristic sheen, stands away from the scalp in multiple directions, and is impossible to comb. This hair shaft disorder occurs in children and improves with age. The hair growth rate can range from slow to normal. The clinical diagnosis of UHS can be confirmed by scanning electron microscopy (SEM) analysis of hair shafts.6, 7, 8 In at least 50% of the hair examined, this reveals a triangular or heart-shaped cross-section, in comparison to the normal circular cross section, as well as longitudinal grooves along the entire length of the hair shaft.9 The hair is not more fragile or brittle than normal hair. No effective therapy is yet available although biotin supplementation was reported to be successful in some cases.10
Until now, no genetic alteration had been linked to UHS although familial occurrence has been well observed. In this study, we report UHS causative mutations in PADI3 (MIM: 606755), TGM3 (MIM: 600238), and TCHH (MIM: 190370), encoding for hair shaft proteins that display sequential interactions with each other. Transfection of cells with constructs encoding for wild-type (WT) and mutant proteins showed that the identified PADI3 and TGM3 mutations have profound effects on enzymatic activity of the respective proteins. The observation of alterations in whiskers and hair coat of Padi3 knockout mice confirms the essential role of this enzyme in hair shaft morphology. Altogether, our findings indicate that UHS occurs when interactions of a structural protein that gives shape and mechanical strength to the hair shaft are impaired by defects either in this protein itself or in others that mediate its interactions.
Material and Methods
Study Participants
Detailed information regarding the clinical descriptions of the individuals included in this study is given in the Supplemental Case Reports. This study was performed according to the principles of the Declaration of Helsinki. Ethical approval was obtained from the ethics committee of the Medical Faculty of the University of Bonn and the participants provided written informed consent prior to blood sampling. Written informed consents of the affected individuals or their legal guardians were obtained for the publication of the case photos included in this manuscript.
Scanning Electron Microscopy
Hair shafts from various individuals with UHS and control subjects were mounted on stubs and sputter coated with either gold or platinum prior to examination in either a FEI Quanta FEG250 or a Philips 505 scanning electron microscope.
Exome Sequencing
Exome sequencing was performed in two different centers. The two affected siblings of the discovery family from UK were exome sequenced by Oxford Gene Technology’s Genefficiency Sequencing Service. Genomic DNA (2 μg) was fragmented and enriched for human exonic sequences using the Human All Exon V5 Agilent Sure Select kit (Agilent Technologies) using the manufacturer’s protocol and sequenced on the Illumina HiSeq 2000 platform using Truseq (v3 Chemistry) (Illumina) to generate 100 base paired-end reads. Fastq files were mapped to the reference human genome (hg19/GRCh37) using the Burrows-Wheeler Aligner (BWA) package (v.0.6.2).11 Local realignment of the mapped reads around potential insertion/deletion (indel) sites was carried out with the Genome Analysis Tool Kit (GATK) v.1.6.12 Duplicate reads were marked using Picard v.1.8 and BAM files were sorted and indexed with SAM tools v.0.1.18.13 Approximately 12 and 14 GB of sequence data was generated for these samples and a minimum of 90.83% and 80.54% of the targeted exome was covered to a depth of at least 20× and 30× coverage, respectively. We filtered the variants for high-quality homozygous or potentially compound heterozygous, novel variants (defined against dbSNP 132 inclusion) that are shared by both siblings and are deleterious based on either of the SIFT, PolyPhen, and Condel predictions.
The affected individuals from Germany and Turkey were exome sequenced at the Cologne Center for Genomics. For whole-exome sequencing, 1 μg of genomic DNA was fragmented with sonication technology (Bioruptor, Diagenode). The fragments were end-repaired and adaptor ligated, including incorporation of sample index barcodes. After size selection, a pool of all five libraries was subjected to an enrichment process with the SeqCap EZ Human Exome Library v.2.0 kit (Roche NimbleGen). The final libraries were sequenced on an Illumina HiSeq 2000 sequencing instrument (Illumina) with a paired-end 2 × 100 bp protocol. Primary data were filtered according to signal purity by the Illumina Realtime Analysis (RTA) software v.1.8. Subsequently, the reads were mapped to the human genome reference build hg19 using the BWA-aln alignment algorithm.11 GATK v.1.6 was used to mark duplicated reads, to do a local realignment around short insertions and deletions, to recalibrate the base quality scores, and to call SNPs and short indels.12 For the Turkish individual, this resulted in 7.5 Gb of unique mapped sequences, a mean coverage of 94×, and 30× coverage of 91% of the target sequences. For the German individual, this resulted in 8.1 Gb of unique mapped sequences, a mean coverage of 106×, and 30× coverage of 92% of the target sequences. The Varbank pipeline v.2.10 and interface developed in-house at the Cologne Center for Genomics were used for data analysis and filtering (unpublished data, H.T., J.A., and P.N.). The GATK UnifiedGenotyper variation calls were filtered for high-quality (DP > 15; AF > 0.25 + VQSLOD > −8 if possible, otherwise: QD > 2; MQ > 40; FS < 60; MQRankSum > −12.5; ReadPosRankSum > −8; HaplotypeScore < 13) rare (MAF ≤ 0.005 based on 1000 Genomes build 20110521 and EVS build ESP6500) variants, predicted to modify a protein sequence or to impair splicing, in homozygous or compound heterozygous state.
Sanger Sequencing
Amplicons were generated under standard polymerase chain reaction conditions by using primers presented in Tables S1–S3. Sanger sequencing was performed using the BigDye Terminator v.1.1 Cycle Sequencing kit (Applied Biosystems) and an ABI 3100 genetic analyzer (Applied Biosystems). The data were analyzed using SeqMan II software (DNASTAR).
Molecular Cloning and Mammalian Cell Cultures
To construct the expression vectors for PADI3 and TGM3, the coding sequences (cDNA) of PADI3 (1,995 bp) and TGM3 (2,082 bp) were amplified from hair follicle cDNA and cloned into the TOPO cloning site of pcDNA 3.1/V5-His-TOPO vector (Invitrogen) according to manufacturer’s protocol. The mutant constructs (PADI3: c.335T>A [p.Leu112His], c.881C>T [p.Ala294Val], c.1813C>A [p.Pro605Thr]; and TGM3: c.1351C>T [p.Gln451∗]) were generated by targeted mutagenesis using QuickChange II Site-Directed Mutagenesis kit (Agilent Technologies) according to manufacturer’s instructions. The constructs were verified by Sanger sequencing. The primers used for cloning and mutagenesis are listed in Table S4. The HaCaT human keratinocyte cell line was established by Boukamp et al.14 The HEK293T cell line was a kind gift from Thomas Zillinger (Institute of Clinical Chemistry and Clinical Pharmacology, University of Bonn). Both HaCaT and HEK293T cell lines were tested for mycoplasma contamination and confirmed to be mycoplasma free. Cells were maintained at 37°C (5% CO2) in DMEM (Lonza) supplemented with 10% FCS (Life Technologies), 1% penicillin-streptomycin (10,000 U mL−1, Life Technologies), and 1% Amphotericin B (250 μg mL−1, Life Technologies). Cells were cultured in 10 cm Petri dishes and on coverslips in 24-well plates for western blotting and immunofluorescence analysis, respectively. Transfections were carried on using the Lipofectamine3000 Transfection Kit (Life Technologies) according to manufacturer’s instructions. For each 10 cm plate, the following amounts of reagents were used: 15 μg plasmid, 1,500 μL Opti-MEM (Life Technologies), 22.5 μL Lipofectamine3000, and 30 μL P3000 reagent. For each well in a 24-well plate, 0.5 μg of plasmid, 50 μL Opti-MEM, 1.5 μL Lipofectamine3000, and 1 μL P3000 reagent were used. Cells were harvested 48 hr after transfection.
