Abstract
Determining the site of a regulatory phosphorylation event is often essential for elucidating specific kinase–substrate relationships, providing a handle for understanding essential signaling pathways and ultimately allowing insights into numerous disease pathologies. Despite intense research efforts to elucidate mechanisms of protein phosphorylation regulation, efficient, large-scale identification and characterization of phosphorylation sites remains an unsolved problem. In this report we describe an application of existing technology for the isolation and identification of phosphorylation sites. By using a strategy based on strong cation exchange chromatography, phosphopeptides were enriched from the nuclear fraction of HeLa cell lysate. From 967 proteins, 2,002 phosphorylation sites were determined by tandem MS. This unprecedented large collection of sites permitted a detailed accounting of known and unknown kinase motifs and substrates.
Keywords: phosphorylation, mass spectrometry, strong cation exchange chromatography
Much of eukaryotic protein regulation occurs when protein kinases add a phosphate moiety in an ATP-dependent manner to a Ser, Thr, or Tyr residue of a substrate protein. Not surprisingly, malfunctions in this critical cellular process have been implicated as causal factors in diseases, such as diabetes, cancer, and Alzheimer's. With >500 identified kinases and thousands of potential substrates, these proteins remain attractive drug targets. Large-scale identification of phosphorylated kinase substrates will certainly enhance our understanding of diverse biological phenomena, potentially leading to targeted intervention in any number of disease paradigms.
The identification of phosphorylation sites is most robustly accomplished by MS (1, 2). With tandem MS (MS/MS), phosphopeptides are fragmented to determine their sequence and to pinpoint the specific Ser, Thr, or Tyr modified by a protein kinase. Despite many reports of thousands of identified proteins from a single biological sample, the large-scale determination of phosphorylation sites is just emerging. To date, the three largest reported repositories of identified sites are from yeast and plant studies [383 (3),125 (4) and ≈200 (5)], whereas the most phosphorylation sites identified from a single human sample stands at 64 (6). Clearly, to study the rich biology relying on protein phosphorylation will require more effective methodologies.
Uninformative fragmentation is a fundamental obstacle to phosphorylation site analysis, regardless of the scale of an experiment. Fragmentation of phosphopeptides by collision-induced dissociation by MS/MS commonly results in the production of a single dominant peak corresponding to a neutral loss of phosphoric acid (H3PO4, 98 Da) from the phosphopeptide (for example, see Fig. 2B). The lack of informative fragmentation at the peptide backbone severely reduces the ability of database searching algorithms to unambiguously identify the phosphopeptide. Furthermore, when a phosphopeptide is identified, it is often not possible to assign the site to a particular Ser, Thr, or Tyr residue because of the lack of informative fragmentation (2).
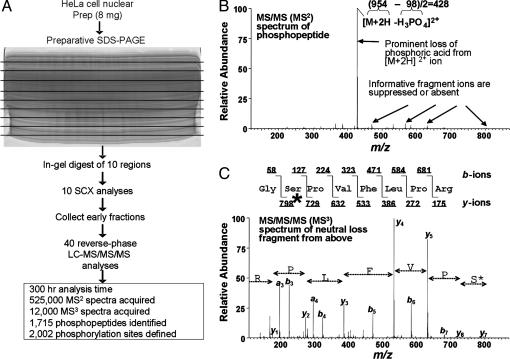
Fig. 2.
Analysis of human nuclear phosphorylation sites by multidimensional LC coupled to MS/MS/MS. (A) Eight milligrams of nuclear extract from asynchronous HeLa cells were separated by SDS/PAGE. The entire gel was excised into 10 regions and proteolyzed with trypsin followed by phosphopeptide enrichment by SCX LC. Early eluting fractions were subjected to amino acid sequence analysis by reverse-phase LC-MS/MS with data-dependent MS/MS/MS acquisition. Phosphorylation sites (n = 2,002) were identified by the sequest algorithm, acquisition of MS/MS/MS spectra, and manual validation. Prep, preparation. (B) Example of a MS/MS spectrum of a phosphopeptide showing a typical extensive neutral loss of phosphoric acid. (C)MS/MS/MS spectrum of the neutral loss precursor ion from B. Abundant peptide bond fragmentation permitted the unambiguous identification of this peptide from the protein cell division cycle 2-related protein kinase 7, with a phosphorylated Ser residue marked by an asterisk.
Further hindering phosphorylation studies, the phosphorylated form of a protein frequently has low stoichiometry relative to its unphosphorylated counterpart. Considering the already low expression levels of most proteins regulated by phosphorylation, it is obvious that “shotgun” sequencing strategies can easily miss these rare peptides. It is essential to employ some type of enrichment strategy to overcome the tremendous complexity of a proteolyzed lysate. Efforts to isolate phosphopeptides in the past have used either chemical modification of phosphate groups (7–9), phosphate-specific MS-based methods (10–13), or affinity-based methods (antibody or metal ion chromatography) (3, 6, 14–16). Regardless of the enrichment procedure, amino acid sequence analysis and site determination were accomplished by MS/MS. Each technique has been successful for the analysis of a few proteins (<30), but only immobilized metal affinity chromatography has shown the potential for the identification of more than a few sites from complex mixtures (3, 6).
