Abstract
Due to recent advances in high-throughput techniques, we and others have generated multiple proteomic and transcriptomic databases to describe and quantify gene expression, protein abundance, or cellular signaling on the scale of the whole genome/proteome in kidney cells. The existence of so much data from diverse sources raises the following question: “How can researchers find information efficiently for a given gene product over all of these data sets without searching each data set individually?” This is the type of problem that has motivated the “Big-Data” revolution in Data Science, which has driven progress in fields such as marketing. Here we present an online Big-Data tool called BIG (Biological Information Gatherer) that allows users to submit a single online query to obtain all relevant information from all indexed databases. BIG is accessible at http://big.nhlbi.nih.gov/.
Keywords: data science, kidney physiology, systems biology, BIG data
in recent years, systems biology-based approaches have taken hold in kidney research. We and others have used proteomics and transcriptomics techniques to quantify certain aspects of gene expression or signaling on the scale of the whole genome (2, 5). Our laboratory alone has produced multiple proteomics and transcriptomics datasets, which have been made available online (https://hpcwebapps.cit.nih.gov/ESBL/Database/). In addition, renal researchers from several other laboratories have produced large-scale datasets, some of which are hosted at the same site. The problem that has arisen is: “How can we efficiently gather information about a given gene product from all of these databases?” Current methods require users to open individual databases separately and search the pages for the gene product of interest, a process that is time consuming and inefficient. The solution that we present in this paper is an online tool called BIG (Biological Information Gatherer) that allows users to easily search all indexed databases with a single query.
METHODS
Data curation.
The datasets used in BIG were from the Kidney Systems Biology Project website (URL: https://hpcwebapps.cit.nih.gov/ESBL/Database/). This consists of large-scale proteomic and transcriptomic datasets from the NHLBI Epithelial Systems Biology Laboratory, as well as various datasets from other laboratories throughout the world. The information was composed into regular English sentences in Microsoft Excel, before transfer into MySQL.
The architecture.
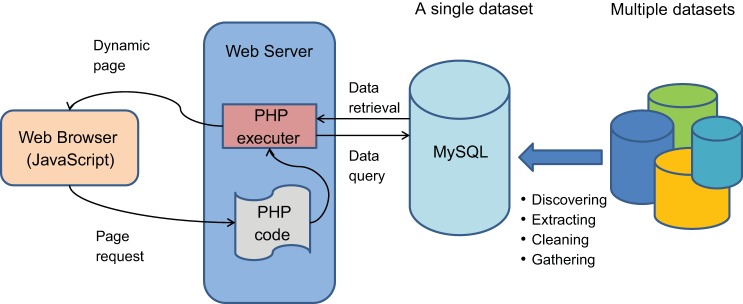
The datasets used in BIG come from various projects and sources. Thus they are originally autonomous, heterogeneous, and distributed. To provide a uniform access to all data in these datasets, we first integrated the data from these various datasets into a single database. The integrating process consisted of five phases (accomplished in Microsoft Excel): discovering the useful data information from the datasets, extracting the useful data from the datasets, cleaning the extracted data, formatting them with uniform format, and gathering all of the cleaned data into a single dataset. (“Cleaning” in this context means removing redundant and irrelevant information.) We created a single dataset using MySQL and uploaded all of the cleaned data to the MySQL database. We created a user input page on the web browser, where the user can input the query key and submit the query request. On the web server side, the scripting language, PHP (http://php.net/), was used to implement the query on the MySQL database. After the targeted data are obtained, a query-results display page is created on the web browser. Figure 1 demonstrates the logic architecture and workflow of BIG.
Fig. 1.
The workflow and logic architecture of Biological Information Gatherer (BIG). The BIG is accessible through a web browser. Users submit the queries from the web browsers, and the embedded PHP codes (a scripting programing language for web site development) are executed to retrieve queried information from the MySQL database. The database was constructed based on a large dataset integrated from multiple individual experimental datasets. These databases are listed at https://hpcwebapps.cit.nih.gov/ESBL/Database/.
RESULTS
The current version of BIG (April 2016) contains 147,236 records gleaned from multiple individual databases listed at https://hpcwebapps.cit.nih.gov/ESBL/Database/ (Fig. 2). Figure 3 shows a screen-shot image of the data entry page of BIG. The user simply enters an official gene symbol into the indicated box, chooses a kidney-structure delimiter, and clicks “submit”. Alternatively, the user can enter a keyword for a protein of interest. BIG then will perform a string search on the protein definitions obtained from UniProt Knowledgebase (www.uniprot.org) to identify a list of possible gene symbols from which the user can select the correct one. A typical output is shown in another screen-shot image (Fig. 4). The first line gives the UniProt “protein name” corresponding to the entered official gene symbol. The information grabbed from all indexed databases is then output as a series of complete sentences in English.
