Abstract
Aims: Nuclear factor erythroid 2 (NF-E2)-related factor 2 (Nrf2) is the master transcriptional regulator of antioxidant gene expression. On increased oxidative stress, an adaptor for Nrf2 degradation, Kelch-like ECH-associated protein 1 (Keap1), is directly modulated by oxidants in the cytoplasm, which results in stabilization and activation of Nrf2. Nrf2 is also constitutively active, to some extent, in the absence of exogenous oxidative stress. We have previously demonstrated that intestinal epithelium-specific TGF-β-activated kinase 1 (TAK1) deletion downregulates the level of Nrf2 protein, resulting in an increase of reactive oxygen species (ROS) in a mouse model. We aim at determining the mechanism by which TAK1 modulates the level of Nrf2.
Results: We found that TAK1 upregulated serine 351 phosphorylation of an autophagic adaptor protein, p62/Sequestosome-1 (SQSTM1), which facilitates interaction between p62/SQSTM1 and Keap1 and subsequent Keap1 degradation. This, ultimately, causes increased Nrf2. Tak1 deficiency reduced the phosphorylation of p62/SQSTM1, resulting in decreased steady-state levels of Nrf2 along with increased Keap1. We also found that this regulation is independent of the canonical redox-mediated Nrf2 activation mechanism. In Tak1-deficient intestinal epithelium, a synthetic phenolic electrophile, butylated hydroxyanisole still effectively upregulated Nrf2 and reduced ROS.
Innovation: Our results identify for the first time that TAK1 is a modulator of p62/SQSTM1-dependent Keap1 degradation and maintains the steady state-level of Nrf2.
Conclusion: TAK1 regulates Nrf2 through modulation of Keap-p62/SQSTM1 interaction. This regulation is important for homeostatic antioxidant protection in the intestinal epithelium. Antioxid. Redox Signal. 25, 953–964.
Keywords: : intestine, TAK1, Nrf2, Keap1, p62/SQSTM
Introduction
Nrf2 is a member of the Cap‘n’ Collar/basic leucine zipper (CNC)/(bZIP) family of proteins and transcriptionally upregulates a number of antioxidant genes (23, 27, 31). Cytoplasmic Nrf2 is constantly associated with Kelch-like ECH-associated protein 1 (Keap1) (20). Keap1 acts as an adaptor molecule for the Cul3 E3 ubiquitin ligase, which targets Nrf2 for degradation via the proteasome pathway (25). Covalent modifications such as oxidation on Keap1 cause conformational changes, which block Cul3-directed ubiquitylation of Nrf2. Consequently, stabilized Nrf2 is accumulated and translocates into the nucleus (3, 21). Nuclear Nrf2 binds to a specific DNA so-called antioxidant responsive element (ARE) and upregulates an array of antioxidant genes, including glutathione S-transferase (GST), NADPH quinone oxidoreductase-1 (NQO1), and heme oxygenase-1 (HO1) (48).
Nrf2-Keap1 complex is primarily regulated by the cellular redox status through modification of specific cysteine residues, including Cys151, Cys273, and Cys288 of mouse Keap1 (9, 10, 53, 55). Increased cellular reactive oxygen species (ROS) oxidize thiols to disulfides of these cysteine residues, or electrophilic oxidative stressors; sulforaphane and quinone compounds such as tert-butylhydroquinone (tBHQ) directly modify these cysteine residues, which cause conformational changes of Keap1 and stabilize Nrf2 (1, 34, 49). Additionally, Nrf2 is degraded by another type of E3 ligase system, βTrCP, in a Keap1-independnt manner in response to Wnt signaling (6, 44, 45). Nrf2 stability is also influenced by cellular autophagic activity, in which a scaffold protein, p62/SQSTM1, binds to and recruits Keap1 into autophagosomes, leading to autophagic degradation of Keap1 and subsequent stabilization of Nrf2 (7, 22, 32, 33). In this regulation, a specific phosphorylation event on serine 351 in p62/SQSTM1 has been recently implicated as a mediator of interaction between p62/SQSMT1 and Keap1 (16).
Innovation.
This study identified a previously uncharacterized nuclear factor erythroid 2 (NF-E2)-related factor 2 (Nrf2) regulation, which is different from the canonical redox-dependent regulation of Nrf2. We found that an inflammatory signaling protein kinase, TGF-β-activated kinase 1 (TAK1), modulates an autophagic adaptor protein p62/SQSTM1 and facilitates downregulation of Keap1, which results in increased Nrf2. The canonical pathway modulates Keap1 on oxidative stress, whereas TAK1 modulates steady-state levels of Nrf2, which is important for normal tissue homeostasis in the intestinal epithelium in vivo. This mechanism may link inflammatory signaling pathways with the redox system.
Collectively, Nrf2 is regulated through multiple different mechanisms. Although stimuli-dependent regulations provide a means to deal with stimuli-dependent acute increase of ROS, the mechanism for maintenance of the steady-state Nrf2 activity is still elusive.
