Abstract
Acanthamoeba myosin IC (AMIC) is a single-headed myosin comprised of one heavy chain (129 kDa) and one light chain (17 kDa). The heavy chain has head, neck (light chain-binding), and tail domains. The tail consists of four subdomains: a basic region (BR) (23 kDa) and two Gly/Pro/Ala-rich (GPA) regions, GPA1 (6 kDa) and GPA2 (15 kDa), flanking an Src homology 3 region (6 kDa). Although the AMIC head is similar in sequence, structure, and function (ATPase motor) to other myosin heads, the organization of the tail has been less clear as has its function beyond an assumed role in binding interaction partners, e.g., the BR has a membrane affinity and the GPA components bind F-actin in an ATP-independent manner. To investigate the spatial arrangement of subdomains in the tail, we have used cryo-electron microscopy and image reconstruction to compare actin filaments decorated with WT AMIC and tail-truncated mutants of various lengths. The BR forms an oval-shaped feature, ≈40 Å long, that diverges obliquely from the head, extending azimuthally around the actin filament and toward its barbed end. GPA2 and GPA1 are located together on the inner (actin-proximal) side of the tail, close enough to act in concert in binding the same or another actin filament. The outer face of the BR is strategically exposed for membrane or vesicle binding.
Members of the myosin superfamily identified to date fall into 18 classes (1, 2). Although they are classified primarily in terms of their head domain sequences, phylogenetic analysis also assigns the neck/tail domains of the heavy chains to the same classes (3). Class I myosins are nonfilamentous proteins with basic tails that bind to phospholipids (4).
Acanthamoeba has three class I myosins: AMIA, AMIB, and AMIC (5). The latter protein, the object of this study, associates with plasma membranes and membranes of large contractile vacuoles that play essential roles in osmoregulation (6–8). More recent localization studies have revealed that the concentration of AMIC around contractile vacuoles and macropinocytosis cups is transient (9, 10). AMIC has an 80-kDa head domain, a 50-kDa tail domain, and one 17-kDa light chain (11) of unknown function binding to a 3-kDa neck domain (Fig. 1A). In this article, we use the terms head domain, tail domain, etc. to refer to previously defined regions of amino acid sequence (Fig. 1 A) and head and tail to denote structural features of our density maps. The tail domain has four subdomains: basic region (BR), Gly/Pro/Ala-rich (GPA)1, GPA2, and Src homology 3 (SH3). The BR binds to acidic vacuole membranes (12). GPA1 and GPA2 are ATP-independent actin-binding sites (13, 14). The bundling of actin filaments by AMIC (unpublished observations) also supports the idea that its tail can bind to another actin filament, as can AMIA and AMIB (15, 16) and cardiac muscle myosin II (17). SH3 domains are found in a number of membrane-cytoskeletal proteins and are thought to engage in protein–protein interactions. Dictyostelium myosin I and AMI bind through their SH3 domains to Pro-rich regions in CARMIL (capping protein, Arp2/3, and myosin I linker) (18, 19), which also binds capping protein and the Arp2/3 complex (18).
Fig. 1.
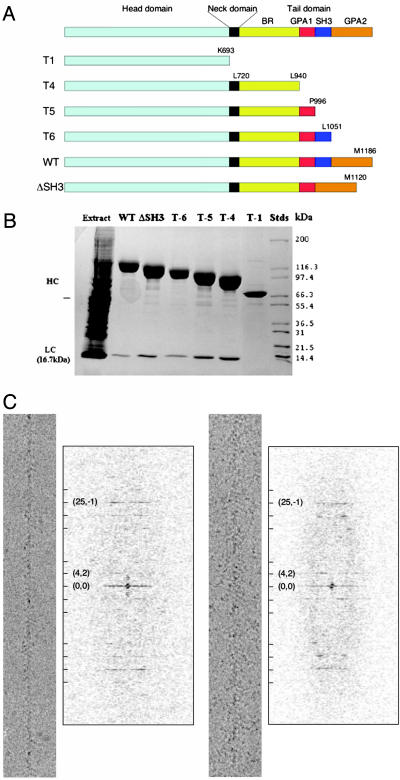
Subdomain organization and actin filament-binding properties of full-length AMIC and truncated variants. (A) Schematic diagram of the domain compositions of the AMIC mutants analyzed in this study. The domain boundaries are from ref. 13. (B) SDS/PAGE of the purified proteins (reproduced from ref. 13). HC, heavy chain; LC, light chain. (C) Typical cryo-electron micrographs and diffraction patterns of AMIC-decorated actin filaments. A few key layer-lines are indexed.
