Abstract
Biotransformation of mycotoxins in animals comprises phase I and phase II metabolisation reactions. For the trichothecene deoxynivalenol (DON), several phase II biotransformation reactions have been described resulting in DON-glutathiones, DON-glucuronides and DON-sulfates made by glutathione-S-transferases, uridine-diphosphoglucuronyl transferases and sulfotransferases, respectively. These metabolites can be easily excreted and are less toxic than their free compounds. Here, we demonstrate for the first time in the animal kingdom the conversion of DON to DON-3-glucoside (DON-3G) via a model system with plant pathogenic aphids. This phase II biotransformation mechanism has only been reported in plants. As the DON-3G metabolite was less toxic for aphids than DON, this conversion is considered a detoxification reaction. Remarkably, English grain aphids (Sitobion avenae) which co-occur with the DON producer Fusarium graminearum on wheat during the development of fusarium symptoms, tolerate DON much better and convert DON to DON-3G more efficiently than pea aphids (Acyrthosiphon pisum), the latter being known to feed on legumes which are no host for F. graminearum. Using a non-targeted high resolution mass spectrometric approach, we detected DON-diglucosides in aphids probably as a result of sequential glucosylation reactions. Data are discussed in the light of an eventual co-evolutionary adaptation of S. avenae to DON.
Fusarium Head Blight (FHB) is an important disease on small grain cereals such as wheat, barley and oats. Although the disease is caused by a species complex, F. graminearum is considered as the most important species in the complex1. The risk associated with infection by this species comes primarily from the toxic secondary metabolites, mycotoxins, that F. graminearum deposits in the host matrix.
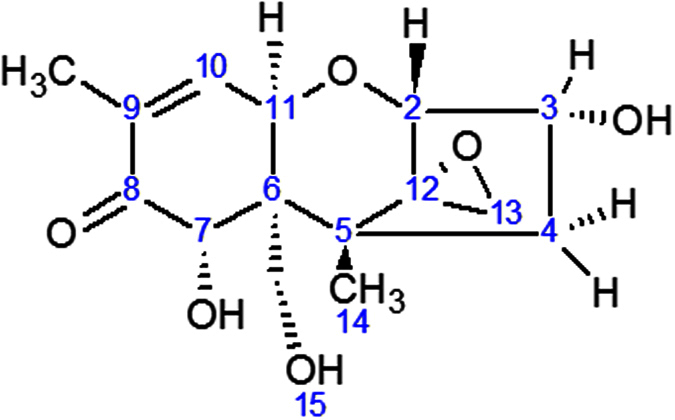
Trichothecenes are a class of mycotoxins produced by several fungal species of the genus Fusarium and related genera in agricultural crops. They belong to the structural group of sesquiterpenoids, all bearing a common tricyclic 12, 13-epoxytrichothec-9-ene core structure. Type A, B, C and D trichothecenes can be distinguished based on substitutions at position C-4, C-7, C-8 and/or C-151. Worldwide the type B trichothecene deoxynivalenol (DON, Fig. 1) is one of the most important mycotoxins because of its omnipresence in many cereal-based matrices1,2,3. DON provokes acute and chronic disease symptoms in humans and animals. Its toxic effects range from diarrhea, vomiting, gastro-intestinal inflammation, necrosis and apoptosis of the intestinal tract, the bone marrow and the lymphoid tissues4,5. DON causes inhibition of the mitochondrial function and has effects on cell division and membrane integrity. Finally, it also inhibits protein, DNA- and RNA synthesis in eukaryotic cells6,7,8. The toxicity of these sesquiterpenes can be explained by their chemical structure containing an epoxide at the C-12 and C-13 position (Fig. 1)9. Although most eukaryotic organisms are susceptible to trichothecenes to a certain level, many of them have developed strategies to arm themselves against the detrimental effects of these mycotoxins, and examples are present throughout the fungal, animal and plant kingdom.
Figure 1. Chemical structure of the mycotoxin deoxynivalenol (DON).
In the yeast Saccharomyces cerevisiae, a spontaneous mutant tolerant to the trichothecene, trichodermin was isolated7. The tolerance was shown to be based on alteration of the target side of trichothecenes. The gene responsible for the trichodermin tolerance was called tcm18 and was suggested to encode for the ribosomal protein L3 (RPL3)10, which is the target of trichothecenes. The DNA sequence of tcm1 was determined11 and a mutation in this gene did not only cause tolerance to trichothecenes, but also affected the maturation of either 40 S or 60 S ribosomal subunits12. Similarly, Mitterbauer et al.13 depicted several mutations in Rpl3 conferring semi-dominant resistance to trichothecenes13. Transgenic tobacco plants expressing a modified Rpl3 cDNA were shown to be able to adapt to DON. However, the tolerance was not consistent because the engineered RPL3 protein which shows a lower affinity to the ribosome assembly factor Rrb1p, was not utilized by the translational machinery in the presence of the native RPL313.
In animals, two major metabolic pathways for detoxification of trichothecenes have been reported. A well-known example is the deepoxidation of the trichothecene DON to deepoxidated DON (DOM-1) in cows and pigs amongst others14. In addition, several type II biotransformation reactions have been reported in which DON is conjugated with glucuronides, sulphonates or glutathione15. In contrast to the vast amount of data on mammals, information on transformation and detoxification strategies in insects remains limited. Nevertheless, insects often live in close proximity of trichothecene-producing fungi and the toxicity has been reported in a few studies. Trichodermin and other 12,13-epoxytrichothecenes have been shown to have larvicidal activity against mosquitoes of Aedes aegypti16. DON has toxic effects on cells of the lepidopteran Spodoptera frugiperda17. In addition, the trichothecenes (type A) diacetoxyscirpenol and neosolaniol are demonstrated to be potent anti-feedants against Galleria mellonella larvae18.
Trichothecenes are also prone to metabolization in plants. The major detoxification process of trichothecenes in plants is through the action of glucosyltransferases which insert a glucose rendering the toxin more water soluble and redirecting it to the vacuoles19. Furthermore, several studies have reported the detection of conjugation products of DON with glutathione (DON-GSH) and the degradation products DON-S-cysteinylglycine (DON-S-cys-gly) and DON-S-cysteine (DON-S-cys)20,21,22.
