Abstract
The transcription factor CREB (cAMP Response Element Binding Protein) is overexpressed in the majority of acute myeloid leukemia (AML) patients, and this is associated with a worse prognosis. Previous work revealed that CREB overexpression augmented AML cell growth, while CREB knockdown disrupted key AML cell functions in vitro. In contrast, CREB knockdown had no effect on long-term hematopoietic stem cell activity in mouse transduction/transplantation assays. Together, these studies position CREB as a promising drug target for AML. To test this concept, a small molecule inhibitor of CREB, XX-650-23, was developed. This molecule blocks a critical interaction between CREB and its required co-activator CBP (CREB Binding Protein), leading to disruption of CREB-driven gene expression. Inhibition of CBP-CREB interaction induced apoptosis and cell cycle arrest in AML cells, and prolonged survival in vivo in mice injected with human AML cells. XX-650-23 had little toxicity on normal human hematopoietic cells and tissues in mice. To understand the mechanism of XX-650-23, we performed RNA-seq, ChIP-seq and Cytometry Time of Flight with human AML cells. Our results demonstrate that small molecule inhibition of CBP-CREB interaction mostly affects apoptotic, cell cycle, and survival pathways, which may represent a novel approach for AML therapy.
Keywords: Acute Myeloid Leukemia, small molecule, CREB, CBP
Introduction
Acute Myeloid Leukemia (AML) is associated with a 5-year overall survival of less than 50% despite the use of intensive chemotherapy regimens and hematopoietic stem cell transplantation 1–3. Treatment for AML is itself associated with significant morbidity and mortality, and most patients who survive experience at least one serious treatment-related long-term complication 4. Therefore, it is critical to identify novel therapeutic targets and develop more effective, less toxic drugs for the treatment of patients with AML.
Previous work has described the transcription factor CREB (cAMP Response-Element Binding Protein) as a critical regulator of the growth and survival of AML cells 5–9. Elevated CREB expression was observed in ~60% of AML patients, and this was associated with a significantly worse prognosis and an increased risk of relapse compared to patients with basal CREB expression 8. CREB overexpression in AML cells augments their growth rate and confers resistance to apoptosis in vitro 6. Conversely, CREB knockdown inhibited AML cell proliferation and induced apoptosis, but had no toxicity to normal hematopoietic stem cells in mouse transplantation assays 10. Together, these observations suggest that CREB is associated with a more aggressive form of AML, yet is not required for normal hematopoietic stem cell function. Therefore, we hypothesized that inhibition of CREB function may represent an effective, targeted approach to AML therapy.
CREB binds genomic DNA at thousands of loci possessing the consensus cAMP Response Element (CRE) 11. Initiation of CREB-driven transcription at these loci requires that CREB recruits and binds a co-activator, the histone acetyltransferase CREB-Binding Protein (CBP) 5. This interaction triggers local histone acetylation and subsequent recruitment of transcriptional machinery to the promoter 12. Thus, our group sought to disrupt the critical interaction between CREB and CBP in an effort to disrupt CREB-driven transcription.
The precise molecular interactions that mediate CBP-CREB binding have been resolved by NMR spectroscopy 13, 14, which facilitated the identification of a small molecule capable of disrupting the CBP-CREB interaction, naphthol AS-E phosphate (KG-501) 15. Although the low potency of KG-501 renders it a poor candidate for potential clinical use (Ki ~90 μM), this molecule provided a molecular backbone for further development 16. Through a series of structure-activity relationship studies, we designed “XX-650-23” [N-(4-cyanophenyl)-3-hydroxy-2-naphthamide], an inhibitor of the CBP-CREB interaction 17–20. The current studies were undertaken to determine the efficacy of targeting the CBP-CREB interaction in AML therapy, and evaluate the toxicity of this approach to normal cells.
Here, we provide evidence that XX-650-23 disrupts the CBP-CREB interaction in AML cells and elicits an array of on-target transcriptional alterations, which leads to cell cycle arrest and apoptosis of AML cells in vitro and significantly increases survival of cell line- and patient-derived xenograft mice with no toxicity to normal hematopoietic cells or animals. These data provide “proof-of-principle” that CREB inhibition represents a potential approach for AML treatment.
Methods
Protein Purification and Biacore
KIX domain mutants were created by standard cloning and mutagenesis methods in the pGEX4T3 vector (GE Healthcare Life Sciences, Pittsburgh, PA, USA). GST-KIX and its mutants were purified with the B-PER GST Fusion Protein Spin Purification Kit (Thermo Scientific/Pierce, Grand Island, NY, USA). Surface Plasmon Resonance analysis was performed on a GE Biacore 3000 surface plasmon resonance instrument in collaboration with the Stanford Protein and Nucleic Acid (PAN) Facility.
AML Cell Lines and Patient Samples
KG-1, HL-60, MOLM-13, MV-4-11, and U937 cell lines were obtained from ATCC and low-passage stocks were used and cultured for less than 3 months maintained. Cells were regularly tested for Mycoplasma and growth characteristics, though no further authentication has been performed by the authors. Cells were plated at a density of 2-4x105 cells/ml, and treated with various doses of XX-650-23. Cell counts and viability were determined using the Vi-CELL XR Cell Viability Analyzer (Beckman Coulter, Brea, CA, USA). HL-60 and KG-1 cells overexpressing CREB were generated using lentiviral gene delivery with subsequent cells sorting for GFP. CREB knockdown was achieved by infecting cells with a lentivirus expressing the shRNA sequence 5′-GCAAATGACAGTTCAAGCCC-3′. For chemotherapy combination experiments, combination index values were calculated using median effects analysis on Calcusyn software as described 21. Human patient bone marrow samples were cultured in DMEM plus 20% FBS and 1x PSG, supplemented with recombinant GM-CSF (20 ng/ml), G-CSF (20 ng/ml), SCF (50 ng/ml), IL-3 (20 ng/ml), and IL-6 (10 ng/ml). Cells (1x105 cells/ml) were cultured with XX-650-23 for up to 72 hours. All samples contained >85% AML blasts and were not sorted prior to performing experiments. Flow cytometry analyses were performed on a DxP10 flow cytometer (Cytek, Fremont, CA, USA). All antibodies were purchased from BD Biosciences (San Jose, CA, USA). Bone marrow from AML patients were collected through voluntary patient participation at University of California, Los Angeles (Los Angeles, California, USA) and Stanford University (Palo Alto, California, USA) in compliance with the Institutional Review Board regulations of each institution. Informed consent was obtained from all human subjects, and all research was conducted in accordance with the statements set forth in the declaration of Helsinki and the Data Protection Directive.
Luciferase Assays
KG-1 cell lines were created to express luciferase in a CREB-dependent or non-CREB-dependent fashion using lentiviral gene delivery. Cells were sorted for mCherry expression. Luciferase activity was measured on a spectrophotometer using the Promega Luciferase Activity Kit (Promega, Madison, WI, USA) per manufacturer’s instructions. The split Renilla luciferase complementation assay has been described previously 20. In this assay, the KID and KIX domains were fused to the N- and C- terminal regions of Renilla luciferase, respectively. Once KIX binds phosphorylated KID, the Renilla luciferase regions were brought together, resulting in luciferase activity.
