Abstract
Mitochondrial fatty acid synthesis (mtFAS) is an evolutionarily conserved pathway essential for the function of the respiratory chain and several mitochondrial enzyme complexes. We report here a unique neurometabolic human disorder caused by defective mtFAS. Seven individuals from five unrelated families presented with childhood-onset dystonia, optic atrophy, and basal ganglia signal abnormalities on MRI. All affected individuals were found to harbor recessive mutations in MECR encoding the mitochondrial trans-2-enoyl-coenzyme A-reductase involved in human mtFAS. All six mutations are extremely rare in the general population, segregate with the disease in the families, and are predicted to be deleterious. The nonsense c.855T>G (p.Tyr285∗), c.247_250del (p.Asn83Hisfs∗4), and splice site c.830+2_830+3insT mutations lead to C-terminal truncation variants of MECR. The missense c.695G>A (p.Gly232Glu), c.854A>G (p.Tyr285Cys), and c.772C>T (p.Arg258Trp) mutations involve conserved amino acid residues, are located within the cofactor binding domain, and are predicted by structural analysis to have a destabilizing effect. Yeast modeling and complementation studies validated the pathogenicity of the MECR mutations. Fibroblast cell lines from affected individuals displayed reduced levels of both MECR and lipoylated proteins as well as defective respiration. These results suggest that mutations in MECR cause a distinct human disorder of the mtFAS pathway. The observation of decreased lipoylation raises the possibility of a potential therapeutic strategy.
Introduction
Childhood dystonia accompanied by bilateral symmetrical basal ganglia signal intensity changes on magnetic resonance imaging (MRI) can be caused by a wide spectrum of genetic and non-genetic causes.1 Striatal degeneration has been described after infection with agents such as mycoplasma or West Nile or Herpes viruses, as well as from dietary depletion of thiamine as seen in Wernicke encephalopathy.2 Examples of genetic metabolic causes of childhood dystonia with specific brain MRI changes include glutaric aciduria type 1 (MIM: 231670) with striatal necrosis and widening of the Sylvian fissures3 and pantothenate kinase-associated neurodegeneration (MIM: 234200) with globus pallidus edema and iron accumulation.4
However, when facing the combination of childhood dystonia with basal ganglia degeneration, one of the major groups of disorders suspected is mitochondrial disease. Commonly, in addition to developing basal ganglia or brain stem MRI lesions causing movement disorders and gaze disturbance, affected individuals suffer cognitive regression and, in some cases, clinical features such as optic atrophy, hearing defects, peripheral neuropathies, or multisystem involvement. Many nuclear and mitochondrial genes have been associated with such mitochondrial disorders. Their common pathogenesis is disturbance in one of the five complexes of the mitochondrial respiratory chain, the pyruvate dehydrogenase enzyme, or one of their cofactors.5
We describe seven individuals from five families that displayed childhood-onset dystonia accompanied by distinctive MRI changes in the basal ganglia and a somewhat later appearance of optic atrophy in all but the youngest one. Contrary to most known mitochondrial disorders, the majority of affected individuals described here remained relatively cognitively intact despite their severe motor and visual impairment. All affected individuals were found to harbor recessive mutations in MECR (MIM: 608205, GenBank: NM_016011.3), encoding the mitochondrial trans-2-enoyl-CoA reductase enzyme, a key protein in mitochondrial fatty acid synthesis (mtFAS).
The mtFAS system is an evolutionarily conserved pathway that is required for respiratory competence in the eukaryotic model organism Saccharomyces cerevisiae. There is ample evidence that mtFAS is involved in the synthesis of lipoic acid (LA) in eukaryotic cells.6, 7, 8 LA is essential for the activity of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGD), branched-chain keto-acid dehydrogenase, 2-oxoadipate dehydrogenase, and the glycine cleavage system, which utilize it as a covalently bound cofactor.9 Disruption of mtFAS leads to loss of lipoylated proteins in yeast and mammals.7, 8, 10 Human MECR rescues the respiratory defect in yeast cells that is caused by deletion of the gene encoding the mtFAS enoyl thioester reductase homolog Etr1,11 providing a system for investigating the functional impact of specific mutations. In this work, we utilize this system to analyze the identified MECR mutations and their resulting cellular changes.
To our knowledge, the mtFAS pathway has not been implicated up to now in human disorders. This inborn error of metabolism resembles typical mitochondrial disorders by the involvement of organs with high energy demands and susceptibility to oxidative stress such as basal ganglia and optic nerve. However, it differs by the relative sparing of cognition and absence of additional organ involvement and typical mitochondrial biomarkers. Understanding its pathogenesis may help explain its unique manifestations and support a possible therapeutic approach through dietary supplementation with LA or its precursor octanoic acid to stop or slow disease progression.
Subjects and Methods
Ethical Approval
The families were recruited at the Oregon Health & Science University (Portland, OR, USA), the Sheba Medical Center (Israel), the IRCCS Foundation Neurological Institute “C. Besta” (Milan, Italy), and the Sydney Children’s Hospitals Network (Westmead, Australia). The Institutional Review Board at each center approved the study and written informed consent was obtained from all participants or their respective legal guardians.
Next-Generation Sequencing
For affected individual 1 (Figure 1, family A, II:1), DNA samples from him and his unaffected sister (family A, II:2) were submitted for whole-exome sequencing (WES) to the University of Washington Center for Mendelian Genomics (for detailed methods, see Web Resources). Pathogenicity of identified variants was evaluated using multiple bioinformatics tools. For each variant, minor allele frequencies were checked in variant databases including the 1000 Genomes, Exome Variant Server (EVS), and Exome Aggregation Consortium (ExAC) databases. The MECR variants were submitted to ClinVar. To check for evolutionary conservation of MECR amino acid residues, the human sequence was aligned with 14 vertebrate sequences.
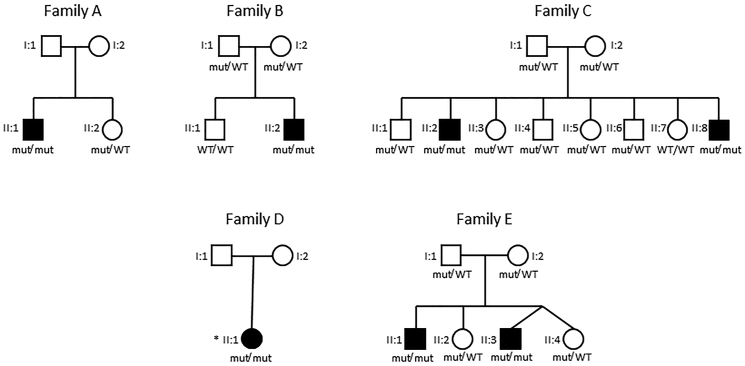
Figure 1.
Pedigrees of the Families Described in This Work
For all tested individuals: Mut/mut refers to compound heterozygous affected individuals with the exception of subject II:1 from family D who is a homozygous affected individual and is specified with an asterisk (∗); Mut/WT refers to heterozygous healthy carriers; and WT/WT refers to healthy non-carriers.
For affected individual 2 (Figure 1, family B, II:2), trio WES was performed on a clinical basis by the company Centogene. Approximately 37 Mb (214,405 exons) of the consensus coding sequences (CCDS) were enriched from fragmented genomic DNA by >340,000 probes designed against the human genome. The samples were then sequenced on the NextSeq Platform (Illumina) with100-bp read paired-end. Analysis was performed in the Hadassah Medical Center; reads alignment and variant calling were performed with DNAnexus software using the default parameters with the human genome assembly hg19 (GRCh37) as a reference. Variants were removed if called fewer than 8 times or were off-target, synonymous, or had a minor allele frequency (MAF) > 1% in ExAC or at the Hadassah in-house databases.
For affected individuals 6 and 7 (Figure 1, family E, II:1 and II:3, respectively), genomic DNA was extracted from the blood of affected individual 6 and his parents and subjected to whole-genome sequencing (WGS) at the Kinghorn Centre for Clinical Genomics (Garvan Institute, Sydney). WGS sequencing libraries were prepared using Illumina TruSeq Nano HT v2.5 sample preparation kits and sequenced one lane per sample, on Illumina HiSeq X sequencers, via 2 × 150 bp reads, with >110 Gb data per lane, >75% bases with at least Q30 base quality, and >30× mean coverage. At this coverage, 96% of the nuclear genome is covered to >15× depth. Reads were aligned to the b37d5 reference genome using BWA MEM v.0.7.10, sorted using novosort v.1.03.01, then realigned around known indels, and base quality scores were recalibrated using GATK v.3.3. Variants were identified using GATK HaplotypeCaller v.3.3 and GenotypeGVCFs, and variant filters established using VQSR. Variants were annotated using VEP v.79 and converted into a database using Gemini v.0.11.0.12 Variants were filtered using Seave, an in-house variant filtration platform. Sanger sequencing was used to confirm variants in all family members.
Bioinformatics and Structural Analysis
Information regarding the exons and domain organization of MECR was taken from the PDB site.13 Structures of MECR were downloaded from the PDB including PDB: 2vcy,14 1guf,15 and 1zsy.
Thermo stability calculations were done using Cupsat,16 duet,17 mCSM,18 MAESTRO,19 SDM,20 I Mutant2,21 and Fold-X.22 Functional predictions of various programs including Sift,23 PolyPhen2,24 MutationTaster,25 MutationAssessor,26 LRT, and PROVEAN27 were collected from the dbNSFP database.28
Structural alignment between MECR human and yeast proteins, in order to borrow the coordinates of NADPH, was done using the TriangleMatch program.29 Structure visualization and images were done with Jmol. Side-chain modeling was performed using SCCOMP.30
Prediction of alternative donor splicing motifs in the vicinity of the splicing variant was done with ASSP.31
PCR Studies
Five milliliters of heparinized blood were drawn from participants, DNA was extracted using the MagNa Pure LC system, and RNA was extracted using Trizol reagent (Ambion). Complementary DNA (cDNA) was generated using random primers (Reverse-iT 1st Strand synthesis kit, ABgene). DNA and cDNA amplification was carried out in a 25 μL reaction containing 50 ng of DNA, 10 μM of each and Red load Taq Master∗5 (LAEOVA). After an initial denaturation of 5 min at 95°C, 30 cycles were performed (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s), followed by a final extension of 10 min at 72°C. Sequencing was performed using an automated ABI Prism 3100 Genetic Analyzer (Perkin Elmer).