AMIC actin-activated ATPase activity is regulated by phosphorylation of Ser-329 in the actin-binding region of the head (20). The structural consequences of a change of nucleotide state, rigor to MgADP, have been examined by cryo-electron microscopy (EM) of actin filaments decorated with constitutively active and inactive mutant AMICs (21). Unlike brush border myosin I (22, 23) and smooth muscle myosin II (24) in which substantial swings of the tail were found to accompany this switch, no such effect was detected with AMIC, an observation that may reflect kinetic differences between the respective systems. That study (21) depicted the AMIC tail as a distal lobe connected at an oblique angle to the neck region. The sequences of the AMIA and AMIB tail domains reveal similar subdomains to AMIC except that SH3 follows a single contiguous GPA region rather than splitting it into GPA1 and GPA2 (25, 26). The AMIB tail has also been visualized by cryo-EM (27).
Although the overall shape of the AMIC tail has been described, the placement of its four subdomains remains unknown. Motivated by the possibility that information of this kind might yield insight into the roles of the various tail subdomains in linking AMIC to potential interaction partners such as membranes and other actin filaments, we have investigated this question by difference imaging. Mutant AMIC molecules truncated by removal of different numbers of tail subdomains were expressed and purified (13). We then used cryo-EM and 3D image reconstruction to systematically compare actin filaments decorated with these mutant myosins.
Materials and Methods
Protein Purification. WT and mutant AMICs were prepared as described (21). Myosins were dissolved in 10 mM Tris (pH 7.5), 100 mM KCl, 2 mM MgCl2, and 1 mM DTT. Rabbit skeletal muscle actin was purchased from Cytoskeleton (Denver). F-actin was dissolved in 50 mM KCl, 10 mM Tris (pH 7.5), 2 mM MgCl2, and 1 mM DTT and stabilized by phalloidin.
Cryo-EM. Drops of actin filament suspension were applied to holey carbon grids to drape actin filaments across holes. Then, the filaments were decorated by incubating the grids on AMIC-containing drops, as described (21). After blotting, the grids were vitrified and imaged in a CM200 FEG electron microscope (FEI, Hillsboro, OR) operated at 120 kV and ×38,000 magnification. Micrographs were recorded, mostly as focus pairs, at defocus values of 1.2–2.5 μm, placing the first contrast transfer function zeros at spacings of 22–29 Å and digitized with a SCAI scanner (Z/I Imaging, Huntsville, AL).
Image Analysis. 3D reconstructions of decorated actin filaments were calculated with the phoelix program (28), assuming helical symmetry with the selection rule, l = 25n + 54m (21). Members of focus pairs were processed separately. Contrast transfer function effects were corrected by flipping phases between zeros on layer-lines (28). For surface renderings, the contour level was chosen such that the motor domain enclosed a volume appropriate for 100% of its molecular weight. unblob (29) was used to calculate volumes and eliminate residual “salt and pepper” noise from the density maps. bsoft (30) was used for general image processing. Resolution was evaluated in terms of the Fourier shell correlation coefficient (31).
Before calculating a difference map, two density maps were aligned and scaled by calculating a relative screw displacement and density normalization to minimize the rms difference between their respective motor domains. For visualization, amira (TGS, San Diego) was used for surface rendering, and contour maps for horizontal sections were drawn with photoshop (Adobe Systems, San Jose, CA).
Molecular Modeling. The atomic model of the Dictyostelium myosin IE head domain (32) was fitted to the reconstruction manually by using o (33). A quasi-atomic model of actin filaments decorated by myosin subfragment 1 from skeletal muscle (34) was used to compare the binding aspect of AMIC to the actin filament with that of myosin II.