In this report, we show that strong cation exchange (SCX) chromatography provides an additional, robust enrichment tool for phosphopeptides. SCX is often used as a primary separation strategy for complex peptide mixtures before analysis by reversephase liquid chromatography (LC)-MS/MS (17), in which peptides elute according to their solution state charge. The most common implementation utilizes an on-line system in which complex mixtures are adsorbed to a bi-phasic column packed with SCX and reverse-phase resins. Several salt “bumps” are then initiated to release discrete peptide fractions to the reverse-phase column for analysis. Although this strategy has been widely successful for the analysis of complex peptide mixtures, the ability of SCX chromatography to enrich phosphopeptides has not been reported likely because of weak retention of most tryptic phosphopeptides.
Here, we describe a SCX-based strategy to enrich for the phosphopeptide component of a proteome and a method to enhance phosphopeptide identification. We demonstrate the technique by isolating and identifying >2,000 phosphorylation sites from HeLa cell nuclei.
Materials and Methods
HeLa Cell Nuclear Preparation, Preparative SDS/PAGE Separation, and in-Gel Proteolysis. HeLa cell nuclear preparation was as described in ref. 18. Protein (8 mg) was separated by a preparative SDS/PAGE gradient (5–15%) gel (15 × 15 × 0.15 cm). The gel was stopped when the buffer front had migrated 4 cm into the gel and stained with Coomassie. The entire gel was then cut into ten regions (≈4 mm × 150 mm) which were subjected to in-gel digestion with trypsin as described but with larger volumes in 15-ml falcon tubes (19).
SCX Chromatography. SCX chromatography was performed on a Surveyor HPLC and PDA detector (Thermo Electron, San Jose, CA). Solvent A (5 mM KH2PO4/30% acetonitrile, pH 2.7), solvent B (solvent A with 350 mM KCl), and solvent C (0.1 M Tris/0.5 M KCl, pH 7.0) were used to develop a salt gradient. Extracted, dried peptides were dissolved in 500 μl of SCX solvent A immediately before analysis. Tryptic peptides were separated at pH 2.7 by SCX chromatography by using a 3.0 mm × 20 cm column (Poly LC, Columbia, MD) containing 5-μm polysulfoethyl aspartamide beads with a 200-Å pore size and a flow rate of 350 μl/min. UV detection was at 220 and 280 nm (20). This column provided the best retention of singly-charged phosphopeptides. A gradient was developed consisting of 5 min at 100% solvent A, 15 min gradient to 15% solvent B, 1 min gradient to 100% solvent B, 15 min at 100% solvent B, 15 min at 100% solvent C, 20 min at 100% solvent A. Fractions were collected every 2 min during the analysis. Four fractions spanning the early eluting peptides were desalted off-line (21) and completely dried.
HeLa Cell Lysate and Synthetic Phosphopeptide Library. Cell lysate was prepared by using unsynchronized HeLa cells lysed by sonication in 8M urea/100 mM NaCl/25 mM Tris, pH 8.05, in the presence of protease (Roche) and phosphatase inhibitors (Sigma). The sample was diluted eightfold, and trypsin was added at 1:50 enzyme to substrate ratio overnight. Acidified proteolyzed samples were desalted by solid-phase extraction (2-ml tC18 cartridges, Waters). Total peptides (300 μg) were then separated by SCX chromatography as described above. A synthetic phosphopeptide library was generated to contain singly charged phosphopeptides at acidic pH with the following sequence, GAPXPXsXFEA(K/R), where X was the amino acid ADEFGLSTV or Y and s denotes a phosphorylated Ser.
MS. Early eluting fractions were analyzed by reverse-phase LC-MS/MS by using 75-μm i.d. × 12 cm self-packed fused-silica C18 capillary columns as described in ref. 22. Peptides were eluted for each analysis by using a 6-hr gradient from 0 to 30% solvent B (0.1% formic acid/98% acetonitrile) in which the ions were detected, isolated, and fragmented in a completely automated fashion on an LCQ DECA XP ion trap mass spectrometer (Thermo Electron). Software for the automatic acquisition of data-dependent third stage of MS (MS/MS/MS) spectra was produced and implemented through a collaboration with Thermo Electron and is available in xcalibur 1.4. An MS/MS/MS spectrum was automatically collected when the most intense peak from the MS/MS spectrum corresponded to a neutral loss event of 98 m/z, 49 m/z ± 0.7 Da.