Mitogen-activated protein kinase kinase kinase 7, also called TGF-β-activated kinase 1 (TAK1) is an intermediate of inflammatory signal transduction pathways, which is commonly activated by a diverse set of inflammatory stimuli, including proinflammatory cytokines, microorganism moieties, and chemical and physical stressors. TAK1 transmits signals from receptor or nonreceptor activated signaling complexes to downstream targets, including, but not limited to, two types of protein kinase cascades: mitogen-activated protein kinase (MAPK) cascade and IκB kinase (IKK)-NF-κB cascade (36, 39). TAK1 is activated by its binding protein TAK1-binding protein 1 (TAB1)-mediated autophosphorylation, or by an adaptor protein, TAK1-binding protein 2 (TAB2)-mediated polyubiquitin-dependent mechanism (36). TAK1, in turn, phosphorylates and activates downstream protein kinases.
Genetic ablation of TAK1 in mouse models has revealed that TAK1 is required for the prevention of oxidative tissue injury in the epidermis, intestinal epithelium, liver, and skin tumors (18, 24, 41, 42). Tak1 deficiency spontaneously upregulates ROS in the absence of any exogenous stimulation, suggesting that TAK1 regulates cellular anti-oxidant levels under steady-state conditions in those tissues. The mechanism by which Tak1 deficiency upregulates ROS has been partly determined. We have demonstrated that the MAPK, JNK, and its target c-Jun are involved in TAK1 regulation of ROS, whereas the IKK-NF-κB pathway is dispensable for TAK1 regulation of ROS (42). In addition, Tak1 deletion is found to reduce the protein level of Nrf2 in the intestinal epithelium (24), suggesting that TAK1 modulates ROS also through the Nrf2-Keap1 pathway under steady-state conditions.
In the current study, we attempted to define the role of TAK1 in the regulation of Nrf2-Keap1, specifically in the absence of exogenous oxidative stress.
Results
TAK1 regulates Nrf2
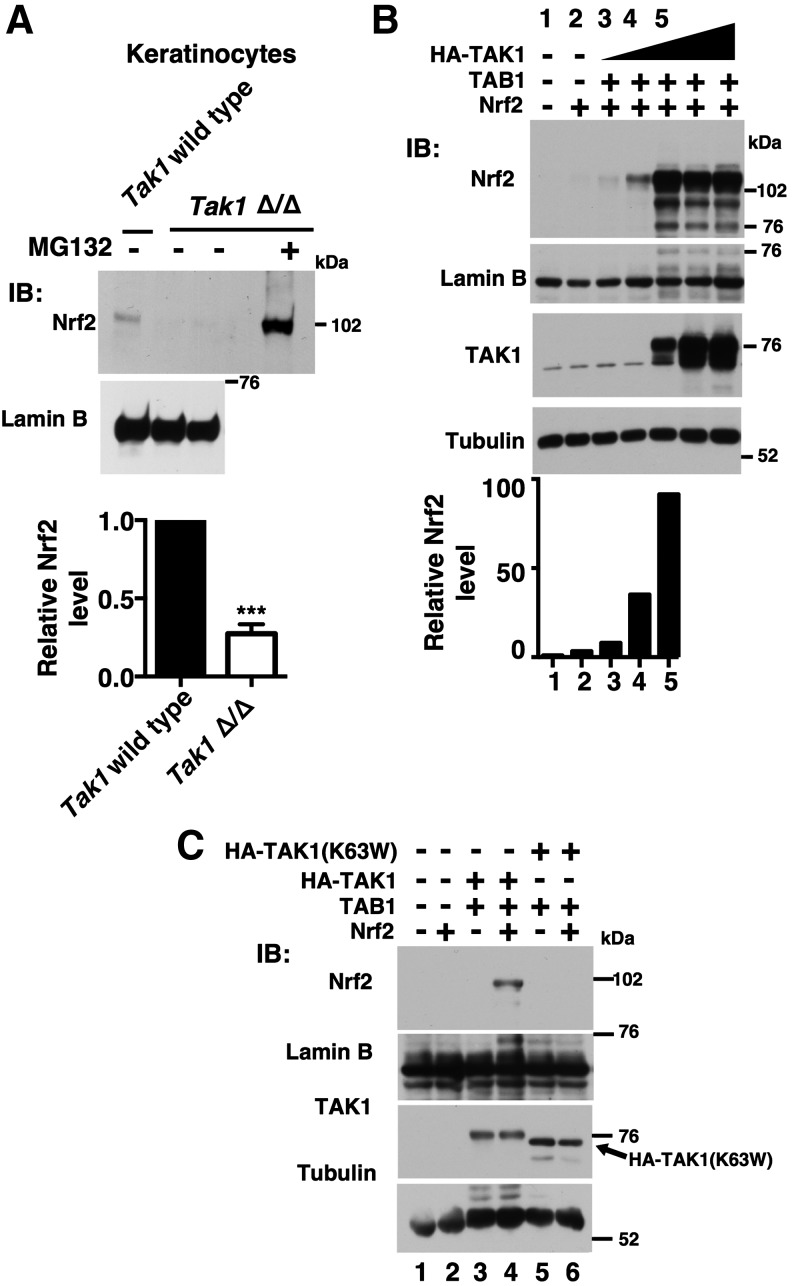
Intestinal epithelial-specific Tak1 deletion reduces the protein amount of Nrf2 in the intestine without any exogenous insults. We previously showed that the Nrf2 mRNA level is not altered (24). Thus, TAK1 regulates Nrf2 at the protein level. In the current study, we began to examine TAK1 regulation of Nrf2 protein level using mouse keratinocytes (40), which are skin epithelial cells and retain characteristics of in vivo epidermal cells. In keratinocytes, Nrf2 is expressed at a certain level and is detectable in the absence of oxidative stress conditions. This steady-state level of Nrf2 protein was diminished in Tak1-deficient keratinocytes, which was greatly upregulated by proteasome inhibition (Fig. 1A). This suggests that Tak1 deficiency may enhance Nrf2 protein degradation, whereas TAK1 may facilitate Nrf2 stabilization.
