Abstract
This article concerns the identification of different types of voltage-gated Na+ channels and of muscarinic and purinergic receptors that are expressed in human erythroid precursor cells and red cell ghosts. We analyzed, by RT-PCR, RNA that was extracted from purified and synchronously growing human erythroid progenitor cells, differentiating from erythroblasts to reticulocytes in 7 to 14 days. These extracts were free of white cell and platelet contamination. Two types of voltage-gated, tetrodotoxin-sensitive Na+ channels were found. These were Nav1.4 and Nav1.7, the former known to be present in skeletal muscle and the latter in peripheral nerve. By using a pan Na+ channel antibody and Western blotting, an immunoreactive channel was detected in ghosts of human red blood cells, consistent with the expression of these two channels. The transcripts for four of the five known subtypes of muscarinic receptors were also identified, including subtypes M2, M3, M4, and M5, whereas subtype M1 was not found. Expression was also detected for the purinergic type receptors P2X1, P2X4, P2X7, and P2Y1 whereas types P2Y2, P2Y4, and P2Y6 were not found. We also searched for but did not find transcripts for hBNP-1, a type 1b human brain sodium phosphate cotransporter, and cystic fibrosis transmembrane conductance regulator (CFTR). Implications regarding the presence of these different types of channels and receptors in human red blood cells and their functional significance are discussed.
This article is the third in a series that exploits the use of human erythroid progenitor cells to identify genes that code for transport proteins and receptors that reside in erythrocyte membranes and their precursors. Our previous work defined the isoform composition of the Na pump (1) and the SK4 isoform (2) responsible for the Ca2+-activated K+ channel (Gardos channel) in human red blood cells. These studies used “β profiling,” a criterion that established that the human erythroid progenitor cell cultures we analyzed were free of white cell and platelet contamination. β profiling was based on the observation that analysis of human reticulocyte RNA by RT-PCR yielded the message for the β2 but not the β1 isoform of the Na pump (3). In sharp contrast, only the β1 and not the β2 isoform of the Na pump was found in a white cell and platelet library (3). Thus, all of the human erythroid progenitor cell preparations used here displayed unambiguous β profiling, by both RT-PCR and Northern analyses (see refs. 1 and 2).
The current effort to identify the voltage-gated Na+ channels in erythroid cells emanates from a report (4) that characterized a tetrodotoxin (TTX)-sensitive Na+ influx in human red blood cells that depended on the combined addition of F– and Ca2+. K+ transport by means of the Gardos channel is known to be activated under this circumstance (5, 6), which can take place without a change in Na+ permeability at low but not at high F– concentrations (4, 7). The TTX-sensitive Na+ flux was also found to be inhibited by pertussis toxin, which implies that the increase in Na+ flux is G protein-mediated (4). There is ample evidence that heterotrimeric G proteins of several types (e.g., Gi, Go, Gq) are present in human red blood cells (8–14); in addition, these cells are also known to contain ≈2.7 μM Al3+/l.rbc (15), which may be necessary for adenylyl cyclase activation (16). It was further shown that, under conditions where the Na+ flux was increased in the presence of added F– and Ca2+, Ca2+ uptake (measured with 45Ca2+) was also increased and could be inhibited with TTX (17). A similar TTX-sensitive Ca2+ influx has been characterized in cardiac myocytes (18).
Based on these observations, we chose to search by RT-PCR our RNA preparations of human reticulocytes and erythroid progenitor cells for the presence of Na+ channel transcripts. Furthermore, because the muscarinic agonist carbamylcholine (CCh) has been reported to induce a TTX-sensitive Na+ uptake in human red blood cells (19), we analyzed our preparations for messages associated with these types of membrane receptors as well. And lastly, because human (20–22) and dog (22, 23) red blood cells are known to respond to extracellularly applied nucleotides, e.g., ATP, we also surveyed our preparations of human erythroid progenitor cells for transcripts identifying various types of purinergic receptors that might be present (24–27, ∥).
Methods
Erythroid Progenitor Cells. All of the samples of RNA used in this study were taken from the same preparations of human erythroid progenitor cells purified and cultured as described (1, 2). The primary erythroid cells used in this study were, with 80–85% synchrony, at basophilic (day 7), polychromatic (day 9), and orthochromatic (day 12) stages of maturation. As stated before, all of these preparations were free of leukocyte and platelet contamination based on the criterion of β profiling (1, 2). Therefore, the results represent the identification of constituents restricted to human erythroid tissue.