Database Correlation. All MS/MS and MS/MS/MS spectra were searched against the human database from the National Center for Biotechnology Information (August 2003) by using the sequest algorithm (23). Modifications were permitted to allow for the detection of oxidized Met (+16), carboxyamidomethylated Cys (+57), and phosphorylated Ser, Thr and Tyr (+80). All peptide matches were initially filtered based on Xcorr and dcorr scores followed by manual validation of all spectra with the aid of in-house software. All phosphopeptide spectra are available upon request from the authors.
Results
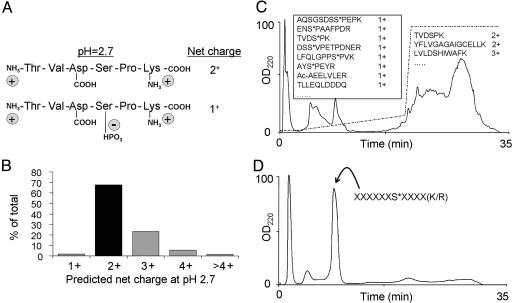
Enrichment of Tryptic Phosphopeptides by Differential Net Solution Charge State. At pH 2.7, only Lys, Arg, His, and the amino terminus of a peptide are charged. Trypsin proteolysis produces peptides with a C-terminal Lys or Arg. Thus, most tryptic peptides carry a net solution charge state of 2+, as shown in Fig. 1A. Because a phosphate group maintains a negative charge at acidic pH values, the net charge state of a phosphopeptide is generally only 1+. An in silico tryptic digest of the human protein database from National Center for Biotechnology Information produced peptides, with 68% predicted to have a net charge of 2+ (Fig. 1B). Any of these peptides would have a net charge state of 1+ after a single phosphorylation event. Because SCX chromatography separates peptides based primarily on ionic charge (24), we expect singly charged species to elute before those with multiple charges. The SCX separation of a complex peptide mixture (300 μg of HeLa cell lysate) at pH 2.7 generated by trypsin proteolysis is shown in Fig. 1C. LC-MS/MS analysis confirmed that early fractions were highly enriched with phosphopeptides that contained a solution charge state of 1+ and had been successfully separated from the more complex multiply charged peptides, which eluted later. The SCX separation of a synthetic library of 2,000 phosphopeptides with a solution charge state of 1+ as shown in Fig. 1D further confirmed that phosphopeptides with a solution charge state of 1+ elute early and can be enriched from more complex multiply charged peptides.
Fig. 1.
Scheme for phosphopeptide enrichment by SCX chromatography. (A) At pH 2.7, most peptides produced by trypsin proteolysis have a solution charge state of 2+, whereas phosphopeptides have a charge state of only 1+. (B) Solution charge state distribution of peptides (5–40 aa) produced by a theoretical digestion of the human protein database with trypsin (n = 6.8 × 108 peptides). Sixty-eight percent of the predicted peptides have a net charge of 2+. Any peptide in this category would shift to a 1+ charge state upon phosphorylation. (C) SCX chromatography separation at pH 2.7 of a HeLa cell lysate after trypsin digestion. The dashed line indicates the salt gradient. Some identified peptides from the collected fractions are shown. Phosphorylation sites are denoted by an asterisk, and N-terminal acetylation is denoted by Ac. (D) SCX chromatography separation at pH 2.7 of a synthetic phosphopeptide library containing 2,000 phosphopeptides with a predicted solution charge state of 1+.
Data-Dependent Acquisition of MS/MS/MS Spectra for Improved Phosphopeptide Identification. In the context of peptide MS, an MS/MS spectrum and MS/MS/MS spectrum represent, respectively, the measurement of fragment ions derived from a single peptide, and fragment ions derived from a single peptide fragment. Thus, if an MS/MS spectrum of a phosphopeptide results in a dominant phosphate-specific fragment ion, an MS/MS/MS spectrum from that dominant fragment ion could result in a more useful fragmentation pattern.
We designed and implemented a strategy to collect an MS/MS/MS spectrum when the following conditions were met**: (i) The MS/MS spectrum revealed a significant loss of phosphoric acid (49 or 98 Da) upon fragmentation; and (ii) the neutral loss event was the most intense peak in the MS/MS spectrum. Meeting these two criteria is often observed for phosphopeptides but is extremely unlikely for nonphosphorylated peptides. In this way, MS/MS/MS spectra were not acquired unless a phosphopeptide was suspected. An example of such a spectrum is shown in Fig. 2B. The primary fragmentation product of this phosphopeptide is a single intense peak 49 Da less than the m/z ratio of the precursor ion. Having met both conditions described above, an MS/MS/MS scan was automatically collected by isolating and fragmenting the neutral loss fragment ion from the MS/MS spectrum. The result in many cases is a richer fragmentation spectrum from which the phosphopeptide sequence could be determined, including the modified residue (a Ser), because the loss of phosphoric acid converted the Ser residue to a dehydroalanine. The amount of time required to collect both the MS/MS and MS/MS/MS spectra was <3 s.