Results
Visualization of Actin Filaments Decorated with WT and Mutant AMICs. In addition to WT AMIC, we expressed and purified four truncation mutants (T1, T4, T5, and T6) and ΔSH3, a deletion mutant that lacks the SH3 region so that GPA1 is spliced directly to GPA2 (Fig. 1 A and B). The mutant myosins increase in length from T1, which lacks the entire tail domain, to T6, which lacks only the distal subdomain, GPA2. All but T1 bind the light chain (ref. 13; Fig. 1B). Cryo-EM (e.g., Fig. 1C) confirmed the observation (13) that in rigor, i.e., in the absence of nucleotide, all of the mutant proteins bind readily to actin filaments. The diffraction patterns of micrographs of the decorated actin filaments showed well defined layer-lines, suggesting full or nearly full occupancy (e.g., Fig. 1C).
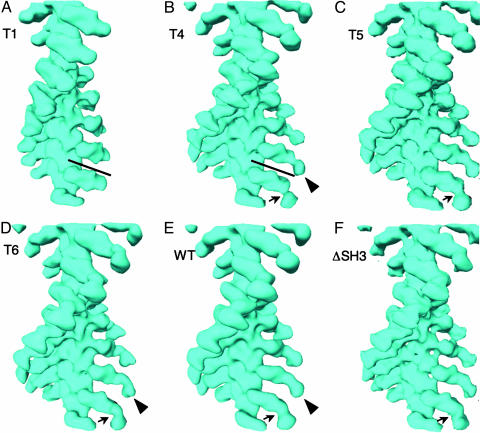
We used the phoelix helical reconstruction program (28) to determine the 3D structures of WT and mutant AMIC molecules (Fig. 2). The resolutions of the resulting density maps range from 22 Å for T4-decorated actin filaments to 33 Å for ΔSH3 (Table 1). The structure obtained for WT AMIC is similar to that calculated previously (21), although slightly more detailed. The most marked change in this series of reconstructions is from T1 to T4 (Fig. 2 A and B), in which the distal lobe appears (Fig. 2B, arrowhead), attached to the head by a narrow connection (Fig. 2B, arrow). Subsequent increases in the size of the tail domain (T5 to T6 to WT) are accompanied by a thickening of this connection and relatively subtle alterations in the distal lobe that are best evaluated in terms of difference maps (see below).
Fig. 2.
Surface renderings of the 3D structures of actin filaments decorated with various AMIC-associated constructs. (A) T1. (B) T4. (C) T5. (D) T6. (E) WT. (F) ΔSH3. Note the very thin connection (arrow) between the head–neck and tail (arrowhead) in the T4 construct (B) and its progressive thickening in the longer tail constructs (C–F, arrows). The two bars denote the difference in length between the T1 and T4 heads. A side-by-side comparison of T1 and WT is given in Fig. 6, which is published as supporting information on the PNAS web site.
Table 1. Resolutions and other reconstruction parameters.