FIG. 1.
TAK1 upregulates Nrf2. (A) Wild-type and Tak1-deficient (Tak1 Δ/Δ) mouse keratinocytes were treated with 10 μM MG132 or vehicle for 5 h, and nuclear proteins were analyzed by immunoblotting. Lamin B is shown as a loading control. The levels of Nrf2 were normalized to lamin B levels, and relative levels to those in wild-type cells are shown in the graph (bottom). Wild type, n = 3; Tak1 Δ/Δ, n = 4. Means ± SEM, ***p < 0.001 (two-tailed unpaired Student's t-test). (B) HEK293 were transfected with expression vectors (CMV promoter) for increasing amounts of HA-TAK1 (0, 1, 10, 100, 500, and 1000 ng from lane 2 to lane 7), and for the same amount of TAB1 (100 ng) and Nrf2 (500 ng). Proteins were isolated at 48 h post-transfection. Nrf2 and TAK1 amounts were analyzed by immunoblotting. Lamin B and tubulin are shown as loading controls. The Nrf2 levels in lane 1–5 that were normalized to the corresponding lamin B levels are shown in the graph (bottom). The data are representative of two independently performed experiments with similar results. (C) HEK293 cells were transfected with expression vectors for HA-TAK1 or HA-tagged kinase-dead form of TAK1 [HA-TAK1(K63W)] together with TAB1 and Nrf2. Proteins were isolated at 48 h post-transfection. HA-TAK1(K63W) migrated faster than wild-type HA-TAK1 (3rd panel, arrow), which is due to lack of autophosphorylation (30). The data are representative of two independently performed experiments with similar results.
We examined whether Nrf2 is upregulated by TAK1 activation in the overexpression system. Indeed, overexpression of an active form of TAK1 (co-expression of TAK1 and TAB1) highly increased the amount of Nrf2 in human embryonic kidney 293 (HEK293) cells (Fig. 1B). A catalytically inactive TAK1, TAK1 (K63W), with TAB1 did not upregulate Nrf2 (Fig. 1C, lane 6), demonstrating that catalytically active TAK1 but not TAK1 protein or TAB1 alone mediates upregulation of Nrf2. Furthermore, activated TAK1 increased the endogenous Nrf2 level, as shown later in Figure 4B, top panel. Thus, activation of TAK1 upregulates Nrf2, presumably by stabilizing Nrf2.
FIG. 4.
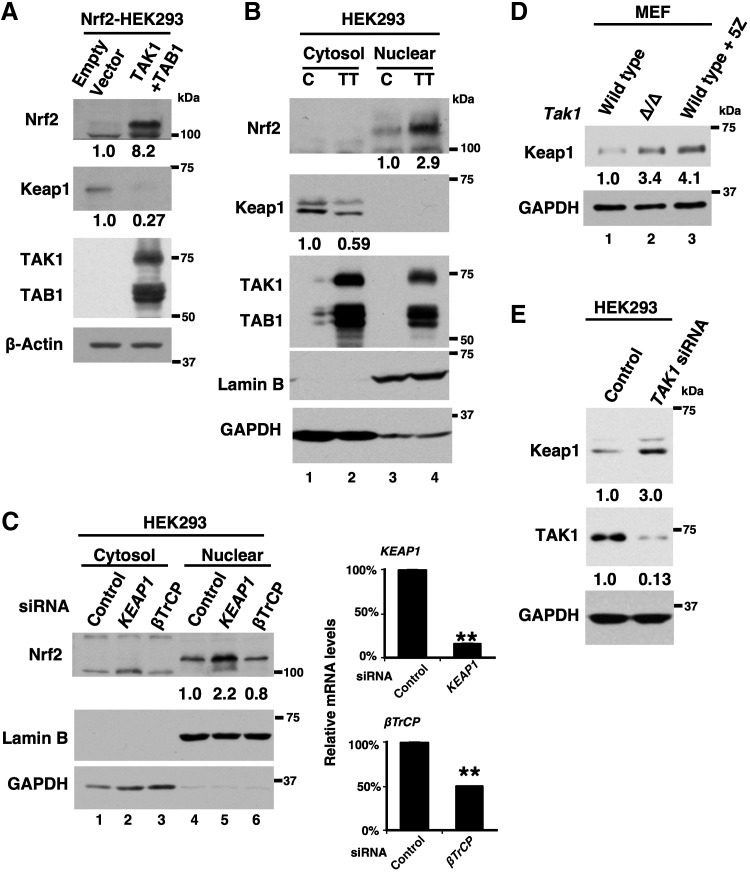
TAK1 downregulates Keap1. (A) Nrf2-HEK293 cells were transfected with an empty vector or expression vectors for TAK1 and TAB1, and proteins in cell lysates at 48 h post-transfection were analyzed by immunoblotting. The levels of Nrf2 and Keap1 were quantified, and the relative values to those of β-actin are shown below the blots. The results are representative of three independently performed experiments with similar results. (B) HEK293 cells were transfected with an empty vector or expression vectors for TAK1 and TAB1 and were harvested at 48 h post-transfection. Cell extracts were fractionated