Finding new detoxification strategies for mycotoxins is a growing field of interest and the first step in order to implement this knowledge in future mycotoxin remediation strategies. New insights on detoxification might come from animals that live in close contact with DON-producing fungi. A well-known example is the English grain aphid (Sitobion avenae) which migrates from the leaves to the developing ear upon its appearance. From that moment, it resides on the same niche as the Fusarium fungus and comes in close contact with its toxic secondary metabolites. The present study aimed to assess the ability of these plant-pathogenic aphids to cope with the trichothecene DON. We demonstrated the presence of type II biotransformation mechanisms linked with DON detoxification in two aphid species. We used a non-targeted high resolution mass spectrometric approach to get an in depth insight into eventual new derived metabolites of DON in animals.
Results
Survival of S. avenae and A. pisum upon exposure to DON
S. avenae and A. pisum aphids were fed for 3 days on a diet containing different concentrations of DON (0, 0.5, 1, and 3 mg l−1) using an aphid feeding apparatus. These concentrations were chosen because they are relevant physiological concentrations in the field: from surveys, it is known that DON levels range from 0 till 5 mg kg−1 at field level (unpublished data). In addition, at the level of a spikelet, these concentrations can mount to 50 mg kg−1 (unpublished data).The survival of S. avenae aphids was not affected by DON up to concentrations of 3 mg l−1 compared to the control when analyzed using a Kruskal-Wallis test (p = 0.337), while survival of A. pisum aphids was significantly reduced. The lowest administered concentration of 0.5 mg l−1 DON significantly reduced (p = 0.024) the survival rate of A. pisum (62% ± 6) compared to the control treatment which was 100% (Table 1). As we wanted to assess the tolerance of S. avenae in more detail, we exposed both aphid species to a concentration of 100 mg l−1 DON for 3 days. Remarkably 43% ± 8 of the S. avenae aphids survived this dose, while for the A. pisum survival rates dropped to 4% ± 2. The LC 50 values of DON for S. avenae and A. pisum were 62.9 mg l−1 and 2.43 mg l−1 respectively.
Table 1. Percentage survival ± S.E. of S. avenae aphids and A. pisum aphids feeding from diet containing different concentrations of deoxynivalenol.
| Concentration | Sitobion avenae | Acyrthosiphon pisum | |
|---|---|---|---|
| Control | 97 ± 3a | 100 ± 0a | |
| DON | 0.5 mg l−1 | 93 ± 3a (p = 1.000) | 62 ± 6bc (p = 0.024) |
| 1 mg l−1 | 93 ± 2a (p = 1.000) | 48 ± 9c (p = 0.006) | |
| 3 mg l−1 | 93 ± 4a (p = 1.000) | 50 ± 11c (p = 0.012) | |
| DON-3G | 0.5 mg l−1 | 88 ± 6a (p = 1.000) | 88 ± 5ab (p = 1.000) |
| 1 mg l−1 | 89 ± 5a (p = 1.000) | 96 ± 2ab (p = 1.000) | |
| 3 mg l−1 | 81 ± 7a (p = 0.160) | 88 ± 6ab (p = 1.000) |
Each treatment consisted of 3 feeding apparatuses each containing 10 nymphs per species. Different letters indicate significant differences (P < 0.05) between treatments using a two-sided non-parametric Kruskall-Wallis test followed by a Dunn’s test for pairwise comparisons. The statistical significances in pairwise comparisons with the control treatments are depicted as a p-value between brackets (this p-value was corrected for multiple pairwise comparisons).
Involvement of RPL3 in the tolerance of S. avenae to DON
Previous research has reported increased tolerance to DON by amino acid modifications in the RPL3 protein which is the target of DON13. For that reason, we sequenced the nucleotide sequence of the gene encoding for RPL3 of S. avenae and A. pisum. After converting the nucleotide sequence to amino acids, no differences were found between the RPL3 sequence of S. avenae when aligned with 60 S RPL3 of A. pisum (NCBI Reference Sequence: XP_001951042.1) and with other insects (Supplementary Figure 1). The typical amino acid changes observed in Saccharomyces cerevisiae which were associated with DON tolerance were not detected in any of the aphid species.
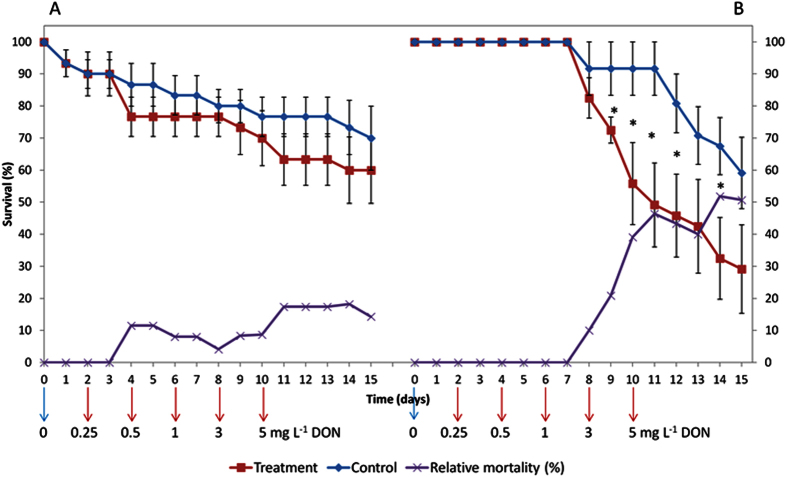
Experiments with transformed plants, have shown that an eventual modified DON insensitive RPL3 protein can be present heterozygously13. In this case, the insensitive RPL3 protein is not used by the translation machinery in the presence of the native RPL3 protein due to a lower affinity of the mutant RPL3 for the ribosome assembly factor Rrb1p. In this scenario, the mutant RPL3 protein only accumulates when organisms are gradually exposed to DON which allows the mutant RPL3 to push out the native RPL3 protein. In order to investigate whether a similar adaptation mechanism was present in A. pisum, aphids were exposed to an increasing concentration of DON in a time-lapse experiment. However, feeding A. pisum with increasing concentrations of DON during a longer period of time did not result in an increased survival (Fig. 2). As expected, the survival of S. avenae was not negatively influenced by increasing DON concentrations.
Figure 2.
Long-term survival of S. avenae aphids (A) and A. pisum aphids (B) (means ± SE) feeding on diet with increasing concentrations of deoxynivalenol (DON) (0 → 5 mg l−1). The purple line indicates the mortality of the aphids relative to the surviving fraction at each time point. *Indicates significant differences (p < 0.05) between treatment and control using one-sided t-tests.
