Single nucleotide substitution played important role in evolutionary expansion of human neocortex.
Keywords: ARHGAP11B, neocortical expansion, human evolution, basal progenitors, neurogenesis, gene duplication, neocortex, brain size, RhoGAP, human-specific gene
Abstract
The gene ARHGAP11B promotes basal progenitor amplification and is implicated in neocortex expansion. It arose on the human evolutionary lineage by partial duplication of ARHGAP11A, which encodes a Rho guanosine triphosphatase–activating protein (RhoGAP). However, a lack of 55 nucleotides in ARHGAP11B mRNA leads to loss of RhoGAP activity by GAP domain truncation and addition of a human-specific carboxy-terminal amino acid sequence. We show that these 55 nucleotides are deleted by mRNA splicing due to a single C→G substitution that creates a novel splice donor site. We reconstructed an ancestral ARHGAP11B complementary DNA without this substitution. Ancestral ARHGAP11B exhibits RhoGAP activity but has no ability to increase basal progenitors during neocortex development. Hence, a single nucleotide substitution underlies the specific properties of ARHGAP11B that likely contributed to the evolutionary expansion of the human neocortex.
INTRODUCTION
Evolutionary enlargement of the human neocortex accompanied the emergence of the cognitive functions that are unique to our species. This enlargement primarily reflects an increase in the number of neurons that are generated during neocortical development (1, 2).
Neocortical neurogenesis involves two main classes of neural progenitor cells that reside in two distinct germinal zones (fig. S1) (3–7). First are apical progenitors (APs), notably apical radial glia, which are anchored to and undergo mitosis at the apical (ventricular) surface of the ventricular zone. Second are basal progenitors (BPs)—comprising basal (or outer) radial glia and basal intermediate progenitors—which originate from a subset of apical radial glia divisions, delaminate from the ventricular surface and migrate basally to the subventricular zone where they undergo mitosis (3–7). BPs are better suited for maximizing neuron production than APs because they are not subjected to the constraint imposed on AP proliferation by the limited ventricular space but rather can make use of the much larger space available in the subventricular zone to undergo mitosis (8–10). Accordingly, the evolutionary expansion of the neocortex is associated with an increase in the generation of BPs and their proliferation before generating neurons, which are reflected by an increase in the BP pool size and correlative thickness of the subventricular zone (fig. S1) (4, 5, 7, 11–15).
Because the size of the BP pool is largely under genetic control, differences in BP abundance among species are likely to be based on species-specific genomic changes. Hence, identifying the elements unique to our genome that contribute to BP amplification during human neocortical neurogenesis is integral to the understanding of human-specific aspects of neocortical expansion (16–18). Recent in vivo functional experiments in mice have mechanistically linked human-specific genomic changes to several uniquely human traits of brain development. Specifically, expression of a humanized version of the forkhead box P2 transcription factor, carrying two human-specific amino acid substitutions, resulted in increased dendrite length and synaptic plasticity of spiny projection neurons in the striatum (19). Expression of SRGAP2C, the product of a human-specific partial duplication of the SRGAP2 gene (20), led to neoteny of spine maturation and increased density of longer spines in the neocortex (21). Furthermore, a few human-specific genomic changes have been experimentally demonstrated to enhance neocortical neurogenesis by increasing the neural progenitor cell proliferation in vivo and thus have been implicated in the evolutionary expansion of the human neocortex. These include the human-accelerated region HARE5, a sequence that serves as enhancer for the Wnt receptor FRIZZLED-8 (22), and the ARHGAP11B gene (23).
ARHGAP11B is the first, and hitherto only, human-specific protein-encoding gene that was shown to increase BP generation and proliferation (23). It arose on the human evolutionary lineage ~1 million years after divergence from the chimpanzee lineage, existed in Neanderthals and Denisovans, and is found in all present-day humans (23–25). ARHGAP11B is the product of a partial duplication of ARHGAP11A (24, 26), which encodes a Rho guanosine triphosphatase–activating protein (RhoGAP) conserved between yeast and humans (27). However, contrary to ARHGAP11A (27, 28), ARHGAP11B does not exhibit RhoGAP activity when expressed in monkey fibroblast-like cells (23). This reflects the fact that the GAP domain of ARHGAP11B is truncated, with the 26 C-terminal amino acid residues being replaced by a 47-residue-long C-terminal sequence that is unique to humans. This, in turn, is due to a shift in the reading frame caused by the absence of 55 nucleotides (nt) in the ARHGAP11B mRNA, which are present at the homologous position in the ARHGAP11A mRNA (23).