Cell Cycle Analysis
KG-1 cells were synchronized at prometaphase using a modified thymidine plus nocodazole block 22. Briefly, KG-1 cells were treated with 2 mM thymidine for 30 h, washed with PBS and released from G1/S block in fresh media for 4 h. The cells were incubated with 300 nM nocodazole (Sigma, St. Louis, MO, USA) for 13 h. XX-650-23 or DMSO was added 3 hours before release. The synchronized cells were washed with PBS and released from the mitotic block in fresh media containing XX-650-23 or DMSO. To analyze DNA content by flow cytometry, cells were harvested, fixed in 70% ice-cold ethanol for at least 1 hour at −20°C, and then stained with propidium iodide. Cells were analyzed on a FACS Calibur flow cytometer (BD Biosciences). Cell-cycle distribution was determined using FlowJo software (TreeStar, Ashland, OR, USA).
Chromatin Immunoprecipitation and High-Throughput Sequencing (RNA-Seq and ChIP-Seq)
For Chip-Seq experiments, KG-1 cells were treated with 5 μM XX-650-23 or DMSO for 6 hours. Cells were cross-linked with 1% formaldehyde at room temperature for 10 min and then incubated with 0.125 mM glycine for 5 min. After cross-linking, chromatin was digested by Micrococcal nuclease and then sonicated using SimpleChIP® Plus Enzymatic Chromatin IP Kit (Cell Signaling, Danvers, MA, USA) following the manufacturer’s protocol. Chromatin immunoprecipitations were carried out with anti-CREB antibody (17–600, Millipore, Billerica, MA, USA), anti-Acetyl-Histone H3 (8173, Lys27) (D5E4) (Cell Signaling Technology) or a control IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The captured immunocomplexes containing bound transcriptional DNA fragments were eluted, with recovered DNA fragments used for PCR amplification. For RNA-Seq experiments, KG-1 cells were treated with XX-650-23 (5μ) or 0.1% DMSO for 12 hours. Sample libraries were run using the Illumina sequencing platform. Two hundred million reads were collected on two biological replicates of the experiment. Libraries were prepared using the Illumina Truseq RNA samples prep kit per manufacturer’s instructions. Fastq files were aligned using TopHat 23. Aligned BAM files were used for CuffDiff calculation of differentially expressed genes 24. CummeRbund R package was used to infer QC of the RNA-seq. RNA-seq and CREB ChIP-Seq data were deposited in the NCBI GEO database (GEO Submission GSE74928). ChIP-seq analysis was performed as previously described 25. BETA analysis was used by inferring differentially expressed genes between DMSO and XX-650-23 treated cells based on a fdr ≤0.01 and gene distance of 100 kb (Galaxy/cistrome). For analysis of RNA-seq data as target genes of known transcription factors, a curated list of all human target genes was extracted from the TRANSFAC Pro database. DNA sequences enriched on ChIP-Seq were defined as previously published26.
RT-PCR
Total RNA was isolated using the Aurum RNA Isolation Mini Kit, and the iScript cDNA Synthesis Kit was used to prepare samples for qPCR (Bio-Rad). Data were analyzed using the Livak method 27. Data were normalized to 7SL scRNA expression values.
Western Blot Analysis
Cell lysates were resolved on SDS PAGE gels, and then transferred onto PVDF membranes (Millipore). Listing of primary antibodies is given in Supplementary Table 1. Secondary antibodies were used at a 1:2500 dilution and purchased from Thermo Scientific/Pierce. Membranes were visualized with enhanced chemiluminescence system. Representative blot of at least three different experiments are shown.
Hematopoietic Cell Colony Assays
Normal human bone marrow cells were resuspended in methylcellulose (2×104 cells/mL, Miltenyi Biotec, Auburn, CA, USA) containing cytokines (GM-CSF, G-CSF, IL-3, IL-6, SCF, erythropoietin), and cultured for 2 weeks. Colonies were scored on the basis of morphology.
Caspase-3 Activity
The ApoTarget Caspase-3 Protease Activity Kit (Invitrogen, Grand Island, NY, USA) was used per manufacturer’s instructions.
Multiparameter Single Cell Mass Cytometry (CyTOF)
Bone marrow from primary AML patients were cultured as above and treated for 48 hours with 2 μM XX-650-23. These were stained for viability using cisplatin as previously described 28. Cells were fixed with 1.6% PFA for 10 min and washed with cell staining media (CSM). Fc receptor block was performed using Human TruStain FcX (Biolegend, San Diego, CA, USA). Cells were stained for surface proteins at room temperature for 30 min, and then cells were washed twice with CSM and permeabilized with methanol pre-cooled to 4°C for 10 min. Cells were then washed twice and stained for intracellular proteins for 30 min at room temperature. Cells were washed and stained with 1 mL of 2000x iridium DNA intercalator (diluted 1:5000 in PBS with 1.6% PFA; DVS Sciences, Sunnyvale, CA, USA) overnight at 4 °C. Data were acquired using internal metal isotope bead standards as previously described 29. Cell events were acquired on a CyTOF I (DVS Sciences/Fluidigm). All antibodies were also purchased from DVS Sciences/Fluidigm. Each patient sample was individually normalized to the internal bead standards prior to analysis. To remove dead cells and debris, cells were gated based on cell length and DNA content as described 30.
AML Xenograft Models of AML
For AML xenograft mouse experiments, HL-60 cells (2x106) expressing firefly luciferase and GFP, or cryopreserved human AML patient cells (5x106), were injected through the tail vein into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (5–8 weeks old). AML cells-injected mice were randomly divided into two groups. Mice were then treated with XX-650-23 or 10% DMSO, injected intravenously (i.v.) daily, until death or an endpoint was reached (moribundity) in accordance with the animal care institutional guidelines. The sample size of AML xenograft mice experiments was determined based on our previous studies as well as the published papers 31,32. Blinding was not used for xenograft mice experiments. All mouse experiments were subject to institutional approval by Stanford University Institutional Animal Care and Use Committee. Leukemia progression in mice at the indicated time points was monitored using an in vivo IVIS 100 bioluminescence/optical imaging system (Xenogen Corporation, Alameda, CA, USA). Analysis was performed on Living Image In Vivo imaging software (Perkin-Elmer, Waltham, MA, USA).
Statistical Analysis
Unless noted, all experiments were performed in triplicate, and unpaired two-tailed Student’s t-test was used to assess experimental mean values for statistically significant differences using Prism software (v5.04) (Graphpad, La Jolla, CA, USA), with p-values of <0.05 deemed statistically significant. Kaplan-Meier plots and statistical significance of differences in mice survival experiments were calculated using a Log-rank (Mantel-Cox) test with Prism. Synergy calculations were performed using CalcuSyn software. IC50s were calculated using Prism.
Results
XX-650-23 is a Competitive Inhibitor of CREB Binding to CBP
The region of CBP critical for binding Ser-133-phosphorylated-CREB is termed the Kinase-Inducible Acceptor domain or ‘KIX’ domain, and spans CBP amino acids 586-666. The small molecule KG-501 (Naphthol AS-E phosphate) was reported to disrupt CREB-CBP binding by interacting with key amino acids in the KIX domain, particularly residues Arg-600 to Valine-608 15. Based on this information and subsequent structure-activity relationship studies, our group developed an optimized molecule targeted to this domain designed to disrupt the CREB-CBP interaction, termed “XX-650-23” (Fig. 1A). Earlier structure-activity relationship studies revealed XX-648-48 as an inactive yet similarly structured compound (N-methyl-(4-chlorophenyl)-3-hydroxy-naphthamide) 20.