Topological Tridimensional Models of PADI3
We constructed, as we had previously published,15 a topological tridimensional model of WT PADI3 using the crystal atomic coordinates of calcium-bound human PADI4 (MIM: 605347; PDB: 1WDA).16 On this basis, topological models of the tridimensional structure of the three mutated enzymes were produced after in silico substitutions of the respective amino acids. All the models were refined by energy minimization (3 × 3,000 iterations) using the MSI insight II modules Biopolymer, CHARMM, and Viewer on an O2 SGI station. Energetic potentials after minimization were all improved and validated. The three models were then validated by analyses of “Anolea data” and Ramachandran plots produced using the Swiss model expasy structure assessment tools and the Swiss-Pdb viewer software v.4.04, respectively. Finally, topological models of mutated PADI3 were individually compared to that of WT PADI3 after a magic fit on Swiss-Pdb viewer. Solid ribbon representations were displayed to show overviews of the tridimensional structures or enlargements focused on either the catalytic sites or the five calcium binding sites.
Immunoblotting of HaCaT and HEK293T Cell Extracts
Cells were collected in ice-cold PBS and centrifuged at 150 × g at 4°C for 10 min. The cell pellets were re-suspended in 40 μL of 10× RIPA buffer (Cell Signaling Technology) and 360 μL Protease Inhibitor Cocktail (Roche), incubated on ice for 15 min, and sonicated 10 times for 10 s, with 10 s breaks on ice. After centrifugation at 10,500 × g for 10 min at 4°C, the supernatants were transferred to clean tubes and purified with Micro Bio-Spin Columns (Bio-Rad Laboratories) according to manufacturer’s instructions. Purified lysates were mixed with 4× Laemmli sample buffer (Bio-Rad Laboratories) diluted in β-mercaptoethanol. After protein separation on TGX stain-free gels 4%–15% (Bio-Rad Laboratories), the proteins were transferred on PVDF membrane (Amersham Biosciences). Western blotting was carried out using the WesternBreeze chemiluminescent kit (Invitrogen) according to manufacturer’s instructions. The following primary antibodies were used with an incubation duration of 1 hr: mouse monoclonal anti-V5 (1:5,000, Sigma Aldrich cat# V8012; RRID: AB_261888) and rabbit monoclonal anti-PADI3/PAD3 (1:400, Abcam cat# ab172959) for PADI3 detection and mouse monoclonal anti-Flag (1:5,000, Sigma Aldrich cat# F1804; RRID: AB_262044) and rabbit polyclonal anti TGM3-C-terminal (1:250, Aviva Systems Biology cat# OAAB12971) for TGM3 detection. Membranes were developed using the ChemiDoc MP imager (Bio Rad) for a maximum of 20 min. Data in Figure 4A are representative of western blotting experiments from three independent transfections of HaCaT cells with PADI3 constructs. Data in Figures 7A and S7 emerge from six independent transfections of HaCaT (1×) and HEK293T (5×) cells with TGM3 constructs. The relative quantities of WT and mutated TGM3 were assessed using Stain-Free technology that is based on total protein normalization (Bio-Rad Laboratories).17
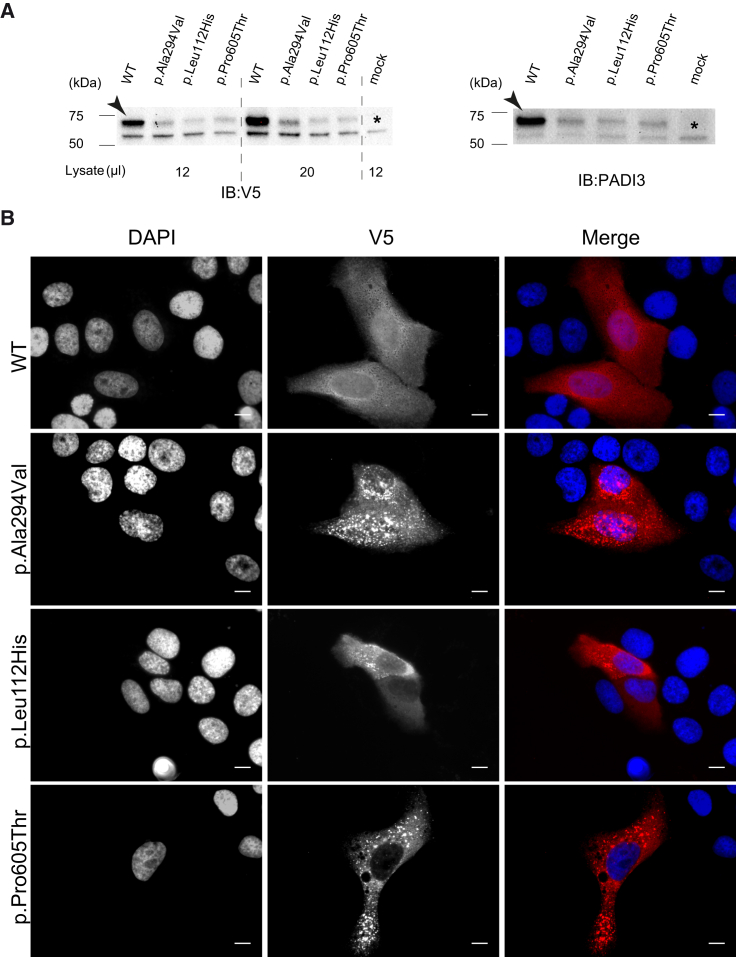
Figure 4.
WT and Mutant Forms of PADI3 Produced in HaCaT Cell Line
(A) Immunoblotting analysis shows the translation of WT and mutant PADI3 in transiently transfected HaCaT cells, which were collected 48 hr after transfection. Immunoblotting was performed with anti-V5 and anti-PADI3 antibodies. Antibody-specific bands are indicated with an arrow and a non-specific cross-reactive band around 55 kDa is indicated with an asterisk. Full-length blot images can be seen in Figure S9.
(B) Immunofluorescence analysis in HaCaT cells transiently transfected with WT and mutant PADI3 encoding plasmids shows the homogeneous cytosolic localization of WT PADI3 whereas the three mutant proteins are observed in the form of large aggregates. Scale bars represent 10 μm.
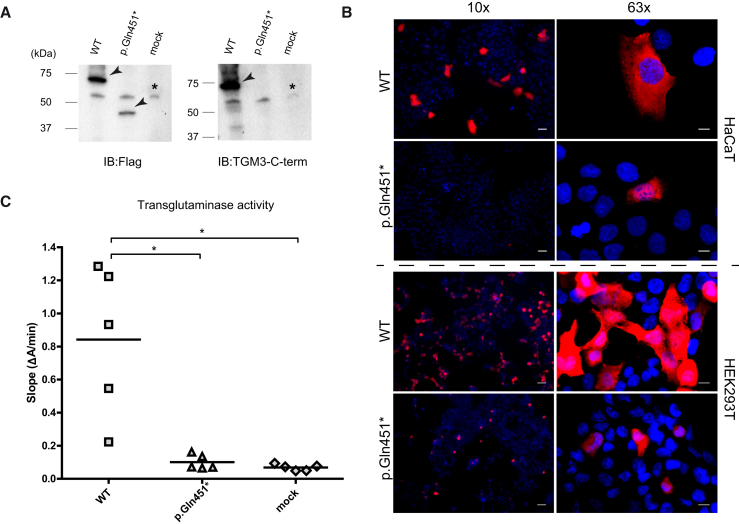
Figure 7.
WT and Mutant Forms of TGM3 Produced in HaCaT and HEK293T Cell Lines
(A) Immunoblotting analysis shows the translation of WT and truncated TGM3 in transiently transfected HaCaT cells, collected 48 hr post-transfection. Immunoblotting was performed with anti-Flag and anti-TGM3 antibodies. Antibody-specific bands showing the WT (∼70 kDa) and truncated TGM3 (∼40 kDa) are indicated with an arrow and a non-specific cross-reactive band around 55 kDa is indicated with an asterisk. The truncated TGM3 can be detected only with the antibody against the Flag epitope fused to the N terminus and not with the antibody against the C terminus of TGM3 as expected. Full-length blot images can be seen in Figure S9. Immunoblotting analysis with HEK293T cells is presented in Figure S7.
(B) Immunofluorescence analyses in HaCaT and HEK293T cells transiently transfected with WT and mutant TGM3 encoding constructs are presented by images captured with 10× and 63× objectives. The analyses show clear differences in the number of cells producing the WT and mutant TGM3 (10× images). Scale bars represent 50 μm and 10 μm for 10× and 63× objectives, respectively.