Restriction Assays
Carrier rates were established using restriction assays. The sequence containing the c.695G>A mutation was amplified with the primers 5′-GAAAAAGCTGGCAGATGTGACCAG-3′ (f) and 5′-TCTAAGAGTCACACATGTGGGCTG-3′ (r), resulting in a 435 bp PCR product. The products were then cut with the GsuI restriction enzyme which cuts the mutant alleles to yield 327 bp and 181 bp products. The c.830+2_830+3insT mutation was amplified with the 5′-TGCACGCAAACAAAGGCATAAACATC-3′ and 5′-CTATGGCAGGCAGCTTAGCAGT-3′ primers. The MseI restriction enzyme cuts the 325 bp fragment to yield 276 bp and 159 bp products for the normal allele and 276 + 104 + 55 bp fragments for the mutant allele. The c.855T>G mutation was amplified with the primers 5′-AGAGCAGGGTTTATAAAGGAACCAG-3′ and 5′- TGAGGGCAAGAACTTCAGGTACC-3′, resulting in a 325 bp PCR product. The products were then cut with the BfaI restriction enzyme which cuts the mutant allele to yield 193 bp and 132 bp products.
cDNA Analysis
Segments of cDNA containing the c.830+2>T splice mutation were amplified with the primers 5′- AAGAGTCTGGGGGCTGAGCAT-3′ and 5′-CAAAAGCCTCGAAGTTTGAGATCC-3′. The PCR products were separated on a 3% agarose gel, extracted using QIAquick Gel extraction kit (QIAGENE), and then sequenced.
Affected Individuals’ Fibroblast Preparation
Affected individual and control fibroblast cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose (4.5 g/L), GlutaMAX, and HEPES (Thermo Fisher Scientific), supplemented with 10% HyClone fetal bovine serum and penicillin-streptomycin (Sigma-Aldrich). Cell lines were cultured at +37°C in 5% CO2 and both subcultured and harvested for experiments at sub-confluent cell density.
Western Blotting Analyses of Affected Individual Fibroblast Cell Extracts
Total cell protein extracts were prepared from the affected individual and control cell lines using M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific). Protein concentrations were measured using Bio-Rad Protein Assay (Bio-Rad) microtiter plate protocol.
SDS-PAGE electrophoresis was done using four control cell lines (control 1–4) and three affected individuals’ cell lines (individuals 1–3). Bio-Rad TGX 4%–20% gradient gels were loaded with ∼40 μg protein per well and run, then immediately blotted on a 0.2 μm nitrocellulose membrane using Bio-Rad Trans-Blot Turbo system and Trans-Blot Transfer Pack. To check the protein loading, the nitrocellulose membrane was stained with Ponceau S and imaged. Nitrocellulose membranes were blocked overnight at 4°C using 1× Casein Blocking Buffer (Sigma-Aldrich). This Casein Blocking Buffer was also used in the western blotting for all antibody dilutions.
To detect MECR, the blocked membrane was incubated for 1 hr at room temperature with a 1:2,000 dilution of rabbit polyclonal anti-Mecr IgG (51027-2-AP, Proteintech). The membrane was washed with TBST buffer (50 mM TRIS, 150 mM NaCl, 0.05% Tween 20) thrice for 5 min. The washed blot was incubated for 1 hr at room temperature with a 1:10,000 dilution of horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG (Bio-Rad) and washed as previously. The signal was detected from the membrane using Clarity ECL reagent (Bio-Rad) with a 4-min treatment time, after which the membrane was imaged using Bio-Rad XRS camera.
The same methods were used to detect lipoic acid from the membrane, using a 1:2,500 dilution of rabbit polyclonal anti-lipoic acid IgG (Merck Millipore) and a 1:5,000 dilution of HRP-conjugated goat anti-rabbit IgG (Bio-Rad). As a loading control, β-actin was detected from the membrane using a 1:2,500 dilution of mouse polyclonal anti-β-actin IgG (Abcam cat# ab8224; RRID: AB_449644) and a 1:2,500 dilution of HRP-conjugated goat anti-mouse IgG (Promega). Bio-Rad Imagelab software was used to quantify band intensities from the imaged western blots. To account for the differences in the protein loading, the observed band intensities of DLAT and DLST lipoic acid antibody signal were normalized using the actin antibody band intensity in the same sample. This was done for each sample by dividing the actin band intensity of control 1 with the actin band intensity for the control or affected individual sample and using this correction as a multiplier for the measured DLAT or DLST band intensity on the lipoic acid western blot. For the presentation of the lipoic acid band intensities in Figure 5, the mean of the four pooled control cell lines was set as 1.0, and the affected individual cell lines are presented as a relative change compared to this value.
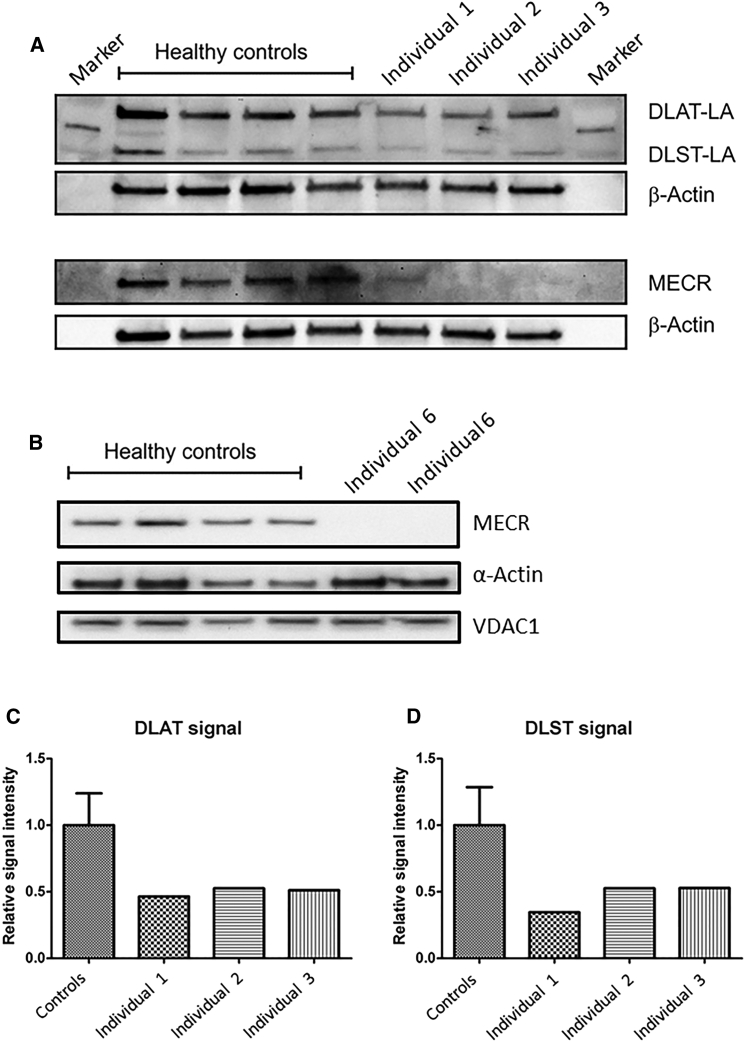
Figure 5.
Western Blotting Analysis for MECR and Lipoylated Protein Content of Cell Extracts from Several Affected Individuals’ Fibroblast Cell Lines
(A) Western blotting of whole-cell extract from cell lines of affected individuals 1–3 (family A, II:1; family B, II:2; family C, II:2) demonstrates reduced lipoylation and decreased level of MECR compared to four control fibroblast cell lines. DLAT-LA: lipoyated E2 subunit of human pyruvate dehydrogenase. DLST-LA: lipoylated E2 subunit of human α-ketoglutarate dehydrogenase. β-Actin: loading control.
(B) Western blotting of whole-cell extract from cell lines of affected individual 6 (family E, II:1) shows decreased level of MECR compared to control fibroblast cell lines.
(C and D) Densitometric quantitation of the DLAT-LA (C) and DLST-LA (D) signals show significantly reduced lipoylation in affected individuals 1–3 fibroblasts compared with controls. For the DLAT/DLST control intensity bars, the average intensity of the luminescence signal of the four samples from healthy controls was calculated. The mean value was defined as 1.0 (100% intensity) and the error bars represent the standard error of the mean (SEM).
For affected individual 6, fibroblast extracts were prepared and immunoblotting performed as previously described32 with the following modifications. 20 μg of fibroblast extract was loaded per lane. For immunoblotting, PVDF membranes were probed with 1:1,000 anti-MECR (Abcam cat# ab180075), 1:2,000 anti-α-actin (Sigma cat# A2172; RRID: AB_476695), or 1:1,000 anti-VDAC1 (Abcam cat# ab14734; RRID: AB_443084) overnight at 4°C or with anti-OXPHOS (ab110411) for 2 hr at room temperature.
Respiration Measurements
Respiratory characteristics of the intact control and affected individuals cell lines were measured using OROBOROS Oxygraph-2k high-resolution respirometry system (Oroboros). In the SUIT (substrate-uncoupler-inhibitor titration) protocol used in the experiment, Oxygraph-2k chamber with the polarographic oxygen sensor was filled with the live cell suspension and closed and warmed to 37°C. After the rate of oxygen consumption in the chamber had equilibrated, the average rate of oxygen consumption was measured as the ROUTINE respiration. Oligomycin (Sigma-Aldrich) was injected to the chamber to a final concentration of 2.5 mM to inhibit respiratory complex V activity, and the average oxygen consumption was measured as LEAK respiration. The maximum capacity of the cellular respiration was measured by titration of an uncoupler carbonyl cyanide m-chlorophenyl hydrazine (CCCP) (Sigma-Aldrich) in 0.5 μM steps until the maximum uncoupled rate of respiration was reached. This maximum value was measured as the electron transport system (ETS) maximum capacity. Finally, antimycin A (Sigma-Aldrich) was injected to a final concentration of 2.5 μM to inhibit complex III, and rotenone was injected to a final concentration of 0.5 μM to inhibit complex I. Average oxygen consumption, resulting from non-respiratory events in the cells, after these titrations was measured as the residual oxygen consumption (ROX).