Application to HeLa Cell Nuclear Phosphoproteins. We applied this strategy to the characterization of phosphoproteins from asynchronous HeLa cells. We examined proteins present in the nuclear fraction and applied a preparative SDS/PAGE separation to allow milligram quantities of starting protein (Fig. 2A). The entire gel was divided into 10 regions and proteolyzed with trypsin followed by phosphopeptide enrichment by SCX chromatography. Early eluting fractions were subjected to reverse-phase LC with on-line sequence analysis by LC-MS/MS. More than 500,000 MS/MS and an additional 12,000 MS/MS/MS spectra were acquired during the course of the experiment. MS/MS spectra were searched with sequest (23) against the human database from the National Center for Biotechnology Information with a variable modification for phosphorylation to Ser, Thr, and Tyr. MS/MS/MS spectra were searched with sequest with a variable modification of -18 to Ser and Thr residues.
Spectral Validation. In total, we confidently assigned 2,002 distinct phosphorylation sites with the sequest algorithm and manual confirmation. Manual confirmation was done by validating the presence of peaks explained by peptide backbone fragmentation and used by sequest (e.g., b- and y-type ions) and peaks unexplained by simple peptide bond fragmentation. Many intense ions can be explained by neutral loss events from either the precursor ion or b- and y-type ions not taken into account by searching algorithms. For example, in addition to the precursor ion, a b- or y-type ion can also show a significant loss of phosphoric acid (see Fig. 4, which is published as supporting information on the PNAS web site). A spectrum that contained ions that were not explained by using this method resulted in the rejection of its sequence assignment. Software was created to automate this process (Fig. 4). Matches were deemed correct when they met rigid criteria, such as the presence of intense Pro-directed fragment ions, possession of the correct net solution charge state, and good agreement in molecular mass of the parent protein and the region excised from the gel. We have provided the entire list of 2,002 sites (Table 3, which is published as supporting information on the PNAS web site).
Classification of Phosphorylation Sites. Kinase specificity typically depends on the primary amino acid sequence surrounding the target phosphorylation site (25). Protein kinases can be separated into Ser/Thr and Tyr kinases, although dual specificity kinases exist (26). The sites detected from our nuclear preparation were exclusively Ser/Thr. Tyr phosphorylation is generally thought to represent <1% of all cellular phosphorylation, but it is not clear whether this proportion can be extrapolated to nuclear proteins.
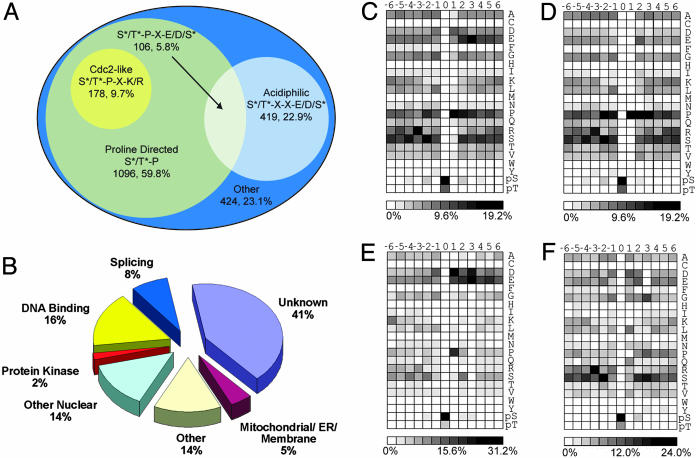
Ser/Thr protein kinases can be subdivided based on substrate specificity, as determined for a number of kinases by in vitro phosphorylation of soluble peptide libraries (27, 28). Major classes include Pro-directed (e.g., extracellular signal-regulated kinase 1, cyclin-dependent kinase 5, and cyclin B/Cdc2, etc.), basophilic (PKA, PKC, and Slk1, etc.) and acidiphilic (casein kinase 1δ, casein kinase 1γ, and casein kinase II) kinases. As shown in Fig. 3A, Pro-directed and acidiphilic sites accounted for 77% of all detected phosphorylation. Furthermore, the detected sites were categorized by their biological function (Fig. 3B). Consistent with our preparation, most sites detected were nuclear in origin or from other organelles known to be present in nuclear preparations (mitochondria and endoplasmic reticulum). Finally, numerous protein kinases and transcription factors were identified that demonstrated the sensitivity of the analysis. Table 1 shows 61 phosphorylation sites from 28 protein kinases detected in this study. Only six of these sites have been described previously (29). Knowledge of these sites will likely provide a framework for piecing together complex phosphorylation regulatory mechanisms.
Fig. 3.