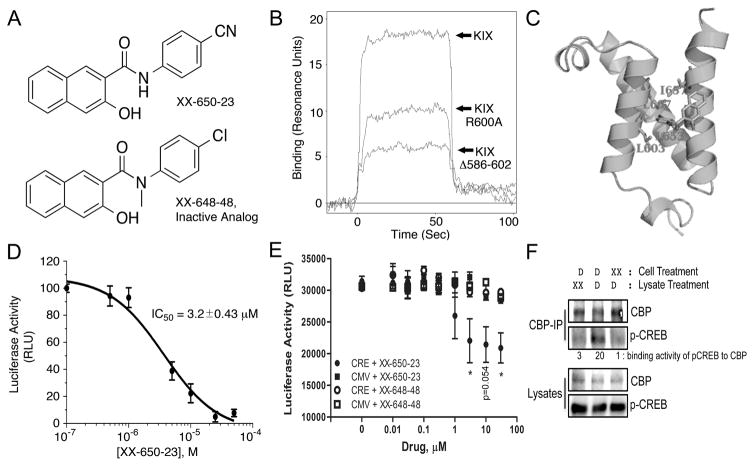
Fig. 1. XX-650-23 Binds to the KIX Domain of CBP and Blocks CREB-Dependent Gene Transcription.
A) The structure of XX-650-23 and its inactive analog XX-648-48. B) The native human KIX domain and two mutant proteins were expressed as a fusion protein with GST and subjected to Biacore analysis to assess XX-650-23 binding characteristics. Mutation of Arginine-600 to Alanine reduced binding of XX-650-23 by ~45% at a concentration of 5μ, while a KIX mutant lacking amino acids 586-602 reduced binding by ~70%. C) Binding model of XX-650-23 to CBP KIX domain. D) Split-Renilla luciferase assays with 293T cells treated with forskolin demonstrated XX-650-23 is a direct inhibitor of CREB-CBP binding, with an IC50 of approximately 3.2 μM. Data are presented as mean ± SD, n=3. E) Two KG-1 cell lines were generated in which luciferase was expressed either under the control of a CREB-driven promoter (CRE) or a CMV-driven promoter (CMV). These two cell lines were each treated with a range of XX-650-23 or XX-648-48 concentrations for 6 hours. Luciferase activity was significantly decreased following XX-650-23 treatment at concentrations of 3, 10 and 30 μM. No statistical difference in luciferase activity was detected following XX-648-48 treatment. Data are presented as mean ± SEM, n=3. *p < 0.05, t-test. F) XX-650-23 inhibits CREB/CBP association. HEK293 cells were transfected with a plasmid expressing CREB. Cells were treated with XX-650-23 (XX, 5 μM, lane 3) or DMSO vehicle (D) for 1 hour. Total lysates of transfected HEK293 cells were immunoprecipitated using anti-CBP antibody. XX-650-23 (XX, 5 μM) was added during anti-CBP Immunoprecipation process (lane 1). Immunoprecipitates (CBP-IP) and total lysates were analyzed by immunoblotting for phospho-CREB (p-CREB) and CBP. Binding activities were calculated by the relative protein amounts of p-CREB bound to CBP normalized against CBP amounts in CBP IP products.
Binding of XX-650-23 to the KIX domain and two KIX domain mutants in which residues critical for CREB binding were altered or removed (Arg-600 mutated to Alanine or deletion of amino acids 586-602) was determined by Surface Plasmon Resonance (Biacore) analysis (Fig. 1B). Mutation of Arginine-600 to Alanine reduced binding of XX-650-23 by ~45%, while a KIX domain mutant lacking amino acids 586-602 reduced binding by ~70%. A hypothetical binding model between XX-650-23 and CBP KIX domain is shown (Fig. 1C). This binding model suggests the major interactions between XX-650-23 and the KIX domain are hydrophobic. The aniline ring of XX-650-23 is predicted to project into a hydrophobic pocket of KIX where Leucine-141 of CREB, an amino acid essential for stable CREB binding, normally docks.
XX-650-23 inhibited the interaction between CREB and CBP with an IC50 of 3.20 ± 0.43 μM in a split Renilla luciferase complementation assay was performed 20 (Fig. 1D). To determine whether XX-650-23 could specifically decrease CREB-driven reporter gene expression in AML cells, KG-1 AML cell lines expressing luciferase under the control of a CREB-driven promoter (two CRE elements placed upstream of an attenuated CMV promoter) or a CMV-driven promoter without CREs, were treated with XX-650-23. CREB-driven luciferase activity was reduced by XX-650-23 in a dose-dependent fashion, whereas CMV-driven luciferase expression was unchanged following treatment (Fig. 1E). The inactive analog XX-648-48 had no significant effect on luciferase activity in either AML cell line. Finally, we demonstrated that treatment of HEK293 lysates or cells with XX-650-23 prevented binding of CREB to CBP (Fig. 1F). Thus, XX-650-23 inhibits CREB function through disruption of CBP-CREB interaction.
XX-650-23 Suppresses AML growth
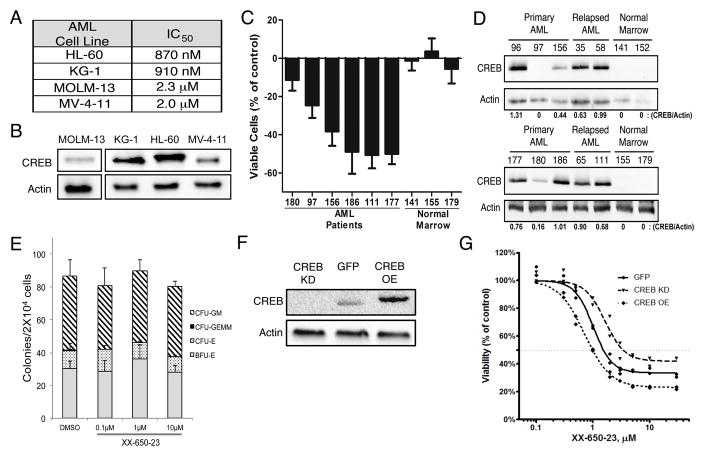
To test the effects of XX-650-23 on AML cells, four AML cell lines were treated with doses ranging from 100 pM to 10 μM for 48 hours. The IC50 of XX-650-23 for these AML cell lines, defined as a 50% reduced viable cell count compared to DMSO-treated cells after 48 hours of culture, ranged from 870 nM to 2.3 μM (Fig. 2A). These four cell lines expressed CREB protein (Fig. 2B). We next tested the effects of XX-650-23 on primary human AML samples, obtained at initial diagnosis or at the time of relapse. AML patient samples treated with 2μM XX-650-23 for 72 hours demonstrated a range of responsiveness (Fig. 2C). AML patient samples with relatively higher CREB expression exhibited a greater loss of viability than those with lower CREB expression (Fig. 2D). Normal bone marrow cells expressed nearly undetectable levels of CREB protein (Fig. 2D), and the viability of the cells did not significantly change by treatment with XX-650-23 (Fig. 2C). These studies were extended to 10 methylcellulose colony assays of normal human hematopoietic cells, which also demonstrated no significant decrease in colony formation when treated with up to 10 μM XX-650-23 (Fig. 2E). These data indicated that higher CREB expression might be associated with greater sensitivity to XX-650-23. Therefore, we modulated CREB expression levels to determine whether we could change sensitivity of cells to XX-650-23. CREB was overexpressed, or knocked down by shRNA, in KG-1 cells (Fig. 2F). CREB knockdown in KG-1 cells decreased their sensitivity to XX-650-23, while CREB overexpression increased their sensitivity (IC50: CREB KD, 3.43 μM; GFP, 1.53 μM; CREB OE, 0.99 μM) (Figure 2G). Experiments were also performed to examine the efficacy of combining XX-650-23 with cytarabine or daunorubicin, standard drugs used in AML therapy. Combination of XX-650-23 with cytarabine or daunorubicin in KG-1 cells showed synergistic effect against KG-1 cell viability (Supplementary Fig. 1).