(C) Transglutaminase activity of WT and mutated TGM3 produced in HEK293T cells. Mock-transfected cell lysates were used as negative control. The transglutaminase activity, represented by the mean slope of 10 min long measurements from technical triplicates (given in Figure S8), is presented as a dot plot. The enzymatic activity assay was performed with samples emerging from five independent transfections. Horizontal lines represent the mean values (mean ± SD; WT, 0.84 ± 0.45; p.Gln451∗, 0.10 ± 0.04; mock, 0.07 ± 0.02). Results demonstrate that the mutated protein did not differ from the mock transfected negative control in terms of activity and WT TGM3 had a significantly higher activity in comparison to both (∗p < 0.05, Dunn’s test).
Immunofluorescence Analysis in HaCaT and HEK293T Cells
Transiently transfected HaCaT and HEK293T cells grown on coverslips were washed with 1× PBS for 5 min, permeabilized for 10 min with 1% Triton X-100, and blocked for 1 hr in PBS containing 1% bovine serum albumin, 10% normal goat serum, and 0.1% Triton X-100. The cells were incubated with mouse monoclonal anti-V5 primary antibody (1:100, Sigma Aldrich cat# V8012; RRID: AB_261888) or mouse monoclonal anti-Flag antibody (1:500, Sigma Aldrich cat# F1804; RRID: AB_262044) for PADI3 and TGM3, respectively, for 3 hr at RT (or overnight at 4°C) and goat anti-mouse-cy3 secondary antibody (1:500, Life Technologies cat# A10521; RRID: AB_2534030) with DAPI (Sigma Aldrich cat# D9542) for 40 min. The mounting was performed with Mowiol 4-88 (Roth). Images were captured with 63× or 10× oil immersion objectives using a Zeiss Axioplan 2 imaging microscope and the Cytovision 7.4 software. ImageJ was used for the analyses by applying the same brightness and contrast thresholds to all data. Data presented in Figures 4B and 7B are representative of analyses from four independent transfections of HaCaT cells with PADI3 constructs and five independent transfections of HaCaT (3×) and HEK293T (2×) cells with TGM3 constructs, respectively. Two to three coverslips were analyzed per construct at each transfection.
Activity of PADI3 in HaCaT Cells
Transiently transfected HaCaT cells grown on coverslips were air-dried. For indirect immunofluorescence, the cells were rehydrated for 15 min in PBS and permeabilized for 10 min in PBS containing 1% Triton X-100, and then non-specific binding sites were blocked with PBS containing 2% fetal bovine serum and 1% Triton X-100. Slides were incubated with the following primary antibodies: mouse monoclonal anti-V5 (1:100, Thermo Fisher Scientific cat# R960-25; RRID: AB_2556564) and ACPA antibodies (6 μg mL−1) purified from a pool of sera of individuals affected by rheumatoid arthritis (MIM: 180300).18, 19 Sera were from informed and consenting individuals attending the Rheumatology Center of Toulouse and have been declared to and approved by the Comité de Protection des Personnes Sud Ouest et Outre-Mer II (Toulouse, France). After incubation with the corresponding secondary antibodies, Alexa Fluor 488 Donkey anti-mouse IgG (1:10,000, Thermo Fisher Scientific cat# A-21202; RRID: AB_2535788), Alexa Fluor 555 Goat Anti-human IgG (H+L) (1:10,000, Thermo Fisher Scientific cat# A21433; RRID: AB_1500626), and DAPI (Sigma-Aldrich cat# D9542), slides were mounted in Mowiol 4-88 (Calbiochem Merck Millipore). Images were captured with 20× dry or 63× oil immersion objectives using a Zeiss apotome microscope (Carl Zeiss). ImageJ was used for the analyses by applying the same brightness and contrast thresholds to all data. Data presented in Figure 5A are representative of analyses from two independent transfections of HaCaT cells with three coverslips for each single PADI3 construct.
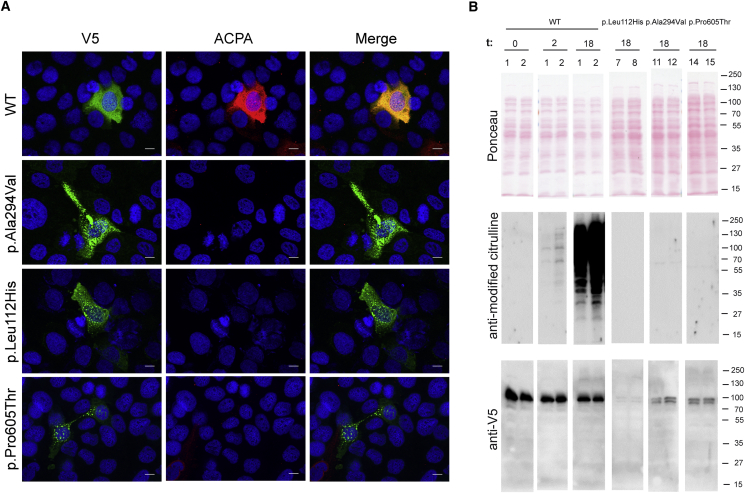
Figure 5.
Consequences of PADI3 Mutations on the Enzyme Activity
(A) Indirect immunofluorescence analysis of transfected HaCaT cells. HaCaT cells were double labeled with anti-V5 monoclonal antibody (green) and ACPA antibodies (red). Nuclei were labeled with DAPI (blue). Scale bars represent 10 μm. The WT PADI3 is clearly active (merge image; yellow color) whereas the mutants display a low activity.
(B) WT PADI3 (bacterial clones 1 and 2) and the mutant forms, p.Leu112His (clones 7 and 8), p.Ala294Val (clones 11 and 12), and p.Pro605Thr (clones 14 and 15), produced in E. coli, were extracted in Tris-HCl buffered salt. Soluble proteins were incubated for the indicated period of time (t; in hours) with calcium, separated by electrophoresis, transferred to membranes, stained with Ponceau Red, and immunodetected with either the anti-modified citrulline rabbit antibodies or anti-V5 antibody. Although citrullinated proteins were detected when the WT-containing extracts have been incubated for 2 hr, no citrullinated proteins were detected in the mutant-containing extracts, even after 18 hr of incubation. The substituted enzymes appear to be cleaved and a V5-reactive doublet is observed. Molecular mass markers are indicated on the right in kDa. Full-length blot images can be seen in Figure S9.
Activity of PADI3 Produced in Escherichia coli
E. coli strain BL21(DE3)-pLysS (Life Technologies) were transformed with 5 ng of the recombinant expression plasmids (pcDNA3.1/V5-His-TOPO-PADI3-WT, pcDNA3.1/V5-His-TOPO-PADI3-p.Leu112His, pcDNA3.1/V5-His-TOPO-PADI3-p.Arg294Val, and pcDNA3.1/V5-His-TOPO-PADI3-p.Pro605Thr) and grown at 37°C overnight on agar-Luria Broth plates (MP Biomedicals) supplemented with ampicillin (50 μg mL−1) and chloramphenicol (34 μg mL−1). Four clones were selected for each plasmid. After selection, the bacterial clones were grown in Luria Broth medium supplemented with ampicillin (50 μg mL−1) at 37°C for 3 hr and then overnight at 30°C. Harvested bacteria were sonicated 4× 8 s (6–10 W) on ice in a Tris-HCl (pH 7.6) buffer containing a cocktail of bacterial protease inhibitors (Sigma Aldrich). After centrifugation at 9,000 × g for 10 min, soluble proteins were recovered in the supernatants (clarified extracts).
To measure an in vitro deimination activity, the clarified extracts were incubated at 37°C in 100 mM Tris-HCl (pH 7.6) buffer containing 10 mM CaCl2 and 5 mM DTT, for either 2 hr or overnight, under agitation at 1,400 rpm.20 The deimination reactions were stopped by boiling for 3 min in Laemmli’s sample buffer. The proteins were then separated by SDS-polyacrylamide (10%) gel electrophoresis and immunodetected with the V5 Epitope Tag monoclonal antibody (1:5,000, Thermo Fisher Scientific cat# R960-25; RRID: AB_2556564) and the anti-modified citrulline antibodies.21, 22 The blots were developed using the ECL prime system (GE Healthcare) as described by the manufacturer. Immunoblotting signals were recorded using a G:Box Chemi XT4 imager and GeneTool analysis software (Syngene) for a maximum of 20 min.