| Construct | Domains in construct | No. of filaments | No. of AMIC molecules* | Resolution,† Å |
|---|---|---|---|---|
| T1 | Head | 14 | 1,820 | 26.4 (22.2) |
| T4 | Head, neck, BR | 30 | 3,900 | 21.8 (20.2) |
| T5 | Head, neck, BR, GPA1 | 65 | 8,450 | 22.8 (18.9) |
| T6 | Head, neck, BR, GPA1, SH3 | 55 | 7,150 | 29.5 (28.9) |
| WT | Head, neck, BR, GPA1, SH3, GPA2 | 43 | 5,590 | 25.1 (18.2) |
| ΔSH3 | Head, neck, BR, GPA1, GPA2 | 28 | 3,640 | 33.3 (31.2) |
Assuming 100% occupancy
Fourier shell correlation coefficient = 0.5 (0.3)
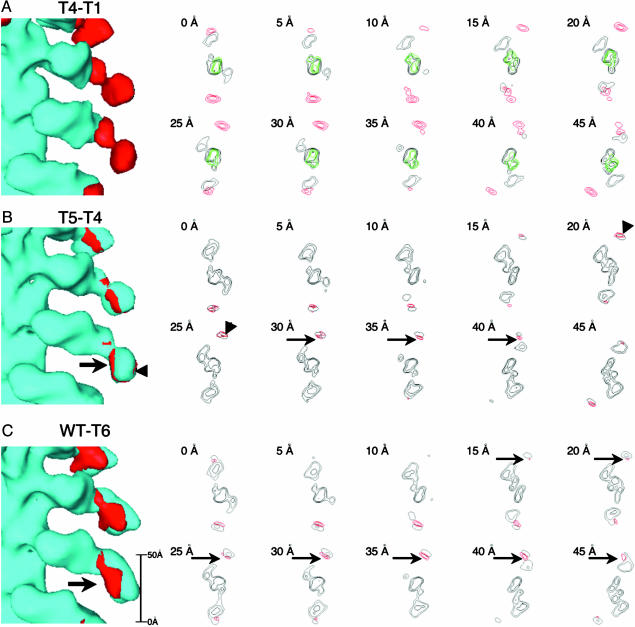
AMIC Head Structure and Binding Aspect. Before addressing the organization of the distal lobe, we examined the head structures. Noting that T1 is perceptibly shorter than T4 (Fig. 2 A and B, bars), we wanted to ascertain where the volume occupied by the head domain stops and that occupied by the neck domain, plus the light chain and the tail domain, starts. The longer T4 structural head registers in the T4–T1 difference map by contributing one lobe of a bilobed feature (red in Fig. 3A). This terminal portion of the head is linked to the proximal portion of the tail by a narrow constriction.
Fig. 3.
Difference densities mapped on surface renderings and in horizontal sections. (A) The difference between T4 and T1 (red) is overlaid on the T1 reconstruction (light blue). The density of actin (34) is shown in green. (B)T5–T4 overlaid on T4. (C)WT–T6 overlaid on T6. In B and C, extra density that we interpret as being GPA1-associated is marked by arrows in B: enhanced density on the other side of BR (arrowheads) is interpreted as an ordering effect.
We performed a more detailed appraisal of the AMIC head by molecular modeling. T1 corresponds to the head domain of chick myosin II (36% sequence identity, 54% similarity), apart from lacking myosin II's 80-residue N-terminal region. As with our previous AMIC reconstruction (21), the crystal structure of the latter domain fits well into the cryo-EM envelope (data not shown), leaving no doubt that the AMIC head domain has essentially the same fold. This fit differs from the previous one by only 0.5–1° in terms of its long axis inclination relative to the actin filament axis.
Recently, a crystal structure for the Dictyostelium myosin IE head domain was reported (32). Because this protein is closer to AMIC (51% identity, 61% similarity up to AMIC residue 667), we repeated the docking with it and again obtained a good fit (Fig. 4). The docked myosin IE head domain is essentially superimposable on the docked myosin II head domain (data not shown). There is, however, a relative rotation of 15° between the rigor binding aspect of AMIC and that of myosin II (Fig. 4, bars), as we ascertained by comparing our fit with a previously published quasi-atomic model (34). This discrepancy is in line with the 10° difference observed between the orientations of the actin-bound heads of AMIB and brush border myosin I (25). It would appear that this property (rigor binding aspect) is specific to the AMIs.
Fig. 4.
Docking of the Dictyostelium myosin IE head (32) into the cryo-EM density map of actin filaments decorated with WT-AMIC. The atomic model of actin is shown in green, and the myosin IE head domain is shown in yellow. The yellow and blue lines illustrate the difference between the rigor actin-binding aspects of the heads of AMIC and myosin II.
Tail Components Visualized by Difference Mapping. Before calculating each difference map, we optimized the relative alignment and normalization of the two reconstructions under comparison.