Fig. 2. Efficacy of CREB Inhibition Depends on CREB Expression.
A) IC50 values for 4 AML cells lines identically treated with XX-650-23 are shown. Cells were cultured with various doses of XX-650-23 for 48 hours. Viable cells were enumerated by Trypan blue exclusion method. B) Western blot of CREB expression levels in 4 AML cell lines, run on non-contiguous lanes. C) Six AML patient and three normal bone marrow samples were treated with 2 μM XX-650-23 for 48 hours, and the percent of viable cells lost or gained compared to DMSO-treated cells is shown. Data are graphed as mean ± SEM (n = 4). D) Western blot of CREB expression in ten AML patient and four normal marrow samples. Relative expression levels of CREB protein are shown. E) Methylcellulose colony assays of normal bone marrow progenitor cells treated with up to 10 μM XX-650-23. Cells were cultured with XX-650-23 for 2 weeks in methylcellulose media and colonies were scored based on morphology. Data are graphed as mean ± SEM (n = 3). F) Western blot of CREB expression levels in KG-1 cells engineered to overexpress CREB (CREB OE) or GFP control (GFP), or in which CREB expression was reduced by shRNA (CREB KD). Representative blots of at least three independent experiments are shown. G) XX-650-23 dose-response data for the three KG-1 cell lines depicted in (F) following 48 hours of treatment.
XX-650-23 Specifically Disrupts CREB-Driven Transcription in AML Cells
To determine the functional specificity and downstream effects of XX-650-23 in AML, the CREB transcriptome was first defined in KG-1 cells using CREB chromatin immunoprecipitation followed by high-throughput sequencing (CREB ChIP-Seq). In parallel, whole transcriptome sequencing (RNA-Seq) of KG-1 cells treated with XX-650-23 was performed to define alterations in the transcription of identified CREB-bound genes. Finally, whole-genomic H3K27 histone acetylation analysis was performed to assess the disruption of CBP-CREB interaction. Together, these three experiments permitted: 1) assembly of a comprehensive catalogue of CREB-bound sites within the AML genome; 2) identification of the set of genes which exhibited altered expression following XX-650-23 treatment, and 3) measurement of alterations in CBP-mediated histone acetylation at genomic CREB binding sites. In this way, the ‘on-target’ effects of XX-650-23 could be assessed.
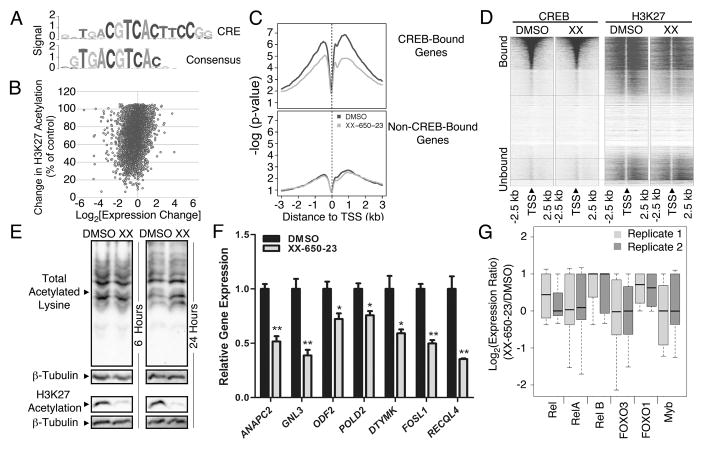
CREB peaks (p-val ≤ 10−5, fdr ≤1%) identified on ChIP-Seq analysis were enriched for the canonical ‘CRE element’ DNA binding sequence (Fig. 3A). CREB binding was detected at 4680 sites in the KG-1 AML cell genome. Nearly half of the CREB-bound genes (1863 of 4213 total CREB-bound genes) in KG-1 cells coincided with the previously reported putative CREB target genes 11. Gene ontology enrichment analysis of the function annotations for regions associated with CREB-bound peaks (Supplementary Table 2) showed that CREB-bound peaks were enriched in clusters of genes regulating cell cycle and apoptosis as well as translation and mRNA processing. Over 90% of occupied CREB-binding sites were within 500 bp of a transcription start site (TSS) (Supplementary Figs. 2A and 2B). Of these CREB-bound 4213 genes, 2787 CREB-bound genes exhibited reduced expression following XX-650-23 treatment and 602 genes exhibited greater than 50% reduced expression. Majority of H3K27 peaks in CREB-bound genes (4052 out of 5282 total peaks) exhibited decreased H3K27 histone acetylation following treatment with XX-650-23 (66.5 ± 17% of control, n=4052 peaks. Fig. 3B & Supplementary Table 3). Histogram analysis of H3K27 acetylation of all CREB-bound genomic loci also showed a relative reduction in overall H3K27 acetylation, represented as a median shift to the left (Supplementary Fig. 2C). Moreover, reductions in H3K27 acetylation were almost exclusively within 1 to 3 kb of these CREB-bound promoter sites. On the contrary the genes that did not show any significant binding site for CREB showed no global detectable changes in acetylation. (Fig. 3C). CREB genomic binding did not change following XX-650-23 treatment (Fig. 3D and Supplementary Fig. 2D). Western blot analysis confirmed that XX-650-23 caused a specific decrease in H3K27 acetylation and is not a general inhibitor of acetyltransferase activity (Fig. 3E).
Fig. 3. Specificity of CREB Inhibition.
A) The consensus sequence obtained following CREB ChIP-Seq, mapped against the canonical CRE element sequence. B) Relationship between H3K27 acetylation and CREB-driven gene expression. RNA-Seq, CREB ChIP-Seq and H3K27 histone acetylation-Seq were performed on KG-1 cells treated with 5 μM XX-650-23 or 0.1% DMSO. Combined analysis defined the set of genes bound by CREB in AML cells, and alterations in transcription and histone acetylation occurring secondary to XX-650-23 at those loci. The percent of histone acetylation remaining following treatment with XX-650-23 is plotted versus the corresponding change in gene expression for all CREB-bound genes identified by CREB ChIP-Seq. C) Changes in H3K27 acetylation for CREB-bound and -unbound genes were averaged and plotted against base-pair distance to transcriptional start sites (TSS) following XX-650-23 or DMSO treatment. Non-CREB-bound genes show no significant change in H3K27 acetylation following XX-650-23 treatment. D) Heatmap of CREB binding and H3K27 acetylation relative to TSS in DMSO and XX-650-23-treated samples. H3K27 signal intensity, but not CREB binding signal intensity, decreased following XX-650-23. E) Western blot of total and H3K27-specific histone acetylation following XX-650-23 treatment following 6 or 24 hours of DMSO or XX-650-23 treatment. Representative blots of at least three independent experiments are shown. F) RT-PCR confirmed downregulation of CREB-bound genes identified on RNA-Seq following 12 hours of XX-650-23 treatment of KG-1 cells for all genes shown. Data are graphed as mean ± SEM (n = 3), *p < 0.05; **p < 0.001, t-test. G) RNA-Seq analysis demonstrated that the activity of six CBP-bound transcription factors remains unchanged following XX-650-23 treatment.