RESULTS
Modern ARHGAP11B contains a novel splice donor site
When analyzing the genomic DNA, however, we noticed that this stretch of 55 nt is, in fact, present in the ARHGAP11B gene. Specifically, we compared modern ARHGAP11A/B orthologous sequences at the genomic DNA, pre-mRNA, mRNA, and protein level (Fig. 1). ARHGAP11B is located ~2 million base pairs (Mbp) 5′ to ARHGAP11A on chromosome 15 (15q13.3) (Fig. 1). ARHGAP11A encompasses 12 exons, the first 8 of which and the subsequent introns are duplicated in ARHGAP11B, with the duplication break point located near the 3′ end of intron 8 (Fig. 1). With the exception of a 593-bp deletion in intron 2 of ARHGAP11B, the two paralogous sequences are highly similar (Fig. 1). The first 220 amino acids of ARHGAP11B are 98% identical to those of ARHGAP11A (216 of 220 residues; for a discussion of the four amino acid changes, see below and the Supplementary Materials) and encompass most of its GAP domain (amino acids 46 to 246) (Fig. 1). However, at the position corresponding to base pair 661 of the ARHGAP11A protein-coding sequence, in exon 5, a single C→G base change in the ARHGAP11B sequence creates a novel GU-purine splice donor site, 55 nt 5′ to the ancestral one (Fig. 1). Therefore, these 55 nt, which, in ARHGAP11A, encode amino acid residues 221 to 238 that are part of the GAP domain (amino acids 46 to 246), are spliced out from the ARHGAP11B pre-mRNA and hence are lacking from the mRNA (Fig. 1 and fig. S2). Thus, the single C→G base change in the ARHGAP11B gene is ultimately the reason for the reading frame shift that leads to the truncation of the GAP domain of ARHGAP11B and its novel C-terminal sequence (amino acids 221 to 267; Fig. 1 and fig. S2), which is unique to ARHGAP11B and not found in any other protein thus far described in the animal kingdom (23).
Fig. 1. ARHGAP11A and ARHGAP11B genomic, pre-mRNA, mRNA, and protein structures.
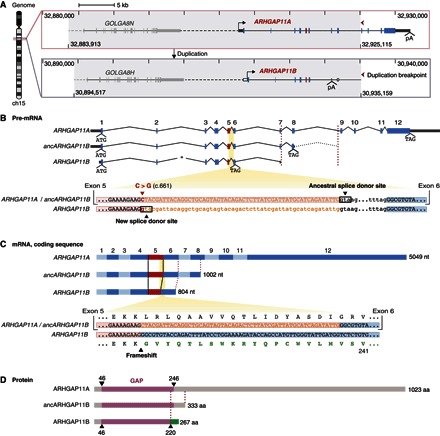
(A) Gene structure and genomic context of human ARHGAP11A (top) and ARHGAP11B (bottom). Gray areas indicate the duplicated genomic region (40.642 Mb), which comprises the GOLGA8 and ARHGAP11 genes. Tick marks and numbers indicate genomic coordinates on chromosome 15 (GRCh37/hg19). Red arrowheads point to the 3′ duplication break point, highlighting the partial nature of the ARHGAP11 duplication. Continuous and dashed horizontal lines indicate intragenic and intergenic regions, respectively. Transcription start sites (forward arrows) and polyadenylation sites (pA) are shown. Rectangles indicate exons; dark gray, untranslated regions; blue and red, protein-coding sequences (red, exon 5); white, exonic sequences of ARHGAP11A that are duplicated but typically untranscribed in modern ARHGAP11B. The duplicated GOLGA8 gene 5′ to ARHGAP11 is depicted in light gray. Image was adapted from the University of California Santa Cruz (UCSC) Genome Browser. (B) ARHGAP11A, predicted ancestral ARHGAP11B (ancARHGAP11B), and modern ARHGAP11B pre-mRNAs (top). Translation start (ATG) and stop (TAG) codons are indicated. Numbers (1 to 12) indicate ARHGAP11A exons, 1 to 8 of which were duplicated. Left red dashed line indicates the position of the pA site in exon 7 of modern ARHGAP11B relative to exon 7 of ARHGAP11A and ancARHGAP11B. Dotted intron line 3′ to the predicted ancARHGAP11B stop codon indicates the duplicated portion of ARHGAP11A intron 8 until the duplication break point (right red dashed line). Asterisk indicates a 593-bp deletion in modern ARHGAP11B intron 2. (B) Alignment of ARHGAP11A/B homologous sequences (bottom) encompassing the 3′ end of exon 5 (red background) and 5′ end of exon 6 (blue background), interspaced by intron 5. Exonic and intronic nucleotide (nt) sequences are displayed in uppercase and lowercase letters, respectively. The C→G base substitution (red, position c.661) produces a new splice donor site in modern ARHGAP11B, 55-nt 5′ to the ancestral one. As a consequence, these 55 nt (orange sequences) become intronic. (C) ARHGAP11A, ancARHGAP11B, and modern ARHGAP11B coding sequences (top). Alternating light-dark blue rectangles indicate exons. Black lines highlight the shortening of modern ARHGAP11B exon 5. (C) Alignment of ARHGAP11A/B coding sequences (bottom) encompassing the 3′ end of exon 5 and the 5′ end of exon 6, with the corresponding amino acid sequences (until residue 241) depicted above and below. The 55-nt sequence corresponding to the 3′ end of ARHGAP11A exon 5 (orange) is spliced out from the modern ARHGAP11B mRNA. The resulting frameshift generates a novel C-terminal amino acid sequence (green) unique to modern ARHGAP11B. (B and C) Note that all RNA sequences are depicted with T instead of U. (D) ARHGAP11A, ancARHGAP11B, and modern ARHGAP11B protein structures showing the conserved portions of the ARHGAP11A GAP domain (purple) and the novel C-terminal domain of modern ARHGAP11B (green) starting at residue 221. aa, amino acids.