A standard multiplex RT-PCR assay was used as before (1, 2) to amplify products from various templates that are present in the cDNA pool from human erythroid progenitor cells cultured from 7–12 days. Table 1 gives the primer sequences used to identify specific isoforms of the Na+ channel whereas those in Table 2 give the primer sequences used to identify purinergic and muscarinic receptors, a sodium phosphate cotransporter and the cystic fibrosis transmembrane conductance regulator (CFTR). As a control in all cases, RT-PCR was performed with each set of primers and plasmids individually encoding each of the channel or receptor isoforms. In every case, a product of the expected size was found with >95% sequence identity. In Table 1, generic forward (F) and reverse (R) primers were designed against highly conserved sequences of Na+ channels, delineated by transmembrane segment 5 of domain 3 and the cytoplasmic loop joining domains 3 and 4 (30, 31). The N terminus of this channel fragment is the sequence MNVLV (M is residue 1161), and the C terminus is the sequence MKKLG (G is residue 1323) of human Nav1.4 sequence (GenBank accession no. M81758). The atypical Na+ channel (Nax) was not included in the current analysis because its function as a voltage-gated Na+ channel remains unconfirmed. Specific primer pairs are optimal for the amplification of a subset of the channels. However, multiple primers may amplify individual channels depending on the stringency of the PCR amplification conditions and the abundance of the channel templates.
Table 1. PCR primer sequences used to identify specific isoforms of the Na channel.
| Symbol | Isoform specificity | Expected size, bp | Primer sequence* |
|---|---|---|---|
| F1 | Nav1.4/1.6 | 484/480 | 5′-CTKGTSTGYCTCATCTTCTGG-3′ |
| F2 | Nav1.3/1.5/1.8 | 471/483/480 | 5′-CTSGTCTGYCTCATCTTCTGG-3′ |
| F3 | Nav1.1/1.2/1.7 | 480 | 5′-CTKGTKTGTCTKATMTTYTGG-3′ |
| F4 | Nav1.9 | 462 | 5′-CTTGTCTGCCTSATKWTCTGG-3′ |
| R1 | All | 5′-CATGGCATTRTAGTAYTTCTTCTG-3′ |
See Methods for details.
K = G or T, S = C or G, Y = T or C, M = A or C, and W = A or T.
Table 2. PCR primer sequences used to search/identify specific isoforms of purinergic and muscarinic receptors, a NaPO4 cotransporter and CFTR.
| Specific primers | Expected size, bp | Primer sequence |
|---|---|---|
| Purinergic receptors | ||
| P2X1 | 463 | 5′-CTGTGAAGACGTGTGAGATCTTTGG-3′ |
| 5′-TTGAAGAGGTGACGGTAGTTGGTC-3′ | ||
| P2X4 | 296 | 5′-GAGATTCCAGATGCGACC-3′ |
| 5′-GACTTGAGGTAAGTAGTGG-3′ | ||
| P2X7 (X7-like) | 675 | 5′-AACATCACTTGTACCTTCC-3′ |
| 5′-TGTGAAGTCCATCGCAGG-3′ | ||
| P2Y1 | 528 | 5′-CGGTCCGGGTTCGTCC-3′ |
| 5′-CGGACCCCGGTACCT-3′ | ||
| P2Y2 | 638 | 5′-CTCTACTTTGTCACCACCAGCG-3′ |
| 5′-TTCTGCTCCTACAGCCGAATGTCC-3′ | ||
| P2Y4 | 425 | 5′-CCACCTGGCATTGTCAGACACC-3′ |
| 5′-GAGTGACCAGGCAGGGCACGC-3′ | ||
| P2Y6 | 365 | 5′-CGCTTCCTCTTCTATGCCAACC-3′ |
| 5′-CCATCCTGGCGGCACAGGCGGC-3′ | ||
| Muscarinic receptors | ||
| M1 | 538 | 5′-CAGGCAACCTGCTGGTACTC-3′ |
| 5′-CGTGCTCGGTTCTCTGTCTC-3′ | ||
| M2 | 654 | 5′-CTCCTCTAACAATAGCCTGG-3′ |
| 5′-GGCTCCTTCTTGTCCTTCTT-3′ | ||
| M3 | 560 | 5′-GGACAGAGGCAGAGACAGAA-3′ |
| 5′-GAAGGACAGAGGTAGAGTGG-3′ | ||
| M4 | 503 | 5′-ATCGCTATGAGACGGTGGAA-3′ |
| 5′-GTTGGACAGGAACTGGATGA-3′ | ||
| M5 | 752 | 5′-ACCACAATGCAACCACCGTC-3′ |
| 5′-ACAGCGCAAGCAGGATCTGA-3′ | ||
| NaPO4 cotransporter | ||
| hBNP-1 | 389 | 5′-GGAGGAGCGCAAGTACATCGAG-3′ |
| 5′-AGTAGCCGACCACCAACAGCAG-3′ | ||
| CFTR | ||
| 4101F | 527 | 5′-GCTCAGATCTGTGATAGAACAGTTTCCT-3′ |
| 4330F | 318 | 5′-TGTGAACACAGGATAGAAGCAATGCT-3′ |
| 4647R | 5′-TTCCATGAGGTGACTGTCC-3′ | |
Primer sequences for the purinergic receptors are taken from ref. 28; for the muscarinic receptors from ref. 29; for the NaPO4 cotransporter as suggested by Robert B. Gunn (Emory University, Atlanta); and for CFTR as suggested by Jackie Zhou (University of North Carolina, Chapel Hill). See Methods for details.