Classification of identified phosphorylation sites and amino acid frequencies surrounding phosphorylated Ser and Thr residues. From the 2,002 detected sites, 1,833 could be localized to a specific Ser or Thr without ambiguity. (A) Venn diagram representation of 1,833 precise sites of phosphorylation with respect to surrounding residues. Seventy-seven percent of the detected phosphorylation sites could be assigned as either Pro-directed or acidiphilic. (B) Phosphorylation sites grouped by protein localization and function. The largest class of proteins detected was “unknown” (uncharacterized or hypothetical). “Other” represents known proteins not in other categories (mostly well characterized cytosolic proteins). (C) Intensity map showing the relative occurrence of residues flanking all phosphorylation sites. (D) Intensity map showing the relative occurrence of residues flanking Pro-directed [(pSer/pThr)-Pro] phosphorylation sites. (E) Intensity map showing the relative occurrence of residues flanking acidiphilic [(pSer/pThr)-Xxx-Xxx-(Asp/Glu/pSer)] sites. (F) Intensity map showing the relative occurrence of residues flanking all other phosphorylation sites. To facilitate comparisons, an intensity gradient of light to dark was used ranging from white (no occurrence) to black (high occurrence).
Table 1. Phosphorylation sites identified from protein kinases detected in this study.
| Gene name | Peptide | Literature |
|---|---|---|
| AF067512† | EYGS*PLKAYT*PVVVTLWYR | No |
| AF162666† | ISDYFEYQGGNGSS*PVR | No |
| AF162667† | ISDYFEFAGGSAPGTS*PGR | No |
| AF387103† | GLSS*GWSSPLLPAPVCNPNK | No |
| AJ297709† | GGDVS*PSPYSSSSWR | No |
| S*PS*PAGGGSSPYSR | No | |
| S*PSYSR | No | |
| SLS*PLGGR | No | |
| AK001247† | EGDPVSLSTPLETEFGSPSELS*PR | No |
| VFPEPTES*GDEGEELGLPLLSTR | No | |
| E54024‡ | DLLSDLQDIS*DSER | No |
| G01025‡ | VPAS*PLPGLER | No |
| T13149‡ | LFQGYS*FVAPSILFK | No |
| ATM_HUMAN§ | SLAFEEGS*QSTTISSLSEK | Yes |
| CDK2_HUMAN§ | IGEGT*YGVVYK | Yes |
| CRK7_HUMAN§ | AIT*PPQQPYK | No |
| GS*PVFLPR | No | |
| NSS*PAPPQPAPGK | No | |
| QDDSPSGASYGQDYDLS*PSR | No | |
| S*PGSTSR | No | |
| SPS*PYSR | No | |
| SVS*PYSR | No | |
| TVDS*PK | No | |
| KPCD_HUMAN§ | NLIDSMDQSAFAGFS*FVNPK | Yes |
| RAB_HUMAN§ | GDGGSTTGLSAT*PPASLPGSLTNVK | No |
| SAS*EPSLNR | No | |
| MATK_HUMAN§ | SAGAPASVSGQDADGSTS*PR | No |
| MPK2_HUMAN§ | LNQPGT*PTR | No |
| PDPK_HUMAN§ | ANS*FVGTAQYVSPELLTEK | Yes |
| PKL1_HUMAN§ | TDVSNFDEEFTGEAPTLS*PPR | No |
| PKL2_HUMAN§ | AS*SLGEIDESSELR | No |
| TST*FCGTPEFLAPEVLTETSYTR | Yes | |
| PR4B_HUMAN§ | DAS*PINRWS*PTR | No |
| EQPEMEDANS*EKS*INEENGEVSEDQSQNK | No | |
| S*LS*PKPR | No | |
| S*PIINESR | No | |
| S*PVDLR | No | |
| S*RS*PLLNDR | No | |
| SINEENGEVS*EDQS*QNK | No | |
| TLS*PGR | No | |
| TRS*PS*PDDILER | No | |
| YLAEDSNMSVPSEPSS*PQSSTR | No | |
| PRKD_HUMAN§ | LTPLPEDNS*MNVDQDGDPSDR | Yes |
| STKA_HUMAN§ | QVAEQGGDLS*PAANR | No |
| WEE1_HUMAN§ | SPAAPYFLGSSFS*PVR | No |
| M4K1_HUMAN§ | DLRS*SS*PR | No |
| M4K4_HUMAN§ | AASSLNLS*NGETESVK | No |
| TTS*RS*PVLSR | No | |
| M4K6_HUMAN§ | LDSS*PVLSPGNK | No |
| KC1E_HUMAN§ | IQPAGNTS*PR | No |
| KPBB_HUMAN§ | QSST*PSAPELGQQPDVNISEWK | No |
Accession numbers were derived from the following sources: †, GenBank; ‡, Protein Information Resource; §, SwissProt. Sites of phosphorylation in the peptides are indicated by an asterisk. A literature search of phosphorylation sites was done by using the Human Protein Reference Database (30).