To validate the RNA-Seq data set, qPCR of XX-650-23 target genes with CREB-bound promoters that exhibited significant (>50%) transcriptional downregulation and reduced H3K27 acetylation was performed in two AML cell lines (KG-1 and HL-60) following treatment with XX-650-23 (Fig. 3F and Supplementary Fig. 2E). XX-650-23 consistently reduced these genes expression in both these cell lines in a statistically significant manner, although we expect that these genes are regulated by other transcription factors in addition to CREB. To assess for potential ‘off-target’ effects of XX-650-23, the RNA-Seq gene expression profiles of other transcription factors that bind CBP, including Rel, RelA, RelB, Foxo3, Foxo1 and Myb, were analyzed. XX-650-23 evoked no significant change in the expression of these other CBP-binding transcription factors target genes (Fig. 3G). These results were confirmed for a set of Myb-driven genes, SP3, FPR1, PRODH, SLC34A2 in both KG-1 and HL-60 cells 33–36 (Supplementary Fig. 2F). Thus, these results provide evidence that XX-650-23 disrupts the interaction of CREB to the KIX domain of CBP but not other KIX binding proteins in AML cells.
XX-650-23 Prolongs Survival in Mouse Models of AML Without Toxicity
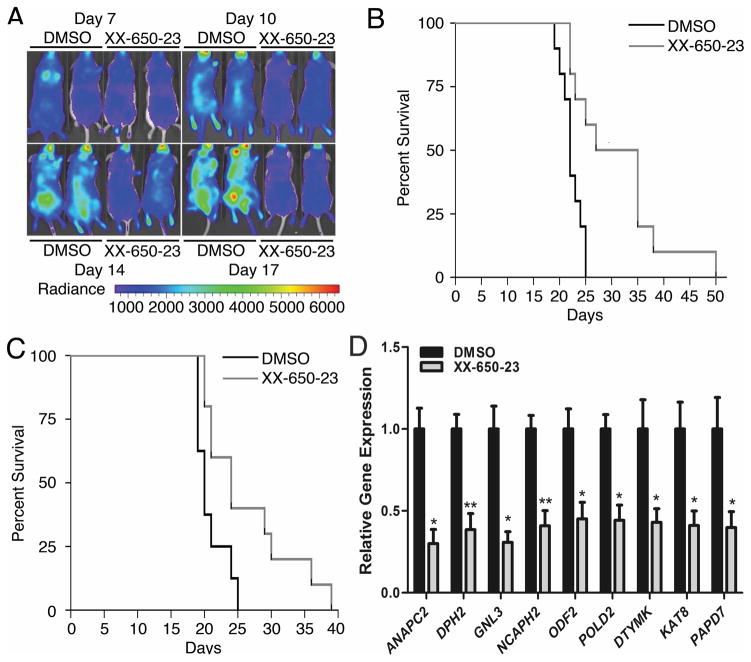
To examine the efficacy of XX-650-23 in an in vivo model of AML, NSG mice were tail-vein injected with HL-60 AML cells expressing Firefly luciferase and GFP. Mice received XX-650-23 (2.3 mg/kg, intravenously) once daily starting the day after cell injection (immediate treatment groups), or starting seven days after AML cell injection (delayed treatment groups). Bioluminescence imaging performed during treatment revealed less disease burden in XX-650-23-treated mice compared to control (average radiance measurements, given in p/sec/cm2/sr: 5.7x105 for the control group versus 2.8x105 for the XX-650-23-treated group at day 17) (Fig. 4A). XX-650-23 significantly prolonged the median survival in Kaplan-Meier analysis in both the immediate (median survival, 22 days versus 31 days, p = 0.0027, long-rank test; mean survival 22.3 days vs. 31.2 days) and delayed treatment groups (median survival, 20 days versus 24 days, p = 0.0211, long-rank test; mean survivals 20.9 vs. 26.4 days) (Fig. 4B and 4C). We also assessed the efficacy of XX-650-23 in a mouse xenograft model using primary AML cell sample (patient sample #186). Mice treated with XX-650-23 demonstrated a significant survival advantage compared to DMSO-treated mice (Supplementary Fig. 3A). Next we evaluated the effects of XX-650-23 on CREB transcriptional activity in AML cells in vivo. This experiment recapitulated in vitro findings (Fig. 4D).
Fig. 4. CREB Inhibition In Vivo.
A) Bioluminescent imaging revealed significantly less disease burden in mice treated with daily intravenous injections of XX-650-23 on treatment days 10, 14 and 17 (DMSO-treated mice, left; XX-650-23-treated mice, right). B) Kaplan-Meier curve analysis demonstrated a significant survival advantage in NSG mice treated with 2.3 mg/kg/day intravenously once a day XX-650-23 (n=10) compared to those treated with vehicle alone (n=10) beginning one day after AML cell injection (p = 0.0027, log-rank tests). C) Kaplan-Meier curve analysis also demonstrated a survival advantage for mice given XX-650-23 (n=10) compared to those treated with vehicle alone (n=8) beginning 7 days after AML cell injection (p=0.0211, log-rank tests). D) RT-PCR showed XX-650-23 elicits the same transcriptional alterations in vivo as observed in vitro. To directly evaluate the effects of XX-650-23 on CREB transcriptional activity in vivo, six NSG mice were injected with 2x106 HL-60 expressing GFP. After a ten-day engraftment period, the mice received three once-daily treatments of either 2.3 mg/kg XX-650-23 or DMSO. The mice were then sacrificed and GFP+ bone marrow cells were sorted and analyzed for transcriptional changes in validated CREB target genes. XX-650-23 treatment significantly reduced expression of all genes. Data are graphed as mean ± SEM (n = 3), *p < 0.05; **p < 0.01, t-test.
Pharmacokinetic studies showed that the half-life of XX-650-23 in plasma is approximately 4.3 hours (Supplementary 3B). XX-650-23- treated mice demonstrated normal complete blood counts, liver function tests and kidney function tests, compared to age-matched NSG mice given no treatment (Supplementary Fig. 3C). Histology demonstrated no microscopic evidence of vital organ damage (Supplementary Fig. 3D).