22Na Efflux. Fresh normal heparinized blood was centrifuged, and the red cells, after washing four times with ≈10 vol of an isotonic solution containing 150 mM NaCl, 20 mM Hepes, 3 mM adenosine, and 5 mM glucose at pH 7.4, were incubated in this solution for ≈3 h at 37°C after the addition of 200 μCi (1 Ci = 37 GBq) of 22Na and chloramphenicol. The suspension was then centrifuged, and the cells were washed four times with a solution containing 145 mM NaCl, 5 mM KCl, and 20 mM Hepes, to remove extracellular 22Na. The 22Na-loaded cells were then suspended (≈2% hematocrit) at 4°C in this solution in the presence and absence of 2.5 mM CaCl2 and 10 mM NaF and in the presence and absence of 3.3 μM TTX. The flasks were then placed at 37°C, and, after a 3-min equilibration, samples were withdrawn at the indicated times and centrifuged, and the supernatants were counted for 22Na. All conditions were performed in duplicate whose variation was <5%. Calculation of the percent 22Na released, outward rate constants, and Na+ efflux was the same as described (32). TTX was purchased from Calbiochem and 22Na from PerkinElmer, and all other chemicals, wherever possible, were of reagent grade.
Western Blotting. Human red blood cell ghosts, prepared as described (33), were used in an immunoblot assay to explore the possible presence of voltage-gated Na+ channel proteins. Two stable HEK 293 cell lines expressing either human Nav1.4 (HEK-Nav1.4) or human Nav1.7 (HEK-Nav1.7) Na+ channels served as a positive control for the assay. HEK-Nav1.4, HEK-Nav1.7, and red cell ghosts were solubilized in 2× NuPAGE sample buffer with reducing agent (Invitrogen) at 50°C for 18 min. Samples (100 μg) were run on a 3–8% NuPAGE Trisacetate SDS-polyacrylamide denaturing and reducing gel system and transferred to a poly(vinylidene difluoride) (PVDF) membrane. The PVDF membrane was blocked with 10% dry milk overnight and then incubated with mouse pan Na+ channel antibody (Sigma), at 1 μg/ml in 5% BSA/TBST, for 3 h at room temperature. The pan Na+ channel antibody recognizes amino acid sequences in the linker between domains 3 and 4 of the channel (TEEQKKYYNAMKKLGSKK), which are highly conserved among all members of the voltage-gated Na+ channel family. Detection of the immunoreactive proteins was carried out by using horseradish peroxidase-conjugated anti-mouse IgG secondary antibody (DAKO), 1/10,000 in 1.25% fetal serum albumin/Tris-buffered saline Triton (FSA/TBST), with Western Lightning, Chemiluminescence Reagent Plus (PerkinElmer).
Results
Our first undertaking concerns the possible presence of voltage-gated TTX-sensitive Na+ channels in human red blood cell membranes. We felt it was important to repeat the initial observation (4) that exposure of human red cells to the combination Ca2+ plus F– turned on a TTX-sensitive Na+ permeability. The results presented in Fig. 1 show that the stimulation induced by Ca2+ plus F– could be inhibited with TTX, thereby confirming the primary observation. It should be noticed that the time course of Na+ efflux reflects first an immediate, initial burst followed by a slower rate paralleling the control and TTX-inhibited effluxes, consistent with the original report (4). Mention should be made that essentially no hemolysis occurred in the experiment reported in Fig. 1, but this was not always the case. Even so, the TTX-sensitive efflux was still apparent in other experiments of a similar type after correction for 22Na that was released from the cells by hemolysis.
Fig. 1.
Stimulation of 22Na efflux from human red blood cells by Ca2+ plus F– and its inhibition by TTX. 22Na-loaded cells suspended in a solution containing 145 mM NaCl, 5 mM KCl, and 20 mM Hepes (pH 7.4) were incubated at 37°Cin the presence and absence (control) of 2.5 mM CaCl2 and 10 mM NaF. The percent 22Na released from the cells was measured at the indicated times. All conditions were carried out in duplicate with <5% variation. It is evident that 3.3 μmol of TTX inhibited the initial burst of stimulation of 22Na efflux that occurred in the presence of Ca2+ plus F–, bringing the remaining efflux to essentially control levels. The effluxes, calculated for the 8-min time point of the control, Ca2+ plus F–, and Ca2+ plus F– plus TTX conditions were, respectively, 3.3, 30.4, and 4.9 mM Na/liter cells per hr. The cells contained 8.2 mmol Na/liter cells and 64.1% H2O. See Methods for details and text for discussion.