into the cytosol and the nuclear fractions, and they were analyzed by immunoblotting. Lamin B and GAPDH were used as the nuclear and cytosol markers, respectively. The levels of Nrf2 and Keap1 were quantified, and the relative values to those of lamin B and GAPDH, respectively, are shown below the blots. The results are representative of two independently performed experiments with similar results. (C) HEK293 cells were transfected with nontargeted (control) siRNA or siRNA targeted against KEAP1, or βTrCP, and the cytoplasmic and nuclear extracts at 48 h post-transfection were analyzed by immunoblotting. The levels of Nrf2 were quantified, and the relative values to those of lamin B from the representative result are shown below the blots. The knockdown efficiency of Keap1 and βTrCP was assessed by the mRNA levels normalized to GAPDH mRNA (left graphs). Three independent experiments were performed. Means ± SEM are shown. **p < 0.01 (two-tailed unpaired Student's t-test). (D) Wild-type and Tak1-deficient (Δ/Δ) MEFs were analyzed by immunoblotting. Cells were either left untreated (left two lanes) or treated with a TAK1 inhibitor, 5z-7oxozeaenol (5Z) for 6 h. GAPDH is shown as a loading control. The levels of Keap1 were quantified, and the relative values to those of GAPDH are shown below the blots. The results are representative of two independently performed experiments with similar results. (E) HEK293 cells were transfected with nontargeted (control) siRNA or siRNA targeted against TAK1, and cell lysates at 48 h post-transfection were analyzed by immunoblotting. The levels of Keap1 were quantified, and the relative values to those of GAPDH are shown below the blots. The results are representative of two independently performed experiments with similar results. MEF, mouse embryonic fibroblast.
Nrf2 is degraded through Keap1-driven proteasome degradation, and this process is initiated by a physical interaction of Nrf2 with Keap1. Given such an Nrf2 degradation mechanism, we postulated several scenarios in which TAK1 could lead to Nrf2 stabilization. TAK1 may inhibit interaction between Nrf2 and Keap1. Alternatively, TAK1 may block the machinery of proteasome degradation of Nrf2 after normal complex formation of Nrf2 and Keap1. Finally, TAK1 may downregulate the protein level of Keap1 that increases the Keap1-unbound (stabilized) form of Nrf2. We tested these possibilities.
TAK1 binds to Keap1 but does not block binding of Nrf2 with Keap1
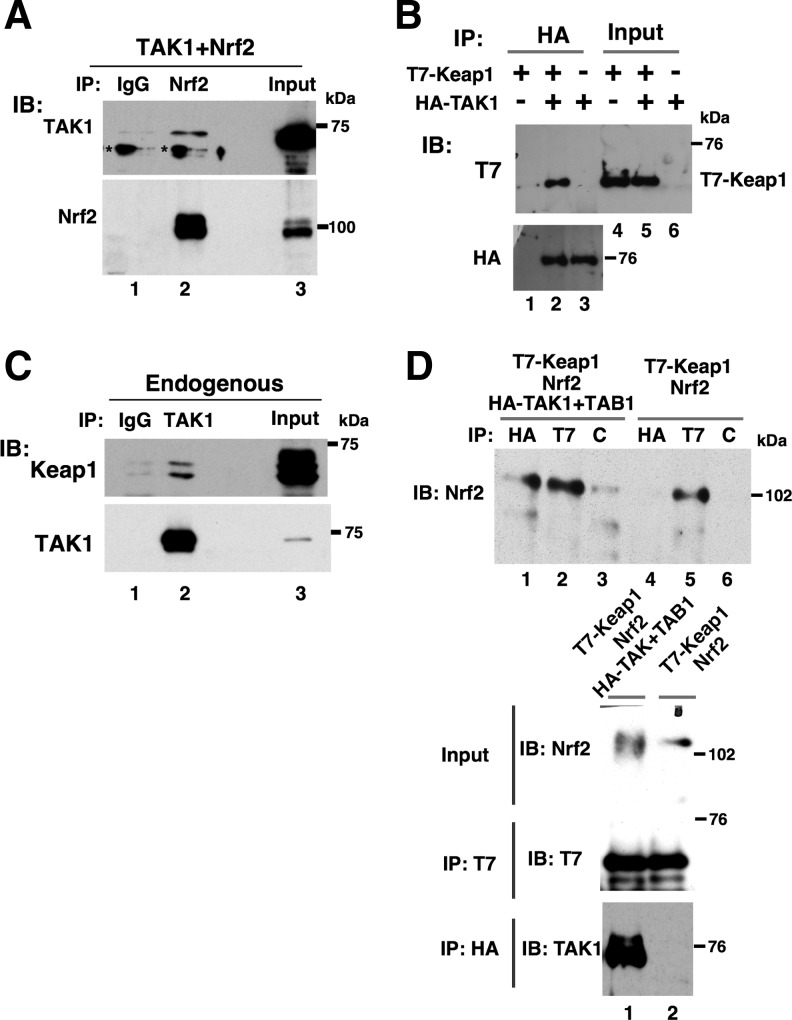