Given that ARHGAP11B arose from ARHGAP11A, an ancient gene (27), the C at base pair 661 of the ARHGAP11A protein-coding sequence is likely to be the ancestral base, and the G at this position in ARHGAP11B presumably is a more recent base substitution. We sought to corroborate this conclusion by determining the respective base at the homologous position in the ARHGAP11A genes of archaic hominins [Neanderthal and Denisova (29, 30), who carry both paralogs] and nonhuman primates (who carry only ARHGAP11A) (23–25). All ARHGAP11A genes analyzed, that is, those of archaic hominins and extant apes, were found to contain a C at this position (fig. S3), indicating that this is the ancestral, conserved base.
In light of this finding, it was important to determine whether the single C→G base substitution in the ARHGAP11B gene that ultimately causes its human-specific C-terminal sequence occurs not only in modern humans but if it was also present in Neanderthals, whose brains were as large as those of modern humans, and Denisovans. The crucial C→G base substitution was also found in Neanderthal and Denisova ARHGAP11B (fig. S3). Moreover, all present-day humans analyzed (31) carry the C→G substitution. Together, these observations indicate that the C→G base substitution, which presumably occurred in the ~5 million years since the ARHGAP11 gene duplication event, took place before the archaic hominins diverged from the modern human lineage >500,000 years ago.
As a corollary, an “ancestral” ARHGAP11B gene presumably originated from partial duplication of ARHGAP11A without the C→G base substitution present in modern ARHGAP11B, thus encoding the full ARHGAP11 GAP domain (Fig. 1 and figs. S4 and S5). We sought to experimentally test whether such an ancestral ARHGAP11B protein may have been functionally distinct from modern ARHGAP11B, bearing a closer resemblance to modern ARHGAP11A.
Reconstructed ancestral ARHGAP11B lacks the novel splice donor site
To this end, we reconstructed, from the ARHGAP11A complementary DNA (cDNA) (23), a cDNA corresponding to the putative ancestral version of the ARHGAP11B mRNA (Fig. 1 and fig. S4). The ancestral ARHGAP11B mRNA lacks the C→G base substitution that creates the novel splice donor site and hence is predicted to exhibit a full-length exon 5 (fig. S4). Consequently, it contains the 55 nt, the lack of which, in modern ARHGAP11B mRNA, causes the reading frame shift and translational stop codon in exon 6 (Fig. 1 and fig. S4). Therefore, translation of ancestral ARHGAP11B mRNA would proceed, like that of ARHGAP11A mRNA, through exon 7 into exon 8 but would terminate there because of a stop codon caused by a 5-bp insertion (fig. S4). These 5 bp, a microduplication of the preceding 5 bp (fig. S4), are present in the modern ARHGAP11B gene in the exon 8–corresponding sequence 3′ to the polyadenylation site in exon 7 (Fig. 1 and fig. S4) and are fixed in the modern human population (31). Notably, the stop codon caused by this 5-bp insertion constitutes the only translational stop predicted in the ancestral ARHGAP11B mRNA transcribed from the duplicated exons 1 to 8 (Fig. 1 and fig. S4). Therefore, in reconstructing the ancestral ARHGAP11B cDNA, we included this 5-bp insertion. The ancestral ARHGAP11B mRNA thus encodes a 333–amino acid protein containing a full-length GAP domain (Fig. 1 and fig. S5).
Ancestral ARHGAP11B exhibits RhoGAP activity
Modern ARHGAP11B lacks RhoGAP activity in intact cells (23) and exhibits only very low RhoGAP activity in a cell-free assay. Therefore, we asked whether ancestral ARHGAP11B exhibits RhoGAP activity, as predicted by the presence of a complete GAP domain. To this end, we performed cell transfection assays in which the readout of RhoGAP activity is a decrease in the level of phosphorylated myosin phosphatase target subunit 1 (MYPT1-pT853) (32). We transfected a monkey cell line with expression plasmids encoding ancestral and modern ARHGAP11B. As a positive control, we transfected a truncated mutant of ARHGAP11A encoding only its GAP domain (ARHGAP11A-250), which was previously shown to exhibit overt RhoGAP activity in the same assay (23). In contrast to modern ARHGAP11B, but similar to ARHGAP11A-250, ancestral ARHGAP11B was found to exhibit RhoGAP activity, as compared to a negative control (empty vector) (Fig. 2). This also corroborates our previous finding (23) that the 26 C-terminal amino acids of the ARHGAP11 GAP domain (residues 221 to 246) are required for full RhoGAP activity.