XX-650-23 Induces Apoptosis in AML cells
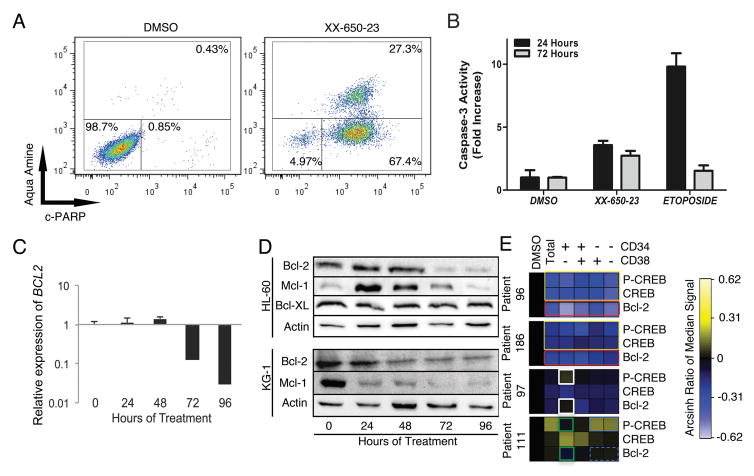
We next examined whether XX-650-23 induced cell death as well as growth inhibition in AML cells. In HL-60 cells treated with XX-650-23, apoptosis was induced in a dose- and time-dependent manner. Over 95% of cells entered early apoptotic or late apoptotic stage after 72 hours of XX-650-23 treatment (Fig. 5A). To confirm that XX-650-23-induced apoptosis is not restricted to HL60 cells, we determined the apoptotic effect of XX-650-23 in additional AML cell lines (KG-1, MV-4-11, and U937 cells). Apoptosis was observed in all these cell lines with XX-650-23 treatment (Supplementary Fig. 4). XX-650-23 elicited apoptosis through the intrinsic apoptosis pathway, with activation of Caspase-3 (Fig. 5B) and detectable Caspase-9 cleavage (data not shown). The balance between pro- and anti-apoptotic protein expression contributes to cell fate decisions 37. CREB regulates the expression of several anti-apoptotic proteins, including BCL2, BCL2L1 and MCL1 38, 39. The transcription of BCL2 decreased following 72 hours of 2 μM XX-650-23 treatment (Fig. 5C), and this was verified by Western blot analysis (Fig. 5D). In parallel, the expression of Mcl-1 initially increased, then decreased at 72 hours, while Bcl-XL expression remained constant. Bcl-2 inhibitor ABT-737 treatment induced in apoptosis, similar to XX-650-23 (Supplementary Figs. 5A and 5B) 40. KG-1 cells also underwent apoptosis following treatment with XX-650-23, and similarly exhibited decreased Bcl-2 expression alongside a pronounced decrease in Mcl-1 (Fig. 5D). Bcl-2 levels were influenced by CREB expression in these AML cell lines (Supplementary Fig. 5C). Together, these data suggest that downregulation of Bcl-2 is involved in XX-650-23-induced apoptosis in AML cell lines.
Fig. 5. CREB Inhibition in AML Cells Induces Apoptosis.
A) HL-60 cells clearly showed early apoptotic (c-PARP+/aqua amine−) or late apoptotic (c-PARP+/aqua amine+) populations following 72 hours of treatment with 2 μ XX-650-23. Representative flow cytometric plots of three independent experiments are shown. B) Caspase-3 activity is activated in response to XX-650-23 treatment in HL60 cells. Data are graphed as mean ± SD (n = 2). C) RT-PCR showed BCL2 expression decreases at 72 hours after XX-650-23 treatment in HL-60 cells. Data are graphed as mean ± SEM (n = 3). D) Western blot analysis shows a decrease in Bcl-2 protein expression following 72 hours of treatment, no change in Bcl-XL expression, and an initial increase followed by a decrease in Mcl-1 expression in HL-60 cells. In KG-1 cells, Mcl-1 and Bcl-2 also showed decreased expression following XX-650-23 treatment. E) Heatmap representing expression of p-CREB (Ser133), total CREB (CREB) and Bcl-2 in four primary AML samples treated with DMSO or XX-650-23 as analyzed by mass cytometry. Expression shown as Arcsin ratio to DMSO control (first column). Gated cell populations based on CD34 and/or CD38 as indicated above heatmap. Patient 96 and 186 demonstrate downregulation of Bcl-2 in all cell populations in response to XX-650-23 (red box) as well as decreases in total CREB and p-CREB (yellow boxes). Patient 97 demonstrates activation of p-CREB in a cell specific manner (white boxes) as well as no effect on Bcl-2 expression in CD34+CD38− population. In contrast, p-CREB and Bcl-2 expression levels were downregulated in CD34+CD38− population (green boxes) in Patient 111, even though p-CREB level was increased (blue box) and Bcl-2 expression level was not changed (blue dashed box) in mature populations.
We examined the relationship between Bcl-2, total CREB, and p-CREB in primary AML patient samples, especially in the CD34+CD38− subpopulation containing the putative ‘leukemia initiating’ cells 41, 42 (see CyTOF gating strategy, Supplementary Fig. 5D). XX-650-23 treatment significantly reduced Bcl-2 expression levels in all cell subpopulations in XX-650-23 hyper-responsive primary AML cells with higher CREB expression (#96 and #186) (Fig. 5E, within red outlined box). This occurred alongside downregulation of total CREB as well as reduced phosphorylation at Serine 133, an activation mark of CREB (Fig. 5E, within yellow outlined box). In contrast, in patient sample 97, which expressed less CREB at baseline and showed less sensitivity to XX-650-23 (Fig. 2C & D), CREB phosphorylation was increased in the CD34+CD38− population without significant Bcl-2 downregulation (Fig. 5E, within white outlined box). However, p-CREB and Bcl-2 expression levels were downregulated in CD34+CD38− population (within green outlined boxes) in a patient 111, even though phosphorylation of CREB was increased (Fig. 5E, within blue outlined box) without significant Bcl-2 downregulation (Fig. 5E, within blue dashed box) in the more mature AML cell populations. These data suggest that Bcl-2 downregulation in CD34+CD38− population may determine the sensitivity to the drug. The relative activation of AKT and ERK was associated with increased or decreased of phosphorylation of CREB (Supplementary Fig. 6A). HL-60 cells also showed an increase in levels of phosphorylated but not total CREB following 24 hours of XX-650-23 treatment (Supplementary Fig. 6B), and this effect can be blocked by validated small molecule inhibitors of the ERK and RSK kinases (Supplementary Fig. 6B), in keeping with previous work describing ERK/RSK-mediated phosphorylation of CREB downstream of the GM-CSF receptor in AML cells 43–45. Blockade of these kinases increased the efficacy of XX-650-23 (Supplementary Fig. 6C).
XX-650-23 Induces AML Cell Cycle Arrest at the G1/S Transition
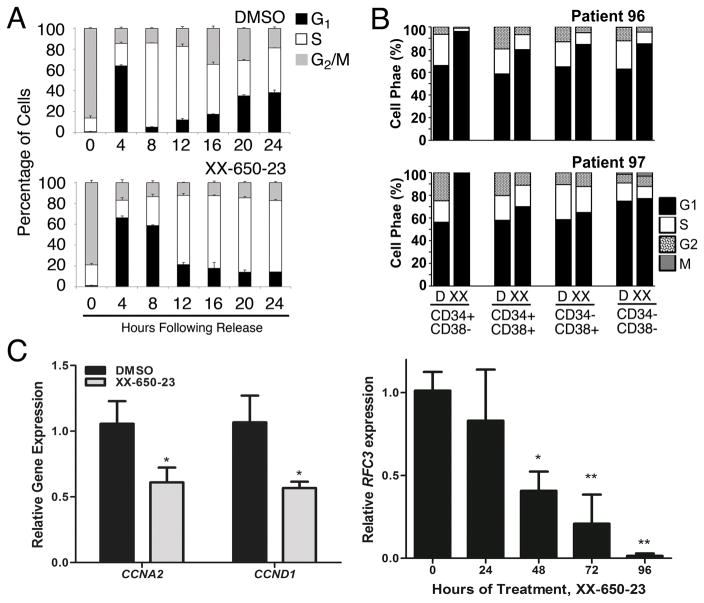
CREB regulates the cell cycle by virtue of transcriptionally regulating the expression of key cell cycle genes. Following release from prometaphase, cell cycle progression was followed by DNA contents for 24 hours in the presence or absence of XX-650-23 in KG-1 cells. XX-650-23 treatment caused aberrant cell cycle progression at the G1/S transition and through S-phase (% cells in G1-phase for DMSO treated cells: 61.18 ± 0.97% and 4.92 ± 0.33% at 4 and 8 hours, respectively, versus XX-650-23-treated cells: 66.20 ± 1.83% and 55.41 ± 0.59% (mean ± SEM, n=3) at 4 and 8 hours following release, respectively.) (Fig. 6A and Supplementary Fig. 7A). CyTOF cell cycle analysis of AML patient bone marrow samples revealed the same perturbation in the G1/S transition following XX-650-23 treatment (Fig. 6B). The percent of cells in both the S and G2/M phases was reduced, and that of G1 was increased.