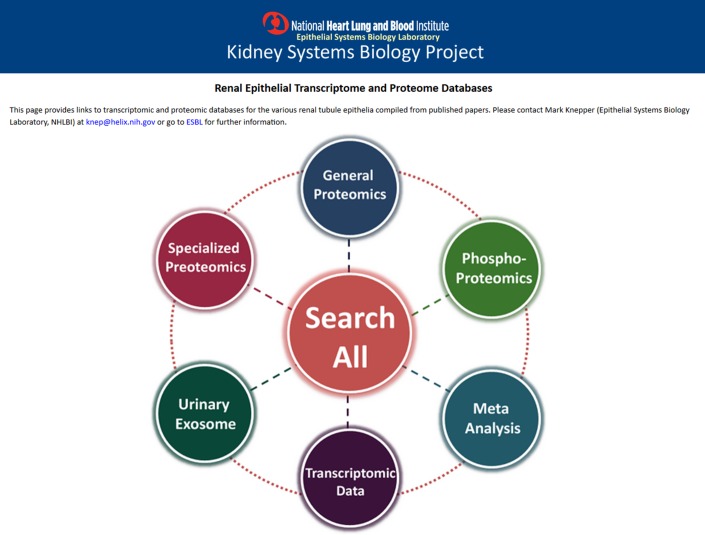
Fig. 2.
A screen-shot image of the web portal for Renal Epithelial Transcriptome and Proteome Databases. BIG can be accessed via center icon “Search All”. Link: https://hpcwebapps.cit.nih.gov/ESBL/Database/.
Fig. 3.
A screen-shot image of the BIG data query page. Queries are based on entry of official gene symbols. User can also use a keyword to search possible gene symbols and then select “Cell Type” or “All”.
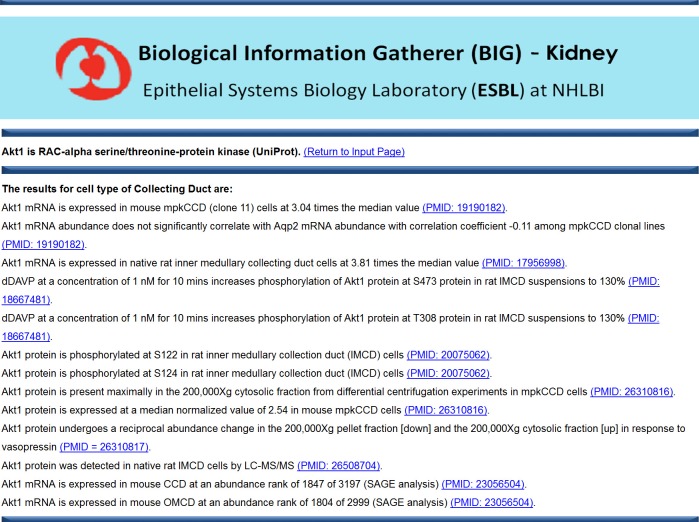
Fig. 4.
A screen-shot image of a typical BIG data output page for query term “Akt1” and cell type “Collecting Duct”.