For the respirometry analysis, the results were normalized using the live cell count of each sample before the measurement. Before filling the chambers, an aliquot of the cell suspension was stained with trypan blue solution and the live cell concentration was counted using Thermo Fisher Scientific Countess II FL Automated Cell Counter. In the data analysis, corrected values for ROUTINE, LEAK, and ETS were generated by subtracting the ROX value of the measurement from these values.
Complex I and IV Activity Measurements
Complex I and complex IV dipstick activity assays were performed on 15 μg of cleared cell lysates of control and affected individuals fibroblasts according to the manufacturer’s protocol (Abcam). The MS1000 was used for measuring the dipstick products.
Statistical Analysis
Respirometry results were analyzed with GraphPad Prism 5 software. Control cell line measurements from four different control cell lines were pooled for the analysis (n = 14), and each affected individual cell line was tested with a two-tailed, unpaired t test against the pooled controls for a statistically significant difference in the group mean. Dipstick assay results from two independent experiments were pooled and analyzed with SPSS 22 software using an independent sample Mann-Whitney U test. Results are presented as the mean ± SEM, n = 6 for each group.
Strains and Plasmids Used in the Yeast Growth Complementation Studies
E. coli Top10F’(F'{lacIq, Tn10(TetR)} mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG) (Invitrogen) was used for molecular cloning. S. cerevisiae strains BJ1991 etr1Δ (MATα, leu2, trp1, ura3-52, pep4-3, prb1-1122, gal2)33 and W1535 8B etr1Δ (MATα, ade2Δ, ade3Δ, leu2-3, leu2-112, his3-11, his3-15,trp1-1, ura3-1, etr1::kanMX)34 have been described previously.
Plasmids YEp35235 and YEplac19536 are E. coli-S. cerevisiae shuttle plasmids used as plasmid backbones in our studies that were generated by different research groups but are identical in all of their relevant features (2μ origin of replication, URA3 marker gene, the bla gene encoding ampicillin resistance, multiple cloning site). Two plasmids containing MECR with the c.695G>A and c.855T>G mutations were generated using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent), using as a template yeast expression plasmid pYE352::HsNRBF-1,11 expressing the H. sapiens MECR ORF from the yeast CTA1 promoter. The following sense and antisense primer pairs specific for each mutation were used (c.695G>A: 5′-AGACTGAAGAGTCTGGAGGCTGAGCATGTCATC-3′ and 5′-GATGACATGCTCAGCCTCCAGACTCTTCAGTCT-3′; c.855T>G: 5′-GAGGAACCATGGTAACCTAGGGGGGGATGG-3′ and 5′-CCATCCCCCCCTAGGTTACCATGGTTCCTC-3′). After the mutagenesis, candidate plasmids were purified using NucleoSpin Plasmid kit (Macherey-Nagel), and the candidate plasmids carrying the mutation allele were sequence verified. For clarity, the plasmids are referred to as YEp352-HsMECR (pYE352::HsNRBF-1, wild-type), YEp352-HsMECR-c.695G>A, or YEp352-HsMECR-c.885T>G henceforth.
To exclude the possibility that lack of complementation was due to the inability of S. cerevisiae cells to efficiently recognize the native mitochondrial targeting signal (MTS) of H. sapiens MECR, we generated plasmids that encode a chimeric fusion protein of the S. cerevisiae COQ3 MTS and HsMECR and its variants (HsMECR-c.695G>A and HsMECR-c.855T>G). Using YEp352-HsMECR or the verified mutant plasmids as the template, HsMECR was amplified with primers that added BspHI (5′-TTCGACTCATGATGTGGGTCTGCAGTACCC-3′) and XhoI (5′-ATATCTCGAGCATGGTGAGAATCTGCTTTG-3′) restriction sites. PCR products were purified using MicroElute Gel Extraction Kit (Omega Bio-Tek). Then, the yeast expression plasmid pYEmtQOR33 was digested with NcoI/XhoI, while the PCR products were digested with BspHI/XhoI, with the digestions of the plasmid vector and the PCR amplified inserts resulting in compatible cohesive ends. Digested and purified vector and inserts were ligated together using Fast-Link DNA Ligation Kit (Epicenter) and transformed into Top 10 cells. Candidates were verified by NcoI/XhoI digestion and sequencing. The resulting plasmids were named MTS-HsMECR, YEp352-MTS-HsMECR-c.695G>A, and YEp352-MTS-HsMECR-c.885T>G.
Yeast Mutation Complementation Studies
The plasmids MTS-HsMECR, YEp352-MTS-HsMECR-c.695G>A, and YEp352-MTS-HsMECR-c.885T>G were used, together with control plasmids, to transform yeast S. cerevisiae strains W1536 8B etr1Δ and BJ1991 etr1Δ, in which yeast 2-enoyl thioester reductase gene ETR1 has been deleted. As positive control plasmids, YEp195-ETR1 containing the yeast ETR1 and YEp352-HsMECR containing wild-type human MECR were used. As a negative control the empty YEp352 expression plasmid was used.
Overnight cultures of the transformed yeast strains were grown in synthetic complete media with glucose lacking uracil (SCD-URA). To ensure that the strains were in a similar phase of growth for the spotting assay, overnight cultures were used to inoculate new SCD-URA cultures to an optical density of ∼0.1, which were grown for ∼4 hr before harvesting and spotting. Cell number was normalized to measured optical density, after which a dilution series of 10−1 to 10−3 was prepared in sterile ddH2O from each culture. Dilution series were spotted on SCD-URA plates, and synthetic complete plates with glycerol (SCG) or lactate (SCL) as the carbon source. Spotted plates were grown in +30°C. SCD-URA plates were imaged after 2 days of growth, SCG and SCL plates after 4 to 5 days.
Western Blotting Analysis of MECR Mutations Modeling Yeast Strains
BJ1991 etr1Δ strain transformed with YEp195-ETR1, YEp352, YEp352-MTS-HsMECR, YEp352-MTS-HsMECR-c.695G>A, or YEp352-MTS-HsMECR-c.885T>G were grown in YPG media (1% yeast extract, 2% peptone, 3% glycerol, and 0.05% glucose) at +30°C for 16 hr.
Whole-cell extracts were collected by TCA precipitation according to Platta et al.37 2.6 mg of total protein was loaded onto 10% polyacrylamide gel (10% mini Protean tgx precast protein gels, Bio-Rad). Proteins were transferred to nitrocellulose membranes and blocked with 2% BSA in TBS-T buffer. Primary antibodies used were Mecr polyclonal Rabbit antibody (Proteintech) 1:3,000 dilution, anti-lipoic acid Rabbit pAb (Calbiochem) 1:3,000 dilution, and Anti-beta actin Mouse antibody (Abcam) 1:2,500 dilution as the loading control. Secondary antibodies used were goat anti-rabbit HRP-conjugate (Immun-star, Bio-Rad) 1:10,000 dilution and anti-mouse IgG HRP conjugate (Promega) in 1:3,333 dilution. All the antibody dilutions were prepared in 2% BSA TBST-buffer. Detection of proteins bound by the antibodies was done using clarity western ECL reagents from Bio-Rad.
Results
Case Descriptions
All seven affected individuals described here (Figure 1, Table 1) presented with involuntary movement disorders, mainly dystonia, during early childhood (15 months–6.5 years). Optic atrophy developed in all but the youngest affected individual, either immediately or within a few years of the appearance of the dystonia, manifesting with reduced visual acuity and in some of the cases abnormal eye movements. In all affected individuals MRI demonstrates bilateral hyperintense T2 signal in either caudate, putamen, or pallidum (Figures 2A–2G) accompanied in some cases with a lactate peak in the spectroscopy (Figures 2H and 2I). Although motor disability (including speech dysarthria) progressed gradually in all affected individuals described here, there was a relative sparing of the cognitive functions in most of them, and no incidence of seizures or disease-related childhood mortality were noted (see detailed clinical descriptions in the Supplemental Data).
Table 1.