Conversion of DON to DON-3G in aphids results in a detoxification
In order to get an insight into the ability of the aphids to detoxify DON, aphids were fed 100 mg l−1 of DON in an aphid feeding apparatus. In the control aphids which fed on artificial diet only, no DON or DON derivatives were detected. In addition, we tested eventual chemical- or photo-degradation of DON in the artificial diet not exposed to aphids. However, in the artificial diet amended with 100 mg l−1 DON, we found 99.37 ± 0.83 mg l−1 DON illustrating that DON was not degraded in the feeding apparatus during the course of the experiment. Using an enzyme based approach we could not demonstrate an involvement of glutathione-S-transferases (GSTs) or cytochrome P450 monooxygenases (P450s) in the detoxification process (data not shown).
In search for another explanation, a targeted liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) approach was used. Aphids which were exposed to DON were analyzed for DON (LOD, 45 μg kg−1; LOQ, 89 μg kg−1), DON-3G (LOD, 34 μg kg−1; LOQ, 67 μg kg−1) 3- and 15-acetyldeoxynivalenol (3-ADON: LOD, 47 μg kg−1; LOQ, 94 μg kg−1) and 15-ADON (LOD, 33 μg kg−1; LOQ, 67 μg kg−1). For structures see Supplementary Figure 2.
Remarkably, DON-3G, a type II-conjugate of DON that is solely reported in plant detoxification pathways, was detected. Moreover, there was a clear difference in the metabolisation efficiency between both aphid species. For DON-3G A. pisum contained 9.9 ± 1.6 mg/kg while in S. avenae 12.5 ± 3.0 mg/kg DON-3G was present. Remarkably, the DON levels in both aphid species differed significantly (p = 0.017): In A. pisum 9.0 ± 1.5 mg/kg DON was recovered while in S. avenae only 2.5 ± 0.4 mg/kg was detected. This result points to a more efficient conversion of DON to DON-3G in S. avenae. This was even more clear when we considered relative amounts of DON. S. avenae aphids (tolerant to DON) efficiently converted DON to DON-3G (82.5% of the total DON titer was present as DON-3G) and both were significantly different (p = 0.000), whereas in A. pisum only 55% of the total DON titer consisted of DON-3G and DON-3G levels did not differ significantly from DON levels (p = 0.812) (Fig. 3). In view of these results, the toxicity of DON-3G in aphids was assessed by exposing both S. avenae and A. pisum to DON-3G concentrations of 0.5 mg l−1, 1 mg l−1, and 3 mg l−1. These experiments clearly demonstrated that DON-3G was no longer toxic for either aphid species (Table 1).
Figure 3. Relative concentrations (%) of deoxynivalenol (DON), deoxynivalenol-3-glucoside (DON-3G) compared to the total DON titer in S. avenae and A. pisum aphids after two days of feeding on artificial diet amended with 100 mg l−1 DON.
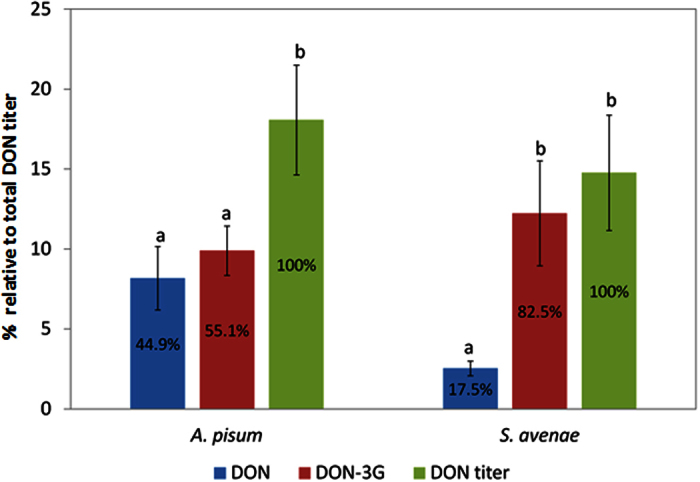
Different letters indicate significant differences (p < 0.05) between treatments using a one-way ANOVA with a post-hoc Tukey test.
DON-3G can be further metabolized: diglucosides of DON
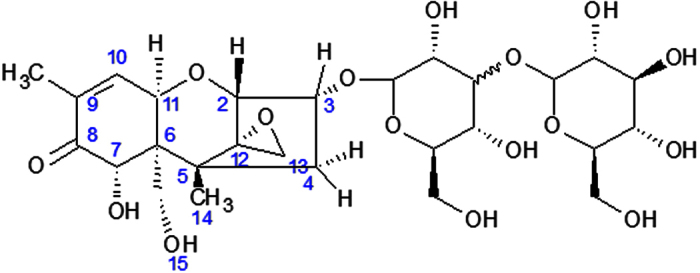
Using a non-targeted high resolution (HR)-MSE approach in the two aphid species fed with DON (100 mg l−1), we were able to recover DON-diglucosides, for which unfortunately reference standards are not available. Other possible conjugates such as DON-GSH (glutathiones) which are known to be produced in other organisms as type II biotransformation products were investigated, however, we were not able to detect DON-GSH in any of the aphid samples confirming the GST enzyme tests. The DON-fed aphid samples were analyzed and discrepancies between aphid species were checked. Three peaks at retention times of 3.98 min, 4.25 min and 4.42 min were observed, corresponding to different structural isomers of DON-diglucoside. For these specific retention times, the measured and theoretical masses were investigated via MassLynxTM software-analysis. The obtained molecular formula was C27H40O16Na+ with a mass of 643.2214 (theoretical) and 643.2202 (measured), resulting in a mass error of −1.9 ppm. The chemical structure of this diglucoside is proposed in Fig. 4. In addition, in view of the presence of DON-3G, we hypothesize here that the presence of the diglucoside form results from a sequential process in which the conversion of DON to DON-3G is the primary step. In order to understand the insertion of the glucose molecules of the three structural isomers, a study via nuclear magnetic resonance (NMR) is interesting. NMR can provide a conclusive structure-elucidation of the complete molecule including spins, alfa- and beta-isomerization, etc. However, it would be limited due to the extremely small amounts that can be isolated from aphid samples.
Figure 4. Proposed chemical structure of deoxynivalenol-diglucoside.
Discussion
The trichothecene DON is a sesquiterpenoid mycotoxin produced by several Fusarium species and is toxic for most eukaryotic cells. In the present study, we assessed the toxicity of DON for two aphid species: the English grain aphid S. avenae and the pea aphid A. pisum. Dietary exposure of both aphids to DON showed that S. avenae were tolerant to DON compared to A. pisum. To explain this unique difference in toxicity of DON between the two aphid species three hypotheses were tested.