Having established the validity of a TTX-sensitive Na+ flux in human red cells, we next considered the identity of the type of Na+ channels underlying this effect. For this study, we used cDNA derived from human erythroid progenitor cells and an RT-PCR reaction assay (30) to determine which, if any, specific voltage-gated Na+ channels were expressed.
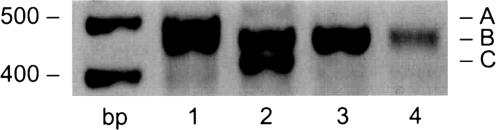
Fig. 2 shows the amplification products in the expected size range of 462–483 bp that were obtained from cDNA templates of cells harvested after 7 days in culture. A signal was obtained in reactions by using the primer pairs (see Table 1) F1/R1 (lane 1), F2/R1 (lane 2), and F3/R1 (lane 3), but only a trace signal in reactions by using primer pair F4/R1 (lane 4). Sequencing of either the PCR products directly or of clones of these products in the vector pTargeT (Promega) identified transcripts of Nav1.4 and Nav1.7 Na+ channels (B in lanes 1–3), and verified that bands A and C (lanes 1 and 2) are artifacts of the amplification conditions. The amplification of Nav1.4 by F1/R1 and that of Nav1.7 by F3/R1 reflect the optimal primer combination for the respective channels (Fig. 2). The amplification of Nav1.4 by F2/R1, however, is likely due to absence of Nav1.3, Nav1.5 and Nav1.8 templates, which normally would be amplified by this primer combination. The fact that only two channels are detected under these conditions supports the conclusion that these channels are specifically expressed in the human erythroid progenitor cells (see ref. 34 for nomenclature and tissue distribution of voltage-gated Na+ channels).
Fig. 2.
Evidence for the expression of voltage-gated Na+ channels in human erythroid progenitor cells. The results were duplicated on cells derived from two different individuals' blood after 7 days in culture. PCR products obtained by using blood reticulocyte cDNA as template and Na+ channel primers (Table 1) were analyzed on 1.6% agarose gel. The mass ladder in base pairs is given on the left. The presence of amplification products in the expected size range suggested the expression of multiple Na+ channels as defined in Table 1. The F1/R1 primer combination is expected to amplify Nav1.4 and Nav1.6; F2/R1 amplifies Nav1.3, -1.5, and -1.8; F3/R1 amplifies Nav1.1, -1.2, and -1.7; and F4/R1 amplifies Nav1.9. Band size and sequence analysis (>98% expected identity) reveals that band B in lanes 1 and 2 corresponds to Nav1.4 and in lane 3 corresponds to Nav1.7 whereas bands A and C are artifacts. The product in lane 4 was not reproducible in multiple amplification reactions and was not further characterized because of the low signal.
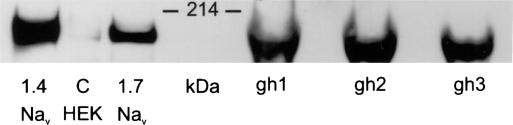
The next question is: are these two Na+ channels expressed in mature human erythrocytes? Thus, by using a monoclonal pan Na+ channel antibody, Western blot analysis shows (see Fig. 3) the presence of an immunoreactive band in three different samples of human erythrocyte ghosts and in HEK 293 cells expressing either human Nav1.4 and/or human Nav1.7 Na+ channel. The protein species migrates in the gel slightly faster than the 214-kDa protein marker (Bio-Rad), consistent with the theoretical molecular mass of Nav1.4 (208 kDa) and Nav1.7 (225 kDa). The HEK 293 cell lines that were used in this assay have been used before to express voltage-gated Na+ currents (35). These results indicate that either or both of the two different types of Na+ channels could reside in human red cell membranes.
Fig. 3.
Western blot showing that Na+ channel proteins are present in human red blood cell ghosts. This analysis was carried out by using a monoclonal pan Na+ channel antibody that, as shown on the left side, detects a single immunoreactive band from HEK 293 cells expressing both the human Nav1.4 and human Nav1.7 Na+ channels. A similar immunoreactive band is detected in three independent preparations of human ghosts, made from the red blood cells of three individuals (labeled gh1 to gh3). To balance the intensity of the immunoreactive blots of the HEK cell line lysates with the human ghosts preparations, 20 μg and 100 μg of the cell lysates and ghost proteins, respectively, were loaded on the gel. The control, C, represents the untransfected HEK 293 cell lysate and shows only a very faint signal, consistent with the presence of very low levels of native Na+ channels. The position of the 214-kDa protein molecular mass marker is indicated.