Fig. 6. CREB Inhibition Induces Cell Cycle Arrest.
A) Cell cycle phase analysis of KG-1 cells treated with XX-650-23 showed G1/S transition block and delayed S-phase progression. Data represent the percentages of cell populations residing at each cell cycle stage and is expressed as mean ± SEM (n = 3). B) CyTOF analysis of AML patient samples also showed a reduction of cells in G2 and S phase following XX-650-23 treatment. C) CREB-regulated genes important for cell cycle progression through G1/S and S were downregulated. CCNA2 and CCND1 expression was decreased following 24 hours of treatment, while RFC3 expression decreased following 48 hours of treatment. Data are graphed as mean ± SEM (n = 3), *p < 0.05; **p < 0.01, t-test.
A number of previously described mediators of the G1/S transition and S-phase progression were downregulated following XX-650-23 treatment, including a set of CREB-bound cyclins, CCNA2 and CCND1, and FOSL1 46–48(Fig. 3F & 6C). We analyzed RNA expression and H3K27 acetylation of CREB-bound genes to identify potential target genes of XX-650-23 in cell cycle perturbation. CREB-bound cell cycle regulators including FOSL1, CDK2, CCNB1, CDC25C, and RAD54L show decreased RNA expression and H3K27 acetylation following XX-650-23 treatment (Supplementary Fig. 7B). Transcriptional alterations of a subset of cell cycle regulators were confirmed by qPCR in two AML cell lines (Supplementary Figs. 7C). In addition, our group has previously reported that Replication Factor C3 (RFC3), a member of the PCNA DNA replication complex, is downregulated following CREB knockdown in AML cells, which is associated with G1/S transition arrest 49. Levels of RFC3 also decrease following XX-650-23 treatment (Fig. 6C).
Discussion
Successful treatment of AML remains a major clinical challenge, and progress in our ability to treat AML patients will depend on the development of effective approaches associated with minimal toxicity. Here, we provide the first evidence that a novel small molecule that inhibits the transcription factor CREB may offer a unique, effective and non-toxic approach to treat AML patients. CREB is an ideal target for AML therapy, given that CREB overexpression is associated with a worse prognosis and knockdown of CREB does not affect normal HSC activity. Previous work facilitated the synthesis of a novel inhibitor of CREB function, XX-650-23, which our data establish as a competitive inhibitor of CBP-CREB interaction. We offer this study as ‘proof of principle’ that CREB inhibition is a viable approach to AML therapy.
Developing ‘targeted therapy’ for AML connotes both that the agent is targeted to a specific function or molecule within AML cells, and that the agent’s toxicity is specific to AML cells. Based on our data, we postulate that the specificity with which XX-650-23 disrupts the CREB-CBP interaction defines its selective toxicity to AML cells. Biochemical evidence supports that XX-650-23 binds CBP and physically disrupts the CREB-CBP interaction. Our whole-transcriptome and genome acetylation studies provide evidence that the CREB-CBP interaction is specifically disrupted. The almost exclusive reduction in H3K27 acetylation at CREB-bound genomic loci reflects the absence of CBP docking at, and only at, these sites. Furthermore, the activity of other transcription factors that also bind the CBP KIX domain was not affected. These data provide evidence that XX-650-23 exerts ‘on-target’ effects. Within the set of genes bound by CREB, the transcription of hundreds of genes was significantly (>50%) down-regulated. While it is not surprising that we did not observe a significant reduction in transcription of all CREB-bound genes, expression of most eukaryotic genes is determined by input from unique sets of diverse transcription factors. Our data demonstrate that key CREB-bound factors that regulate the cell cycle and apoptosis are downregulated, and sufficient to cause AML cell cycle arrest and apoptosis in vitro and in vivo.
Targeting the activity of specific transcription factors for the treatment of leukemia has begun to show promise in a number of pre-clinical studies. The interaction of CBP with β- and γ-catenin has recently been targeted using a small molecule, and this strategy was effective against both primary and relapsed ALL 31. Another group recently demonstrated the efficacy of targeting the mutant fusion transcription factor CBPβ-SMMHC, which drives inv(16)+ AML 32, and the critical interaction between menin and MLL fusion proteins, which drives subtypes of both AML and ALL, has also been successfully targeted 50. The data presented here similarly provide “proof of principle” that CREB can be targeted for the treatment of AML, and lay the groundwork for advancing this strategy to the clinical arena.
Supplementary Material
Acknowledgments
We thank Kai Fu and Jing Lu for analysis of ChIP-seq and RNA-seq data. In vivo serum measurements of XX-650-23 were performed in collaboration with Ludmila Alexandrova, Ph.D., at the Stanford University Mass Spectrometry (SUMS) core facility. Surface Plasmon Resonance (Biacore) analysis was performed in collaboration with Michael Eckart, Ph.D., at the Stanford Protein and Nucleic Acid (PAN) facility. In vivo animal imaging was performed in collaboration with the Stanford Small Animal Imaging (SCI3) facility, directed by Timothy Doyle, Ph.D.
Financial support: This research was supported by NIH R01 HL75826, Maxfield Foundation, SPARK program and Child Health Research Institute Lucile Packard Foundation for Children’s Health, Leukemia and Lymphoma Society of America (SLP-8009-15), Pediatric Cancer Research Foundation, Hyundai Hope on Wheels (#02500CA) (K.M.S.), NIH R01 GM087305 (X.X.), and by the American Cancer Society Greeley & Seattle Gala/Friends of Rob Kinas Postdoctoral Fellowship, Bear Necessities and Jane C. Ventura Charitable Trust, Harvey Cohen Endowment, and the Stanford University Dean’s PostDoctoral Fellowship program (B.M.).
Footnotes
Conflict of Interest Statement: G. N. has personal financial interest in the company DVS Sciences, the manufacturers that produced some of the reagents and instrumentation used in this manuscript.
Author contributions
BM and HDC designed research, performed research, analyzed data and wrote the paper. KH, RD, BCT, AK, FX, BXL and BXL performed research. GAD, RF, KD performed research and analyzed data. GD, GN, MRB, EML, GN, MP and XX analyzed data. NL collected and provided primary cells from AML patients. KMS designed research, analyzed data and wrote the paper.