First, we examined the amino acid sequence of the gene encoding RPL3, the target molecule of DON, in both aphid species. Trichodermin and other sesquiterpenoids of the same group are known inhibitors of the peptidyltransferase center of eukaryotic ribosomes, and thereby block protein synthesis23,24. Research showed that RPL3 plays an essential role in the formation of this peptidyltransferase center10,25,26. One of the resistance mechanisms to DON identified in yeast is the modification of this ribosomal target by amino acid changes in RPL327. Mitterbauer et al.13 used yeast as a model system to identify several mutations in the gene encoding RPL3 (e.g. W255C, a change of tryptophan into cysteine at position 255), which confer resistance to trichothecenes, in particular to DON. However, the amino acid sequence of S. avenae’s RPL3 showed none of these mutations. In addition, no functional aberrations were observed between the amino acid sequence of RPL3 from S. avenae and the predicted RPL3 from A. pisum. Amino acids at places 190 and 382 are valine (V) for A. pisum, but isoleucine (I) for S. avenae. However, when comparing the chemical structure of these two amino acids we concluded that this cannot explain the improved survival of S. avenae in presence of DON compared to A. pisum. Not the whole amino acid sequence of the S. avenae RPL3 was picked up, leaving seven amino acids undetermined at the end. As our sequencing data of the RPL3 of S. avenae did not show the mutations reported in yeast, we can conclude that the target of DON, the gene encoding RPL3, is not the reason of the tolerance in grain aphids.
Second, we investigated the hypothesis of Mitterbauer et al.13 stating that organisms might be heterozygous for the RPL3 locus. In this hypothesis, native ribosomes originating from one allele could be preferentially dismantled and degraded in vivo upon DON exposure. In consequence, the remaining fraction of resistant ribosomes on the second allele could allow the synthesis of new ribosomal proteins, eventually leading to a higher and steady-state level of modified RPL3 protein in the ribosomes. This hypothesis was validated by these researchers27 via an engineered tomato RPL3 containing the mutations of the yeast RPL3, which resulted in an adaptation but not in a constitutive tolerance against DON pointing to the semi-dominant nature of this tolerance. Expression of this aberrant gene in tobacco showed that the aberrant RPL3 protein was not utilized when the wild-type RPL3 protein was present, unless the transgenic plants were challenged with sub-lethal amounts of DON. Indeed, after toxin treatment in a dose-dependent manner, they noticed an accumulation of the modified protein due to the selection pressure in the presence of DON13. We investigated this hypothesis with our two aphid species in an experimental setup where we fed the aphids with increasing DON-concentrations over a period of 2 weeks. However, we did not detect any augmented tolerance especially in A. pisum when gradually exposed to increasing DON doses.
Finally, we investigated whether DON was subject to a type II biotransformation process in aphids. Metabolisation of the trichothecene monoacetoxyscirpenol in insects has been reported once before in literature although the type of metabolisation nor the detoxification was addressed in detail at that time28,29. In general, detoxification of xenobiotics is well-known in insects. Some major groups of genes encoding metabolic enzymes to detoxify xenobiotics like insecticides and plant derived metabolites have been described i.e., glucosyltransferases esterases, P450s, and GSTs30,31,32.
We were able to demonstrate the presence of DON-3G in both aphid species pointing to a glucosyltransferase involved in the detoxification of DON in aphids. Moreover, the tolerant S. avenae species converted DON to DON-3G more efficiently than the susceptible A. pisum. To our knowledge, this is the first time that the conversion of DON to DON-3G is reported in animal species. To date, DON glucosylation has solely been reported in plant cells. In plants, a vast number of genes that code for putative UDP-glycosyltransferases (UGTs) has been revealed19. In barley and Brachypodium distachyon, genes from the UGT family with potential relevance for DON tolerance have functionally been characterized33,34. Although DON glucosylation has never been reported in animals, genes encoding for UGTs are known to be present in insects. They catalyze the conjugation of a range of diverse small lipophilic compounds with polar compounds (i.e. carbohydrates) to produce glucosides, and as such they play an important role in type II detoxification processes of xenobiotics in insects35. However, the presence of these UGTs have never been linked with mycotoxin glucosylation. It has been shown that the insect UGT enzymes typically use UDP-glucose as the main sugar donor unlike vertebrate UGTs which mainly utilize UDP-glucuronic acid36. In aphids, up-regulated ecdysteroid UDP-glucosyltransferases have been associated with tolerance of cotton aphids to the neonicotinoid insecticide thiamethoxam. In addition, they have been shown to be expressed upon migration of aphids from primary to secondary hosts37. These examples illustrate the plethora of biological processes in which UGT enzymes are involved.
Although we provide valuable evidence for a role of glucosylation in DON-detoxification in aphids, several other detoxification enzymes have been described in aphids for coping with xenobiotics (e.g. secondary compounds from plants or insecticides); including P450s, GSTs, esterases and oxidoreductases38,39,40,41,42,43. Some of these enzymes are known to be involved in the detoxification of mycotoxins20. In honeybees Apis mellifera, the larvae of Trichoplusia ni and corn earworms Helicoverpa zea, it has been demonstrated that P450s play a role in bio-activation or detoxification of aflatoxin B1 produced by Aspergillus spp44,45. However, in this study we showed that GSTs and P450s were not involved.
Using a non-targeted HR-MSE approach, we were able to detect DON-diglucosides (via HR-MSE) in both S. avenae and A. pisum aphids. To date, the only report on the presence of DON-diglucosides was in beer, where oligoglucosylated DONs with up to four bound hexose units were present46. Remarkably, although detoxification of DON through conjugation with glutathione (DON-GSH) has been observed in plants20,21 and in many animal species15 no glutathione derivatives of DON were observed during the HR-MSE analyses of our aphid samples.
The question remains why S. avenae is able to convert DON to DON-3G more efficiently than A. pisum. Insights might come from the knowledge that S. avenae occurs on cereal ears which are often colonized by Fusarium spp. producing DON, while A. pisum occurs on plant species that are no hosts for DON producing Fusarium spp47. It is remarkable that A. pisum although it disposes of a very large arsenal of UGTs compared to other insects35, does not convert DON efficiently to DON-3G which points to the substrate specificity of these enzyme. Consequently, we might speculate on adaption by co-evolution in S. avenae. It has been reported before that insects are capable to develop tolerance when exposed to a toxin over many generations. Drosophila melanogaster larvae which were exposed to Aspergillus nidulans over 26 generations displayed higher survival rates in the presence of A. nidulans and a higher tolerance to the mycotoxin sterigmatocytin (i.e. an aflatoxin precursor) compared to control lines48.