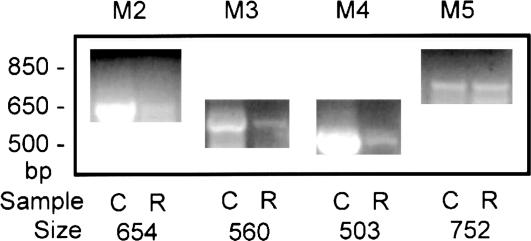
Exposure of human red blood cells to Ca2+ plus F– is not the only way to induce a TTX-sensitive Na+ permeability because the muscarinic agonist CCh has also been reported (19) to have the same effect. The parallel is striking in that the time course of CCh activation is similar to that seen in Fig. 1 and also seems to depend on the presence of Ca2+. That transcripts for muscarinic receptors were also expressed in our preparations of human erythroid progenitor cells is evident from the results presented in Fig. 4, where C is the positive control (human brain) and R represents the erythroid cells. Here, it is clear that four of the five known subtypes of muscarinic receptors are present in these cells. By using the primers listed in Table 2, we found that the products obtained for M2-M5 were of the expected size and showed >97% identity with the expected sequences. We found no evidence for the presence of the M1 subtype.
Fig. 4.
Evidence that the transcripts for four different isoforms of muscarinic receptors are present in human erythroid progenitor cells cultured for 7 days. The positive control blot for each of the four muscarinic receptors is represented by the C lane, with single-stranded cDNA derived from human brain tissue used as template (Clontech). R represents the results obtained with single-stranded cDNA derived from the progenitor cells used as templates (see Methods). The mass ladder (in base pairs) is shown on the left (Life Technologies, Grand Island, NY). For each of the four muscarinic isoforms M2 through M5, PCR products of approximately the expected sizes were found. Each product was sequenced and in every case was found to have >97% identity with its expected sequence. These same results were also obtained on another individual's set of human erythroid progenitor cells processed after 7 days in culture. See text for discussion.
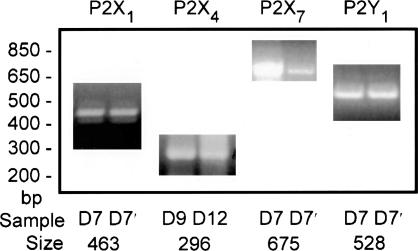
As mentioned earlier, human and other types of red blood cells respond to extracellularly applied nucleotides. This finding led us to survey our preparations of human erythroid progenitor cells for possible types of purinergic receptors that might be present. The results depicted in Fig. 5, by using the primers defined in Table 2, provide clear evidence that the message for purinergic types P2X1, P2X4, P2X7 (or P2X7-like), and P2Y1 are present. Not only are the RT-PCR products of the expected size, but the expected sequence identity was >99% for P2X1, P2X4, and P2Y1; 97% for P2X7; and >88% for P2X7-like. Other purinergic receptor types that we looked for but did not find were P2X2, P2Y4, and P2Y6.
Fig. 5.
Evidence that the transcripts for four different types of purinergic receptors are present in two different samples of human erythroid progenitor cells in culture for 7 (and 7′), 9, or 12 days. PCR products were obtained with the isoform-specific primers as defined in Table 2. Single-stranded cDNA derived from human erythroid progenitor cells was used as template (see Methods). The mass ladder (in base pairs) is given on the left. The approximate expected size product was found (see Table 2) for each of the four types of purinergic receptors presented in this figure. In each case, the product was sequenced and in every instance was found to have >96% identity with the expected sequence. See text for discussion.
Two other types of transcripts that we searched for in the cDNAs prepared from our human erythroid progenitor cells but did not find were hBNP-1 and CFTR (data not shown). hBNP-1 is an Na+-phosphate cotransporter that has been identified and functionally characterized in human red blood cells (36). There is accumulating evidence that CFTR protein resides in human red blood cell membranes (37–39) although we did not detect its message in the progenitor cells, but see Note Added in Proof.
Discussion
Human red blood cells are normally thought to be inexcitable, yet, as shown in this article, they harbor two types of voltage-gated, TTX-sensitive Na+ channels. The type Nav1.4 (also denoted as SkM1 or SCN4A) is known to be primarily present in skeletal muscle whereas the other type, Nav1.7 (also denoted as PN1 or SCN9A), is found primarily in peripheral nervous tissue. These channels are evidently cryptic in normal red blood cells and require, for activation along with Ca2+, either F– (4) or a muscarinic agonist such as CCh (19). In both cases, an Na+ permeability is turned on at least for a brief time (see refs. 4 and 19) that seems to be G protein-mediated. The number of copies of either type of Na+ channels that are resident in human red blood cell membranes is not known. It is also not known in either case what role the membrane potential plays in the permeability change. A further caveat that should be emphasized is that we have no information that directly relates the identified Na+ channel types to the observed changes in Na+ permeability.