References
- 1.Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood. 2005;106:1154–1163. doi: 10.1182/blood-2005-01-0178. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 3.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012;120:3187–3205. doi: 10.1182/blood-2012-03-362608. [DOI] [PubMed] [Google Scholar]
- 4.Schultz KA, Chen L, Chen Z, Kawashima T, Oeffinger KC, Woods WG, et al. Health conditions and quality of life in survivors of childhood acute myeloid leukemia comparing post remission chemotherapy to BMT: a report from the children's oncology group. Pediatr Blood Cancer. 2014;61:729–736. doi: 10.1002/pbc.24881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conkright MD, Montminy M. CREB: the unindicted cancer co-conspirator. Trends in cell biology. 2005;15:457–459. doi: 10.1016/j.tcb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Pigazzi M, Ricotti E, Germano G, Faggian D, Arico M, Basso G. cAMP response element binding protein (CREB) overexpression CREB has been described as critical for leukemia progression. Haematologica. 2007;92:1435–1437. doi: 10.3324/haematol.11122. [DOI] [PubMed] [Google Scholar]
- 7.Pigazzi M, Manara E, Bresolin S, Tregnago C, Beghin A, Baron E, et al. MicroRNA-34b promoter hypermethylation induces CREB overexpression and contributes to myeloid transformation. Haematologica. 2013;98:602–610. doi: 10.3324/haematol.2012.070664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer cell. 2005;7:351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Shankar DB, Cheng JC, Sakamoto KM. Role of cyclic AMP response element binding protein in human leukemias. Cancer. 2005;104:1819–1824. doi: 10.1002/cncr.21401. [DOI] [PubMed] [Google Scholar]
- 10.Cheng JC, Kinjo K, Judelson DR, Chang J, Wu WS, Schmid I, et al. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008;111:1182–1192. doi: 10.1182/blood-2007-04-083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 13.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J Mol Biol. 1999;287:859–865. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 14.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 15.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, et al. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uttarkar S, Dukare S, Bopp B, Goblirsch M, Jose J, Klempnauer KH. Naphthol AS-E Phosphate Inhibits the Activity of the Transcription Factor Myb by Blocking the Interaction with the KIX Domain of the Coactivator p300. Mol Cancer Ther. 2015;14:1276–1285. doi: 10.1158/1535-7163.MCT-14-0662. [DOI] [PubMed] [Google Scholar]
- 17.Jiang M, Li BX, Xie F, Delaney F, Xiao X. Design, synthesis, and biological evaluation of conformationally constrained analogues of naphthol AS-E as inhibitors of CREB-mediated gene transcription. J Med Chem. 2012;55:4020–4024. doi: 10.1021/jm300043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li BX, Yamanaka K, Xiao X. Structure-activity relationship studies of naphthol AS-E and its derivatives as anticancer agents by inhibiting CREB-mediated gene transcription. Bioorg Med Chem. 2012;20:6811–6820. doi: 10.1016/j.bmc.2012.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie F, Li BX, Broussard C, Xiao X. Identification, synthesis and evaluation of substituted benzofurazans as inhibitors of CREB-mediated gene transcription. Bioorg Med Chem Lett. 2013;23:5371–5375. doi: 10.1016/j.bmcl.2013.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li BX, Xiao X. Discovery of a small-molecule inhibitor of the KIX-KID interaction. Chembiochem : a European journal of chemical biology. 2009;10:2721–2724. doi: 10.1002/cbic.200900552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bijnsdorp IV, Giovannetti E, Peters GJ. Analysis of drug interactions. Methods Mol Biol. 2011;731:421–434. doi: 10.1007/978-1-61779-080-5_34. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrari R, Su T, Li B, Bonora G, Oberai A, Chan Y, et al. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012;22:1212–1221. doi: 10.1101/gr.132308.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res. 2012;40:e31. doi: 10.1093/nar/gkr1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81:467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gang EJ, Hsieh YT, Pham J, Zhao Y, Nguyen C, Huantes S, et al. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene. 2013 doi: 10.1038/onc.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFbeta-SMMHC delays leukemia in mice. Science. 2015;347:779–784. doi: 10.1126/science.aaa0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapias A, Ciudad CJ, Noe V. Transcriptional regulation of the 5'-flanking region of the human transcription factor Sp3 gene by NF-1, c-Myb, B-Myb, AP-1 and E2F. Biochim Biophys Acta. 2008;1779:318–329. doi: 10.1016/j.bbagrm.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Xu H, Inouye M, Hines ER, Collins JF, Ghishan FK. Transcriptional regulation of the human NaPi-IIb cotransporter by EGF in Caco-2 cells involves c-myb. Am J Physiol Cell Physiol. 2003;284:C1262–1271. doi: 10.1152/ajpcell.00456.2002. [DOI] [PubMed] [Google Scholar]
- 35.Pattabiraman DR, Gonda TJ. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 36.Miettinen HM. Regulation of human formyl peptide receptor 1 synthesis: role of single nucleotide polymorphisms, transcription factors, and inflammatory mediators. PLoS One. 2011;6:e28712. doi: 10.1371/journal.pone.0028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portt L, Norman G, Clapp C, Greenwood M, Greenwood MT. Anti-apoptosis and cell survival: a review. Biochim Biophys Acta. 2011;1813:238–259. doi: 10.1016/j.bbamcr.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Wang JM, Chao JR, Chen W, Kuo ML, Yen JJ, Yang-Yen HF. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19:6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eliseev RA, Vanwinkle B, Rosier RN, Gunter TE. Diazoxide-mediated preconditioning against apoptosis involves activation of cAMP-response element-binding protein (CREB) and NFkappaB. J Biol Chem. 2004;279:46748–46754. doi: 10.1074/jbc.M406217200. [DOI] [PubMed] [Google Scholar]
- 40.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 41.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 42.Majeti R, Weissman IL. Human acute myelogenous leukemia stem cells revisited: there's more than meets the eye. Cancer Cell. 2011;19:9–10. doi: 10.1016/j.ccr.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon EM, Raines MA, Blenis J, Sakamoto KM. Granulocyte-macrophage colony-stimulating factor stimulation results in phosphorylation of cAMP response element-binding protein through activation of pp90RSK. Blood. 2000;95:2552–2558. [PubMed] [Google Scholar]
- 44.Sakamoto KM, Fraser JK, Lee HJ, Lehman E, Gasson JC. Granulocyte-macrophage colony-stimulating factor and interleukin-3 signaling pathways converge on the CREB-binding site in the human egr-1 promoter. Mol Cell Biol. 1994;14:5975–5985. doi: 10.1128/mcb.14.9.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong A, Sakamoto KM. Granulocyte-macrophage colony-stimulating factor induces the transcriptional activation of egr-1 through a protein kinase A-independent signaling pathway. J Biol Chem. 1995;270:30271–30273. doi: 10.1074/jbc.270.51.30271. [DOI] [PubMed] [Google Scholar]
- 46.Boulon S, Dantonel JC, Binet V, Vie A, Blanchard JM, Hipskind RA, et al. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol Cell Biol. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desdouets C, Matesic G, Molina CA, Foulkes NS, Sassone-Corsi P, Brechot C, et al. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burch PM, Yuan Z, Loonen A, Heintz NH. An extracellular signal-regulated kinase 1- and 2-dependent program of chromatin trafficking of c-Fos and Fra-1 is required for cyclin D1 expression during cell cycle reentry. Mol Cell Biol. 2004;24:4696–4709. doi: 10.1128/MCB.24.11.4696-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chae HD, Mitton B, Lacayo NJ, Sakamoto KM. Replication factor C3 is a CREB target gene that regulates cell cycle progression through the modulation of chromatin loading of PCNA. Leukemia. 2015;29:1379–1389. doi: 10.1038/leu.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.