To predict the kinase/substrate relationships from the data set, the computer algorithm scansite (27) can be used. scansite makes use of soluble peptide library phosphorylation data to predict substrates recognized by specific kinases. Table 2 shows the results of correlating the amino acid sequences surrounding the sites identified by this study against known matrices at the highest stringency level (0.002) and a lower stringency level (0.01). At the highest stringency, scansite predicted several phosphorylation sites within our data set from each of the Pro-directed kinases, the basophilic kinases (AKT, PKA, and Clk2), the acidiphilic kinase casein kinase 2, and the DNA damage-activated kinases ataxia telangiectasia mutated kinase and DNA-dependent protein kinase. It is also possible to use scansite matrices to predict sites that require phosphorylation to become suitable binding domains. Our data set included several known 14–3-3 binding sites and two known phosphoinositide-dependent protein kinase 1 binding sites from PKC-δ and p90RSK. Because only a fraction of the total number of detected sites could be assigned with high confidence by scansite, it seems likely that many more kinase motifs are present in our data set.
Table 2. scansite prediction at highest stringency (0.2%) and medium stringency (1.0%) for kinase phosphorylation and binding motifs from this data set.
| Type | Hits (0.2%) | Hits (1.0%) | |
|---|---|---|---|
| Kinase | |||
| Casein kinase 2 | Acidiphilic | 65 | 172 |
| Glycogen synthase kinase 3 | Pro-directed | 64 | 206 |
| CDC2 | Pro-directed | 55 | 262 |
| AKT | Basophilic | 53 | 122 |
| Extracellular signal-regulated kinase 1 | Pro-directed | 51 | 235 |
| Cyclin-dependent kinase 5 | Pro-directed | 49 | 260 |
| P38 mitogen-activated protein kinase | Pro-directed | 33 | 160 |
| PKA | Basophilic | 17 | 48 |
| Clk2 | Basophilic | 11 | 72 |
| DNA-dependent protein kinase | Gln-directed | 8 | 62 |
| Calmodulin-dependent kinase 2 | Basophilic | 7 | 21 |
| Ataxia telangiectasia mutated kinase | Gln-directed | 6 | 23 |
| PKC-δ | Basophilic | 2 | 9 |
| PKC-α/β/γ | Basophilic | 1 | 7 |
| PKC-ε | Basophilic | 1 | 8 |
| Casein kinase 1 | Other | 0 | 23 |
| Protein kinase D | Basophilic | 0 | 5 |
| Motif | |||
| 14-3-3 binding motif | Pro-directed | 31 | 85 |
| Phosphoinositide-dependent kinase 1 binding motif | Pro-directed | 2 | 3 |
Bioinformatic Analysis of Phosphorylation Sites. The magnitude of this data set made possible a preliminary classification of identified phosphorylation sites into specific motifs. The relative occurrence of each amino acid (including pSer/pThr) flanking the site of phosphorylation was calculated and plotted by using intensity maps (Fig. 3). Considering the entire data set (Fig. 3C), it is clear that a Pro at the +1 position and/or a Glu at position +3 were favored. To further elucidate significant flanking residues, similar maps were generated considering data that conformed to either pSer/pThr-Pro-containing sites (Fig. 3D), pSer/pThr-Xxx-Xxx-Glu/Asp/pSer-containing sites (Fig. 3E), or the subset of all data that did not conform to either general classification (Fig. 3F).
Several further insights into kinase motifs can be made from the plots. For example, when the acidic residue is fixed at position +3, it can be seen that an aspartic acid residue is highly favored at position +1(Fig. 3E). Although this result was not predicted by the soluble peptide libraries (30), a propensity for aspartic acid at the +1 position of casein kinase 2 sites has been reported (31). In the Pro-directed subset (Fig. 3C), additional Pro at the +2 and +3 positions as well as Ser at -3 and Arg at -2 are favored. We have constructed a web site that returns peptides from our data set conforming to a user-specified motif (http://gygi.med.harvard.edu/pubs/phosphorylation/phosmotif.html).
Discussion
The strategy described here exploits the difference between the solution charge state of most tryptic phosphopeptides when compared with their unphosphorylated counterparts. Because SCX chromatography separates peptides primarily based on charge, phosphopeptides containing a single basic group elute first and are highly enriched. After this off-line enrichment, sequence analysis of the phosphopeptides is accomplished by reverse-phase LC-MS/MS. If the tandem mass spectrum of a phosphopeptide is dominated by phosphate-associated losses, an MS/MS/MS spectrum is collected from the dominant fragment ion after the neutral loss. In this way, large numbers of phosphopeptides can be isolated, separated, and sequence-analyzed in an automated fashion. The identification of 2,002 phosphorylation sites from a HeLa cell nuclear preparation is provided to demonstrate the technique. To our knowledge, this is the largest data set of posttranslational modifications determined to date.