Summary of Affected Individuals’ Mutations and Main Clinical Features
| Affected Individuals | Origin | MECR Mutations | Age at Onset of Dystonia | Age at Onset of Optic Atrophy | Intellect | MRI Involvement | Age at Last Assessment |
|---|---|---|---|---|---|---|---|
| 1 (Family A, II:1) | Ashkenazi Jewish | c.695G>A, c.855T>G | early childhood | mid childhood | preserved | bilateral hyperintense T2 signal in the putamen | 48 y |
| 2 (Family B, II:2) | mixed Jewish origin | c.695G>A, c.830+2InsT | 15 m | – | preserved | bilateral hyperintense pallidal T2 signal | 24 m |
| 3 (Family C, II:2) | Ashkenazi Jewish | c.695G>A, c.830+2InsT | 2–3 y | 8 y | preserved | bilateral hyperintense T2 signal in the dorsal striatum | 27 y |
| 4 (Family C, II:8) | Ashkenazi Jewish | c.695G>A, c.830+2InsT | 5 y | 12 y | preserved | NA | 45 y |
| 5 (Family D, II:1) | Tunisian | homozygous c.854A>G | 6.5 y | 6.5 y | relatively preserved (mild concentration difficulties) | bilateral hyperintense T2 signal in the dorsal putamen | 7 y |
| 6 (Family E, II:1) | Anglo-Saxon | c.772C>T, c.247_250del | 23 m | 6 y | deterioration of linguistic skills and executive functions to extremely low range at 9 y | bilateral hyperintense pallidal T2 signal with cavitation and lactate peak on MRS | 16 y |
| 7 (Family E, II:3) | Anglo-Saxon | c.772C>T, c.247_250del | 3 y | 5 y | low average verbal comprehension with extremely low function on the other WISC IV indices | bilateral hyperintense pallidal T2 signal with lactate peak on MRS | 12 y |
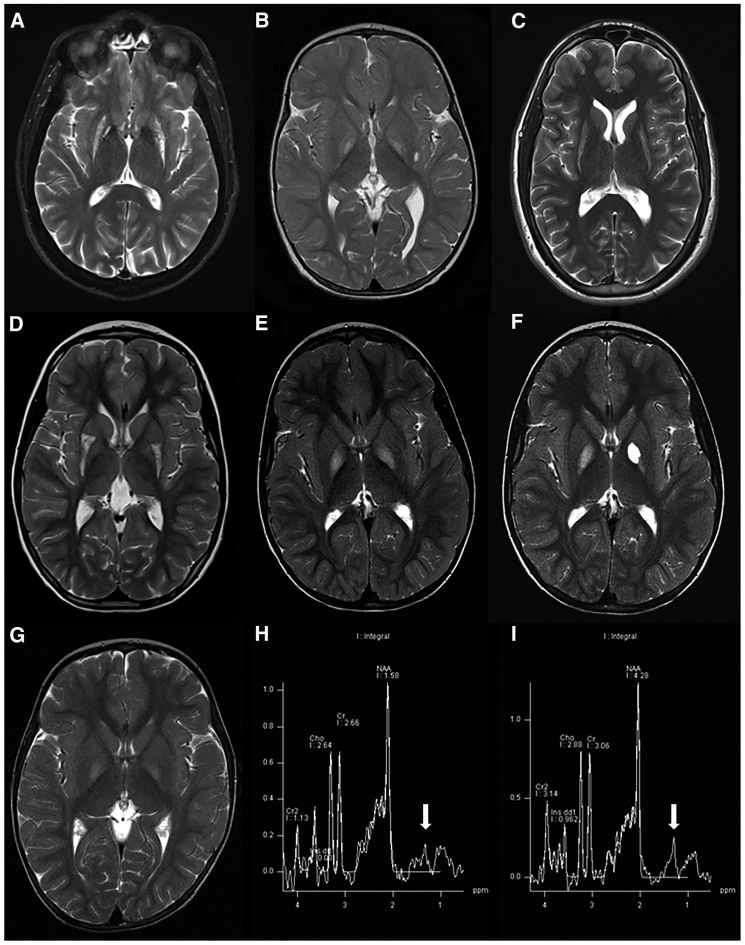
Figure 2.
T2 Axial MRI Sections of Affected Individuals Are Suggestive of Basal Ganglia Necrosis
(A) Hyper intense signal in the putamen and mild pallidal hypo intense signal in affected individual 1 (family A, II:1).
(B) Hyper intense pallidal signal in affected individual 2 (family B, II:2).
(C) Hyper intense signal in both putamen and caudate in affected individual 4 (family C, II:8).
(D) Selective damage to posterior putamen with apparent preservation of the anterior portion in affected individual 5 (family D, II:1).
(E) Hyper intense pallidal signal in affected individual 6 (family E, II:1).
(F) Follow-up MRI 2 years later in the same affected individual demonstrates cystic left pallidal changes.
(G) Hyper intense pallidal signal in affected individual 7 (family E, II:3).
(H and I) A lactate peak (see arrows) is demonstrated in the spectroscopy of affected individuals 6 and 7 (family E, II:1 and II:3), respectively.
Sequencing
WES performed in affected individual 1 (family A, II:1) revealed two compound heterozygous variations in MECR (GenBank: NM_016011.3): a missense variant c.695G>A (p.Gly232Glu) and a stop gain nonsense variant c.855T>G (p.Tyr285∗). WES performed in affected individual 2 (family B, II:2) also revealed the same c.695G>A (p.Gly232Glu) MECR missense variant as in affected individual 1 and a c.830+2_830+3insT splice site variant. It is noteworthy that this individual was also found heterozygous for a potentially deleterious c.327G>C (p.Lys109Asn) variant in NDUFAF5 (MIM: 612360). WGS performed in affected individual 6 (family E, II:1) revealed compound heterozygous variations in MECR, a c.772C>T (p.Arg258Trp) missense variant, and a c.247_250del (p.Asn83Hisfs∗4) frameshift variant. The clustering of the aforementioned next-generation sequencing results was enabled through the GeneMatcher tool.38
After the diagnosis of affected individuals 1 and 2, and due to similarity of their clinical and radiologic features, the three MECR variations located in these individuals were sequenced in affected individuals 3 and 4 (family C, II:2 and II:8), and both brothers were found to harbor the same two MECR variations as in affected individual 2 (c.695G>A; p.Gly232Glu and c.830+2_830+3insT).
In affected individual 5 (family D, II:1), Sanger sequencing of the entire coding region and splice site of MECR (performed as part of a specific population screening) demonstrated that she is homozygous for a c.854A>G (p.Tyr285Cys) missense variant involving the same residue as the nonsense variant found in affected individual 1.
All suspected mutations found by WES and WGS were confirmed with Sanger sequencing and segregation in all five families was consistent with an autosomal-recessive inheritance (Figure 1, Table 1).
cDNA Analysis
To test the c.830+2_830+3insT splice site mutation effects on MECR mRNA, we synthesized cDNA from total RNA of blood cells from affected individual 2 (family B, II:2), his parents, and a healthy control subject. A PCR primer set designed to amplify 284 bp fragment revealed that the c.830+2_830+3insT mutation produced two mutant fragments: one of 180 bp indicating mRNA transcript lacking the whole of exon 7, and a second transcript of 356 bp created by activation of a donor splice site within intron 7, resulting in addition of 108 bp from intron 8 to the mRNA sequence (Figure S1).
Bioinformatics and Population Screening
The suspected mutations detected in this study are all predicted to be damaging and were found to be extremely rare in the ExAC database (Table 2, Table S1).
Table 2.
Summary of the MECR Mutations and Their Prevalence
| Genomic Change (hg19) | Transcript Change (NM_016011.3) | Type of Variant | Protein Change | Allele Frequency in ExAC | Allele Frequency in Ashkenazi Genome Project | Allele Frequency in In-House Ashkenazi Jewish Controls |
|---|---|---|---|---|---|---|
| chr1: g.29522746A>C | c.855T>G | nonsense | p.Tyr285∗ | 0/120,182 | 0/256 | 0/200 |
| chr1: g.29528516C>T | c.695G>A | missense | p.Gly232Glu | 5/121,408 | 0/256 | 0/206 |
| chr1: g.29527026InsA | c.830+2_830+3insT | splice site | – | 10/120,664 | 1/256 | 0/210 |
| chr1: g.29522747T>C | c.854A>G | missense | p.Tyr285Cys | 1/120,182 | 0/256 | NA |
| chr1: g.29527086G>A | c.772C>T | missense | p.Arg258Trp | 10/121,102 | 0/256 | NA |
| chr1: g.29543124_29543127del | c.247_250del | frameshift | p.Asn83Hisfs∗4 | 0/121,412 | 0/256 | NA |
The p.Tyr285∗ nonsense variant is expected to remove significant parts of both the catalytic and the cofactor binding domains (Figures 3A and 3F) and give rise to a non-functional protein product. The early frameshift four nucleotide deletion (p.Asn83Hisfs∗4) creates a new reading frame with a stop codon located four amino acids downstream. It is very likely that this transcript is degraded as a result of the nonsense-mediated mRNA decay (NMD) process39 and in any case gives rise to a non-functional product (Figure 3D).
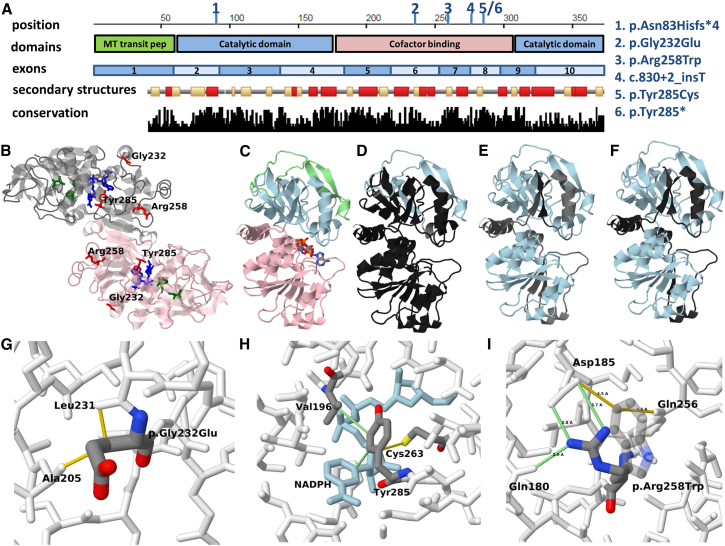
Figure 3.
Structural Context of the Variants Detected in This Study
(A) Sequence presentation that includes the domain organization, exon boundaries, secondary structures, and conservation data. In the secondary structure, track helices are indicated in dark color and strands in brighter color. The variants detected in this study are indicted on the coordinates ruler by the numbers 1–6, with the corresponding variant represented by each number given on the side.
(B) Global view of the homodimer (PDB: 2vcy). The NADPH cofactor-binding site is shown in blue and the suspected fatty acid binding site is shown in dark green. The p.Gly232Glu, p.Tyr285Cys, and p.Arg258Trp missense mutations are shown in red. Positions 232 and 258 are close to the surface and although located in the cofactor binding domain, are not parts of the binding site itself while position 285 is part of the cofactor binding pocket.
(C) Structure of the monomer colored by structural domains. The cofactor-binding domain is shown in pink, the catalytic domain in blue, and the transition peptide in green.
(D) The region expected to be missing (black) or altered (gray) due to the p.Asn83Hisfs∗4 frameshift deletion variant (if the transcript survives decay and is translated).
(E) The region expected to be altered (gray) or missing (black) due to the c.830+2_830+3insT splicing variant assuming no alternative splicing donor site is utilized.
(F) The region expected to be missing (black) due to p.Tyr285∗ nonsense variant.
(G) Detailed view of the region around residue 232. The side chain introduced by the p.Gly232Glu mutation creates severe steric clashing with residues 231 and 205 (shown in yellow), resulting in reduced stability and/or significant local conformation changes in this region.