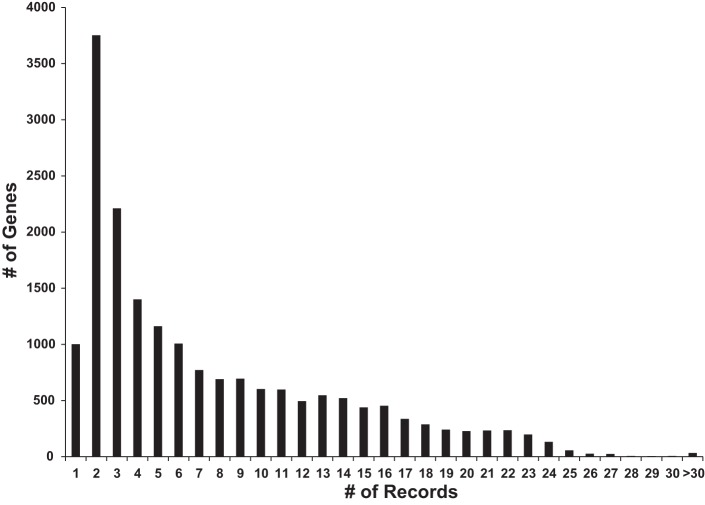
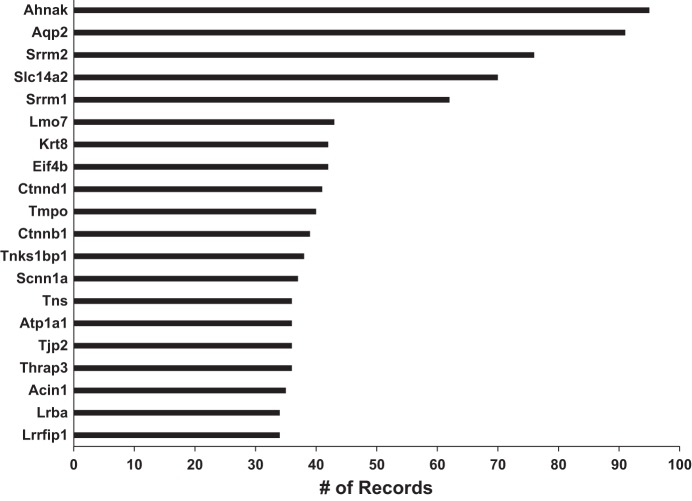
Figure 5 is a histogram showing the current distribution of the number of records for each official gene symbol from the HUGO Gene Nomenclature Committee (http://www.genenames.org/) in BIG. The distribution resembles a Poisson distribution with λ value of 2. Figure 6 shows the 20 gene symbols with the most records. The gene with the most records is Ahnak, a very large protein containing almost 6,000 amino acids. Most of the records relate to its many phosphorylation sites, many of which are regulated by vasopressin in the renal collecting duct. Interestingly among the top 20 genes, nuclear proteins dominate, having the Gene Ontology Cellular Component term “nucleus” (Lrrfip1, Acin1, Thrap3, Tjp2, Tnks1bp1, Ctnnb1, Tmpo, Ctnnd1, Krt8, Srrm1, Srrm2, Ahnak). This may reflect the large number of proteins present in nuclear proteomes relative to the cytoplasm (8).
Fig. 5.
The histogram of current BIG record distribution. The numbers of records for each official gene symbol in BIG were calculated. X-axis shows bins corresponding to numbers of records; Y-axis shows the numbers of official gene symbols within each bin.
Fig. 6.
The 20 official gene symbols with the most records. X-axis is the numbers of records; Y-axis shows the official gene symbols.
To illustrate the use of BIG, we present two case studies as follows.
Case study 1. Mining information about a particular gene product from a published paper.
A paper was recently published by Klein et al. (4) on the role of AMP-activated kinase (AMPK) on phosphorylation of aquaporin-2 and the vasopressin-regulated urea channel Slc14a2. We wanted to find out more about this protein kinase using BIG. The first step is to identify the relevant official gene symbol for the kinase. Using UniProt (http://www.uniprot.org/), we found that there are two separate genes that code for AMPK catalytic subunits, i.e., Prkaa1 and Prkaa2. Based on RNA-seq data (6), it appears that the isoform expressed in collecting duct is Prkaa1. When “Prkaa1” is entered into BIG using the “Collecting Duct” delimiter, several pieces of information are revealed. Based on single-tubule RNA-seq, the mRNA level for Prkaa1 protein is highest among all renal tubule segments in the connecting tubule (6). Prkaa1 mRNA is expressed in both native rat inner medullary collecting duct (IMCD) cells (17) and cultured mouse murine immortalized cortical collecting duct (mpkCCD) cells (14). The corresponding Prkaa1 protein has been detected proteomically in both IMCD cells (8) and in mpkCCD cells (16). In mpkCCD cells, the relative protein abundance is much higher than the relative transcript abundance, suggesting a long protein half-life. Indeed, the half-life of Prkaa1 protein in mpkCCD cells is 34.4 h in the absence of vasopressin and 35.5 h in the presence of vasopressin (10). Prkaa1 protein is present maximally in the 1,000 g pellet fraction from differential centrifugation experiments in mpkCCD cells (16). In other studies, Prkaa1 protein was identified by mass spectrometry in nuclear fractions isolated from mouse mpkCCD cells (11). In addition, Prkaa1 was found to be attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (7). Furthermore, Prkaa1 protein is phosphorylated at S485 in rat IMCD cells (1), although there is no evidence that this phosphorylation site is altered in abundance in response to vasopressin. Interestingly, Prkaa1 mRNA abundance significantly correlates negatively with Aqp2 mRNA abundance (correlation coefficient −0.89) among mpkCCD clonal lines derived from the original mpkCCD cell line (17), suggesting that Prkaa1 expression may have some effect on Aqp2 expression or vice versa. Thus, by entering a gene symbol into BIG, we can obtain a significant amount of baseline information about a particular protein without performing new laboratory experiments.