The data-dependent MS/MS/MS strategy described here provided the capability to collect additional information when a peptide demonstrated significant phosphate-associated losses. MS/MS/MS spectra were only successfully used for phosphopeptide identification when relatively abundant ions were selected. When a phosphopeptide ion of lower intensity was selected, the resulting MS/MS/MS spectra contained no fragment ions. This result is due to insufficient trapping of ions over several stages of MS. Approximately 50% of all MS/MS/MS spectra contained no useful information because there was an insufficient number of ions isolated. This problem resulted in only 96 of 2,002 phosphorylation sites being determined only from an MS/MS/MS spectrum. However, in hundreds of cases, the MS/MS/MS spectra were useful to aid in site localization and in confirming the identification of matching MS/MS spectra. For MS/MS–MS/MS/MS pairs with lower-scoring MS/MS spectra (Xcorr < 2.5), average MS/MS/MS scores were increased by 10%. When the Xcorr for the MS/MS was >2.5, the average MS/MS/MS score showed no improvement. The greater ion capacity of the newer linear ion traps (32) should make the acquisition of MS/MS/MS spectra even more useful for phosphorylation analysis.
This data set provides new bioinformatic opportunities to study and predict kinase–substrate relationships. The intensity maps in Fig. 3 provide some insight into sequence-specific trends surrounding each phosphorylation site. Pro-directed and acidiphilic kinases make up a large fraction of our data set. A more comprehensive bioinformatic analysis should help discover and validate known and novel kinase consensus motifs.
The SCX isolation method has the caveat that some sites are not amenable to analysis. Specifically, a His-containing phosphopeptide would elute as a 2+ peptide and not be selected by the strategy described here. Similarly a multiply phosphorylated tryptic peptide with only two basic sites would have a net charge state of 0 and therefore might fail to be captured by the SCX stationary phase. In essence, any phosphorylated peptide with a charge state other than ++ would not be detected by the method as implemented. It is important to note that phosphopeptides not amenable to this enrichment represent only a fraction of all phosphopeptides (Fig. 1B). Further exploration into new methods and combinations of existing technologies is required to provide a truly comprehensive view of the dynamic phosphoproteome.
In conclusion, this methodology significantly enhances the ability to routinely discover large numbers of phosphorylated species within complex protein mixtures by exploiting peptide solution charge states generated by tryptic digests. Enrichment by off-line SCX chromatography increased the likelihood of selecting phosphorylated peptides for sequencing in the mass spectrometer, whereas data-dependent MS/MS/MS software aided in confirming sequence and phosphorylation site location. Finally, the combination of stable isotope labeling (33, 34) with the methods described here would allow for a large-scale comparative phosphorylation analysis of different cell states with which several hundred phosphorylation sites could be simultaneously profiled.
Supplementary Material
Acknowledgments
We thank I. Jardine, I. Mylchreest, J. Shofstahl, J. Schwartz, and E. Hemenway (Thermo Electron) for integrating the MS/MS/MS algorithm idea and providing early access. We also thank past and present members of the Gygi lab for useful discussions and critical reading, R. Duarte (Harvard Medical School) for producing phosphorylation intensity maps, J. Rush (Cell Signaling Technology, Beverly, MA) for synthetic peptides, and S. Gerber (Harvard Medical School) for HeLa cell lysate. This work was supported in part by National Institutes of Health Grants HG00041 (to S.P.G.), GM67945 (to S.P.G.), and GMS6203 (to L.C.C.).
Abbreviations: SCX, strong cation exchange; MS/MS, tandem MS; MS/MS/MS, third stage of MS; LC, liquid chromatography.
Footnotes
Tomaino, R., Rush, J., Steen, H., Licklider, L., Hemenway, E., Shofstahl, J., Schwartz, J., Mylchreest, I. & Gygi, S. P., 50th American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics, June 2–8, 2002, Orlando, FL, abstr. ThOEpm.