BR. In Fig. 3A, the difference density between the T4 and T1 mutants is shown in red, superimposed on the T1-actin surface rendering (Left), and is also mapped in red on a transverse section (Right) together with T1/actin reconstruction in black and actin from the quasi-atomic model (34) in green. The difference density is bilobed, with the two lobes being of roughly equal size. The most straightforward interpretation is that the proximal lobe, which is the terminal portion of the AMIC head, is contributed by the light chain (149 residues) plus the neck domain (residues 694–720), whereas the distal lobe represents the major, C-terminal portion of the BR (residues 721–940).
GPA1. Fig. 3B shows the difference density between T5 and T4, presumably caused by GPA1, overlaid on T4 and in transverse sections. Extra density appears on both the inner (actin-proximal) and outer sides of the T4 tail (arrows and arrowheads, respectively). We interpret the inner part as GPA1 both because it is larger and because it represents space not occupied in T4 (see below). In contrast, the outer part is in a region already occupied by density in T4 and its apparent increase most likely represents an improvement in order or a slight outward movement of the BR in this mutant.
GPA2. Extra density that can be interpreted as GPA2 shows up clearly in the difference map between WT and T6 (Fig. 3C). This density locates on the inner side of the tail and extends up into the neck. Despite the lower resolution of the ΔSH3 reconstruction, the difference map between it and T5 also maps GPA2 to the same location (data not shown), confirming this assignment. Taken together, the above data indicate that GPA1 and GPA2 are in mutual proximity on the inner surface of the tail (arrows in Fig. 3 B and C).
SH3. In principle, SH3 should account for extra density seen in T6 but not in T5 and in a difference between WT and ΔSH3. However, we found these difference maps (data not shown) to be noisier than those described above. They are consistent with SH3 being either at the tip of the tail or at the neck region but did not allow a conclusive localization. To explain the marginal visibility of SH3, we suspect that its attachment to the GPAs is somewhat flexible.
Discussion
Volume Recovery, Flexibility, and Disorder. In cryo-EM reconstructions of protein complexes, some density may be poorly visible on account of disorder. Accordingly, there is ambiguity as to the most appropriate contour value to use for surface rendering. For this reason, we tried two values. One, based on fitting the myosin IE head domain (Fig. 4), is expected to underestimate the size of the tail on account of both global disorder (the outer portions are relatively remote from the stabilizing attachment point on the actin filament) and local disorder. However, it enclosed a volume corresponding to 135 kDa, or 95% of the expected mass (125-kDa heavy chain plus 16.7-kDa light chain). A second contour level, chosen to enclose the whole AMIC mass, might overestimate the overall dimensions, effecting some global swelling to replace density missing as a result of local disorder. However, the perceived size and shape of the AMIC tail were little affected by these variations in procedure (Fig. 7, which is published as supporting information on the PNAS web site).
Which subdomains are most likely to be affected by disorder? To address the potential concern that the truncated tails might not fold correctly, we subjected the purified proteins to digestion with trypsin, because unfolded proteins tend to be acutely sensitive to proteolysis. However, we found them to be no more sensitive than full-length AMIC (Fig. 8, which is published as supporting information on the PNAS web site). The inference that tails with partial sets of subdomains fold and assemble correctly is consistent with observations on expressed tail subdomains of AMIA (26).
The volume recovery of subdomains in difference maps provides an indicator of their order/disorder with the caveats that difference maps are affected by residual noise from both contributing density maps, and small features may be diminished or even reduced below the detection threshold by limited resolution. By this criterion, GPA2 and BR are well localized in the WT–T6 and T4–T1 difference maps, with ≈100% and ≈82% recovery, respectively. GPA1 is less evident in the T5–T4 difference maps (≈50% recovery), taking only the inner density into account. GPA1 is unusually rich in Gly (35%) and contains several tracts of tandem Gly repeats: as such, it may be flexible, in line with the properties attributed to “glycine-loop” proteins (35). SH3 is also marginally visible. This small domain is likely to have the canonical SH3 fold (26) but to be coupled loosely to GPA1/GPA2.