In human red blood cells, low concentrations of F– together with Ca2+ are known to activate a Ca-activated K+ channel (Gardos channel) with a concomitant hyperpolarization of the membrane (5, 40). But the value of the membrane potential that occurs at higher F– concentrations, when the Na+ permeability transiently increases (e.g., Fig. 1), is not known, nor is its time course known relative to the time course of the hyperpolarization due to the Gardos channel activation. It would be interesting, for instance, to study the activation of this process in high K+ media and/or with the Gardos channel inhibited. The time course of the increase in Na+ permeability that occurs with exposure of human red blood cells to Ca2+ plus CCh correlates with the time course (initial increase) of changes in Ca2+ and cGMP content (41). Whether the pattern of these changes applies to the Ca2+ plus F– case is not known although the transient increase in a TTX-sensitive Ca2+ uptake has been correlated with the Na+ permeability changes (17). The implication here is that activation of muscarinic receptors by means of G proteins such as Gi, Gs, and Gq, which are known to be present in human red blood cell membranes (8–14), uses the same Na+ channels as does the activation by the combination Ca2+ plus F–. More studies are needed to evaluate this relationship. By means of [3H]quinuclidinyl benzilate binding, the density of muscarinic receptors in human red blood cells has been estimated (42, 43) to be 23 fmol/mg membrane protein (≈7 sites per cell). It should be emphasized that, whereas we identified the transcripts for only four different muscarinic receptors, we do not know which subtypes are expressed in the mature red blood cell membrane although it is likely that subtypes M2 and M4 are preferred. This is because subtypes M2 and M4, involving Gi-type G proteins, are pertussis toxin-sensitive in contrast to subtypes M1, M3, and M5, which are coupled to Gq-type G proteins and are pertussis toxin-insensitive (44, 45).
It is of interest to speculate about the functional significance of cholinergic (muscarinic) receptors on human red blood cells. Acetylcholinesterase known to be present on the surfaces of human red blood cells (see ref. 46) can be considered, by its activity, to ensure localized action, say on vagal stimulation of the heart, and thereby reducing the distal effectiveness of acetylcholine in the peripheral circulation. This type of activity might also function to prevent red cell receptor-associated Na+ channel activation, whose consequences have yet to be determined.
Mention should be made that there is evidence that Cav2.1-type Ca2+ channels are present in human red blood cells (47), but these channels are not known to be TTX-sensitive. It has been reported that CCh plus Ca2+ increases the “rigidity” of human red blood cell membranes and that TTX inhibits this effect in the absence of Ca2+ (48, 49). The significance of these results relative to the present work is not clear. We could not repeat the claimed effects of TTX on changes in cation permeability of human red cells treated with lipid vesicles (50).
As pointed out before, human red blood cells respond to the application of externally applied nucleotides and were thought to express purinergic receptors on their surfaces. We identified (Fig. 5) four kinds of mRNAs representing two classes of purinergic receptors in human erythroid progenitor cells. P2X types are known to be ATP/ADP-gated in contrast to P2Y types, which are PLC/adenylyl cyclase (G protein)-mediated. We do not know the extent to which any type that we identified is expressed on the mature human red blood cell membrane. Whereas human red blood cells are known to display ecto-ATPase (ectonucleotidase) type activity (e.g., ref. 51), these proteins are thought to be different from those contained in purinergic receptors (see ref. 52).
Acknowledgments
We thank Drs. Cathy Berlot, Maria Donoghue-Velleca, Cecilia Canessa, and Michael Caplan for their helpful advice during the course of this work. We also thank Drs. Richard Boucher (University of North Carolina, Chapel Hill), Jackie Zhou, and Robert Gunn for providing primer sequences and Lynda Tyrrell for technical assistance. This work was supported in part by National Institutes of Health Grant HL 09906 (to J.F.H.) and the Rehabilitation and Medical Research Services, Department of Veterans Affairs (S.D.D.-H.).
Abbreviations: TTX, tetrodotoxin; CCH, carbamylcholine; CFTR, cystic fibrosis transmembrane conductance regulator.
Note Added in Proof. Immunoblots for CFTR were carried out by J. R. Riordan (Samuel C. Johnson Medical Research Center, Scottsdale, AZ) on human red blood cell ghosts prepared by us as described in Methods. By use of a panel of different specific antibodies (53–55), CFTR protein could not be detected in the ghosts from four different individuals (J. R. Riordan, personal communication).
Footnotes
Schillers, H., Stumpf, A., Wenners-Epping, K., Waite, M. & Oberleithner, H. (2004) Deutsche Physiologische Gesellschaft Congress, March 14–17, 2004, Leipzig, Germany, topic category 101, Session 029 (abstr.).
References
- 1.Hoffman, J. F., Wickrema, A., Potapova, O., Milanick, M. & Yingst, D. R. (2002) Proc. Natl. Acad. Sci. USA 99, 14572–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman, J. F., Joiner, W., Nehrke, K., Potapova, O., Foye, K. & Wickrema, A. (2003) Proc. Natl. Acad. Sci. USA 100, 7366–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stengelin, M. K. & Hoffman, J. F. (1997) Proc. Natl. Acad. Sci. USA 94, 5943–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varečka, L., Peterajová, E. & Ševčik J. (1995) Biochem. Biophys. Res. Commun. 217, 286–291. [DOI] [PubMed] [Google Scholar]
- 5.Wilbrandt, W. (1937) in The Properties and Functions of Membranes, Natural and Artificial, Transactions of the Faraday Society (Gurney and Jackson, London), pp. 956–959.