We first tested the possibility that TAK1 modulates interaction between Nrf2 and Keap1. Such modulation may be through the physical interaction of TAK1 with Nrf2 and/or Keap1. We examined whether TAK1 binds to Nrf2 and/or Keap1. TAK1 was found to marginally interact with Nrf2 when both TAK1 and Nrf2 were overexpressed (Fig. 2A, lane 2). In contrast, Keap1 was effectively co-precipitated with TAK1 in the co-overexpression system (Fig. 2B, lane 2). Furthermore, we found that Keap1 was co-precipitated with TAK1 at the endogenous protein levels (Fig. 2C, lane 2). Thus, TAK1 seems to bind to Keap1, but Nrf2 interaction with TAK1 may be through Keap1.
FIG. 2.
TAK1 binds to Keap1 but does not inhibit binding of Nrf2 with Keap1. (A) HEK293 stably expressing Nrf2 cells (Nrf2-HEK293) were transfected with an expression vector for FLAG-tagged TAK1, and cell lysates at 48 h post-transfection were immunoprecipitated with control IgG or anti-Nrf2. The precipitates were analyzed by immunoblotting. Inputs (1/200 or 1/20 used for immunoprecipitation in TAK1 immunoblot or Nrf2 immunoblot, respectively) are also shown. Asterisks indicate nonspecific bands. (B) HEK293 cells were transfected with expression vectors for HA-TAK1 and T7-Keap1 as indicated, and cell lysates at 48 h post-transfection were immunoprecipitated with anti-HA. Input immunoblotting used 1/200 proteins for immunoprecipitation. (C) Protein extracts from nontransfected HEK293 cells were immunoprecipitated with control IgG or anti-TAK1, and immunoprecipitates were analyzed by immunoblotting. Inputs (1/200) are also shown. (D) HEK293 cells were transfected with expression vectors for HA-TAK1, TAB1, T7-Keap1, and Nrf2 as indicated, and cell lysates at 48 h post-transfection were immunoprecipitated with anti-HA (HA), anti-T7 (T7), or control IgG (C). Inputs (1/200) of Nrf2 are also shown (2nd panel). All the data are representative of two or more independently performed experiments with similar results.
We then examined the interaction between Nrf2 and Keap1 in the presence and absence of an active form of TAK1 (TAK1+TAB1) in the co-overexpression system (Fig. 2D). Consistent with a number of earlier studies (reviewed in ref. (20)), Nrf2 was effectively co-precipitated with Keap1 (Fig. 2D, lane 5), and their interaction was not altered by additional co-overexpression of TAK1+TAB1 (Fig. 2D, lane 2). Nrf2 was also co-precipitated with TAK1 when all the proteins were co-expressed (lane 1). Thus, TAK1 can bind to Nrf2 and Keap1, but it does not prevent interaction between Nrf2 and Keap1. We note here that co-expression of activated TAK1 with Nrf2 increased differentially migrating forms of Nrf2 on SDS-PAGE, which was detected as a smeared band (Fig. 2D lane 1, 2nd panel). This suggests that TAK1 may modulate Nrf2 protein. However, the importance of TAK1 modification of Nrf2 is not clear at this point. These results demonstrate that TAK1 stabilizes Nrf2 through a mechanism other than inhibition of interaction between Nrf2 and Keap1.