After one bacteria transformation, two independent clones for p.Leu112His, three for p.Pro605Thr, and four for WT and p.Ala294Val were analyzed, with identical results; only those corresponding to two clones are illustrated. Data are representative of two technical replicates. The V5-antibody detections of recombinant PADI3 were confirmed using an anti-PADI3 antibody.22
Padi3-Deficient Mice
All experiments with animals were approved by a local ethic committee (INSERM US006 CEEA-122) and carried out according to our institution guidelines and EU legislation. Padi3-deficient mice (B6NCrl;B6N-Atm1BrdPadi3tm1a(KOMP)Wtsi/Or, abbreviated to Padi3tm1a) were generated by the Phenomin Program at the ICS Laboratory (Strasbourg, France). A promoter-less LacZ-reporter cassette containing a neomycin-resistance gene and flanked by two flippase recognition target sites was inserted between exons 4 and 5 of Padi3, with loxP sites flanking the exons 5 and 6. (See International Mouse Phenotyping Consortium website in the Web Resources for more details.) Mice were maintained in the TAAM animal facility (CNRS UPS44, Orléans, France) under pathogen-free conditions. They were killed by cervical dislocation. All efforts were made to minimize suffering. The following oligonucleotides were used for genotyping: Padi3 forward (5′-CTTTATTGATAAACACAGGCAGGGAGC-3′), Padi3 reverse (5′-CAATGGAATCCCTCTGTCCCTCACC-3′), and LacZ reverse (5′-CCAACAGCTTCCCCACAACGG-3′). A wild-type PCR product of 241 bp and a tm1a allele product of 365 bp were produced. Genotyping was confirmed using the Padi3 forward (5′-CCCTCTTTGAGGACCACAGGCTTATC-3′) and reverse (5′-GCACTCAAGAAGCAGAGGCAGGC-3′) primers, a wild-type PCR product of 369 bp, and a tm1a allele product of 421 bp were produced. No randomization was used. The SEM observations of whiskers and hair coat were done in blind, by two independent scientists.
Transglutaminase Activity of TGM3
In order to compare the enzymatic activity of WT and mutant TGM3 in cell extracts of transiently transfected HEK293T cells, we adapted the transglutaminase assay described by Aufenvenne et al.23 This assay is based on the incorporation of monodansylcadaverine into casein by transglutaminase, which causes an augmentation of fluorescence and an emission wavelength shift. Lysates of cells transiently transfected with WT or mutant TGM3 constructs were prepared 48 hr after transfection as described in the immunoblotting section. Lysates of mock-transfected cells were used as a negative control. The translation of the target proteins was always confirmed by immunoblotting. For the assay, the lysates were incubated for 15 min in pre-warmed assay buffer (50 mM Tris/HCl, 10 mM CaCl2, 10 mM reduced glutathione, 2.5% glycerol, 2.5% DMSO, 25 μM monodansylcadaverine, 20 mM N,N-dimethylcasein [pH 8]) at 37°C. Measurements were then made for 10 min at 37°C using a LS55 fluorescence spectrometer (Perkin Elmer) with excitation and emission wavelengths of 332 nm and 500 nm, respectively, and a 5.0 nm slit. The linear slopes of the measurements from technical triplicates of samples emerging from five independent transfections were used as a measure of the transglutaminase activity.23 The activity data were not normally distributed (p < 0.050, Shapiro-Wilk) and analyzed by a Kruskal-Wallis test (one-way ANOVE on Ranks) with post hoc Dunn’s pairwise multiple comparisons test.
Results
Identification of Mutations in PADI3, TGM3, and TCHH
In this study, we identified UHS-causative mutations in three functionally related genes in a total of 11 individuals/families (Figure 1, Table 1; Supplemental Case Reports). The first family originated from the UK and had two affected and two unaffected siblings. The affected individuals reported typical hair problems in childhood with improvement when growing older (Figure 1J); gross and scanning electron microscopy observations were in line with an UHS phenotype (Figures 1K and 1L). In order to elucidate the genetic background of UHS in this family, we performed whole-exome sequencing (WES) in both of the affected siblings. We filtered the data for novel and deleterious homozygous or potentially compound heterozygous variants that are shared by the siblings and identified a homozygous missense variant c.881C>T (p.Ala294Val) within PADI3 (GenBank: NM_016233.2). The mutation co-segregated with the disease phenotype in the family (Figures 2A and S1).
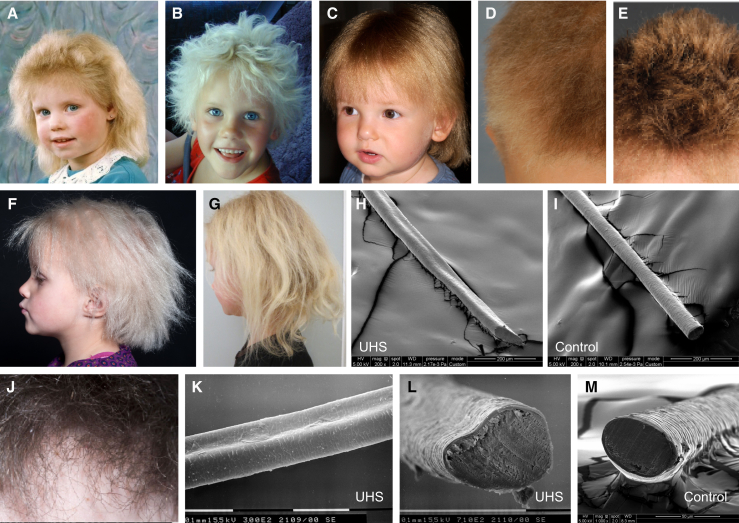
Figure 1.
Clinical Appearance of UHS
(A–G) Clinical presentation of UHS. Typical signs of UHS can be observed in three German girls (A–C), a German boy (D), a Swiss boy (E), and a Danish girl (F and G), who were included in the study. The improvement of the phenotype with aging can be observed in the Danish girl (G).
(H and I) SEM image of the Danish girl’s hair shaft showing the longitudinal groove (H) in comparison to a normal (I) hair shaft.
(J–M) The improved hair phenotype at age 15 (J) and the SEM findings (K and L) of the male sibling from the UK family. Shown are longitudinal grooves (K) and heart-shaped cross section (L) of altered hairs, both indicative of UHS, in comparison to the circular cross section from a control hair shaft (M).
Detailed information regarding the clinical histories can be found in the Supplemental Case Reports.
Table 1.
Individuals Carrying PADI3, TGM3, and TCHH Mutations
| Country | Gene | Mutation | Consequence | Clinical Description |
|---|---|---|---|---|
| United Kingdom | PADI3 | c.881C>T, homozygous | p.Ala294Val | this study |
| Denmark | PADI3 | c.881C>T, homozygous | p.Ala294Val | Nissen and Svendsen24 |
| Germany | PADI3 | c.335T>A, homozygous | p.Leu112His | this study |
| Spain | PADI3 | c.881C>T, c.335T>A | p.Ala294Val, p.Leu112His | Novoa et al.25 |
| Germany | PADI3 | c.881C>T, c.335T>A | p.Ala294Val, p.Leu112His | this study |
| Germany | PADI3 | c.881C>T, c.335T>A | p.Ala294Val, p.Leu112His | this study |
| Germany | PADI3 | c.881C>T, c.1813C>A | p.Ala294Val, p.Pro605Thr | this study |
| Germany | PADI3 | c.335T>A, c.1813C>A | p.Leu112His, p.Pro605Thr | this study |
| Switzerland | PADI3 | c.881C>T, c.1732A>T | p.Ala294Val, p.Lys578∗ | this study |
| Turkey | TGM3 | c.1351C>T, homozygous | p.Gln451∗ | Kilic et al.26 |
| Germany | TCHH | c.991C>T, homozygous | p.Gln331∗ | this study |
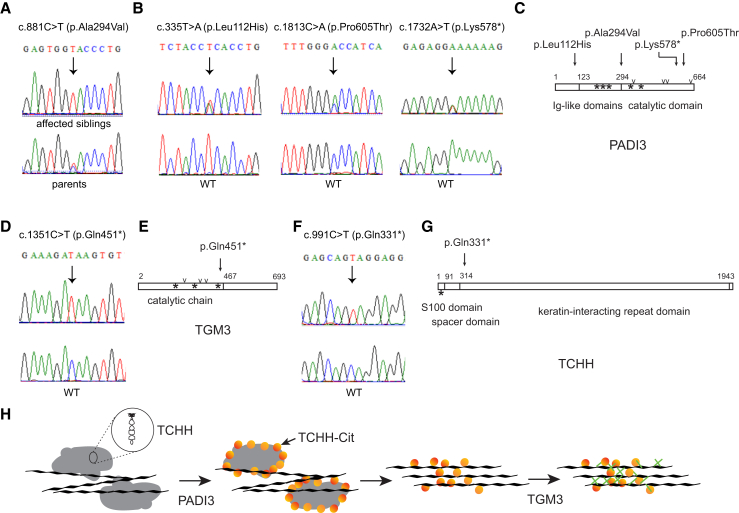
Figure 2.