- 6.Gárdos, G. (1989) Acta Physiologica (Hungary) 15, 121–125. [PubMed] [Google Scholar]
- 7.Passow, H. (1964) in The Red Blood Cell, eds. Bishop, C. & Surgenn, D. M. (Academic, New York), pp. 71–145.
- 8.Birnbaumer, L., Codina, J., Mattera, R., Yatani, A., Graf, R., Olate, J., Sanford, J. & Brown, A. M. (1988) Cold Spring Harbor Symp. Quant. Biol. 53, 229–238. [DOI] [PubMed] [Google Scholar]
- 9.Damonte, G., Sdraffa, A., Zocchi, E., Guida, L., Polvani, C., Tonetti, M., Benatti, U., Boquet, P. & De Flora, A. (1990) Biochem. Biophys. Res. Commun. 166, 1398–1405. [DOI] [PubMed] [Google Scholar]
- 10.Graf, R., Mattera, R., Codina, J., Evans, T., Ho, Y.-K., Estes, M. K. & Birnbaumer, L. (1992) Eur. J. Biochem. 210, 609–619. [DOI] [PubMed] [Google Scholar]
- 11.English, D., Akard, L. P., Taylor, G. S., Rizzo, M. T., Dominguez, J. & Garcia, J. G. N. (1992) J. Lab. Clin. Med. 119, 87–98. [PubMed] [Google Scholar]
- 12.Johnson, G. J., Leis, L. A. & Dunlop, P. C. (1996) Biochem. J. 318, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olearczyk, J. J., Stephenson, A. H., Lonigro, A. J. & Sprague, R. S. (2001) Med. Sci. Monit. 7, 669–674. [PubMed] [Google Scholar]
- 14.Olearczyk, J. J., Stephenson, A. H., Lonigro, A. J. & Sprague, R. S. (2004) Am. J. Physiol. 286, H940–H945. [DOI] [PubMed] [Google Scholar]
- 15.Kehoe, R. A., Cholak, J. & Story, R. V. (1940) J. Nutrition 19, 579–592. [Google Scholar]
- 16.Sternweis, P. C. & Gilman, A. G. (1982) Proc. Natl. Acad. Sci. USA 79, 4888–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varečka, L., Peterajová, E. & Pišová, E. (1998) FEBS Lett. 433, 157–160. [DOI] [PubMed] [Google Scholar]
- 18.Santana, L. F., Gómez & Lederer, W. J. (1998) Science 279, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 19.Szász, I. & Gárdos, G. (1974) FEBS Lett. 44, 213–216. [DOI] [PubMed] [Google Scholar]
- 20.Kashket, S. & Denstedt, O. F. (1958) Can. J. Biochem. Physiol. 36, 1057–1064. [PubMed] [Google Scholar]
- 21.Trams, E. G. (1980) J. Theor. Biol. 87, 609–621. [DOI] [PubMed] [Google Scholar]
- 22.Romualdez, A., Volpi, M. & Sha'afi, R. I. (1976) J. Cell. Physiol. 87, 297–306. [DOI] [PubMed] [Google Scholar]
- 23.Parker, J. C. & Snow, R. L. (1972) Am. J. Physiol. 223, 888–893. [DOI] [PubMed] [Google Scholar]
- 24.Bernie, C. O., Hawkins, P. T., Stephens, L. R., Harden, T. K., Downes, C. P. (1989) Mol. Pharmacol. 35, 526–532. [PubMed] [Google Scholar]
- 25.Boyer, J. L., Downes, C. P., Harden, T. (1989) J. Biol. Chem. 264, 884–890. [PubMed] [Google Scholar]
- 26.Di Virgilio, F., Chiozzi, P., Ferrari, D., Falzoni, S., Sanz, J. M., Morelli, A., Torboli, M., Bolognesi, G. & Baricordi, O. R. (2001) Blood 97, 587–600. [DOI] [PubMed] [Google Scholar]
- 27.Light, D. B., Dahlstrom, P. K., Gronau, R. T., Baumann, N. L. (2001) J. Membr. Biol. 182, 193–202. [DOI] [PubMed] [Google Scholar]
- 28.Adrian, K., Bernhard, M. K., Breitinger, H.-G. & Ogilvie, A. (2000) Biochim. Biophys. Acta 1492, 127–138. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J., Krysiak, P. S., Laurier, L. G., Sims, S. M. & Preiksaitis, H. G. Am. J. Physiol. 279, G1059–G1069. [DOI] [PubMed]