Finally, it is tempting to argue on the origin of the glucosyltransferase in aphids. Although we do not provide evidence in the present study, one of the possibilities of acquiring this specific glucosyltransferase is through horizontal gene transfer. Horizontal transfer of a bacterial gene encoding an enzyme which detoxifies the plant toxin hydrogen cyanide to plant-feeding spider mites (Tetranychus urticae) has previously been shown in a model system using Phaseolus lunatus. This event resulted in an increased survival upon exposure of the insects to hydrogen cyanide49. In our model system using aphids, a potential source of bacterial enzymes degrading the mycotoxin DON are endosymbionts. Aphids are insects known to contain many obligatory (e.g. Buchnera aphidicola) and facultative symbionts50. The latter group is mutualistic in the context of various ecological interactions. Although it has been suggested that they are involved in detoxification of various toxic metabolites, solid evidence for this feature is still lacking. Due to this, the direct or indirect (e.g. though horizontal gene transfer) role of symbionts in detoxification processes remains enigmatic50,51. Nevertheless, this discussion illustrates a possible new aspect of the chemical warfare between plants and insects.
Methods
Insects and chemicals
Laboratory stock cultures of cereal aphids S. avenae and pea aphids A. pisum were maintained on wheat seedlings and young broad bean plants, respectively, under standard conditions of 22 °C and a photoperiod of 16 h light with a relative humidity ranging from 50% to 70%, stimulating parthenogenesis52.
DON was kindly provided by M. Lemmens (BOKU, Vienna, Austria). Purity of the provided stock standard was >99%. A stock solution was prepared by dissolving 5 mg DON in 5 ml (1 mg ml−1) sterile water and stored at −20 °C. DON-3G (50 μg ml−1) was purchased at Sigma-Aldrich (Diegem, Belgium).
Survival of S. avenae and A. pisum when feeding from DON and DON-3G
To determine the effect of DON and DON-3G on the survival of S. avenae and A. pisum, DON and DON-3G were added to the artificial aphid diet based on formulation A from Prosser and Douglas53 to a final concentration of 0.5, 1 or 3 and 100 mg l−1. Sterile water was added to the artificial diet as control (0 mg l−1 DON or DON-3G). For both aphid species, there were three aphid feeding apparatuses prepared as described by Sadeghi, et al.54 for all treatments and control. Each apparatus contained ten randomly picked nymphs who could feed on a parafilm sachet containing 200 μl of the diet. Over a period of three days the surviving nymphs were counted. Abbott’s formula55 was used to correct the survival rates: (nTa/nCa)* 100, nTa represents the number of survivors after treatment and nCa the number of survivors in the control treatment. Statistical differences (P < 0.05) between aphid survival when feeding from different DON concentrations were analyzed by using non-parametric Kruskal-Wallis analysis followed by a Dunn’s test to perform pairwise comparisons using IBM SPSS (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp.). These experiments were repeated at least two times with 4 samples per treatment.
To determine the long-term effect of DON on the survival of S. avenae and A. pisum, aphids were gradually exposed to increasing concentrations: 0 → 0.5 → 1 → 3 → 5 mg l−1 DON in aphid feeding apparatus54. For both aphid species, there were six aphid feeding apparatus prepared for the treatment and for the control (0 mg l−1 DON). Each apparatus contained five nymphs produced by adult aphids within 24 h (day 0). In these experiments, the survival of the nymphs was checked once every day and the diet was changed every two days. Statistical different survival percentages (P < 0.05) between treatments at different time points in the long-term survival experiment were analyzed using one-sided t-tests (SPSS Statistics 22).
For non-targeted and targeted LC-MS/MS analyses S. avenae and A. pisum aphids (in different developmental stages) were taken from the laboratory stock cultures and put in aphid feeding apparatuses containing 0 or 100 mg l−1 DON. After 40 h the surviving aphids were stored at −20 °C until analysis. For each aphid species we performed five replications of aphids that were fed with 0 and 100 mg l−1 DON. In addition, after 40 h of feeding the artificial diet of all repeats were analyzed.
Analysis of the ribosomal protein L3
RNA from S. avenae and A. pisum aphids was extracted using TRI reagent (Sigma-Aldrich) according to the manufacturer’s instructions. The extracted RNA was quantified using a Quantus Fluorometer (Promega, Madison, WI, USA). With a GoScript Reverse Transcription System (Promega), first-strand cDNA was synthesized. The PCR reactions were performed in a total reaction volume of 25 μl, consisting of 0.125 μl GoTaq DNA polymerase (Promega), 5 μl of 5× GoTaq buffer colorless (Promega), 1.25 μl dNTPs, 1 μl of each primer (5 μM), 14.625 μl nuclease-free water (Promega) and 2 μl of the cDNA. The RPL3 sequence was picked up in two parts (p1 and p2) using following primers (5′-3′): p1_F GCACATCCACTTTCGTCAAG, p1_R CTAGGATGCCATGCTCCAAT, p2_F ACCAAGGGTCGTGGATACAA and p2_R CGCTGTGGCTTTCTCTTCTT. PCR analysis was performed with a Bio-Rad T100 Thermal Cycler, the thermocycler profile used was: 5 min at 95 °C followed by 35 cycles at 95 °C for 30 s, 59.7 °C (p1) or 60 °C (p2) for 20 s and 72 °C for 60 s, followed by 72 °C for 10 min and cooled down to 15 °C. The remaining product was purified using the E.Z.N.A. Cycle-Pure Spin kit (VWR, Leuven, Belgium) and send to LGC Genomics (Berlin, Germany) for single sample DNA sequencing. Sequence alignment between S. avenae and A. pisum was done with ClustalW Multiple alignment in Bioedit.