Architecture of the AMIC Tail. AMIC bends sharply at the head–tail junction (arrows in Fig. 2). Adding the GPA and SH3 subdomains to the T4 construct (T5, T6, and WT) has little effect on the length of the tail (Fig. 2): rather, density accretes on its inner surface and there is a marked thickening of the neck. It follows that the distal portion of the tail folds back along this surface into the neck region where a considerable amount of density is added. It is possible that a second fold takes place there, bringing the end of GPA2 back toward the tip of the distal lobe (Fig. 5).
Fig. 5.
Schematic representation of the relative positions of the various subdomains of AMIC as bound to F-actin in the rigor conformation. The GPA regions are positioned where they might readily engage the same filament (on a conformational change in the neck or on release of the head) or another parallel and adjacent actin filament. The BR, facing outward, is appropriately placed to allow ready access to membranes.
Comparison with the AMIB and AMIA Tails. Visualized by cryo-EM (25), the shape and actin-binding aspect of the AMIB head are similar to those of AMIC, but its tail appears much longer and broader, which is surprising because the AMIB tail domain is shorter than that of AMIC (434 vs. 466 residues). (The AMIB light chain is larger than that of AMIC, 27 vs. 16.7 kDa, but it should bind in the neck region and not affect tail length.) It may be that the dimensions of the AMIB tail were exaggerated somewhat in contouring. On the other hand, the AMIA tail in solution is estimated to be ≈160 Å long (26) and that of AMIB may have similar properties (but see below).
AMIC also differs from AMIB in the position of its SH3 sequence. However, it is possible that, in both molecules, GPA1 and GPA2 form a single integrated domain to which SH3 is attached at different sites, inserted into a surface loop in AMIC and appended to the C terminus in AMIB. This might contribute to a relative lengthening of the AMIB tail. There is a precedent for the grafting of additional domains onto different sites on a conserved framework in the Clp ATPases (36). The high sequence similarity of the GPA2 regions of AMIB and AMIC (70% identity) supports this notion, as does our observation that AMIC GPA1 and GPA2 are in close proximity. Moreover, this scenario is consistent with phylogenetic arguments that the SH3 domains of Acanthamoeba myosins were acquired relatively late (26).
The AMIA tail domains have been studied by hydrodynamic methods (26) from which it was concluded that the TH2/3 domain (GPA plus SH3) is highly extended, with an axial ratio of ≈8:1 if prolate, and that this domain is arranged side by side with TH1 (BR) in the tail. Our findings on AMIC are consistent with the latter conclusion with the proviso that TH2/3 should be considerably compacted on binding TH1 unless the organization of AMIA departs radically from that of AMIC.
Strategic Positioning of the GPA Region and BR. GPA1 and GPA2 form the second, ATP-independent actin-binding site of AMIC (13). One hypothetical function for this site is that it may serve as an anchor for actin-based movement. In rigor, GPA1 and GPA2 are not far (≈60 Å) from the actin filament to which the head binds and could approach it with an appropriate pivoting in the neck region. The brush border myosin 1 tail, which swings through 31°, gives a precedent for such a movement (23). A swing of this magnitude in AMIC would approximately halve the separation of the tail from the actin filament. It might, in fact, come closer, depending on the pivot point and because we are not seeing all of the GPA density that is on the actin-proximal side of the tail. This position would be close enough to make contact likely when the head is released, or the putative swing of the AMIC tail might be >31°. At present, however, this possibility is moot, as the processivity or otherwise of AMIC remains to be determined. In this context, kinetic studies of the T2 mutant of AMIC, which lacks the entire tail, show it to have a short duty cycle, which is inconsistent with processivity (37), as do AMIA and AMIB (38). However, the presence of an ATP-insensitive actin-binding site in the tail might overcome the short duty cycle by keeping the myosin attached to the actin filament, thus allowing processivity. The proposition that AMIC might be a processive monomeric motor is reminiscent of KIF1A, a kinesin superfamily member that can slide on microtubules (39) and has a Lys-rich loop at a position accessible from the microtubule (40), comparable with the situation of GPA on AMIC. However, it is still controversial whether KIF1A slides processively as a monomer in vivo (41).