TAK1 does not inhibit the Nrf2 degradation machinery
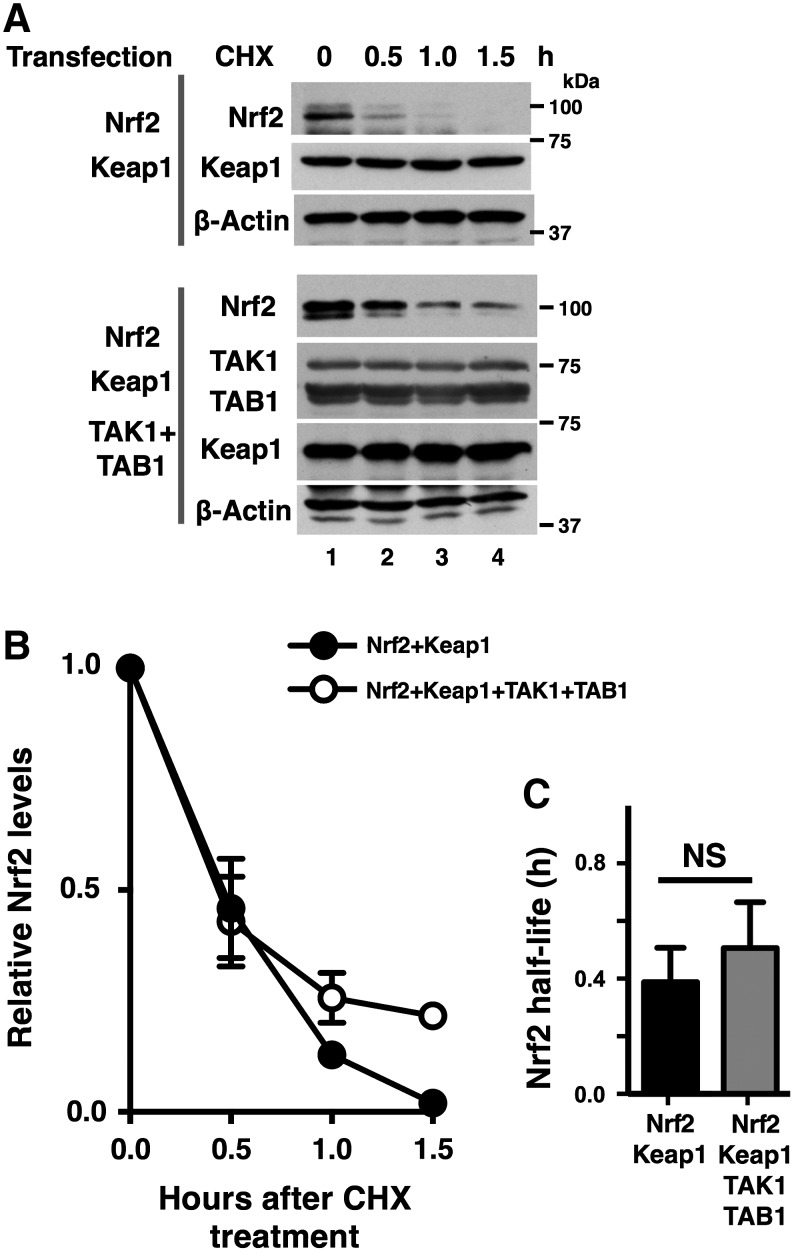
We then examined the possibility that TAK1 inhibits the Nrf2 degradation machinery after normal binding of Nrf2 with Keap1. To this end, we co-overexpressed Nrf2 and Keap1 with or without an active form of TAK1 (TAK1+TAB1). We observed that exogenously introduced Keap1 protein was stable both with and without TAK1+TAB1 (Fig. 3A). Under such experimental conditions, the canonical Keap1-dependent proteasome degradation of Nrf2 should occur as previously reported (8). We determined the Nrf2 degradation rate by a cycloheximide chase assay. Consistent with earlier studies (21, 31), Nrf2 was quickly degraded with a half-life around 0.5 h in our experimental settings. The degradation rate of Nrf2 was not pronouncedly altered by TAK1 activation (Fig. 3A, B), in which the half-lives of Nrf2 were 0.38 h (95% confidence interval = 0.31–0.50) and 0.50 h (95% confidence interval = 0.41–0.66) in the absence and presence of an active form of TAK1, respectively (Fig. 3C). Thus, TAK1 does not seem to directly modulate the process of canonical Nrf2 degradation.
FIG. 3.
TAK1 does not inhibit Nrf2 degradation machinery. (A) HEK293 cells were transfected with expression vectors for Nrf2, Keap1, TAK1, and TAB1 as indicated, and at 48 h post-transfection, 100 μg/ml cycloheximide (CHX) was added to the culture medium. Cells were harvested at 0, 0.5, 1.0, and 1.5 h after CHX addition (lanes, 1–4). Cell lysates were analyzed by immunoblotting. A stable protein, β-actin, is used as a loading control. (B, C) The immunoblotting band intensity of Nrf2 was quantified by ImageJ software and normalized to the intensity of the β-actin band. Three independent experiments were performed. Means ± SEM are shown (B). Nrf2 protein half-lives were calculated by GraphPad software using an exponential decay equation model. The bars depict the 95% confidence intervals (C). NS, not significant (two-tailed unpaired Student's t-test).