Sample preparation and targeted LC-MS/MS analysis
Individual mycotoxin solid standards (1 mg) of DON, 3-ADON, 15-ADON and DOM-1 (internal standard) were purchased from Sigma-Aldrich NV/SA (Bornem, Belgium). DON-3G (50.2 ng μl−1 in acetonitrile) was obtained from Biopure Referenzsubstanzen GmbH (Tulln, Austria). All mycotoxin solid standards were dissolved in methanol (1 mg ml−1), and were stored at −18 °C. Working solutions of DON, 3-ADON, 15-ADON and DOM (10 ng μl−1) were prepared in methanol and stored at −18 °C, while DON-3G (50.2 ng μl−1) was dissolved in acetonitrile and stored at 4 °C. The targeted LC-MS/MS analysis was performed using a Waters Acquity UPLC system coupled to a Quattro Premier XE mass spectrometer (Waters, Milford, MA, USA) equipped with an electrospray interface in positive mode (ESI+). Following MRM-traces were monitored: DON (297 > 203.3; 249.4), 3-ADON (339.2 > 231.2; 203.1), 15-ADON (339.1 > 137.1; 321.2) and DON-3G (476.1>248.6 ; 296.9). LC-MS/MS parameters are described in detail by De Boevre, et al.56. MassLynxTM version 4.1. and QuanLynx® version 4.1. software (Waters, Milford, MA, USA) were used for data acquisition and processing.
Aphid samples were collected, crushed and individually weighed in recipients. According to their weight, 500 ng g−1 of DOM internal standard (10 ng μl−1) was added. A matrix-matched calibration curve with a linear range of 0 ng g−1 to 1500 ng g−1 for DON, 3-ADON, 15-ADON and DON-3G with non-contaminated S. avenae aphids was prepared. The reference standards were allowed to equilibrate for 15 min. An extraction with 1.5 ml acetonitrile/water/acetic acid (79/20/1, v/v/v) was performed, and the samples were vigorously vortexed for 1 min. The sample extract was centrifuged at 4,307 g for 1 min and the supernatant was collected in a small test tube using a glass Pasteur pipette with a bulb. This process was repeated twice. The organic mycotoxin-mixture was evaporated until dry under N2 at 60 °C using the TurboVap® LV (Biotage, Dusseldorf, Germany), and redissolved in 150 μl of injection solvent (50/50 v/v, H2O/MeOH (95/5, v/v), 0.1% of HCOOH + 10 mM of HCOONH4 [solvent A]; MeOH/H2O (95/5, v/v), 0.1% of HCOOH + 10 mM of HCOONH4 [solvent B]). Finally, the redissolved sample was vortexed for 3 minutes, collected in an Ultrafree-MC centrifugal device (0.22 μm, Millipore, Bedford, MA, USA) and centrifuged for 10 minutes at 10,000 g.
To confirm the presence of DON, 3-ADON, 15-ADON and DON-3G, two transitions between precursor and fragments were monitored. According to the Commission Decision of the Council Directive 96/23/EC in August 12, 2002 concerning the performance of analytical methods and the interpretation of results (2002/657/EC, 2002), a system of identification points was applied to interpret the data57. The first criterion indicates that the relative retention time, relative to the internal standard DOM-1, should not exceed 2.5%. The second identification point involved that the relative abundance of both transitions should not exceed the range of 20% to 50%, depending on the relative intensity between the transitions. Also, all MRM-transitions should possess a signal-to-noise (s/n) ratio higher than 3:1 (2002/657/EC, 2002).
Statistical differences (P < 0.05) between concentrations of DON, DON-3G and total titer retrieved in aphids were analyzed by using a one-way ANOVA with a post-hoc Tukey test (SPSS Statistics 22).
Sample preparation and non-targeted LC-MSE analysis
To an exact amount of the aphid sample (individually checked), 750 μl of extraction solvent acetonitrile/water/acetic acid (79/20/1, v/v/v) was added. Using a glass spatula, the aphids samples were crushed until a homogeneous mass was obtained. The spatula was rinsed with 750 μl of extraction solvent. The organic mixture was vigorously vortexed for 1 minute, followed by centrifugation at 4,307 g for 1 minute. The obtained supernatant was transferred into a small test tube. To extract the maximum amount of mycotoxins, 1.5 ml of extraction solvent was added to the centrifuged residue. The vortex and centrifugation step were repeated, and the remaining supernatant was transferred into the same test tube. The organic mycotoxin-mixture was evaporated until dryness under N2 at 60 °C using the TurboVap® LV (Biotage, Dusseldorf, Germany). The residue was redissolved with 150 μl of MeOH/CAN/H2O (30/30/40, v/v/v) and centrifuged in an Ultrafree®-MC centrifugal device (0.22 μm) for 5 minutes at 14,000 g.
DON and its derivatives (DON-3G, 3-ADON, 15-ADON, DON-GSH, DON-diglucosides, DON-triglucosides and DON-tetraglucosides) were investigated using UPLC/Q-TOF-MS with the MSE data acquisition strategy. The LC instrument used was an Acquity UPLCTM system (Waters Milford, MA, USA) with a ZORBAX RRHD Eclipse Plus C18 (1.8 μm, 2.1 × 100 mm) from Agilent Technologies (Diegem, Belgium). The mobile phase consisted of H2O/MeOH (95/5, v/v) containing 0.1% of HCOOH and 10 mM of HCOONH4 [solvent A] and MeOH/H2O (95/5, v/v) containing 0.1% of HCOOH and 10 mM of HCOONH4 [solvent B]. The following gradient elution program was applied: 0–0.5 min: 0% B, 0.5–20 min: 0–99% B, 20–21 min: 99% B, 21–24 min: 0% B, 24–28 min: 0% B. The flow rate was 0.4 ml min−1. The column temperature was set at 30 °C, and the temperature of the autosampler was 10 °C. Five μl of the sample was injected. Instrument control and data processing were carried out by MassLynxTM version 4.1. software (Waters, Milford, MA, USA). The Q-TOF MS instrument used was a Synapt G2-Si MS system (Waters, Milford, MA, USA). The data acquisition mode was TOF MSE in ESI+ mode. The data acquisition ranged from 50 Da to 1200 Da with a 0.1 s scan time. The MS source temperature was set at 150 °C, and the desolvation temperature was set at 500 °C with a desolvation gas flow set at 800 l h−1 and a cone gas flow at 100 l h−1. The capillary voltage was 2.8 kV and the sampling cone voltage was 30 V. The collision energy was set as 45 eV–60 eV ramp (trap) for the high-energy scan. Data was collected in continuum mode and the mass was corrected to ensure accuracy during the MS analysis after acquisition using leucine enkephaline (200 pg μl−1) at a flow rate of 100 μl min−1 as lock mass compound. HRMS data were processed using MassLynxTM software and compounds were identified after applying lockspray correction, extracting the chromatogram and generating the molecular formula from the exact mass.