Mutations Causing UHS and Interplay of PADI3, TGM3, and TCHH
(A) The homozygous c.881C>T (p.Ala294Val) mutation in PADI3 identified by exome sequencing in the UK siblings was verified by Sanger sequencing. The mutation co-segregates with the UHS phenotype in the pedigree as shown in Figure S1.
(B) Electropherograms show the PADI3 mutations c.335T>A (p.Leu112His), c.1813C>A (p.Pro605Thr), and c.1732A>T (p.Lys578∗) in comparison to the wild-type sequences.
(C) Schematic representation of PADI3 showing the various domains of the protein and the positions of calcium-binding sites (∗) and of major amino acids involved in the catalytic sites (v). The positions of the substitutions responsible for UHS are indicated by an arrow.
(D) Electropherograms show the homozygous TGM3 mutation c.1351C>T (p.Gln451∗) identified in the Turkish male in comparison to the wild-type sequence.
(E) Schematic representation of TGM3.
(F) Electropherograms show the homozygous TCHH mutation c.991C>T (p.Gln331∗) identified in the German female in comparison to the wild-type sequence.
(G) Schematic representation of TCHH.
(H) Cartoon depicting the cascade of interactions between TCHH, PADI3, and TGM3 in hair shaft biology. In trichohyalin granules (gray areas), TCHH appears as a dimer with a long rod-shaped domain and a globular end domain. After deimination by PADI3, TCHH is less structured, and the granules progressively dissolved. Citrullinated-TCHH (TCHH-Cit) interacts with keratin intermediate filaments (black lines) organizing them, and becomes a substrate for TGM3. Cross-links of TCHH to itself (intra- or inter-chains), to keratins and between keratins and the cornified cell envelope components are then catalyzed (green dash).
Then, we Sanger sequenced PADI3 in 17 additional case subjects and detected the above-mentioned mutation as well as two other recurrent missense mutations, c.335T>A (p.Leu112His) and c.1813C>A (p.Pro605Thr), in seven other individuals/families (Table 1, Figures 2B, 2C, and S2).24, 25 We also identified an individual who carries c.881C>T (p.Ala294Val) and a nonsense mutation, c.1732A>T (p.Lys578∗), that occurred only once in our UHS cohort (Table 1, Figures 2B, 2C, and S2). These mutations were observed either in homozygous state or in compound heterozygosity as confirmed by parental DNA sequencing in three of the families (Figure S1). The allele frequencies of the PADI3 substitutions from Exome Aggregation Consortium (ExAC) data are presented in Table S5. The nonsense mutation was not observed in ExAC.
As a next step, we performed WES in four further UHS-affected case subjects with no PADI3 mutations. We identified the nonsense mutation c.1351C>T (p.Gln451∗) in TGM3 (GenBank: NM_003245.3) in a Turkish individual26 and the nonsense mutation c.991C>T (p.Gln331∗) in TCHH (GenBank: NM_007113.3) in a German individual (Figures 2D–2G; Table S5). No homozygous loss-of-function mutations were observed in TGM3 or TCHH in ∼60,000 sequenced individuals of the ExAC database.
PADI3, TGM3, and TCHH Interplay in Hair Shaft Formation
PADI3, a gene of the PADI family (PADI1-4 and 6), encodes the 664-amino acid peptidylarginine deiminase type III (Enzyme Commission: EC.3.5.3.15).27 This posttranslational modification enzyme converts positively charged L-arginine residues of proteins into neutral citrulline residues in the presence of calcium ions. The process is called deimination or citrullination. PADI3 is mainly detected in skin, including hair follicles where it modifies hair shaft proteins.28, 29 Although the enzyme has already been a focus of interest in hair biology, it could not be linked to any disorder until now. TGM3 encodes transglutaminase 3, a member of the transglutaminase family (Enzyme Commission: EC.2.3.2.13), which catalyzes the calcium-dependent formation of isopeptide bonds between glutamine and lysine residues in various proteins including the archetypal hair shaft protein trichohyalin, encoded by TCHH. TCHH is a structural protein co-localized with PADI3 in the inner root sheath of the hair follicle and in the medulla of the hair shaft. Deimination by PADI3 reduces the overall charge of TCHH, and that enables its association with the keratin intermediate filaments (KIF). Then, TCHH and KIF are crosslinked together by TGM3. KIFs are then stabilized, hardened, and linked to cornified envelopes through further crosslinking by transglutaminases, particularly by TGM3 (Figure 2H).30, 31, 32 Thereby, TCHH and its sequential modifications by PADI3 and TGM3 have a very important role in shaping and mechanical strengthening of the hair. Of note, the TCHH mutation we identified leads to the synthesis, if any, of a very short protein, probably without any function in KIF interaction, as the KIF interacting domain would be almost entirely missing (Figure 2G).
Missense Mutations in PADI3 Affect the 3D Enzyme Structure
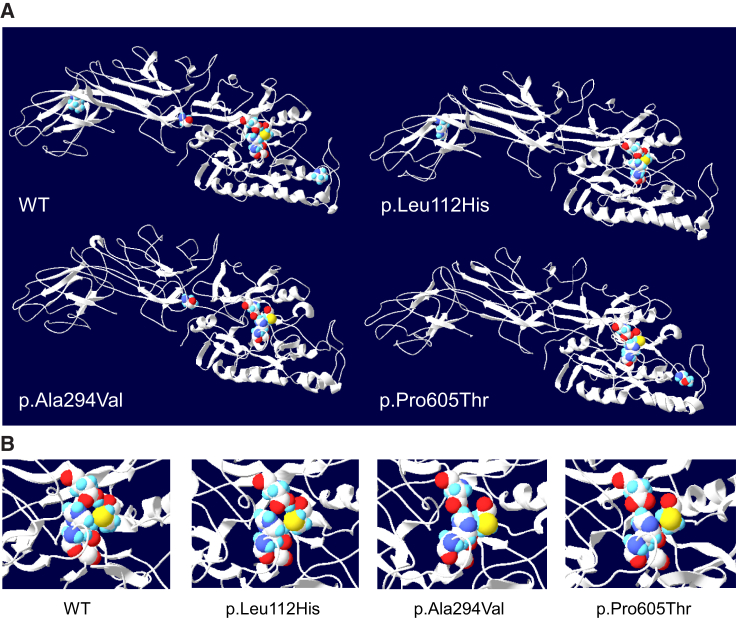
We next investigated the consequences of PADI3 mutations on the corresponding proteins. The p.Lys578∗ is expected to induce the synthesis of a truncated protein lacking the 87 amino acids of the carboxyl terminus, in particular the Cys646 (Figure 2C, Table S6) absolutely necessary for the enzyme activity.28 Intriguingly, we observed that the three PADI3 amino acids substituted as a result of the missense mutations correspond to residues that are conserved in the five human PADI proteins, and also in the PADI3 from other species, suggesting that they have an important role (Figure S3). However, none of them is directly involved in the catalytic site or in one of the five calcium-binding sites of PADI3 (Table S6). Nevertheless, when we analyzed the effect of the PADI3 missense mutations on the predicted three-dimensional structure of the enzyme,15 the mutations p.Ala294Val and p.Pro605Thr were shown to induce clear modifications of β sheets and α helices, in particular in the immunoglobulin-like NH2 domains, and also around the catalytic site and the calcium-binding sites. The effects of the p.Leu112His substitution were less drastic (Figures 3, S4, and S5).
Figure 3.