(H) Detailed view of the region around residue 285. The NADPH is shown in blue. Favorable interactions formed with Tyr285 are shown in green lines. Putative disulfide bond formed between the Cys285 mutant residue and Cys263 is shown in yellow line.
(I) Detailed view of the region around residue 258. The p.Arg258Trp mutation abolishes several salt bridges and hydrogen bonds (shown in green lines) with side chains of residues Asp185 and Gln180. The tryptophan side chain (shown in semitransparent display) makes unfavorable interactions with polar groups in the area (yellow lines).
The splicing variant, a T insertion within the GT splicing donor consensus, is more difficult to interpret. The first nucleotide following the GT donor dinucleotide is changed from A to T. This position is not part of the core GT donor site but is almost always occupied in complex eukaryotes by a purine base (usually G but A in the current instance). A pyrimidine in this position is likely to significantly weaken the site and prevent splicing in most transcripts. If there is no alternative donor site utilized downstream, the frame is shifted and a new sequence of 89 amino acids is introduced before a stop codon in the intron between exons 7 and 8 (Figure 1E). Even if an alternative donor site from one of the two adjacent sites (predicted by ASSP) is utilized, it also eventually results in a shift of the reading frame. Thus, this splicing variant most likely causes most of the mature product to be non-functional.
The three missense mutations are located within the cofactor-binding domain and are evolutionarily conserved, demonstrating their functional importance (Figure 3A). Gly232 and Tyr285 are completely conserved through evolution from yeast to human while Arg258 is partially conserved, with most of the exceptions being lysine (another positively charged residue), but never tryptophan or other aromatic residues. This is well reflected in their functional prediction scores obtained by a variety of tools like Sift, PolyPhen, MutationTaster, MutationAssessor, LRT, and PROVEAN (see Table S1).
Due to the Ashkenazi Jewish origin of three of the five families, we assessed the frequency of their mutations in an Ashkenazi Jewish population (using the Ashkenazi Jewish genome project and in-house controls). Only the c.830+2_830+3insT splice site mutation was found in 1 out of 256 alleles of the Ashkenazi Jewish genome project; the other mutations were not detected in either the Ashkenazi Jewish genome project or the in-house controls (Table 2).
A cohort of 150 individuals with bilateral striatal necrosis that were suspected for NBIA but found negative for mutations in the known genes were screened by either next-generation sequencing or Sanger sequencing of the entire coding region of MECR. Only 1 of these 150 individuals was found to harbor mutations in MECR and is described here as affected individual 5 (family D, II:1).
Structural Analysis
Two structures of human MECR are available in the protein data bank (PDB):13 a structure from 2005 (PDB: 1zsy)13 with 1.75 Å resolution and a structure from 2007 with a 2.41 Å resolution (PDB: 2vcy).14 The protein is considered to be a homodimer (Figure 3B). Each monomer is composed of two functional units, the catalytic domain and a cofactor-binding domain. The catalytic domain includes residues in both N-terminal and C-terminal sides of the cofactor-binding domain. The location of the NADPH cofactor-binding domain is not known in the human protein but can be inferred from the structure of Candida tropicalis protein (PDB: 1guf), as the human and yeast proteins share 40% sequence identity.
The NADPH binding is coordinated by loops of the Rossmann fold of the cofactor-binding domain on the boundary with the catalytic domain (Figure 3C) while the location of the fatty acid substrates (which vary in length from 2 to 16 carbons) is assumed to be in a deep pocket that extends from the active site into the catalytic domain (Figure 3C).
Both Gly232 and Arg258 are partially exposed to the solvent and located relatively close to the protein surface (Figure 3B). Although they are located within the cofactor-binding domain, they are not part of the NADPH binding site itself, the first shell of residues that are predicted to be part of the fatty acyl binding site, or the dimer interface. In contrast, Tyr285 is part of the cofactor binding pocket. We modeled the side-chain conformation of the mutants in order to analyze the changes in atomic contacts in the vicinity of the mutations. The p.Gly232Glu mutation gives rise to a very unfavorable conformation with severe interference between the glutamate side chain and residues Ala215 and Leu231 (Figure 3G). Specifically, it appears that any amino acid with a side chain in position 232 will interfere with the carbonyl of residue 231. Such interference is predicted to cause a very unstable conformation or result in formation of a stable non-functional conformation. Since position 232 is quite close to the protein surface, the p.Gly232Glu variant may affect interaction with an unknown factor. The p.Tyr285Cys substitution is predicted to affect the binding affinity of the NADPH because both share extensive contacts (Figure 3H), especially between the aromatic rings. Tyr285 also creates favorable interactions with hydrophobic side chains such as Val196, which are predicted to be abolished in the p.Tyr285Cys mutants. Interestingly, an additional cysteine residue (Cys263) is very close and might form disulfide bond with p.Tyr285Cys. The formation of a disulfide bond also depends on the oxidizing state of the environment (the mitochondrial intermembrane space is rather supportive for such bonds). If this bond is formed, it is expected to dramatically affect the shape and dynamics of the region and also abolish contacts that are formed in the wild-type protein between the Cys263 and the NADPH. With regard to the p.Arg258Trp variant, it is predicted to cause a destabilizing effect, as three salt bridges with residues Gln180 and Asp185 in the wild-type structures are abolished (Figure 3I). These salt bridges may be important for the integrity of subdomains in the structure. Instead, new unfavorable contacts between polar groups of adjacent side chains and the aromatic ring of Trp258 are formed.
Mutation Modeling in Yeast
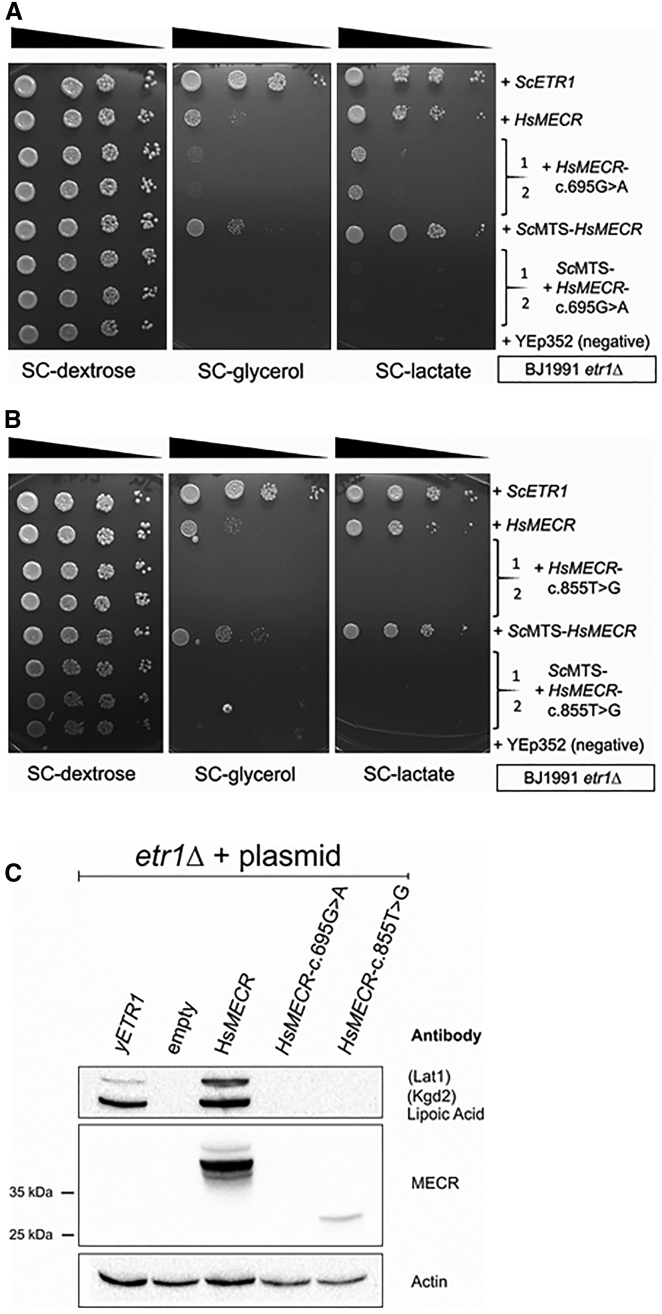
In order to analyze the effect of the mutations in an in vivo system, we turned to a yeast complementation assay. It has been previously demonstrated that MECR is capable of complementation of the respiratory-deficient phenotype of the etr1Δ mutation in S. cerevisiae.11 The cDNAs of the alleles harboring the c.695G>A and c.855T>G mutations were cloned individually on yeast expression plasmids under control of the yeast CTA1 promoter and the plasmids transformed into etr1Δ strains. Respiratory growth of the transformants was tested on plates containing only a non-fermentable carbon source (lactate or glycerol) (Figures 4A and 4B). As in previous experiments, MECR complemented the respiratory deficiency of the etr1Δ strains. The c.695G>A MECR allele rescued growth on lactate only poorly compared to wild-type MECR, while the c.855T>G mutation variant, predicted to encode a truncated MECR protein lacking the cofactor binding site, did not rescue growth on respiratory media at all. These results indicated that c.695G>A MECR encodes a diminished function allele, while the c.855T>G mutation results in a complete loss of function. Because we have previously found that yeast is apparently sometimes unable to efficiently recognize human mitochondrial targeting signals (MTSs),40 we constructed versions of MECR and the c.695G>A and c.855T>G mutation variants that expressed proteins N terminally appended with a yeast MTS. This modification visibly improved complementation of the etr1Δ strains by wild-type MECR but did not positively affect growth rescue by the c.695G>A variant. Likewise, the allele encoding the truncated form of MECR did not rescue any growth even when this variant was appended with yeast MTS. As the extent of growth rescue of etr1Δ strains by MECR is strain dependent (our unpublished observation), these experiments were also conducted in a different yeast strain background, with similar results (Figures S2A and S2B).
Figure 4.