Case study 2. Mining information about a particular gene product found in a large-scale proteomic study.
Proteomics has the ability to identify a protein not previously under consideration with regard to the physiological process under study. An example from our laboratory's recent paper published in Am J Physiol Cell Physiol (15) is Rap1a, which appeared to undergo a reciprocal abundance change in the 200,000 g pellet fraction (down) and the 200,000 g cytosolic fraction (up) in response to vasopressin. Rap1a is a target for cAMP-mediated regulation via Rapgef3 (Epac1) and Rapgef4 (Epac2) and thus is highly relevant to vasopressin signaling. We entered “Rap1a” into the BIG entry page and selected “Collecting Duct”. The output revealed the following information about Rap1a in the collecting duct. Rap1a mRNA is expressed in mouse mpkCCD (clone 11) cells at 3.93 times the median value (17) and in native rat inner medullary collecting duct cells at 5.74 times the median value (14) and hence is highly abundant in collecting duct cells. 1-Desamino-8-d-arginine vasopressin at a concentration of 0.1 nM for 1 h was found to decrease the abundance of Rap1a protein in a nuclear extract of mouse mpkCCD to 60% (11). Consistent with its presence in the nucleus, Rap1a protein was found to be present maximally in the 1,000 g pellet fraction from differential centrifugation experiments in mpkCCD cells (16). Rap1a protein was found to have a half-life of 31.7 h in mouse mpkCCD cells treated with vasopressin (10). Rap1a was found to be attached to the apical plasma membrane by surface biotinylation in cultured mouse mpkCCD cells (7). Overall, these data paint a picture of Rap1a as a very dynamic protein with potential roles in the regulation of aquaporin-2 gene expression and trafficking in renal collecting duct cells.
DISCUSSION
In this article, we present a new information-access tool called BIG that uses the “big data” concept to obtain information from multiple large-scale databases in the knowledge domain “Kidney Physiology”. Big data is a burgeoning subject area within the field of data science. It has been heavily utilized in recent years in the areas of national security surveillance and marketing (3, 12, 13). The general idea underlying the big data concept is to look for information about specific objects or groups of objects across multiple large datasets. In the present paper, objects are particular genes and the gene products for which they code. To search for information, we utilize strings corresponding to official gene symbols as a primary key. The search can be narrowed to information about a particular renal tubule segment (or glomerulus) using an appropriate secondary key. Thus, to enter a search using BIG, the user needs only to enter the primary key (official gene symbol) and the secondary key (renal tubule segment) and click “submit”. The output then reports in English sentences all information available that has been mapped to the selected primary and secondary keys. The databases used were manually curated from the outputs of various transcriptomic and proteomics studies, as well as several reductionist studies of the action of vasopressin in the kidney. We have previously used the big data concept in devising a software program called AbDesigner (https://hpcwebapps.cit.nih.gov/AbDesigner/), which accesses information about a given protein from various sources to identify optimal peptide sequences to be used to make synthetic peptides for antibody production (9).
How can BIG be used for the study of renal physiology? In the results section, we have presented two examples of the use of BIG. In one example, we have taken an interesting protein studied in the context of the physiology of the renal collecting duct, namely the AMP-activated kinase (4), and inserted its official gene symbol into BIG. The output provides a number of interesting facts about this protein in the renal collecting duct that could be used to design new studies on its function. In the other example, we have selected a protein found in a recent published study (15) that appears to translocate from one fraction to another, namely Rap1a, a regulatory target for the cyclic AMP-dependent small-GTPase activing protein Rap1gef3 (Epac1). We inserted its gene symbol into BIG to obtain further information about its function in the renal collecting duct.
The current version of BIG was designed in part to illustrate the use of the big data concept in physiological research. The accessible information is only a fraction of the total information potentially available, and, as new information and large-scale data sets become available, it will be important to continue updating BIG. Users with datasets that they would like to integrate into BIG are invited to contact the authors.