Topological Tridimensional Models of WT and Mutant PADI3
(A) Overall view of the tridimensional solid-ribbon representation of calcium-bound WT and three mutant PADI3 models. The tridimensional solid residues Leu112, Ala294, and Pro605, with their respective lateral chains, are reported on the proposed model of the WT enzyme, as well as the corresponding substituted amino acids on the model of each mutant, p.Leu112His, p.Ala294Val, and p.Pro605Thr. The four amino acids involved in the catalytic site are also shown. According to these models, the p.Ala294Val and p.Pro605Thr substitutions induce a profound disorganization of the predicted immunoglobulin-like domains, with disappearance of several β sheets and modification of some α helices of the catalytic domain as compared to the WT. The effect of the p.Leu112His substitution is more discrete.
(B) Zoomed view of the major amino acids involved in the catalytic site. Its proposed structure is clearly modified after p.Ala294Val and p.Pro605Thr substitutions. It should be noted that none of the substituted amino acids are directly involved in this site.
Aggregation and Reduced Enzymatic Activity of PADI3 Mutants
We then cloned the WT PADI3 cDNA into a mammalian expression vector in order to induce the translation of a C-terminally V5-tagged PADI3 protein. Mutant constructs with the three recurrent missense mutations were generated by targeted mutagenesis. HaCaT cells were transiently transfected (5%–10% transfection efficiency) with constructs encoding for WT and mutant forms of PADI3. Immunoblotting of cell extracts showed that all of the constructs led to translation of a protein of about 70 kDa (Figure 4A). The subcellular location of WT and mutated proteins was determined by immunofluorescence analyses that showed, as expected, a diffuse homogeneous cytoplasmic distribution of the WT PADI3,22 whereas in all three mutants the proteins were observed to form large aggregates throughout the cytoplasm (Figure 4B). To assess the enzymatic activity of WT and mutated PADI3, we performed double immunostaining with an anti-V5 monoclonal antibody and with human anti-citrullinated protein autoantibodies (ACPA) from individuals with rheumatoid arthritis, these antibodies specifically detecting citrullinated proteins.18, 33 Although translation of the WT PADI3 resulted in a strong labeling with the ACPA antibodies, the signal in the cells producing the mutant proteins was barely above background (Figure 5A). We also produced the WT and mutant PADI3 in E. coli. After incubation of the WT PADI3-containing bacterial extracts with calcium for 2 hr, deiminated proteins were detected. By contrast, when the extracts containing the mutated PADI3 were incubated for 2 hr, and even up to 18 hr, no deiminated proteins were detected (Figures 5B and S6). Altogether, the results suggested that the mutated forms of PADI3 are either not or only weakly active.
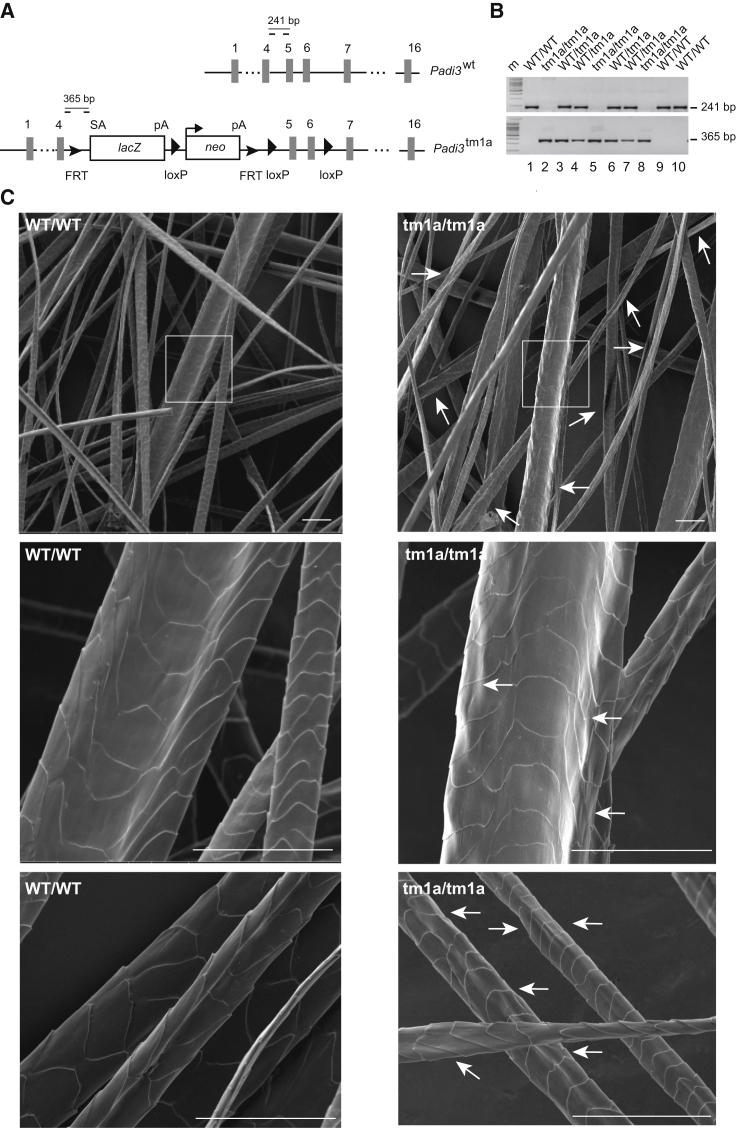
Padi3 Knockout Mice Show Whisker and Hair Anomalies
In order to demonstrate the importance of PADI3 in hair shaft formation and structure, we generated Padi3 knockout mice (Figures 6A and 6B). Breeding mice heterozygous for the Padi3tm1a mutation produced offspring with the three possible genotypes at the expected Mendelian ratios, showing that absence of Padi3 is compatible with life. Grossly, the skin of 7-week-old null mice appeared normal. A further characterization by SEM revealed alterations in the morphology of hair coat (Figure 6C) and, less markedly, of whiskers. The surface of the lower (proximal) part of the vibrissae and the hairs on their entire length were irregular and rough and appeared as if hammered.
Figure 6.
Padi3 Gene Disruption in Mice
(A) Schematic representation of Padi3 and the targeting vector. The exons are shown by numbered gray rectangles. A cassette containing the beta-galactosidase (LacZ) and neomycin (neo) genes is inserted between exons 4 and 5. This results in the production of a truncated Padi3 protein corresponding to amino acids 1–136 fused to β-galactosidase. The position of the inserted loxP and flippase recognition target (FRT) sites are indicated, as well as the splicing acceptor site (SA) and the polyadenylation sites (pA) of LacZ and neo. The positions of the oligonucleotides (dashes) used for genotyping are shown, as well as the length of the amplified PCR products in wild-type (Padi3wt) and targeted (Padi3tm1a) alleles.
(B) PCR on genomic DNA from tail biopsies of ten mice from the same littermate (five females [1–5] and five males [6–10]) showing amplification of the 241 bp fragment for the Padi3wt allele and the 365 bp fragment for the Padi3tm1a allele.
(C) Representative pictures of SEM analysis of back hair coat from the three WT and three Padi3tm1a/tm1a mice (7 weeks old). Most hairs of the knockout mice are rough, with an irregular surface (white arrows). This is evident for thick hairs (top and middle micrographs) but was also observed for thin hairs (bottom). In some cases the hairs are twisted. The boxed areas are enlarged in the middle. At least 6 vibrissae and 200 hairs were analyzed per animal. Scale bars represent 50 μm.
Nonsense Mutation in TGM3 Leads to Reduced Enzymatic Activity
We cloned the WT TGM3 cDNA into a mammalian expression vector in order to induce the translation of an N-terminally FLAG-tagged TGM3 protein. Mutant construct was generated by targeted mutagenesis. HaCaT and HEK293T cell lines were transiently transfected with WT and mutant TGM3 encoding constructs. WT TGM3 construct led to translation of a protein of about 70 kDa, while the nonsense mutation resulted in a truncated protein of around 40 kDa with a lower detection level (Figures 7A and S7). Immunofluorescence analysis showed that TGM3 is located in the cytoplasm and that the truncated form is present in a dramatically lower number of cells (Figure 7B) in accordance with the western blot results. Generally these cells had a smaller cytosolic surface area in comparison to those producing the WT TGM3 (Figure 7B). We performed a transglutaminase activity assay23 with HEK293T cell lysates containing WT and mutated TGM3. The analysis results revealed that the WT had a significantly higher transglutaminase activity in comparison to the truncated protein and the latter did not differ from the mock transfected negative control (Figures 7C and S8).