References
- 1.Aebersold, R. & Goodlett, D. R. (2001) Chem. Rev. 101, 269-295. [DOI] [PubMed] [Google Scholar]
- 2.McLachlin, D. T. & Chait, B. T. (2001) Curr. Opin. Chem. Biol. 5, 591-602. [DOI] [PubMed] [Google Scholar]
- 3.Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F. & White, F. M. (2002) Nat. Biotechnol. 20, 301-305. [DOI] [PubMed] [Google Scholar]
- 4.Peng, J., Schwartz, D., Elias, J. E., Thoreen, C. C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D. & Gygi, S. P. (2003) Nat. Biotechnol. 21, 921-926. [DOI] [PubMed] [Google Scholar]
- 5.Nuhse, T. S., Stensballe, A., Jensen, O. N. & Peck, S. C. (2003) Mol. Cell Proteomics 2, 1234-1243. [DOI] [PubMed] [Google Scholar]
- 6.Ficarro, S., Chertihin, O., Westbrook, V. A., White, F., Jayes, F., Kalab, P., Marto, J. A., Shabanowitz, J., Herr, J. C., Hunt, D. F. & Visconti, P. E. (2003) J. Biol. Chem. 278, 11579-11589. [DOI] [PubMed] [Google Scholar]
- 7.McLachlin, D. T. & Chait, B. T. (2003) Anal. Chem. 75, 6826-6836. [DOI] [PubMed] [Google Scholar]
- 8.Zhou, H., Watts, J. D. & Aebersold, R. (2001) Nat. Biotechnol. 19, 375-378. [DOI] [PubMed] [Google Scholar]
- 9.Oda, Y., Nagasu, T. & Chait, B. T. (2001) Nat. Biotechnol. 19, 379-382. [DOI] [PubMed] [Google Scholar]
- 10.Steen, H., Kuster, B., Fernandez, M., Pandey, A. & Mann, M. (2001) Anal. Chem. 73, 1440-1448. [DOI] [PubMed] [Google Scholar]
- 11.Shou, W., Verma, R., Annan, R. S., Huddleston, M. J., Chen, S. L., Carr, S. A. & Deshaies, R. J. (2002) Methods Enzymol. 351, 279-296. [DOI] [PubMed] [Google Scholar]
- 12.Annan, R. S., Huddleston, M. J., Verma, R., Deshaies, R. J. & Carr, S. A. (2001) Anal. Chem. 73, 393-404. [DOI] [PubMed] [Google Scholar]
- 13.Schlosser, A., Pipkorn, R., Bossemeyer, D. & Lehmann, W. D. (2001) Anal. Chem. 73, 170-176. [DOI] [PubMed] [Google Scholar]
- 14.De Corte, V., Demol, H., Goethals, M., Van Damme, J., Gettemans, J. & Vandekerckhove, J. (1999) Protein Sci. 8, 234-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann, M., Ong, S. E., Gronborg, M., Steen, H., Jensen, O. N. & Pandey, A. (2002) Trends Biotechnol. 20, 261-268. [DOI] [PubMed] [Google Scholar]
- 16.Gronborg, M., Kristiansen, T. Z., Stensballe, A., Andersen, J. S., Ohara, O., Mann, M., Jensen, O. N. & Pandey, A. (2002) Mol. Cell Proteomics 1, 517-527. [DOI] [PubMed] [Google Scholar]
- 17.Washburn, M. P., Wolters, D. & Yates, J. R. (2001) Nat. Biotechnol. 19, 242-247. [DOI] [PubMed] [Google Scholar]
- 18.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- 20.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J. & Gygi, S. P. (2003) J. Proteome Res. 2, 43-50. [DOI] [PubMed] [Google Scholar]
- 21.Rappsilber, J., Ishihama, Y. & Mann, M. (2003) Anal. Chem. 75, 663-670. [DOI] [PubMed] [Google Scholar]
- 22.Peng, J. & Gygi, S. P. (2001) J. Mass Spectrom. 36, 1083-1091. [DOI] [PubMed] [Google Scholar]
- 23.Eng, J., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- 24.Alpert, A. J. & Andrews, P. C. (1988) J. Chromatogr. 443, 85-96. [DOI] [PubMed] [Google Scholar]
- 25.Manning, B. D. & Cantley, L. C. (2002) Sci. STKE 2002, PE49. [DOI] [PubMed] [Google Scholar]
- 26.Dhanasekaran, N. & Premkumar Reddy, E. (1998) Oncogene 17, 1447-1455. [DOI] [PubMed] [Google Scholar]
- 27.Obenauer, J. C., Cantley, L. C. & Yaffe, M. B. (2003) Nucleic Acids Res. 31, 3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Neill, T., Dwyer, A. J., Ziv, Y., Chan, D. W., Lees-Miller, S. P., Abraham, R. H., Lai, J. H., Hill, D., Shiloh, Y., Cantley, L. C. & Rathbun, G. A. (2000) J. Biol. Chem. 275, 22719-22727. [DOI] [PubMed] [Google Scholar]
- 29.Peri, S., Navarro, J. D., Amanchy, R., Kristiansen, T. Z., Jonnalagadda, C. K., et al. (2003) Genome Res. 13, 2363-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Songyang, Z., Lu, K. P., Kwon, Y. T., Tsai, L. H., Filhol, O., Cochet, C., Brickey, D. A., Soderling, T. R., Bartleson, C., Graves, D. J., et al. (1996) Mol. Cell. Biol. 16, 6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meggio, F. & Pinna, L. A. (2003) Faseb J. 17, 349-368. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, J. C., Senko, M. W. & Syka, J. E. (2002) J. Am. Soc. Mass Spectrom 13, 659-669. [DOI] [PubMed] [Google Scholar]
- 33.Oda, Y., Huang, K., Cross, F. R., Cowburn, D. & Chait, B. T. (1999) Proc. Natl. Acad. Sci. USA 96, 6591-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen, H., Pandey, A. & Mann, M. (2002) Mol. Cell Proteomics 1, 376-386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.