Additional Information
How to cite this article: Zutter, N. D. et al. Aphids transform and detoxify the mycotoxin deoxynivalenol via a type II biotransformation mechanism yet unknown in animals. Sci. Rep. 6, 38640; doi: 10.1038/srep38640 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Nathalie De Zutter is holder of a Ph.D. grant by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT). The authors would like to thank Maarten Ameye, Jolien Scheerlinck and Tom Devos for their contributions to this article. The authors also acknowledge the Hercules infrastructure funding AUGE/13/13 and the Special Research Fund of Ghent University.
Footnotes
The authors declare no competing financial interests.
Author Contributions N.D.Z., K.A., G.H. and G.S. conceived and designed the experiments. N.D.Z. planned and performed the experiments, analyzed and interpreted the data and wrote the manuscript. N.A-M., M.D.B. and C.V.P. performed the (non)targeted LC-MS/MS experiments. M.D.B., K.A., S.D.S., G.H. and G.S. contributed to the writing the manuscript, supervised the study and helped interpreting the data. All authors reviewed the manuscript.
References
- Goswami R. S. & Kistler H. C. Heading for disaster: Fusarium graminearum on cereal crops. Molecular Plant Pathology 5, 515–525 (2004). [DOI] [PubMed] [Google Scholar]
- De Boevre M. et al. Natural occurrence of mycotoxins and their masked forms in food and feed products. World Mycotoxin Journal 5, 207–219 (2012). [Google Scholar]
- Bottalico A. & Perrone G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. European Journal of Plant Pathology 108, 611–624 (2002). [Google Scholar]
- Bennett J. W. & Klich M. Mycotoxins. Clinical Microbiology Reviews 16, 497–516 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka J. J. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin Journal 3, 323–347 (2010). [Google Scholar]
- Rocha O., Ansari K. & Doohan F. M. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Additives & Contaminants 22, 369–378 (2005). [DOI] [PubMed] [Google Scholar]
- Jimenez A., Sanchez L. & Vazquez D. Simultaneous ribosomal resistance to trichodermin and anisomycin in Saccharomyces cerevisiae mutants. Biochimica Et Biophysica Acta 383, 427–434 (1975). [DOI] [PubMed] [Google Scholar]
- Grant P. G., Schindler D. & Davies J. E. Mapping of trichodermin resistance in Saccaromyces cerevisiae genetic locus for a component of 60S ribosomal subunit. Genetics 83, 667–673 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A. E., Hohn T. M. & McCormick S. P. Trichothecene biosynthesis in Fusarium species - Chemistry, genetics and significance. Microbiological Reviews 57, 595–604 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M. & Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences 78, 238–242 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D. & Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccaromyces cerevisiae. Journal of Bacteriology 155, 8–14 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lobato M., Cannon M., Mitlin J. A., Mount R. C. & Jimenez A. Characterization of Saccharomyces cerevisiae strains displaying high-level or low-level resistance to trichothecene antibiotics. Biochemical Journal 267, 709–713 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterbauer R. et al. Toxin-dependent utilization of engineered ribosomal protein L3 limits trichothecene resistance in transgenic plants. Plant Biotechnology Journal 2, 329–340 (2004). [DOI] [PubMed] [Google Scholar]
- Wu D. H. et al. Metabolic pathways of trichothecenes. Drug Metabolism Reviews 42, 250–267 (2010). [DOI] [PubMed] [Google Scholar]
- Wen J. K., Mu P. Q. & Deng Y. Q. Mycotoxins: cytotoxicity and biotransformation in animal cells. Toxicolology Research 5, 377–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J. F. & Hosken M. Larvicidal activity of some 12,13-epoxytrichothec-9-enes. Biochemical Pharmacology 24, 959–962 (1975). [DOI] [PubMed] [Google Scholar]
- Fornelli F., Minervini F. & Mule G. Cytotoxicity induced by nivalenol, deoxynivalenol, and fumonisin B, in the SF-9 insect cell line. In Vitro Cellular and Developmental Biology - Animal 40, 166–171 (2004). [DOI] [PubMed] [Google Scholar]
- Mule G., Dambrosio A., Logrieco A. & Bottalico A. Toxicity of mycotoxins of Fusarium sambucinum for feeding in Galleria mellonella. Entomologia Experimentalis Et Applicata 62, 17–22 (1992). [Google Scholar]
- Berthiller F. et al. Masked mycotoxins: A review. Molecular Nutrition and Food Research 57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner S. A. et al. Transcriptome analysis of the barley-deoxynivalenol interaction: evidence for a role of glutathione in deoxynivalenol detoxification. Molecular Plant-Microbe Interactions 23, 962–976 (2010). [DOI] [PubMed] [Google Scholar]
- Kluger B. et al. Stable isotopic labelling-assisted untargeted metabolic profiling reveals novel conjugates of the mycotoxin deoxynivalenol in wheat. Analytical and Bioanalytical Chemistry 405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder P., Scheer C. E., Diekmann F. & Stampfl A. How plants cope with foreign compounds - Translocation of xenobiotic glutathione conjugates in roots of barley (Hordeum vulgare). Environmental Science and Pollution Research 14, 114–122 (2007). [DOI] [PubMed] [Google Scholar]
- Barbacid M. & Vazquez D. Binding of acetyl-C-14 trichodermin to peptidyl transferase center of eukaryotic ribosomes. European Journal of Biochemistry 44, 437–444 (1974). [DOI] [PubMed] [Google Scholar]
- Carrasco L., Barbacid M. & Vazquez D. Trichodermin group of antibiotics, inhibitors of peptide-bond formation by eukaryotic ribosomes. Biochimica Et Biophysica Acta 312, 368–376 (1973). [DOI] [PubMed] [Google Scholar]
- Schulze H. & Nierhaus K. H. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. Embo Journal 1, 609–613 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskas A. & Dinman J. D. Ribosomal protein L3: Gatekeeper to the a site. Molecular Cell 25, 877–888 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterbauer R. & Adam G. Saccharomyces cerevisae and Arabidopsis thaliana: Useful model systems for the identification of molecular mechanisms involved in resistance of plants to toxins. European Journal of Plant Pathology 108, 699–703 (2002). [Google Scholar]