GRANTS
This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute (project ZIA-HL001285 and ZIA-HL006129, M. A. Knepper).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Y.Z., C.-R.Y., and M.A.K. conception and design of research; Y.Z. and C.-R.Y. performed experiments; Y.Z., C.-R.Y., and M.A.K. analyzed data; Y.Z., C.-R.Y., V.R., J.P., and M.A.K. interpreted results of experiments; Y.Z., C.-R.Y., and J.P. prepared figures; Y.Z., C.-R.Y., and M.A.K. drafted manuscript; Y.Z., C.-R.Y., V.R., J.P., and M.A.K. edited and revised manuscript; Y.Z., C.-R.Y., V.R., J.P., and M.A.K. approved final version of manuscript.
REFERENCES
- 1.Bansal AD, Hoffert JD, Pisitkun T, Hwang S, Chou CL, Boja ES, Wang G, Knepper MA. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J Am Soc Nephrol 21: 303–315, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huling JC, Pisitkun T, Song JH, Yu MJ, Hoffert JD, Knepper MA. Gene expression databases for kidney epithelial cells. Am J Physiol Renal Physiol 302: F401–F407, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim GH, Trimi S, Chung JH. Big-data applications in the government sector. Commun ACM 57: 78–85, 2014. [Google Scholar]
- 4.Klein JD, Blount MA, Frohlich O, Denson CE, Tan X, Sim JH, Martin CF, Sands JM. Phosphorylation of UT-A1 on serine 486 correlates with membrane accumulation and urea transport activity in both rat IMCDs and cultured cells. Am J Physiol Renal Physiol 298: F935–F940, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knepper MA. Systems biology in physiology: the vasopressin signaling network in kidney. Am J Physiol Cell Physiol 303: C1115–C1124, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo CS, Chen CW, Wang PJ, Chen PY, Lin SY, Khoo KH, Fenton RA, Knepper MA, Yu MJ. Quantitative apical membrane proteomics reveals vasopressin-induced actin dynamics in collecting duct cells. Proc Natl Acad Sci U S A 110: 17119–17124, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickering CM, Grady CR, Medvar B, Emamian M, Sandoval PC, Zhao Y, Yang CR, Jung HJ, Chou CL, Knepper MA. Proteomic profiling of nuclear fractions from native renal inner medullary collecting duct cells. Physiol Genomics 48: 154–166, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pisitkun T, Hoffert JD, Saeed F, Knepper MA. NHLBI-AbDesigner: an online tool for design of peptide-directed antibodies. Am J Physiol Cell Physiol 302: C154–C164, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandoval PC, Slentz DH, Pisitkun T, Saeed F, Hoffert JD, Knepper MA. Proteome-wide measurement of protein half-lives and translation rates in vasopressin-sensitive collecting duct cells. J Am Soc Nephrol 24: 1793–1805, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenk LK, Bolger SJ, Luginbuhl K, Gonzales PA, Rinschen MM, Yu MJ, Hoffert JD, Pisitkun T, Knepper MA. Quantitative proteomics identifies vasopressin-responsive nuclear proteins in collecting duct cells. J Am Soc Nephrol 23: 1008–1018, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seref S, Sinanc D. Big data: a review. In: The 2013 International Conference on Collaboration Technologies and Systems, CTS 2013, San Diego, CA, May 20–24, 2013. New York: IEEE, 2013, p. 42–47. [Google Scholar]
- 13.Tirunillai S, Tellis GJ. Mining marketing meaning from online chatter: strategic brand analysis of big data using latent dirichlet allocation. J Mark Res 51: 463–479, 2014. [Google Scholar]
- 14.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang CR, Raghuram V, Emamian M, Sandoval PC, Knepper MA. Deep proteomic profiling of vasopressin-sensitive collecting duct cells. II. Bioinformatic analysis of vasopressin signaling. Am J Physiol Cell Physiol 309: C799–C812, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang CR, Tongyoo P, Emamian M, Sandoval PC, Raghuram V, Knepper MA. Deep proteomic profiling of vasopressin-sensitive collecting duct cells. I. Virtual Western blots and molecular weight distributions. Am J Physiol Cell Physiol 309: C785–C798, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu MJ, Miller RL, Uawithya P, Rinschen MM, Khositseth S, Braucht DW, Chou CL, Pisitkun T, Nelson RD, Knepper MA. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc Natl Acad Sci U S A 106: 2441–2446, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]