Fig. 2. Ancestral but not modern ARHGAP11B shows RhoGAP activity.
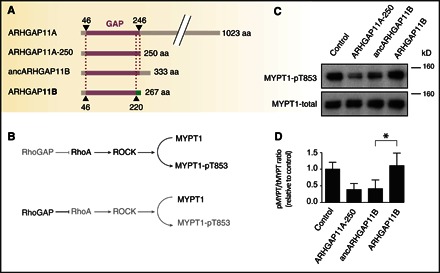
(A) Domain structures of ARHGAP11A, ARHGAP11A-250, ancestral ARHGAP11B (ancARHGAP11B), and modern ARHGAP11B. Purple, GAP domain; green, C-terminal domain unique to ARHGAP11B. (B) RhoGAP activity assay in transfected COS-7 cells based on the lack of phosphorylation of MYPT1 at Thr853 (pT853) by inhibition of Rho-associated protein kinase (ROCK/Rho-kinase). Gray indicates reduced activity/presence. (C) Immunoblots of COS-7 cells transfected with empty vector (control), ARHGAP11A-250, ancARHGAP11B, or modern ARHGAP11B using anti-MYPT1-pT853 (top) or anti-MYPT1 (bottom) antibodies; the position of a 160-kDa marker is indicated. (D) Quantification of immunoblots as shown in (C). The ratio of MYPT1-pT853 (pMYPT) to total MYPT1 (tMYPT) was determined, the mean value obtained for the control was set to 1.0, and the mean values of the other three conditions were expressed relative to this. Data are the means of three independent transfections; error bars, SD; *P < 0.05 (paired t test).
Ancestral ARHGAP11B does not amplify BPs
Given that ancestral ARHGAP11B exhibits RhoGAP activity but lacks the human-specific C-terminal sequence of modern ARHGAP11B due to the absence of the novel splice donor site, we finally examined the effect of ancestral ARHGAP11B on BP pool size during cortical development. To this end, we expressed ancestral and modern ARHGAP11B in mouse embryonic day (E) 13.5 neocortex by in utero electroporation and analyzed the electroporated neocortex at E14.5 for an increased abundance of targeted mitotic BPs, a previously established readout for ARHGAP11B function (23). Confirming our previous results (23), modern ARHGAP11B expression resulted in an increase of approximately twofold in targeted mitotic BPs, as compared to control (Fig. 3). However, similar to ARHGAP11A (23), ancestral ARHGAP11B did not exert any effect on mitotic BP abundance (Fig. 3). Consistent with these findings, modern ARHGAP11B, but not ancestral ARHGAP11B, increased the proportion of targeted progenitors that were positive for the BP marker T-box brain protein 2 (Tbr2) (fig. S6). Neither modern nor ancestral ARHGAP11B affected the abundance of mitotic APs (fig. S7).
Fig. 3. Modern but not ancestral ARHGAP11B increases BPs in mouse developing neocortex.
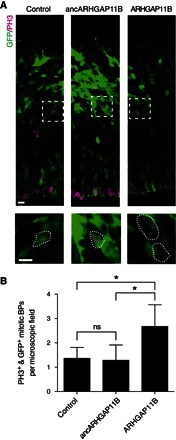
In utero electroporation of pCAGGS–green fluorescent protein (GFP) together with empty vector (control), ancestral ARHGAP11B (ancARHGAP11B), and modern ARHGAP11B expression plasmids in E13.5 mouse neocortex followed by analysis at E14.5. (A) GFP (green) and phosphohistone H3 (PH3, magenta) immunofluorescence labeling targeted and mitotic cells, respectively. Areas in dashed boxes are shown at higher magnification at the bottom. Scale bars, 10 μm. (B) Quantification of targeted mitotic BPs, identified by the presence of both GFP and PH3 immunofluorescence at a basal location. Data are the means of four (control), six (ancARHGAP11B), and five (ARHGAP11B) embryos from three litters, expressed per 100-μm-wide field of cortical wall; error bars, SD; *P < 0.05; ns, not significant [analysis of variance (ANOVA) with Bonferroni’s post hoc test for multiple comparisons].
In addition to the C→G base substitution, ARHGAP11B exhibits a small number of nucleotide changes in exons 1 to 7 when compared to ARHGAP11A (fig. S4). For three of the four amino acid residues where ARHGAP11B differs from ARHGAP11A in residues 1 to 220, ARHGAP11A carries the ancestral base, including an A at base pair 649 instead of the G in ARHGAP11B (fig. S4). In light of the significance of the single C→G base substitution at base pair 661 of the ancestral ARHGAP11B cDNA (fig. S4) for the function of ARHGAP11B, we examined whether this A→G base substitution in modern ARHGAP11B (fig. S4), which leads to the loss of a predicted threonine phosphorylation site (fig. S5), is of functional relevance. However, this was not the case as reintroducing Thr217, and thus, the predicted phosphorylation site into modern ARHGAP11B did not affect its lack of RhoGAP activity and its ability to amplify BPs (fig. S8).