- Karlovski P. Biological detoxification of the mycotoxin deoxynivalenol and its use in genetically engineered crops and feed additives. Applied Microbiology and Biotechnology 91, 491–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd P. F. & Vanmiddlesworth F. L. In vitro metabolism of the trichothecene 4-monoacetoxyscirpenol by fungus-feeding and non-fungus-feeding insects. Experientia 45, 292–395 (1989). [Google Scholar]
- Schuler M. A. P450s in plant-insect interactions. BBA-Proteins Proteomics 1814, 36–45 (2011) [DOI] [PubMed] [Google Scholar]
- Schuler M. A. & Berenbaum M. R. Structure and function of P450s in insect adaptation to natural and synthetic toxins: insights gained from molecular modeling. Journal of Chemical Ecology 39, 1232–1245 (2013). [DOI] [PubMed] [Google Scholar]
- Li X. C., Schuler M. A. & Berenbaum M. R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics Annual Review of Entomology 52, 231–251 (2007). [DOI] [PubMed] [Google Scholar]
- Schweiger W. et al. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Molecular Plant-Microbe Interactions 26, 781–792 (2013). [DOI] [PubMed] [Google Scholar]
- Schweiger W. et al. Validation of a candidate deoxynivalenol-inactivating UDP-glucosyltransferase from barley by heterologous expression in yeast. Molecular Plant-Microbe Interactions 23, 977–986 (2010). [DOI] [PubMed] [Google Scholar]
- Ahn S. J., Vogel H. & Heckel D. G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochemistry and Molecular Biology 42, 133–147 (2012). [DOI] [PubMed] [Google Scholar]
- Bock K. W. Vertebrate UDP-glucuronosyltransferases: functional and evolutionary aspects. Biochemical Pharmacology 66, 691–696 (2003). [DOI] [PubMed] [Google Scholar]
- Lukasik I. Effect of host plant alternation on some adaptive enzymes of the bird cherry oat aphid, Rhopalosiphum padi (L.) Journal of Pest Science 82, 203–209 (2009). [Google Scholar]
- Zhang M. X. et al. Xenobiotic metabolism of plant secondary compounds in the English grain aphid, Sitobion avenae (F.) (Hemiptera: Aphididae). Pesticide Biochemistry and Physiology 107, 44–49 (2013). [DOI] [PubMed] [Google Scholar]
- Cai Q. N. et al. Detoxification of gramine by the cereal aphid Sitobion avenae. Journal of Chemical Ecology 35, 320–325 (2009). [DOI] [PubMed] [Google Scholar]
- Lu Y. & Gao X. Multiple mechanisms responsible for differential susceptibilities of Sitobion avenae (Fabricius) and Rhopalosiphum padi (Linnaeus) to pirimicarb. Bulletin of Entomological Research 99 (2009). [DOI] [PubMed] [Google Scholar]
- Castaneda L. E., Figueroa C. C. & Nespolo R. F. Do insect pests perform better on highly defended plants? Costs and benefits of induced detoxification defences in the aphid Sitobion avenae. Journal of Evolutionary Biology 23, 2474–2483 (2010). [DOI] [PubMed] [Google Scholar]
- Castaneda L. E., Figueroa C. C., Fuentes-Contreras E., Niemeyer H. M. & Nespolo R. F. Energetic costs of detoxification systems in herbivores feeding on chemically defended host plants: a correlational study in the grain aphid. Sitobion avenae. Journal of Experimental Biology 212, 1185–1190 (2009). [DOI] [PubMed] [Google Scholar]
- Figueroa C. C., Koenig C., Araya C., Santos M. J. & Niemeyer H. M. Effect of dimboa, a hydroxamic acid from cereals, on peroxisomal and mitochondrial enzymes from aphids: Evidence for the presence of peroxisomes in aphids. Journal of Chemical Ecology 25, 2465–2475 (1999). [Google Scholar]
- Sen Zeng R., Wen Z. M., Niu G. D. & Berenbaum M. R. Aflatoxin B1: Toxicity, bioactivation and detoxification in the polyphagous caterpillar Trichoplusia ni. Insect Science 20, 318–328 (2013). [DOI] [PubMed] [Google Scholar]
- Niu G. D., Johnson R. M. & Berenbaum M. R. Toxicity of mycotoxins to honeybees and its amelioration by propolis. Apidologie 42, 79–87 (2011). [Google Scholar]
- Zachariasova M., Vaclavikova M., Lacina O., Vaclavik L. & Hajslova J. Deoxynivalenol oligoglycosides: new “masked” Fusarium toxins occurring in malt, beer, and breadstuff. J. Agricultural and Food Chemistry 60, 9280–9291 (2012). [DOI] [PubMed] [Google Scholar]
- Ferrari J. et al. Population differentiation and genetic variation in host choice among pea aphids from eight host plant genera. Evolution 60, 174–1584 (2006). [PubMed] [Google Scholar]
- Trienens M. & Rohlfs M. Experimental evolution of defense against a competitive mold confers reduced sensitivity to fungal toxins but no increased resistance in Drosophila larvae. Bmc Evolutionary Biology 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw N. et al. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. Elife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen A. K. & Moran N. A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Molecular Ecology 23, 1473–1496 (2014). [DOI] [PubMed] [Google Scholar]
- Oliver K. M., Degnan P. H., Burke G. R. & Moran N. A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annual Review of Entomology 55, 247–266 (2010) [DOI] [PubMed] [Google Scholar]
- De Zutter N., Audenaert K., Haesaert G. & Smagghe G. Preference of cereal aphids for different varieties of winter wheat. Arthropod-Plant Interactions 6, 345–350 (2012). [Google Scholar]
- Prosser W. A. & Douglas A. E. A test of the hypotheses that nitrogen is upgraded and recycled in an aphid (Acyrthosiphon pisum) symbiosis. Journal of Insect Physiology 38, 93–99 (1992). [Google Scholar]
- Sadeghi A., Van Damme E. J. M. & Smagghe G. Evaluation of the susceptibility of the pea aphid, Acyrthosiphon pisum, to a selection of novel biorational insecticides using an artificial diet. Journal of Insect Science 9, 1–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott W. S. A method for computing the effectiveness of an insecticide. Journal of Economical Entomology 18, 265–267 (1925). [Google Scholar]
- De Boevre M. et al. Development and validation of an LC-MS/MS method for the simultaneous determination of deoxynivalenol, zearalenone, T-2-toxin and some masked metabolites in different cereals and cereal-derived food. Food Additives and Contaminants Part A-Chemistry 29, 819–835 (2012). [DOI] [PubMed] [Google Scholar]
- European Commission (Official Journal European Communities L221, 8–36., 2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.