TAK1 downregulates Keap1
Keap1 is a critical adaptor of Nrf2 and targets Nrf2 for canonical proteasome degradation. Thus, increased availability of endogenous Keap1 should enhance Nrf2 degradation, whereas decreased availability of Keap1 should increase Keap1-unbound Nrf2 and limit Nrf2 degradation. The results in Figure 3 showed that the stability of overexpressed Keap1 was not altered by TAK1 activation. However, it is still possible that TAK1 regulates the endogenous level of Keap1, thereby modulating the level of Nrf2. To test this possibility, we utilized HEK293 cells stably expressing exogenous Nrf2, because the amount of Nrf2 is very low in HEK293 cells. We transfected the cells with an active form of TAK1 (TAK1+TAB1) (Fig. 4A). Endogenous Keap1 protein was diminished by expression of an activated TAK1, whereas Nrf2 protein was increased.
To further assess the protein levels of Keap1 and Nrf2, we fractionated cell lysates from HEK293 cells (with no exogenous Nrf2 expression) expressing either an empty vector or TAK1+TAB1 into the nuclear and cytosol fractions. Endogenous Nrf2 was enriched in the nuclear fraction, and we could detect it on a long-exposed immunoblotting (Fig. 4B, top panel). Nrf2 was increased by activation of TAK1, which was accompanied by the reduction of cytoplasmic Keap1 (Fig. 4B). These results suggest that TAK1 downregulates Keap1, raising the possibility that reduction of Keap1 is the cause of increased Nrf2. However, although Keap1-cullin 3 is the major pathway for degradation of Nrf2, βTrCP has also been implicated in Nrf2 degradation under some circumstances (6, 44, 45). Importantly, the βTrCP-dependent mechanism is Keap1-independent. Thus, the steady-state Nrf2 may also be regulated by βTrCP. To test whether βTrCP is involved in maintenance of the steady-state Nrf2 level, we knocked down βTrCP. We found that although Keap1 knockdown effectively increased the steady-state Nrf2, about 50% reduction of βTrCP did not alter the level of Nrf2 (Fig. 4C). Thus, TAK1 is likely to regulate the steady-state Nrf2 level, primarily through modulating Keap1.
We then asked whether ablation of TAK1 can upregulate Keap1. We measured the level of Keap1 in wild-type and Tak1-deficient mouse embryonic fibroblasts (MEFs) (Fig. 4D). The higher level of Keap1 was observed in Tak1-deficient MEFs compared with wild-type MEFs (Fig. 4C, lanes 2). In addition, a selective inhibitor of TAK1, 5z-7oxozeaenol, treatment upregulated Keap1 in wild-type MEFs (Fig. 4C, lane 3). Similarly, TAK1 knockdown upregulated Keap1 protein in HEK293 cells (Fig. 4E). Collectively, these results demonstrate that TAK1 downregulates Keap1.
TAK1 facilitates binding between Keap1 and p62/SQSTM1
We sought to determine the mechanism by which TAK1 downregulates Keap1. Keap1 is known to be regulated through several binding proteins such as sestrins and p62/SQSTM1 (2, 7, 22, 32, 33). Among them, p62/SQSTM1 binds to and targets Keap1 for autophagic degradation (32). p62/SQSTM1 is an important regulator of innate immune signaling by binding and modulating TRAF6 (TNF receptor-associated factor 6) complex (37). TAK1 is also a critical signaling intermediate of TNF and IL-1 pathways, and it interacts with TRAF6 (36, 39). These lead us to test the possibility that TAK1 modulates Keap1 through p62/SQSTM1.
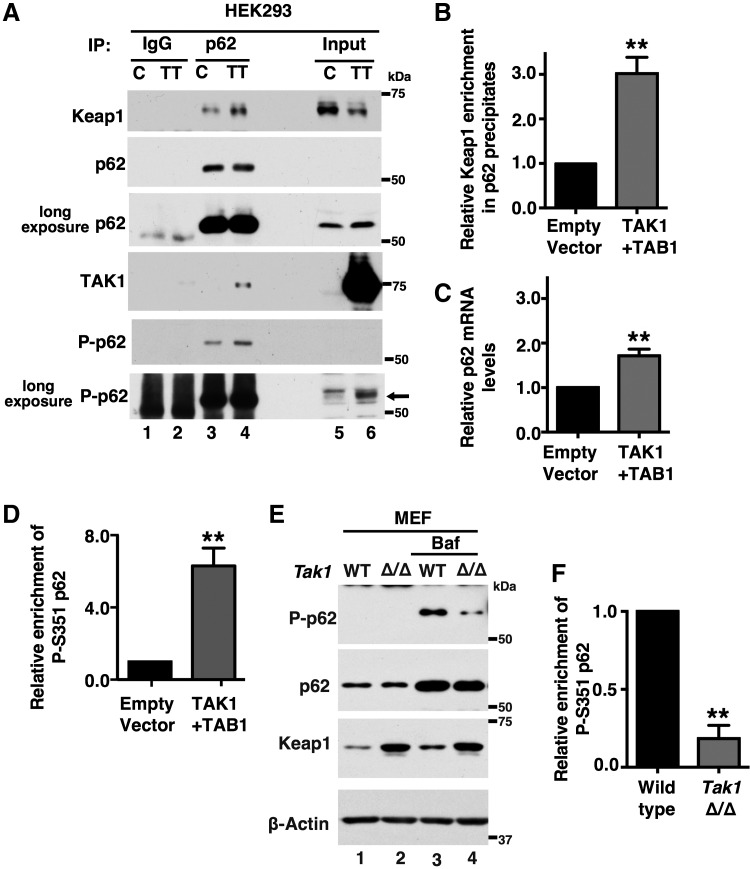
We examined whether the interaction between Keap1 and p62/SQSTM1 was altered by TAK1. An active form of TAK1 (TAK1+TAB1) was expressed in HEK293 cells, and immuneprecipitates with anti-p62/SQSTM1 were analyzed (Fig. 5A). Activation of TAK1 reduced the protein level of Keap1 (top panel, lane 6), as also shown in Figure 4. Interestingly, even though Keap1 was reduced by TAK1, the amount of Keap1 that co-precipitated with p62/SQSTM1 was rather increased by TAK1 activation (top panel, lane 4). We quantified the percentages of p62/SQSTM1-bound Keap1 in total Keap1 protein, and we found that they were about three-fold higher in TAK1+TAB1 expressing cells compared with cells with empty vectors (Fig. 5B).