DISCUSSION
In conclusion, our previous (23) and present data together indicate that ARHGAP11 proteins that exhibit RhoA-GAP activity (ARHGAP11A and ancestral ARHGAP11B) do not promote BP amplification. Moreover, modern ARHGAP11B, which lacks RhoA-GAP activity in vivo but does promote BP amplification, does not appear to do so by exerting a dominant-negative effect on the GAP activity of ARHGAP11A toward RhoA (23). Rather, we hypothesize that the novel, human-specific C-terminal sequence of modern ARHGAP11B has a key role in the mechanism by which this protein promotes BP amplification. In this regard, the present data show that this sequence is due to a single C→G base substitution, which creates a novel splice donor site that results in the replacement of the ancestral C-terminal sequence of the ARHGAP11B GAP domain. The present data also demonstrate that this single C→G base substitution underlies the ARHGAP11B-mediated BP amplification implicated in neocortex expansion.
Hence, it is not the ARHGAP11 partial gene duplication event ~5 million years ago, as such, that impacted human neocortex evolution. Presumably, ARHGAP11A and ancestral ARHGAP11B coexisted as functionally similar proteins for some time after the gene duplication event. The ability of ARHGAP11B to amplify BPs likely arose more recently from a change that is tiny on a genomic scale but substantial in its functional and evolutionary consequences.
MATERIALS AND METHODS
Mice
All experimental procedures were designed and conducted in agreement with the German Animal Welfare Legislation. Institutional review board guidelines were followed. Animals used for this study were kept pathogen-free at the Biomedical Services Facility of the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG). All mice used were C57BL/6JOlaHsd wild-type congenic. E0.5 was set at noon of the day on which the vaginal plug was observed.
Sequence alignments and prediction of ancestral nucleotides
Nonhuman primate, Neanderthal, and modern human ARHGAP11A and ARHGAP11B sequences were obtained from the Ensembl (release 84) database. Denisovan ARHGAP11A and ARHGAP11B sequences were obtained from the UCSC Genome Browser. Sequence alignments were performed using Kalign (European Molecular Biology Laboratory) or CLUSTAW (http://www.genome.jp/tools/clustalw/). Ancestral nucleotide residues were predicted from multiple sequence alignments.
Mapping of RNA-sequencing reads corresponding to the modern ARHGAP11B C-terminal sequence
RNA-sequencing (RNA-seq) reads aligning to ARHGAP11B were obtained from a previous RNA-seq data set produced from purified human basal radial glia (23). Reads covering ARHGAP11B from 5′ to the new splice donor site to 3′ of the translational stop were extracted using chr15:30927281–30927833 hg19 genomic coordinates and mapped to the modern ARHGAP11B cDNA Ensembl reference sequence using the Geneious software (version 9. 0. 4; http://www.geneious.com).
Ancestral ARHGAP11B and ARHGAP11B-A217T cDNAs
Ancestral ARHGAP11B (ancARHGAP11B) cDNA expression plasmids were generated as follows. The ancARHGAP11B coding sequence (anc11B_short; see fig. S4) was amplified by polymerase chain reaction (PCR) using pCAGGS-ARHGAP11A (23) as a template and the following primers: 5′-GGGGATCCGCCACCATGTGGGATCAGAGGCTGGTGAGGTTGGCC-3′ (forward) and 5′-CCCGGATCCCTACTGGCATGGCAATGTACGCTTAGCATTTGGTGT-3′ (reverse).
The reverse primer contained an additional 9-nt-long overhang sequence (underlined in the reverse primer sequence above), encompassing the 3′ end of ancARHGAP11B, including the 5-bp insertion and the resulting translational stop codon (see fig. S4). The PCR product was inserted into the pCR-bluntII-topo vector (Invitrogen), and the ancARHGAP11B coding sequence was then subcloned into either pCAGGS (33) or pCAGGS-HA-KK1 (K. Kaibuchi, Nagoya University).
The ARHGAP11B-A217T cDNA expression plasmids were generated as follows: a pCR-bluntII-topo-ARHGAP11B-A217T was produced by site-directed mutagenesis (QuikChange Lightning Multi Site-Directed Mutagenesis Kit, Agilent) using pCR-bluntII-topo-ARHGAP11B as a template, and the ARHGAP11B-A217T was then subcloned into either pCAGGS or pCAGGS-HA-KK1. All constructs were verified by DNA sequencing.
RhoGAP activity assay and immunoblotting
RhoGAP activity was determined as previously described (32). Briefly, cDNA expression constructs encoding full-length, truncated, or mutant versions of human ARHGAP11A and ARHGAP11B, all hemagglutinin (HA)–tagged, were generated either previously (23) or as described above. COS-7 cells were transfected with pCAGGS (control, empty vector), pCAGGS-HA-ARHGAP11A-250, pCAGGS-HA-ancARHGAP11B, pCAGGS-HA-ARHGAP11B-A217T, and pCAGGS-HA-ARHGAP11B. One day after transfection, total cell lysates were subjected to SDS–polyacrylamide gel electrophoresis, followed by immunoblot analyses with rabbit anti-MYPT1 (34), rabbit anti-MYPT1-pT853 (Cell Signaling Technology), and rabbit anti-HA (Cell Signaling Technology) antibodies. Immunoreactive bands were quantified from densitometric scans of x-ray films using Fiji.