Alternatively, the tail site could bind to another actin filament, increasing cortical tension as shown for Dictyostelium myosin I (42). This hypothesis is supported by the fact that F-actin is readily bundled by AMIC (unpublished observations), and by AMIA and AMIB (15, 16). In this scenario, the second actin filament would be as close to and essentially on the same side of AMIC as the first filament, at least for simultaneous binding in the rigor conformation, unless the tail unfolded when it bound to the second filament.
In our current model (Fig. 5), BR, which is charged for membrane binding (12), faces outward, away from the actin filament. This position allows it free access to a vacuolar membrane or potentially sizable vesicle, which may thereby be coupled by AMIC to the actin filament.
Supplementary Material
Acknowledgments
We thank Dr. David Belnap for advice on image analysis and providing coordinates from his earlier fit, Dr. Bridget Carragher for making phoelix available, and Dr. Jenny Hinshaw and Mr. Michael Johnson for help in installing the program. This work was supported in part by a fellowship from the Japan Society for the Promotion of Science (to T.I.).
Abbreviations: AMI, Acanthamoeba myosin I; BR, basic region; EM, electron microscopy; GPA, Gly/Pro/Ala-rich; SH3, Src homology 3.
References
- 1.Sellers, J. R. (1999) Myosins (Oxford Univ. Press, Oxford), 2nd Ed.
- 2.Berg, J. S., Powell, B. C. & Cheney, R. E. (2001) Mol. Biol. Cell 12, 780-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korn, E. D. (2000) Proc. Natl. Acad. Sci. USA 97, 12559-12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zot, H. G., Doberstein, S. K. & Pollard, T. D. (1992) J. Cell Biol. 116, 367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maruta, H., Gadasi, H., Collins, J. H. & Korn, E. D. (1978) J. Biol. Chem. 253, 6297-6300. [PubMed] [Google Scholar]
- 6.Baines, I. C., Brzeska, H. & Korn, E. D. (1992) J. Cell Biol. 119, 1193-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines, I. C., Corigliano-Murphy, A. & Korn, E. D. (1995) J. Cell Biol. 130, 591-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doberstein, S. K., Baines, I. C., Wiegand, G., Korn, E. D. & Pollard, T. D. (1993) Nature 365, 841-843. [DOI] [PubMed] [Google Scholar]
- 9.Ostap, E. M., Maupin, P., Doberstein, S. K., Baines, I. C., Korn, E. D. & Pollard, T. D. (2003) Cell Motil. Cytoskel. 54, 29-40. [DOI] [PubMed] [Google Scholar]
- 10.Kong, H.-H. & Pollard, T. D. (2003) J. Cell Sci. 115, 4993-5002. [DOI] [PubMed] [Google Scholar]
- 11.Wang, Z. Y., Sakai, J., Matsudaira, P. T., Baines, I. C., Sellers, J. R., Hammer, J. A., III, & Korn, E. D. (1997) J. Muscle Res. Cell Motil. 18, 395-398. [DOI] [PubMed] [Google Scholar]
- 12.Doberstein, S. K. & Pollard, T. D. (1992) J. Cell Biol. 117, 1241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, X., Brzeska, H. & Korn, E. D. (2000) J. Biol. Chem. 275, 24886-24892. [DOI] [PubMed] [Google Scholar]
- 14.Lynch, T. J., Brzeska, H., Miyata, H. & Korn, E. D. (1989) J. Biol. Chem. 264, 19333-19339. [PubMed] [Google Scholar]
- 15.Fujisaki, H., Albanesi, J. P. & Korn, E. D. (1985) J. Biol. Chem. 260, 11183-11189. [PubMed] [Google Scholar]
- 16.Lynch, T. J., Albanesi, J. P., Korn, E. D., Robinson, E. A., Bowers, B. & Fujisaki, H. (1986) J. Biol. Chem. 261. 17156-17162. [PubMed] [Google Scholar]