Discussion
In this study we identified disease-causative mutations for UHS in PADI3, TGM3, and TCHH, with the former two genes encoding for posttranslational modification enzymes that act on the structural hair protein trichohyalin encoded by the latter gene. Our cell culture data show that the identified mutations in PADI3 and TGM3 lead to reduced or no enzymatic activity and the phenotype of the Padi3 knockout mice we generated show structural alterations in the whiskers and hair coat morphology. Our findings are also supported by the phenotype of the already existing Tgm3 knockout mice. These mice exhibit irregular, twisted whiskers and, at birth, have a wavy hair coat, which improves 4 weeks after birth.34 Scanning and transmission electron microscopy analyses of hairs from adult Tgm3−/− mice reveal alterations such as irregular torsions, deformed grooves, abnormal cuticle, and shorter KIF.34 It is also convincing that both nonsense and missense mutations in Tgm3 are responsible for the wellhaarig mouse phenotype, named for the curly whiskers and wavy coat of the mutant animals.35 Based on the phenotype of the Tgm3−/− mice, John et al. have suggested that alterations in TGM3 or its substrates could be related to recessive forms of pili torti (MIM: 261900) or similar hair phenotypes, which improve with age.34 Our findings validate this hypothesis and furthermore identify UHS as the hair phenotype related to alterations not only in TGM3 but also in the other proteins of the TCHH-PADI3-TGM3 cascade. In some affected individuals, TCHH would not be produced, or produced as a truncated form unable to interact with KIF; in others, because of a PADI3 activity defect, TCHH would remain insoluble, preventing its interaction with KIF; in the last group, TCHH/KIF interaction would not be stabilized by TGM3-mediated crosslinks. Taken together, this suggests that one of the particular processes in hair shaft formation that leads to UHS when disturbed might be the interaction of TCHH and KIF. This interaction is known to be crucial for shaping and mechanical strengthening of the hair shaft. These findings also show that the compromise of any one of the proteins that play a role in this process would be enough to lead to the same phenotype.
It is intriguing that, despite the defects affecting a structural component of the hair shaft, the phenotype in UHS is commonly reported to improve with age, similar to the hair coat phenotype of Tgm3−/− mice. Interestingly, improvement with age has also been observed in other hair shaft disorders (e.g., pili annulati, pili torti). The improvement in UHS might be due to the compensatory expression of another isoform of peptidylarginine deiminase, transglutaminase, or other structural hair shaft components. On the other hand, aging-related changes in hair follicles such as increase in diameter and length can have mechanistic influences that might account for this improvement.
TGM3 and PADI3 are strongly detected in the upper epidermis. Nevertheless, no anomalies have been reported in the interfollicular epidermis of individuals with isolated UHS. Similarly, Tgm3−/− mice show no obvious skin defects.34 This is probably due to the fact that other isoforms of these enzyme families are present in the epidermis, which can compensate for the loss of PADI3 and TGM3 activity, whereas these two are the only isoforms detected in the hair cuticles and medulla.27, 28, 30
Up to now, both simplex and familial cases of UHS have been reported with suggested autosomal-dominant and -recessive inheritance with variable levels of penetrance. In this study we consistently observed an autosomal-recessive inheritance pattern in a total of 11 familial and simplex cases. Based on the pedigrees reported in literature, it is most likely that autosomal-dominant forms of this condition exist7, 36, 37 which might be underlined by defects in other genes involved in hair shaft formation and maintenance related processes. It would be of interest then to investigate closely whether differences could be observed in the phenotypes of these individuals in comparison to the individuals who carry recessive mutations in PADI3-TCHH-TGM3 cascade.
An interesting observation was that 5 out of 60,659 individuals in ExAC are homozygous for c.881C>T (p.Ala294Val). UHS is a non-debilitating phenotype that resolves with age and therefore, it is likely that these presumably affected individuals had participated in the ExAC project as “healthy” controls. These data also indicate that UHS is a more common phenotype than estimated.
In summary, we elucidated the molecular genetic background of UHS by identifying recessive mutations in PADI3, TCHH, and TGM3. The three genes encode for proteins that play an essential role during hair shaft formation through their sequential interactions. This finding, in combination with the data describing the functional effects of the mutations in PADI3 and TGM3, provides valuable information regarding the pathophysiology of UHS. Furthermore, it contributes to a better understanding of the protein interaction cascades in molecular histogenesis of the hair. This could be of further value for cosmetic and pharmaceutic industries paving the way for development of novel products.
Acknowledgments
P.N. is a founder, CEO, and shareholder of ATLAS Biolabs GmbH, which is a service provider for genomic analyses. We would like to thank the individuals and their families for participating in this study. We would like to acknowledge Dr. Marianne Chabod, the technical assistance of Carole Pons and Géraldine Offer, the Toulouse Rio Imagerie imaging (INSERM U1043) and electron microscopy (University of Toulouse) facilities, and the TAMM animal facility, in particular Laëtitia Trottereau. We would like to thank Dr. Thomas Zillinger for providing us with HEK293T cells. We would like to acknowledge Dr. Thomas Butterbrodt from Bio-Rad Laboratories GmbH and Dr. Kourosh Zolghadr from Leica Biosystems for their technical assistance. S.S. is supported by the Deutsche Forschungsgemeinschaft (DFG, SFB1089), the German Ministry of Research and Education (BMBF, 01GQ0806), and the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement number 602102 (EPITARGET). V.O. is supported by the German Research Foundation (DFG) (OJ 53/3-1). M.S. and G.S. are supported by The University of Toulouse, the Centre National de la Recherche Scientifique, and the Institut National de la Santé et de la Recherche Médicale. This work was further supported by local funding (BONFOR to R.C.B. and S.S.). R.C.B. is a member of the DFG-funded Excellence Cluster ImmunoSensation and was a recipient of a Heisenberg Professorship of the DFG (BE 2346/4-2).
Published: November 17, 2016
Footnotes
Supplemental Data include nine figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.10.004.