Immunoblotting of COS-7 cells transfected with pCAGGS (control and empty vector), pCAGGS-HA-ancARHGAP11B, or pCAGGS-HA-ARHGAP11B was also performed to compare the immunoreactive bands detected by the anti-HA antibody with that detected by a new monoclonal antibody (to be described in detail elsewhere) raised against the 21 C-terminal amino acid residues of modern ARHGAP11B (KALKKVNMKLLVNIREREDNV).
In utero electroporation
In utero electroporation of E13.5 embryos was performed as previously described (23). Briefly, littermate embryos were injected intraventricularly with pCAGGS-GFP (0.5 μg/μl) and either pCAGGS-empty-vector (control), pCAGGS-ARHGAP11B, pCAGGS-ancARHGAP11B, or pCAGGS-ARHGAP11B-A217T (1 μg/μl) in 1× phosphate-buffered saline (PBS) containing 0.1% fast green (Sigma). Electroporations were performed with six 50-ms pulses of 30 to 33 V at 1-s intervals. The peritoneal cavity was then surgically closed, and the embryos were left to develop for 24 hours. Mothers were then killed, and embryonic brains were dissected in 1× PBS. Electroporated brains were dissected 24 hours after electroporation, fixed overnight at 4°C in 4% paraformaldehyde dissolved in 120 mM phosphate buffer (pH 7.4), cryoprotected overnight at 4°C in 30% sucrose in 1× PBS, embedded in Tissue-Tek OCT (Sakura), and stored at −20°C.
Immunofluorescence
Immunofluorescence was performed on 25-μm cryosections, as previously described (23). Antigen retrieval was performed in 0.01 M citrate buffer for 1 hour at 70°C. Sections were then quenched with 0.1 M glycine in 1× PBS for 30 min, followed by blocking, and primary and secondary antibody incubations in 1× PBS containing 0.2% gelatin, 150 mM NaCl, and 0.3% Triton X-100. Primary antibodies used were rat anti-histone H3 (1:500; phospho-S10, ab10543, Abcam), chicken anti-Tbr2 (1:300; ab15894, Millipore), and goat anti-EGFP (1:500; MPI-CBG). Secondary antibodies used were donkey anti-goat Alexa Fluor 488 (1:500; Thermo Fisher) and donkey anti-rat and donkey anti-chicken Cy3 (1:500; The Jackson Laboratories). Primary antibodies were incubated overnight at 4°C, and secondary antibodies, together with DAPI (1:1000; Sigma), were incubated for 2 hours at room temperature. Sections were mounted in Mowiol 4-88 (Sigma).
Image acquisition
All images were acquired and analyzed from coronal sections of the prefrontal and parietal dorsolateral neocortex. Confocal images were acquired using an LSM 700 inverted microscope (Zeiss). Z-stacks of 3 to 10 2-μm optical sections, 1024 × 1024 pixels each, were acquired. Images were analyzed and processed with ImageJ (http://imagej.nih.gov/ij/).
Quantification of mitotic APs, mitotic BPs, and Tbr2+ cells
Numbers of phosphohistone H3 and GFP–double-positive cells were quantified throughout the entire radial dimension of the cortical wall in electroporated areas within standardized microscopic fields spanning 100 μm in width and 10 μm in depth. Typically, three to seven microscopic fields per embryo were quantified. All mitotic figures located within the apical-most 30 μm of the ventricular surface were scored as apical mitoses, and all other mitotic figures were scored as basal mitoses. Numbers of GFP-positive cells, and of Tbr2 and GFP–double-positive cells, were quantified throughout the entire radial dimension of the cortical wall in electroporated areas within standardized microscopic fields spanning 50 μm in width and 2 to 3 μm in depth. Typically, three to seven microscopic fields per embryo were quantified.
Statistics
Cell counts were compiled using Excel (Microsoft), and results were analyzed and plotted using Prism (GraphPad Software). Statistical significance was determined using repeated-measures ANOVA with Bonferroni’s post hoc test for multiple comparisons.