Modeling of the c.695G>A and c.855T>G Mutated MECR Alleles in the Respiratory-Deficient Yeast BJ1991etr1Δ mtFAS Defective Strain
(A and B) Analysis of rescue of respiratory growth of the etr1Δ mutant by the MECR c.695G>A and c.855T>G allele variants, respectively. Yeast cells transformed with plasmids carrying the indicated constructs were grown on liquid media, normalized according to cell density, and serial diluted (1×, 1/10×, 1/100×, and 1/1,000×). For each mutation two independent clones transformed with mutation plasmids were tested. Equal volumes of cell suspensions were spotted on synthetic media containing only a non-fermentable carbon source (glycerol or lactate) or plates containing the fermentable carbon source glucose as growth control, and grown for several days. Growth on lactate or glycerol indicates respiratory competence.
(C) Western blotting analyses of yeast extracts from BJ1991etr1Δ cells carrying YEp195-ETR1(yETR1), YEp352 (empty), YEp352-MTS-HsMECR (HsMECR), YEp352-MTS-HsMECR-c.695G>A (HsMECR-c.695G>A), or YEp352-MTS-HsMECR-c.885T>G (HsMECR-c.885T>G). Whole yeast cell extract was separated by SDS-page, transferred to a solid support, and probed for the relevant antigens. Antisera used for protein detection are indicated. Lat1: E2 subunit of yeast pyruvate dehydrogenase; Kgd2: E2 subunit of yeast α-ketoglutarate dehydrogenase. Actin: Loading control. An additional loading control (Ponceau S staining of the blotting membrane) can be found in the Supplemental Data.
We analyzed MECR expression and protein lipoylation in the strains transformed with the allele variants (Figure 4C). Extract of cells carrying the wild-type MECR construct displayed ample amounts of MECR as judged by western blotting. No signal corresponding to MECR was found in the lane containing extract from the yeast strain carrying the c.695G>A missense variant, indicating that the mutant protein may not be stable. A weak band cross-reacting with anti-MECR serum was visible in the lane with extract from etr1Δ cells expressing the premature stop codon mutation, consistent with the production of a truncated protein from this allele. In the MECR-expressing cells, the Lat1 and Kgd2 subunits of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, respectively, were lipoylated at even higher levels than in the wild-type yeast control. In contrast, no lipoylation was visible in the lanes containing extract from cells harboring the c.695G>A and c.855T>G variants. These data provide additional evidence that these variants manifest diminished function or loss-of-function alleles.
Analysis of Affected Individuals’ Fibroblasts
We obtained skin samples from four of the affected individuals for establishment of fibroblast cell lines. The cells were analyzed for MECR and lipoic acid content. Cellular extracts of fibroblast samples from affected individuals 1–3 and 6 exhibited strongly reduced levels of MECR protein (Figures 5A and 5B). Because of the reported role of MECR in the production of the lipoic acid precursor octanoic acid and the requirement of mtFAS for cellular respiratory competence, protein lipoylation levels, respiratory activity, and mitochondrial morphology were investigated. Consistent with the decrease in MECR, protein lipoylation levels were reduced by about 50% in affected individuals’ samples that were tested compared to healthy control fibroblasts (Figures 5A, 5C, and 5D). The reduction of MECR and protein lipoylation was accompanied by diminished respiratory activity in some of the affected individuals’ cell lines (Figures 6A–6C).
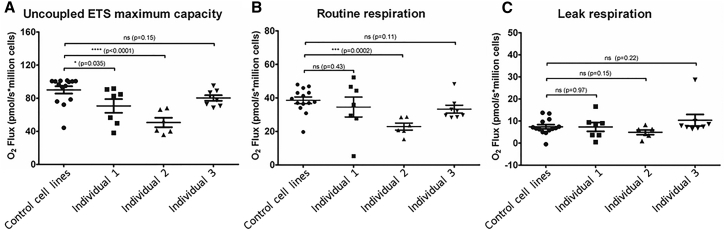
Figure 6.
Respirometry Analysis of Affected Individuals’ Fibroblast Cell Lines
Analysis was carried out measuring oxygen consumption using the Oroboros 2k respirometer; (A) ETS (electron transport system activity); (B) routine level respiration; (C) leak level respiration.
A statistically significant reduction of the ETS (electron transport system) maximum capacity is seen in affected individuals 1 and 2 cell lines (family A, II:1, and family B, II:2, respectively), while the mean measurement in the affected individual 3 (family C, II:2) cell line is lower than that of the control group, but not significantly so (Figure 6A). ETS maximum capacity is a state where proton carriers capable of transporting protons across the inner mitochondrial membrane uncouple the ETS from the normal cellular processes that pump protons inside the mitochondrial matrix. When the uncoupler is carefully titrated, this uncoupled state allows the ETS to function at its maximum capacity. A significant difference in routine respiration was seen only in the cell line of affected individual 2 (Figure 6B). This routine respiration describes the respiratory behavior of the cells under normal culture conditions. No significant differences between the control subjects and affected individuals were seen in the leak respiration measurement, an oligomycin-inhibited state reflecting proton leak from the complexes of the ETS (Figure 6C). No increase in such events was observed in the intact cell respirometry.
Previous studies examining the effects of mtFAS dysfunction in both yeast33, 41 and mammalian cells (unpublished) revealed changes in mitochondrial morphology upon inactivation of mtFAS; however, we did not observe any appreciable differences between affected individual and control cells (data not shown).
Affected individual 6 (family E, II:1) fibroblast extracts exhibited strongly reduced amounts of MECR protein relative to controls on immunoblotting (Figure 5B). No effect of reduced MECR on OXPHOS complexes I-V levels was observed (Figure S3A). Similarly, the activities of complex I and complex IV in his fibroblasts was only modestly reduced to 65% and 75% of controls, respectively (Figure S3B).
Discussion
The fatty acid de novo synthesis (FAS) pathway consists of several steps by which an acetyl group is elongated and the growing acyl chains are bound to an acyl carrier protein (ACP). The existence of a bacterial type fatty acid synthesis pathway in mitochondria that is distinct from the cytosolic FAS apparatus has only recently entered public awareness.42 Enzymes of the mitochondrial pathway accept in vitro substrates with up to C16–18 in carbon chain lengths, but an important product is the octanoyl group, which serves as the precursor of LA synthesis43 by a separate pathway. The mtFAS is essential for respiratory activity in the yeast S. cerevisiae, where defects in components of this pathway lead to loss of respiratory complexes due to faults in mitochondrial RNA processing and translation and respiratory complex assembly.34 A second hallmark phenotype of mtFAS-defective yeast mutants is LA deficiency and concomitant reduced activity of the LA-dependent enzyme complexes.7, 34, 42
A metabolomics study in HeLa cells in which the mtFAS was either downregulated or upregulated demonstrated that despite no significant change was seen in mitochondrial fatty acid composition, several metabolic pathways were affected.44 Among these were glycolysis and sorbitol pathways, tricarboxylic acid (TCA) cycle and amino acid anaplerosis, pentose phosphate pathway, redox status, and polyamine synthesis. The most striking changes observed in these experiments were in the bioactive lipid levels, which are important determinants of cell survival.
The knockout of the LA synthase gene (Lias) in mouse leads to early embryonic death.45 A conditional tamoxifen-induced knockout of MCAT, encoding mitochondrial malonyl-CoA transferase, resulted in a lipoylation deficiency, respiratory complex defects, and a premature aging phenotype46 similar to mouse models of other mitochondrial defects47 and to the mitochondrial phenotype found in yeast mtFAS mutants.
Human disorders of LA synthesis or attachment defects (i.e., through mutations in LIAS [MIM: 607031] or LIPT1 [MIM: 610284]) have been reported.48, 49 Affected individuals present with neonatal-onset epilepsy, muscular hypotonia, and severe psychomotor retardation, often with death in infancy. Metabolite analysis in these infants typically reveals elevated plasma lactate and pyruvate levels and, with exception of individuals with LIPT1 defect, non-ketotic hyperglycinemia.
Human MECR encodes the mitochondrial trans-2-enoyl-CoA reductase, which catalyzes the last step of the mtFAS.11 Deletion of the Mecr homolog in mouse leads to early embryonic death (unpublished), while myocardial overexpression causes cardiac dysfunction.50 The mitochondria in the hearts of these overexpressing mice are dramatically enlarged, a phenotype reminiscent of overexpressing the homologous Etr1 yeast enoyl thioester reductase in S. cerevisiae.33 Recent reports indicate a role of MECR in peroxisome-proliferator-activator-receptor (PPAR)-dependent signaling, although it is debated whether a product of mtFAS or a function of MECR as a transcriptional coactivator is responsible for this regulatory role.51, 52
The individuals described here are less severely affected than individuals with LA synthesis defects and do not display cardiac damage as in the Mecr overexpression murine model. This could be explained by potential alternative sources of substrate for the intact LA synthesis enzymes in cases of defective mtFAS. The first is the parallel cytosolic FAS pathway that might partially compensate in the cases of underactivity of mtFAS enzymes (but not overactivity as in the overexpressing model). The second is the fatty acid β-oxidation pathway in which octanoyl-CoA is an obligatory intermediate. The abundance of β-oxidation in myocytes compared to neurons, where β-oxidation is essentially absent, might account for the predominantly neurologic manifestations and lack of cardiac and other muscle involvement in the affected individuals described here. These potential compensatory mechanisms might also explain why, unlike many mitochondrial diseases, in the individuals with MECR mutations there is relative sparing of cognition and lack of additional multisystem involvement and typical mitochondrial biomarkers. It is possible that partial endogenous compensation is sufficient to prevent damage in most tissues except for those most susceptible to energy depletion or oxidative stress, such as the basal ganglia and optic nerve.53, 54 If this is the case, it raises the possibility of a therapeutic strategy through supplementation to protect vulnerable tissues from further damage. Two strong candidates for therapeutic use in alleviating mtFAS defect clinical features exist: LA and octanoic acid. LA supplementation has been shown to exert positive effects in individuals with mitochondrial defects and is used as a standard therapeutic treatment in mitochondrial disease.55 A recent report describes beneficial effects of LA supplementation on mitochondrial function in human cells where mtFAS has been inhibited.10 However, randomized clinical trials or formal confirmation of utilization of supplemented LA as substrate for attachment as a cofactor to keto-acid reductases are lacking. The enzyme required for this transfer reaction, LIPT1, lacks the LA-activating function of its E. coli lplA homolog which is capable of adenylation of free LA and transfer thereof to target proteins.56 Rather than serving as free LA transferase, LIPT1 is more likely to act as octanoyl-CoA transferase like the more similar yeast Lip3 protein.57 Octanoyl-CoA is present in mitochondria under physiological conditions as an intermediate of β-oxidation, but also free octanoic acid can easily enter into mammalian mitochondria in a non-protonated state where it is then activated to octanoyl-CoA. Octanoic acid has been shown to provide an energy source in mitochondria58, 59 and could also ultimately be utilized as a precursor for mitochondrial LA synthesis. The question of whether exogenous octanoic and/or lipoic acid can be activated in mitochondria and are able to serve there as cofactor to ketoacid dehydrogenase complexes clearly requires more attention.