- 17.Miyanishi, T., Ishikawa, T., Hayashibara, T., Wakabayashi, T. & Maita, T. (2002) Biochemistry 41, 5429-5438. [DOI] [PubMed] [Google Scholar]
- 18.Jung, G., Remmert, K., Wu, X., Volosky, J. M. & Hammer, J. A., III (2001) J. Cell Biol. 153, 1479-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zot, H. G., Bhaskara, V. & Liu, L. (2000) Arch. Biochem. Biophys. 375, 161-164. [DOI] [PubMed] [Google Scholar]
- 20.Brzeska, H. & Korn, E. D. (1996) J. Biol. Chem. 271, 16983-16986. [DOI] [PubMed] [Google Scholar]
- 21.Carragher, B. O., Cheng, N., Wang, Z.-Y., Korn, E. D., Reilen, A., Belnap, D. M., Hammer, J. A., III, & Steven, A. C. (1998) Proc. Natl. Acad. Sci. USA 95, 15206-15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jontes, J. D., Wilson-Kubalek, E. M. & Milligan, R. A. (1995) Nature 378, 751-753. [DOI] [PubMed] [Google Scholar]
- 23.Jontes, J. D. & Milligan, R. A. (1997) J. Cell Biol. 139, 683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittaker, M., Wilson-Kubalek, E. M., Smith, J. E., Faust, L., Milligan, R. A. & Sweeney, H. L. (1995) Nature 378, 748-751. [DOI] [PubMed] [Google Scholar]
- 25.Lee, W. L., Ostap, E. M., Zot, H. G. & Pollard, T. D. (1999) J. Biol. Chem. 274, 35159-35171. [DOI] [PubMed] [Google Scholar]
- 26.Jung, G., Saxe, C. L., III, Kimmel, A. R. & Hammer, J. A., III (1989) Proc. Natl. Acad. Sci. USA 86, 6186-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jontes, J. D., Ostap, E. M., Pollard, T. D. & Milligan, R. A. (1998) J. Cell Biol. 141, 155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carragher, B. O., Whittaker, M. & Milligan, R. A. (1996) J. Struct. Biol. 116, 107-112. [DOI] [PubMed] [Google Scholar]
- 29.Conway, J. F., Trus, B. L., Booy, F. P., Newcomb, W. W., Brown, J. C. & Steven, A. C. (1996) J. Struct. Biol. 116, 200-208. [DOI] [PubMed] [Google Scholar]
- 30.Heymann, J. B. (2001) J. Struct. Biol . 133, 156-169. [DOI] [PubMed] [Google Scholar]
- 31.Saxton, W. O. & Baumeister, W. (1982) J. Microsc. 127, 127-138. [DOI] [PubMed] [Google Scholar]
- 32.Kollmar, M., Durrwang, U., Kliche, W., Manstein, D. J. & Kull, F. J. (2002) EMBO J. 21, 2517-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjieldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 34.Mendelson, R. & Morris, E. P. (1997) Proc. Natl. Acad. Sci. USA 94, 8533-8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinert, P. M., Mack, J. W., Korge, B. P., Gan, S. Q., Haynes, S. R. & Steven, A. C. (1991) Int. J. Biol. Macromol. 13, 130-139. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa, T., Maurizi, M. R. & Steven, A. C. (2004) J. Struct. Biol. 146, 180-188. [DOI] [PubMed] [Google Scholar]
- 37.Ostap, E. M., Lin, T., Rosenfeld, S. S. & Tang, N. (2002) Biochemistry 41, 12450-12456. [DOI] [PubMed] [Google Scholar]
- 38.Ostap, E. M. & Pollard, T. D. (1996) J. Cell Biol. 132, 1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada, Y. & Hirokawa, N. (1999) Science 283, 1152-1157. [DOI] [PubMed] [Google Scholar]
- 40.Kikkawa, M., Sablin, E. P., Okada, Y., Yajima, H., Fletterick, R. J. & Hirokawa, N. (2001) Nature 411, 439-445. [DOI] [PubMed] [Google Scholar]
- 41.Al-Bassam, J., Cui, Y., Klopfenstein, D., Carragher, B. O., Vale, R. D. & Milligan, R. A. (2003) J. Cell Biol. 163, 743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai, J., Ting-Beall, H. P., Hochmuth, R. M., Sheetz, M. P. & Titus, M. A. (1999) Biophys. J. 77, 1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.