Supplementary Material
Acknowledgments
We apologize to all researchers whose work could not be cited due to space limitation. We are grateful to the services and facilities of the MPI-CBG for the outstanding support provided, notably J. Helppi and his team from the Animal Facility and Holger Brandl of the Bioinformatics Facility. We thank A. Goffinet (University of Louvain) for advice on genomics, K. Kaibuchi and M. Amano (Nagoya University) for donating pCAGGS-HA-KK1 and anti-MYPT1 antibody, and members of the Huttner and Pääbo laboratories for critical discussion. We are particularly indebted to K. Prüfer and B. Vernot for support with evolutionary genomics. Funding: M.F. was a member of the International Max Planck Research School for Cell, Developmental and Systems Biology and a doctoral student at Technische Universität Dresden. S.P. was supported by the Paul G. Allen Family Foundation. W.B.H. was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) (SFB 655, A2) and the European Research Council (250197), by the DFG-funded Center for Regenerative Therapies Dresden, and by the Fonds der Chemischen Industrie. Author contributions: M.F. and W.B.H. conceived the study. M.F. performed the genomic analyses. M.F., T.N., and W.B.H. designed the experiments. M.F. and T.N. performed the experiments and analyzed the data. S.P., M.H., and W.B.H. provided intellectual guidance in the analysis and interpretation of data. M.F. and W.B.H. wrote the manuscript with contributions from all co-authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Correspondence for reagents should be addressed to W.B.H.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1601941/DC1
fig. S1. Cartoon illustrating APs and BPs in embryonic mouse and fetal human neocortex.
fig. S2. Presence of the novel, human-specific C-terminal sequence in modern but not ancestral ARHGAP11B.
fig. S3. Reconstruction of the ancestral splice donor site.
fig. S4. Ancestral ARHGAP11B nucleotide sequence.
fig. S5. Ancestral ARHGAP11B protein sequence.
fig. S6. Modern but not ancestral ARHGAP11B increases Tbr2+ progenitors in mouse developing neocortex.
fig. S7. Neither modern nor ancestral ARHGAP11B affects the abundance of mitotic APs in mouse developing neocortex.
fig. S8. ARHGAP11B-A217T mutant, like modern ARHGAP11B, lacks GAP activity and increases BPs in the mouse developing neocortex.
REFERENCES AND NOTES
- 1.G. F. Striedter, Principles of Brain Evolution (Sinauer Associates Inc., Sunderland, 2005). [Google Scholar]
- 2.Rakic P., Evolution of the neocortex: A perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fietz S. A., Huttner W. B., Cortical progenitor expansion, self-renewal and neurogenesis—A polarized perspective. Curr. Opin. Neurobiol. 21, 23–35 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Lui J. H., Hansen D. V., Kriegstein A. R., Development and evolution of the human neocortex. Cell 146, 18–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrell V., Reillo I., Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 72, 955–971 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Taverna E., Götz M., Huttner W. B., The cell biology of neurogenesis: Toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 30, 465–502 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Dehay C., Kennedy H., Kosik K. S., The outer subventricular zone and primate-specific cortical complexification. Neuron 85, 683–694 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Smart I. H., Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: A pilot study based on recording the number, location and plane of cleavage of mitotic figures. J. Anat. 116, 67–91 (1973). [PMC free article] [PubMed] [Google Scholar]
- 9.Fish J. L., Dehay C., Kennedy H., Huttner W. B., Making bigger brains–The evolution of neural-progenitor-cell division. J. Cell Sci. 121, 2783–2793 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Wilsch-Bräuninger M., Florio M., Huttner W. B., Neocortex expansion in development and evolution — From cell biology to single genes. Curr. Opin. Neurobiol. 39, 122–132 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Kriegstein A., Noctor S., Martínez-Cerdeño V., Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 7, 883–890 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Borrell V., Götz M., Role of radial glial cells in cerebral cortex folding. Curr. Opin. Neurobiol. 27, 39–46 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Florio M., Huttner W. B., Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Sun T., Hevner R. F., Growth and folding of the mammalian cerebral cortex: From molecules to malformations. Nat. Rev. Neurosci. 15, 217–232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montiel J. F., Vasistha N. A., Garcia-Moreno F., Molnár Z., From sauropsids to mammals and back: New approaches to comparative cortical development. J. Comp. Neurol. 524, 630–645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geschwind D. H., Rakic P., Cortical evolution: Judge the brain by its cover. Neuron 80, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae B.