- 30.Dib-Hajj, S. D., Tyrrell, L., Black, J. A. & Waxman, S. G. (1998) Proc. Natl. Acad. Sci. USA 95, 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catterall, W. A. (2000) Neuron 26, 13–25. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman, J. F. (1962) J. Gen. Physiol. 45, 837–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proverbio, F. & Hoffman, J. F. (1977) J. Gen. Physiol. 69, 605–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldin, A. L., Barchi, R. L., Caldwell, J. H., Hofmann, F., Howe, J. R., Hunter, J. C., Kallen, R. G., Mandel, G., Meisier, M. H., Netter, Y. B., Noda, M., Tamkin, M. M., Waxman, S. G., Wood, J. N. & Catterall, W. A. (2000) Neuron 28, 365–368. [DOI] [PubMed] [Google Scholar]
- 35.Cummins, T. R., Howe, J. R. & Waxman, S. G. (1998) J. Neurosci. 18, 9607–9619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmer, R. T. & Gunn, R. B. (2000) J. Gen. Physiol. 116, 363–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sprague, R. S., Ellsworth, M. L., Stephenson, A. H., Kleinhenz, M. E. & Lonigro, A. J. (1998) Am. J. Physiol. 275, H1726–H1732. [DOI] [PubMed] [Google Scholar]
- 38.Sprague, R. S., Stephenson, A. H., Ellsworth, M. L., Keller, C. & Lonigro, A. J. (2001) Exp. Biol. Med. 226, 434–439. [DOI] [PubMed] [Google Scholar]
- 39.Abraham, E. H., Sterling, K. M., Kim, R. J., Salikhova, A. Y., Huffman, H. B., Crockett, M. A., Johnston, N., Parker, H. W., Boyle, W. E., Jr., Hartov, A., et al. (2001) Blood Cells Mol. Dis. 27, 165–180. [DOI] [PubMed] [Google Scholar]
- 40.Bennekou, P. & Christophersen, P. (2003) Red Cell Membrane Transport in Health and Disease, eds. Bernhardt, I. & Ellory, J.C. (Springer, Berlin).
- 41.Tang, L. C., Schoomaker, E. & Wiesmann, W. P. (1984) Biochim. Biophys. Acta 772, 235–238. [DOI] [PubMed] [Google Scholar]
- 42.Tang, L. C. (1986) Gen. Pharmacol. 17, 281–285. [DOI] [PubMed] [Google Scholar]
- 43.Aronstam, R. S., Abood, L. G. & MacNeil, M. K. (1977) Life Sci. 20, 1175–1180. [DOI] [PubMed] [Google Scholar]
- 44.Offermanns, S., Wieland, T., Homann, D., Sandmann, J., Bombien, E., Spicher, K., Schultz, G. & Jakobs, K. H. (1994) Mol. Pharmacol. 45, 890–898. [PubMed] [Google Scholar]
- 45.Bräuner-Osborne, H. & Brann, M. R. (1996) Eur. J. Pharmacol. 295, 93–102. [DOI] [PubMed] [Google Scholar]
- 46.Augustinsson, K. B. (1948) Acta Physiol. Scand. 15, Suppl. 52, 1–182. [Google Scholar]
- 47.Andrews, D. A., Yang, L. & Low, P. S. (2002) Blood 100, 3392–3399. [DOI] [PubMed] [Google Scholar]
- 48.Huestis, W. H. & McConnell, H. M. (1974) Biochem. Biophys. Res. Commun. 57, 726–732. [DOI] [PubMed] [Google Scholar]
- 49.Huestis, W. H. (1976) J. Supramol. Struct. 4, 355–365. [DOI] [PubMed] [Google Scholar]
- 50.Huestis, W. H. (1977) J. Biol. Chem. 252, 6764–6768. [PubMed] [Google Scholar]
- 51.Herbert, E. (1956) J. Cell. Comp. Physiol. 47, 11–36. [DOI] [PubMed] [Google Scholar]
- 52.Zimmerman, H. (1999) Trends Pharmacol. Sci. 20, 231–236. [DOI] [PubMed] [Google Scholar]
- 53.Gentzsch, M., Cui, L., Mengos, A., Chang, X., Chen, J. H. & Riordan, J. R. (2003) J. Biol. Chem. 278, 6440–6449. [DOI] [PubMed] [Google Scholar]
- 54.Mall, M., Kreda, S. M., Mengos, A., Jensen, T. J., Hirtz, S., Seydewitz, H. H., Yankaskas, J., Kunzelmann, K., Riordan, J. R. & Boucher, R. C. (2004) Gastroenterology 126, 32–41. [DOI] [PubMed] [Google Scholar]
- 55.Gentzsch, M., Chang, X. B., Cui, L., Wu, Y., Ozols, V. V., Choudhury, A., Pagano, R. E. & Riordan, J. R. (2004) Mol. Biol. Cell 15, 2684–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]