Web Resources
Enzyme Commission, http://www.chem.qmul.ac.uk/iubmb/enzyme/
ExAC Browser (accessed 5 March 2015), http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
International Mouse Phenotyping Consortium, http://www.mousephenotype.org/data/genes/
OMIM, http://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
varbank, https://varbank.ccg.uni-koeln.de
UCSC Human Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
Supplemental Data
References
- 1.Dupré A., Rochiccioli P., Bonafé J.L. Cheveux incoiffables: anomalie congénitale des cheveux. Bull. Soc. Fr. Dermatol. Syphiligr. 1973;80:111–112. [Google Scholar]
- 2.Stroud J.D., Mehregan A.H. New York: Medcom Press; 1973. The First Human Hair Symposium. [Google Scholar]
- 3.Calderon P., Otberg N., Shapiro J. Uncombable hair syndrome. J. Am. Acad. Dermatol. 2009;61:512–515. doi: 10.1016/j.jaad.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Trüeb R.M., Spycher M.A., Schumacher F., Burg G. [Pili torti et canaliculi in ectodermal dysplasia] Hautarzt. 1994;45:372–377. doi: 10.1007/s001050050086. [DOI] [PubMed] [Google Scholar]
- 5.Navarini A.A., Kaufmann F., Kaech A., Trüeb R.M., Weibel L. Picture of the month. Uncombable hair (pili trianguli et canaliculi) Arch. Pediatr. Adolesc. Med. 2010;164:1165–1166. doi: 10.1001/archpediatrics.2010.228-a. [DOI] [PubMed] [Google Scholar]
- 6.Ferrando J., Fontarnau R., Gratacos M.R., Mascaro J.M. [Pili canaliculi (uncombable hair syndrome or spun glass hair syndrome). A scanning electron microscope study of ten new cases (author’s transl)] Ann. Dermatol. Venereol. 1980;107:243–248. [PubMed] [Google Scholar]
- 7.Hebert A.A., Charrow J., Esterly N.B., Fretzin D.F. Uncombable hair (pili trianguli et canaliculi): evidence for dominant inheritance with complete penetrance based on scanning electron microscopy. Am. J. Med. Genet. 1987;28:185–193. doi: 10.1002/ajmg.1320280126. [DOI] [PubMed] [Google Scholar]
- 8.Mallon E., Dawber R.P., De Berker D., Ferguson D.J. Cheveux incoiffables--diagnostic, clinical and hair microscopic findings, and pathogenic studies. Br. J. Dermatol. 1994;131:608–614. doi: 10.1111/j.1365-2133.1994.tb04970.x. [DOI] [PubMed] [Google Scholar]
- 9.Hicks J., Metry D.W., Barrish J., Levy M. Uncombable hair (cheveux incoiffables, pili trianguli et canaliculi) syndrome: brief review and role of scanning electron microscopy in diagnosis. Ultrastruct. Pathol. 2001;25:99–103. [PubMed] [Google Scholar]
- 10.Shelley W.B., Shelley E.D. Uncombable hair syndrome: observations on response to biotin and occurrence in siblings with ectodermal dysplasia. J. Am. Acad. Dermatol. 1985;13:97–102. doi: 10.1016/s0190-9622(85)70150-8. [DOI] [PubMed] [Google Scholar]
- 11.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., Fusenig N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Méchin M.C., Coudane F., Adoue V., Arnaud J., Duplan H., Charveron M., Schmitt A.M., Takahara H., Serre G., Simon M. Deimination is regulated at multiple levels including auto-deimination of peptidylarginine deiminases. Cell. Mol. Life Sci. 2010;67:1491–1503. doi: 10.1007/s00018-010-0262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arita K., Hashimoto H., Shimizu T., Nakashima K., Yamada M., Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 17.Posch A., Kohn J., Oh K., Hammond M., Liu N. V3 stain-free workflow for a practical, convenient, and reliable total protein loading control in western blotting. J. Vis. Exp. 2013;82:50948. doi: 10.3791/50948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masson-Bessière C., Sebbag M., Girbal-Neuhauser E., Nogueira L., Vincent C., Senshu T., Serre G. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J. Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 19.Anquetil F., Clavel C., Offer G., Serre G., Sebbag M. IgM and IgA rheumatoid factors purified from rheumatoid arthritis sera boost the Fc receptor- and complement-dependent effector functions of the disease-specific anti-citrullinated protein autoantibodies. J. Immunol. 2015;194:3664–3674. doi: 10.4049/jimmunol.1402334. [DOI] [PubMed] [Google Scholar]
- 20.Méchin M.C., Enji M., Nachat R., Chavanas S., Charveron M., Ishida-Yamamoto A., Serre G., Takahara H., Simon M. The peptidylarginine deiminases expressed in human epidermis differ in their substrate specificities and subcellular locations. Cell. Mol. Life Sci. 2005;62:1984–1995. doi: 10.1007/s00018-005-5196-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senshu T., Akiyama K., Kan S., Asaga H., Ishigami A., Manabe M. Detection of deiminated proteins in rat skin: probing with a monospecific antibody after modification of citrulline residues. J. Invest. Dermatol. 1995;105:163–169. doi: 10.1111/1523-1747.ep12317070. [DOI] [PubMed] [Google Scholar]
- 22.Nachat R., Méchin M.C., Takahara H., Chavanas S., Charveron M., Serre G., Simon M. Peptidylarginine deiminase isoforms 1-3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J. Invest. Dermatol. 2005;124:384–393. doi: 10.1111/j.0022-202X.2004.23568.x. [DOI] [PubMed] [Google Scholar]
- 23.Aufenvenne K., Oji V., Walker T., Becker-Pauly C., Hennies H.C., Stöcker W., Traupe H. Transglutaminase-1 and bathing suit ichthyosis: molecular analysis of gene/environment interactions. J. Invest. Dermatol. 2009;129:2068–2071. doi: 10.1038/jid.2009.18. [DOI] [PubMed] [Google Scholar]
- 24.Nissen C.V., Svendsen M.T. [Uncombable hair syndrome] Ugeskr. Laeger. 2013;175:2878. [PubMed] [Google Scholar]
- 25.Novoa A., Azon A., Grimalt R. Síndrome del pelo impeinable. Uncombable hair syndrome. Ann. Pediatr. (Paris) 2012;77:139–140. doi: 10.1016/j.anpedi.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Kiliç A., Oğuz D., Can A., Akil H., Gürbüz Köz O. A case of uncombable hair syndrome: light microscopy, trichoscopy and scanning electron microscopy. Acta Dermatovenerol. Croat. 2013;21:209–211. [PubMed] [Google Scholar]
- 27.Kanno T., Kawada A., Yamanouchi J., Yosida-Noro C., Yoshiki A., Shiraiwa M., Kusakabe M., Manabe M., Tezuka T., Takahara H. Human peptidylarginine deiminase type III: molecular cloning and nucleotide sequence of the cDNA, properties of the recombinant enzyme, and immunohistochemical localization in human skin. J. Invest. Dermatol. 2000;115:813–823. doi: 10.1046/j.1523-1747.2000.00131.x. [DOI] [PubMed] [Google Scholar]
- 28.Méchin M.C., Sebbag M., Arnaud J., Nachat R., Foulquier C., Adoue V., Coudane F., Duplan H., Schmitt A.M., Chavanas S. Update on peptidylarginine deiminases and deimination in skin physiology and severe human diseases. Int. J. Cosmet. Sci. 2007;29:147–168. doi: 10.1111/j.1467-2494.2007.00377.x. [DOI] [PubMed] [Google Scholar]
- 29.Takahara H.S.G., Simon M. Deimination in skin and regulation of peptidyl-arginine deiminase expression in keratinocytes. In: Nicholas A., editor. Protein Deimination in Health and Disease. Springer; New York: 2014. pp. 113–128. [Google Scholar]
- 30.Steinert P.M., Parry D.A., Marekov L.N. Trichohyalin mechanically strengthens the hair follicle: multiple cross-bridging roles in the inner root shealth. J. Biol. Chem. 2003;278:41409–41419. doi: 10.1074/jbc.M302037200. [DOI] [PubMed] [Google Scholar]
- 31.Tarcsa E., Marekov L.N., Andreoli J., Idler W.W., Candi E., Chung S.I., Steinert P.M. The fate of trichohyalin. Sequential post-translational modifications by peptidyl-arginine deiminase and transglutaminases. J. Biol. Chem. 1997;272:27893–27901. doi: 10.1074/jbc.272.44.27893. [DOI] [PubMed] [Google Scholar]
- 32.Westgate G.E., Botchkareva N.V., Tobin D.J. The biology of hair diversity. Int. J. Cosmet. Sci. 2013;35:329–336. doi: 10.1111/ics.12041. [DOI] [PubMed] [Google Scholar]
- 33.Vincent C., Nogueira L., Clavel C., Sebbag M., Serre G. Autoantibodies to citrullinated proteins: ACPA. Autoimmunity. 2005;38:17–24. doi: 10.1080/08916930400022582. [DOI] [PubMed] [Google Scholar]
- 34.John S., Thiebach L., Frie C., Mokkapati S., Bechtel M., Nischt R., Rosser-Davies S., Paulsson M., Smyth N. Epidermal transglutaminase (TGase 3) is required for proper hair development, but not the formation of the epidermal barrier. PLoS ONE. 2012;7:e34252. doi: 10.1371/journal.pone.0034252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennan B.M., Huynh M.T., Rabah M.A., Shaw H.E., Bisaillon J.J., Radden L.A., 2nd, Nguyen T.V., King T.R. The mouse wellhaarig (we) mutations result from defects in epidermal-type transglutaminase 3 (Tgm3) Mol. Genet. Metab. 2015;116:187–191. doi: 10.1016/j.ymgme.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garty B., Metzker A., Mimouni M., Varsano I. Uncombable hair: a condition with autosomal dominant inheritance. Arch. Dis. Child. 1982;57:710–712. doi: 10.1136/adc.57.9.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Luna M.M., Rubinson R., de Kohan Z.B. Pili trianguli canaliculi: uncombable hair syndrome in a family with apparent autosomal dominant inheritance. Pediatr. Dermatol. 1985;2:324–327. doi: 10.1111/j.1525-1470.1985.tb00475.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.