Modeling of the c.695G>A and c.855T>G mutations in yeast revealed several interesting features. The c.695G>A missense variant was able to partially complement the respiratory deficiency in two different yeast etr1Δ strain backgrounds, while growth of the etr1 mutants transformed with a plasmid carrying the c.855T>G premature stop codon variant did not exhibit any growth improvement. On the other hand, although the missense allele supported some yeast growth, no mutant MECR protein corresponding to the c.695G>A allele variant accumulated in these yeast cells, while a faint band corresponding to the truncated MECR form could be detected. Nonetheless, neither of the mutant variants was capable of restoring lipoylation in the yeast model. The poor or absent respiratory growth rescue of the yeast etr1Δ by these MECR variants strongly supports their pathogenicity, since the wild-type allele, when appended with a yeast MTS, dramatically improves growth of this strain on non-fermentable carbon sources and restores lipoic acid to near wild-type levels. Thus, although not all MECR mutations were modeled in yeast, we believe that the conclusive results from yeast complementation with both a missense as well as a nonsense mutation, accompanied by the structural prediction modeling of all six mutations, are sufficient to implicate this gene and protein in the human disease.
Analysis of affected individual fibroblasts indicated a strong reduction of mutant MECR protein in their cell lines, ranging from the MECR band being barely visible to undetectable. These data are congruent with the yeast model data and indicate production of an unstable protein. In line with these results, we were unable to purify any appreciable amounts of the c.695G>C-variant encoded mutant MECR protein expressed in E. coli, although we are able to obtain ample amounts of the wild-type protein using the same purification procedure (not shown). However, lipoylation levels in affected individuals’ cells are not reduced as dramatically as in the yeast cells carrying the MECR mutations. A potential explanation of this discrepancy could be a possibly stabilizing effect of the compound heterozygous state of the affected individuals, in a way that the C-terminally truncated inactive enzyme stabilizes the labile missense protein variant. This possibility may be explored in the future in cell lines of homozygous individuals or in a compound heterozygous mouse model. Another explanation could again be the contribution of octanoyl-CoA from the β-oxidation in the fibroblasts, which is nonexistent in the yeast.
Measurement of respiratory activity of several of the affected individuals’ cell lines indicated a mild respiratory defect in two out of the three lines. This was most pronounced for the ETS activity, where the difference was significant for two of three cell lines, with the third following the trend. There is also an indication of differences in the routine respiration level. The effect of mtFAS deficiency on the mammalian respiratory chain has not been systematically investigated, but yeast cells carrying deletions of mtFAS components suffer from a general defect in mitochondrial biogenesis.34 Our data imply that there may be a similar effect in human cells. In spite of the rather severe reduction of MECR in the affected individuals’ cells, and in accord with the lipoylation data obtained from the fibroblast extracts, the respiratory defects detected in these cells appears to be quite mild.
The somewhat more pronounced respiratory defect in affected individual 2 (family B, II:2) specifically might be related to his additional NDUFAF5 heterozygous potentially deleterious variant.
To conclude, we present evidence that mutations in MECR cause a mtFAS-related human disorder that manifests as childhood-onset basal ganglia degeneration and optic atrophy. The clinical and radiographic features of this disease are distinctive and recognizable and it differs from most classical mitochondrial diseases by the relatively preserved cognition and lack of additional multisystem involvement and typical mitochondrial biomarkers. We propose a name for this disorder, MEPAN (mitochondrial enoyl CoA reductase protein-associated neurodegeneration), and a rational therapeutic of LA or octanoic acid supplementation, which should be investigated for safety and efficacy in this condition.
Consortia
The members of the University of Washington Center for Mendelian Genomics are Michael J. Bamshad, Suzanne M. Leal, Deborah A. Nickerson, Peter Anderson, Marcus Annable, Elizabeth Marchani Blue, Kati J. Buckingham, Jennifer Chin, Jessica X. Chong, Rodolfo Cornejo, Jr., Colleen P. Davis, Christopher Frazar, Zongxiao He, Gail P. Jarvik, Guillaume Jimenez, Eric Johanson, Tom Kolar, Stephanie A. Krauter, Daniel Luksic, Colby T. Marvin, Sean McGee, Daniel J. McGoldrick, Karynne Patterson, Marcos Perez, Sam W. Phillips, Jessica Pijoan, Peggy D. Robertson, Regie Santos-Cortez, Aditi Shankar, Krystal Slattery, Kathryn M. Shively, Deborah L. Siegel, Joshua D. Smith, Monica Tackett, Gao Wang, Marc Wegener, Jeffrey M. Weiss, Riana I. Wernick, Marsha M. Wheeler, and Qian Yi.
Acknowledgments
The authors would like to acknowledge the families for their collaboration. The Israeli group was supported by funds from the Israel Science Foundation (grant number 2023/14) and the Pinchas Borenstein Talpiot Medical Leadership Program. The Finnish group was supported by the Sigrid Juselius Foundation and the Academy of Finland. The American group was funded by the NBIA Disorders Association and by philanthropic support. Sequencing for the American group was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant 2UM1HG006493 to Drs. Debbie Nickerson, Michael Bamshad, and Suzanne Leal. The Australian group was supported by a New South Wales Office of Health and Medical Research Council Sydney Genomics Collaborative grant (J.C. and L.G.R.), NHMRC project grant 1026891 (J.C.), Cancer Institute NSW fellowship 13/ECF/1-46 (M.J.C.), and generous financial support from the Kinghorn Foundation. J.C. is also grateful to the Crane and Perkins families for their generous financial support. The Italian group was supported by Pierfranco and Luisa Mariani Foundation. They also thank the Cell lines and DNA Bank of Paediatric Movement Disorders and Mitochondrial Diseases of the Telethon Genetic Biobank Network, which is supported by TELETHON Italy (project no. GTB09003) and the Bank for the Diagnosis and Research of Movement Disorders (MDB) of the EuroBiobank.
Published: November 3, 2016
Footnotes
Supplemental Data include detailed case descriptions, three figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.09.021.
Contributor Information
Bruria Ben-Zeev, Email: bruria.benzeev@sheba.health.gov.il.
Susan J. Hayflick, Email: hayflick@ohsu.edu.
University of Washington Center for Mendelian Genomics:
Michael J. Bamshad, Suzanne M. Leal, Deborah A. Nickerson, Peter Anderson, Marcus Annable, Elizabeth Marchani Blue, Kati J. Buckingham, Jennifer Chin, Jessica X. Chong, Rodolfo Cornejo, Jr., Colleen P. Davis, Christopher Frazar, Zongxiao He, Gail P. Jarvik, Guillaume Jimenez, Eric Johanson, Tom Kolar, Stephanie A. Krauter, Daniel Luksic, Colby T. Marvin, Sean McGee, Daniel J. McGoldrick, Karynne Patterson, Marcos Perez, Sam W. Phillips, Jessica Pijoan, Peggy D. Robertson, Regie Santos-Cortez, Aditi Shankar, Krystal Slattery, Kathryn M. Shively, Deborah L. Siegel, Joshua D. Smith, Monica Tackett, Gao Wang, Marc Wegener, Jeffrey M. Weiss, Riana I. Wernick, Marsha M. Wheeler, and Qian Yi
Web Resources
1000 Genomes, http://browser.1000genomes.org
Alternative Splice Site Predictor, http://wangcomputing.com/assp/
Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/
CUPSAT, http://cupsat.tu-bs.de
DbNSFP, http://varianttools.sourceforge.net/Annotation/DbNSFP
DNAnexus, https://www.dnanexus.com/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
FoldX, http://foldxsuite.crg.eu/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
I-Mutant, http://folding.biofold.org/i-mutant/i-mutant2.0.html
MAESTRO, https://biwww.che.sbg.ac.at/
Mutation Assessor, http://mutationassessor.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PROVEAN, http://provean.jcvi.org
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
SCCOMP, http://www.sheba-cancer.org.il/cgi-bin/sccomp/sccomp1.cgi
TriangleMatch, http://bioinfo3d.cs.tau.ac.il/TriangleMatchBeta/
University of Washington Center for Mendelian Genomics detailed sequencing methods, http://uwcmg.org/docs/Exome_Genome_Sequencing/uwcmg_Detailed_Methods_Sequencing.docx
Supplemental Data
References
- 1.Schneider S.A., Bhatia K.P. Secondary dystonia--clinical clues and syndromic associations. Eur. J. Neurol. 2010;17(Suppl 1):52–57. doi: 10.1111/j.1468-1331.2010.03051.x. [DOI] [PubMed] [Google Scholar]
- 2.Zuccoli G., Yannes M.P., Nardone R., Bailey A., Goldstein A. Bilateral symmetrical basal ganglia and thalamic lesions in children: an update (2015) Neuroradiology. 2015;57:973–989. doi: 10.1007/s00234-015-1568-7. [DOI] [PubMed] [Google Scholar]
- 3.Kölker S., Christensen E., Leonard J.V., Greenberg C.R., Boneh A., Burlina A.B., Burlina A.P., Dixon M., Duran M., García Cazorla A. Diagnosis and management of glutaric aciduria type I--revised recommendations. J. Inherit. Metab. Dis. 2011;34:677–694. doi: 10.1007/s10545-011-9289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer E., Kurian M.A., Hayflick S.J. Neurodegeneration with brain iron accumulation: genetic diversity and pathophysiological mechanisms. Annu. Rev. Genomics Hum. Genet. 2015;16:257–279. doi: 10.1146/annurev-genom-090314-025011. [DOI] [PubMed] [Google Scholar]