-I., Jayaraman D., Walsh C. A., Genetic changes shaping the human brain. Dev. Cell 32, 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silver D. L., Genomic divergence and brain evolution: How regulatory DNA influences development of the cerebral cortex. Bioessays 38, 162–171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enard W., Gehre S., Hammerschmidt K., Hölter S. M., Blass T., Somel M., Brückner M. K., Schreiweis C., Winter C., Sohr R., Becker L., Wiebe V., Nickel B., Giger T., Müller U., Groszer M., Adler T., Aguilar A., Bolle I., Calzada-Wack J., Dalke C., Ehrhardt N., Favor J., Fuchs H., Gailus-Durner V., Hans W., Hölzlwimmer G., Javaheri A., Kalaydjiev S., Kallnik M., Kling E., Kunder S., Moßbrugger I., Naton B., Racz I., Rathkolb B., Rozman J., Schrewe A., Busch D. H., Graw J., Ivandic B., Klingenspor M., Klopstock T., Ollert M., Quintanilla-Martinez L., Schulz H., Wolf E., Wurst W., Zimmer A., Fisher S. E., Morgenstern R., Arendt T., de Angelis M. H., Fischer J., Schwarz J., Pääbo S., A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Dennis M. Y., Nuttle X., Sudmant P. H., Antonacci F., Graves T. A., Nefedov M., Rosenfeld J. A., Sajjadian S., Malig M., Kotkiewicz H., Curry C. J., Shafer S., Shaffer L. G., de Jong P. J., Wilson R. K., Eichler E. E., Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charrier C., Joshi K., Coutinho-Budd J., Kim J.-E., Lambert N., de Marchena J., Jin W.-L., Vanderhaeghen P., Ghosh A., Sassa T., Polleux F., Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd J. L., Skove S. L., Rouanet J. P., Pilaz L.-J., Bepler T., Gordân R., Wray G. A., Silver D. L., Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol. 25, 772–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florio M., Albert M., Taverna E., Namba T., Brandl H., Lewitus E., Haffner C., Sykes A., Wong F. K., Peters J., Guhr E., Klemroth S., Prüfer K., Kelso J., Naumann R., Nüsslein I., Dahl A., Lachmann R., Pääbo S., Huttner W. B., Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Sudmant P. H., Kitzman J. O., Antonacci F., Alkan C., Malig M., Tsalenko A., Sampas N., Bruhn L., Shendure J., 1000 Genomes Project, Eichler E. E., Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antonacci F., Dennis M. Y., Huddleston J., Sudmant P. H., Steinberg K. M., Rosenfeld J. A., Miroballo M., Graves T. A., Vives L., Malig M., Denman L., Raja A., Stuart A., Tang J., Munson B., Shaffer L. G., Amemiya C. T., Wilson R. K., Eichler E. E., Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 46, 1293–1302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riley B., Williamson M., Collier D., Wilkie H., Makoff A., A 3-Mb map of a large segmental duplication overlapping the α7-nicotinic acetylcholine receptor gene (CHRNA7) at human 15q13–q14. Genomics 79, 197–209 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Zanin E., Desai A., Poser I., Toyoda Y., Andree C., Moebius C., Bickle M., Conradt B., Piekny A., Oegema K., A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev. Cell 26, 496–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J., Zhou X., Wang J., Li Z., Kong X., Qian J., Hu Y., Fang J.-Y., RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep. 3, 1526–1538 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F., Mallick S., Schraiber J. G., Jay F., Prüfer K., de Filippo C., Sudmant P. H., Alkan C., Fu Q., Do R., Rohland N., Tandon A., Siebauer M., Green R. E., Bryc K., Briggs A. W., Stenzel U., Dabney J., Shendure J., Kitzman J., Hammer M. F., Shunkov M. V., Derevianko A. P., Patterson N., Andrés A. M., Eichler E. E., Slatkin M., Reich D., Kelso J., Pääbo S., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P. H., de Filippo C., Li H., Mallick S., Dannemann M., Fu Q., Kircher M., Kuhlwilm M., Lachmann M., Meyer M., Ongyerth M., Siebauer M., Theunert C., Tandon A., Moorjani P., Pickrell J., Mullikin J. C., Vohr S. H., Green R. E., Hellmann I., Johnson P. L. F., Blanche H., Cann H., Kitzman J. O., Shendure J., Eichler E. E., Lein E. S., Bakken T. E., Golovanova L. V., Doronichev V. B., Shunkov M. V., Derevianko A. P., Viola B., Slatkin M., Reich D., Kelso J., Pääbo S., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium, Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McVean G. A., Abecasis G. R., A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kagawa Y., Matsumoto S., Kamioka Y., Mimori K., Naito Y., Ishii T., Okuzaki D., Nishida N., Maeda S., Naito A., Kikuta J., Nishikawa K., Nishimura J., Haraguchi N., Takemasa I., Mizushima T., Ikeda M., Yamamoto H., Sekimoto M., Ishii H., Doki Y., Matsuda M., Kikuchi A., Mori M., Ishii M., Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLOS ONE 8, e83629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niwa H., Yamamura K.-i., Miyazaki J.-i., Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., Iwamatsu A., Kaibuchi K., Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/12/e1601941/DC1
fig. S1. Cartoon illustrating APs and BPs in embryonic mouse and fetal human neocortex.
fig. S2. Presence of the novel, human-specific C-terminal sequence in modern but not ancestral ARHGAP11B.
fig. S3. Reconstruction of the ancestral splice donor site.
fig. S4. Ancestral ARHGAP11B nucleotide sequence.
fig. S5. Ancestral ARHGAP11B protein sequence.
fig. S6. Modern but not ancestral ARHGAP11B increases Tbr2+ progenitors in mouse developing neocortex.
fig. S7. Neither modern nor ancestral ARHGAP11B affects the abundance of mitotic APs in mouse developing neocortex.
fig. S8. ARHGAP11B-A217T mutant, like modern ARHGAP11B, lacks GAP activity and increases BPs in the mouse developing neocortex.