- 5.Lake N.J., Compton A.G., Rahman S., Thorburn D.R. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol. 2016;79:190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- 6.Wada H., Shintani D., Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody S., Oh C., Hoja U., Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- 8.Feng D., Witkowski A., Smith S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J. Biol. Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayr J.A., Feichtinger R.G., Tort F., Ribes A., Sperl W. Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 2014;37:553–563. doi: 10.1007/s10545-014-9705-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen C., Han X., Zou X., Li Y., Yang L., Cao K., Xu J., Long J., Liu J., Feng Z. 4-methylene-2-octyl-5-oxotetrahydrofuran-3-carboxylic acid (C75), an inhibitor of fatty-acid synthase, suppresses the mitochondrial fatty acid synthesis pathway and impairs mitochondrial function. J. Biol. Chem. 2014;289:17184–17194. doi: 10.1074/jbc.M114.550806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miinalainen I.J., Chen Z.J., Torkko J.M., Pirilä P.L., Sormunen R.T., Bergmann U., Qin Y.M., Hiltunen J.K. Characterization of 2-enoyl thioester reductase from mammals. An ortholog of YBR026p/MRF1’p of the yeast mitochondrial fatty acid synthesis type II. J. Biol. Chem. 2003;278:20154–20161. doi: 10.1074/jbc.M302851200. [DOI] [PubMed] [Google Scholar]
- 12.Paila U., Chapman B.A., Kirchner R., Quinlan A.R. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput. Biol. 2013;9:e1003153. doi: 10.1371/journal.pcbi.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Z.J., Pudas R., Sharma S., Smart O.S., Juffer A.H., Hiltunen J.K., Wierenga R.K., Haapalainen A.M. Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new insights into its substrate recognition properties. J. Mol. Biol. 2008;379:830–844. doi: 10.1016/j.jmb.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Lappalainen I., Vihinen M. Structural basis of ICF-causing mutations in the methyltransferase domain of DNMT3B. Protein Eng. 2002;15:1005–1014. doi: 10.1093/protein/15.12.1005. [DOI] [PubMed] [Google Scholar]
- 16.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pires D.E., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2015;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pires D.E., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laimer J., Hofer H., Fritz M., Wegenkittl S., Lackner P. MAESTRO--multi agent stability prediction upon point mutations. BMC Bioinformatics. 2015;16:116. doi: 10.1186/s12859-015-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worth C.L., Preissner R., Blundell T.L. SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 23.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 26.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nussinov R., Wolfson H.J. Efficient detection of three-dimensional structural motifs in biological macromolecules by computer vision techniques. Proc. Natl. Acad. Sci. USA. 1991;88:10495–10499. doi: 10.1073/pnas.88.23.10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyal E., Najmanovich R., McConkey B.J., Edelman M., Sobolev V. Importance of solvent accessibility and contact surfaces in modeling side-chain conformations in proteins. J. Comput. Chem. 2004;25:712–724. doi: 10.1002/jcc.10420. [DOI] [PubMed] [Google Scholar]
- 31.Wang M., Marín A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–227. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Riley L.G., Cooper S., Hickey P., Rudinger-Thirion J., McKenzie M., Compton A., Lim S.C., Thorburn D., Ryan M.T., Giegé R. Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia--MLASA syndrome. Am. J. Hum. Genet. 2010;87:52–59. doi: 10.1016/j.ajhg.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torkko J.M., Koivuranta K.T., Miinalainen I.J., Yagi A.I., Schmitz W., Kastaniotis A.J., Airenne T.T., Gurvitz A., Hiltunen K.J. Candida tropicalis Etr1p and Saccharomyces cerevisiae Ybr026p (Mrf1’p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence. Mol. Cell. Biol. 2001;21:6243–6253. doi: 10.1128/MCB.21.18.6243-6253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kursu V.A., Pietikäinen L.P., Fontanesi F., Aaltonen M.J., Suomi F., Raghavan Nair R., Schonauer M.S., Dieckmann C.L., Barrientos A., Hiltunen J.K., Kastaniotis A.J. Defects in mitochondrial fatty acid synthesis result in failure of multiple aspects of mitochondrial biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 2013;90:824–840. doi: 10.1111/mmi.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill J.E., Myers A.M., Koerner T.J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 36.Gietz R.D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 37.Platta H.W., Girzalsky W., Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z., Kastaniotis A.J., Miinalainen I.J., Rajaram V., Wierenga R.K., Hiltunen J.K. 17beta-hydroxysteroid dehydrogenase type 8 and carbonyl reductase type 4 assemble as a ketoacyl reductase of human mitochondrial FAS. FASEB J. 2009;23:3682–3691. doi: 10.1096/fj.09-133587. [DOI] [PubMed] [Google Scholar]
- 41.Kastaniotis A.J., Autio K.J., Sormunen R.T., Hiltunen J.K. Htd2p/Yhr067p is a yeast 3-hydroxyacyl-ACP dehydratase essential for mitochondrial function and morphology. Mol. Microbiol. 2004;53:1407–1421. doi: 10.1111/j.1365-2958.2004.04191.x. [DOI] [PubMed] [Google Scholar]
- 42.Hiltunen J.K., Autio K.J., Schonauer M.S., Kursu V.A., Dieckmann C.L., Kastaniotis A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta. 2010;1797:1195–1202. doi: 10.1016/j.bbabio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Hiltunen J.K., Schonauer M.S., Autio K.J., Mittelmeier T.M., Kastaniotis A.J., Dieckmann C.L. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J. Biol. Chem. 2009;284:9011–9015. doi: 10.1074/jbc.R800068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clay H.B., Parl A.K., Mitchell S.L., Singh L., Bell L.N., Murdock D.G. Altering the mitochondrial fatty acid synthesis (mtFASII) pathway modulates cellular metabolic states and bioactive lipid profiles as revealed by metabolomic profiling. PLoS ONE. 2016;11:e0151171. doi: 10.1371/journal.pone.0151171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi X., Maeda N. Endogenous production of lipoic acid is essential for mouse development. Mol. Cell. Biol. 2005;25:8387–8392. doi: 10.1128/MCB.25.18.8387-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith S., Witkowski A., Moghul A., Yoshinaga Y., Nefedov M., de Jong P., Feng D., Fong L., Tu Y., Hu Y. Compromised mitochondrial fatty acid synthesis in transgenic mice results in defective protein lipoylation and energy disequilibrium. PLoS ONE. 2012;7:e47196. doi: 10.1371/journal.pone.0047196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 48.Mayr J.A., Zimmermann F.A., Fauth C., Bergheim C., Meierhofer D., Radmayr D., Zschocke J., Koch J., Sperl W. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am. J. Hum. Genet. 2011;89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tort F., Ferrer-Cortès X., Thió M., Navarro-Sastre A., Matalonga L., Quintana E., Bujan N., Arias A., García-Villoria J., Acquaviva C. Mutations in the lipoyltransferase LIPT1 gene cause a fatal disease associated with a specific lipoylation defect of the 2-ketoacid dehydrogenase complexes. Hum. Mol. Genet. 2014;23:1907–1915. doi: 10.1093/hmg/ddt585. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z., Leskinen H., Liimatta E., Sormunen R.T., Miinalainen I.J., Hassinen I.E., Hiltunen J.K. Myocardial overexpression of Mecr, a gene of mitochondrial FAS II leads to cardiac dysfunction in mouse. PLoS ONE. 2009;4:e5589. doi: 10.1371/journal.pone.0005589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parl A., Mitchell S.L., Clay H.B., Reiss S., Li Z., Murdock D.G. The mitochondrial fatty acid synthesis (mtFASII) pathway is capable of mediating nuclear-mitochondrial cross talk through the PPAR system of transcriptional activation. Biochem. Biophys. Res. Commun. 2013;441:418–424. doi: 10.1016/j.bbrc.2013.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D.G., Yoo J.C., Kim E., Lee Y.S., Yarishkin O.V., Lee D.Y., Lee K.H., Hong S.G., Hwang E.M., Park J.Y. A novel cytosolic isoform of mitochondrial trans-2-enoyl-CoA reductase enhances peroxisome proliferator-activated receptor α activity. Endocrinol. Metab. (Seoul) 2014;29:185–194. doi: 10.3803/EnM.2014.29.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gubellini P., Picconi B., Di Filippo M., Calabresi P. Downstream mechanisms triggered by mitochondrial dysfunction in the basal ganglia: from experimental models to neurodegenerative diseases. Biochim. Biophys. Acta. 2010;1802:151–161. doi: 10.1016/j.bbadis.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Rasool N., Lessell S., Cestari D.M. Leber hereditary optic neuropathy: bringing the lab to the clinic. Semin. Ophthalmol. 2016;31:107–116. doi: 10.3109/08820538.2015.1115251. [DOI] [PubMed] [Google Scholar]
- 55.Parikh S., Goldstein A., Koenig M.K., Scaglia F., Enns G.M., Saneto R., Anselm I., Cohen B.H., Falk M.J., Greene C. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet. Med. 2015;17:689–701. doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cronan J.E. Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol. Mol. Biol. Rev. 2016;80:429–450. doi: 10.1128/MMBR.00073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hermes F.A., Cronan J.E. The role of the Saccharomyces cerevisiae lipoate protein ligase homologue, Lip3, in lipoic acid synthesis. Yeast. 2013;30:415–427. doi: 10.1002/yea.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell S.E., Tandon N.N., Woldegiorgis G., Luiken J.J., Glatz J.F., Bonen A. A novel function for fatty acid translocase (FAT)/CD36: involvement in long chain fatty acid transfer into the mitochondria. J. Biol. Chem. 2004;279:36235–36241. doi: 10.1074/jbc.M400566200. [DOI] [PubMed] [Google Scholar]
- 59.Lee S.M., Bahl J.J., Bressler R. Prevention of the metabolic effects of 2-tetradecylglycidate by octanoic acid in the genetically diabetic mouse (db/db) Biochem. Med. 1985;33:104–109. doi: 10.1016/0006-2944(85)90132-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.