FIG. 5.
TAK1 facilitates interaction between Keap1 and p62/SQSTM1. (A) HEK293 cells were transfected with expression vectors for TAK1 and TAB1 or an empty vector, and cell lysates were isolated at 48 h post-transfection. Proteins were immunoprecipitated with anti-p62/SQSTM1 or control IgG, and immunoprecipitates were analyzed by immunoblotting. Inputs (1/20) are also shown. Arrow indicates the position of p62/SQSTM1 (bottom panel). (B) The immunoprecipitated and input Keap1 band intensities were quantified by ImageJ software. Four independent experiments were performed, and the percentage of Keap1 protein in p62/SQSTM1 immunoprecipitates was calculated. The percentages without TAK1+TAB1 were designated as 1. Mean ± SEM is shown. **p < 0.01 (two-tailed unpaired Student's t-test). (C) p62/SQSTM1 mRNA levels were measured by real-time PCR. Three independent experiments were performed. Mean ± SEM is shown. **p < 0.01 (two-tailed unpaired Student's t-test). (D) The immunoblot intensity of phospho-S351 p62/SQSTM1 in whole cell lysates was quantified and normalized to the total amount of p62/SQSTM1. The values in empty vector were designated as 1. Three independent experiments were performed. Mean ± SEM is shown. **p < 0.01 (two-tailed unpaired Student's t-test). (E) Wild-type and Tak1-deficient (Tak1 Δ/Δ) MEFs were either left untreated (lanes 1 and 2) or treated with Bafilomycin A1 (2.5 mg/ml) for 12 h (lanes 3 and 4). (F) The immunoblot intensity of phospho-S351 p62/SQSTM1 in whole cell lysates was quantified and normalized to the total amount of p62/SQSTM1. Three independent experiments were performed. Relative values to those in wild-type cells are shown (mean ± SEM). **p < 0.01 (two-tailed unpaired Student's t-test).
We also observed that TAK1 activation slightly increased the amount of p62/SQSTM1 (Fig. 5A). This is likely due to TAK1-dependent activation of NF-κB, which is the major activator of p62/SQSTM1 (56). We found that the p62/SQSTM1 mRNA level was upregulated about 1.7-fold (Fig. 5C). This increase may contribute to the increased binding of Keap1 with p62/SQSTM1; however, the increased p62/SQSTM1 protein level is not pronounced compared with the increased binding between p62/SQSTM1 and Keap1. Thus, we sought an additional mechanism for the increased binding. Binding affinity between Keap1 and p62/SQSTM1 is known to be modulated by phosphorylation of p62/SQSTM1 at a specific amino-acid residue, serine 351 (S351) in mice (S349 in humans) (16). Given that, TAK1 may mediate S351 phosphorylation of p62/SQSTM1.
We examined the S351 phosphorylation status of p62/SQSTM1 (Fig. 5A). S351-phosphorylated p62/SQSTM1 was highly increased by activation of TAK1. We note here that TAK1 was also co-precipitated with p62/SQSTM1 (Fig. 5A). TAK1 may make a complex with p62/SQSTM1 and Keap1. If TAK1 mediates S351 phosphorylation of p62/SQSTM1, Tak1 deficiency should reduce it. To visualize S351 phosphorylation of p62/SQSTM1 under basal conditions, we treated cells with Bafilomycin A1, which inhibits fusion of autophagosomes with lysosomes and blocks degradation of p62/SQSTM1 (52). Bafilomycin A1 highly upregulated p62/SQSTM1 in both wild-type and Tak1-deficient MEF cells (Fig. 5E). However, S351 phosphorylation of p62/SQSTM1 was profoundly lower in Tak1-deficient MEF cells compared with wild-type cells (Fig. 5E, F). These results demonstrate that TAK1 mediates S351 phosphorylation of p62/SQSTM1, which facilitates the binding between Keap1 and p62/SQSTM1 and induces autophagic degradation of Keap1.
TAK1 regulation of Keap1 is independent of oxidative stress-induced modification
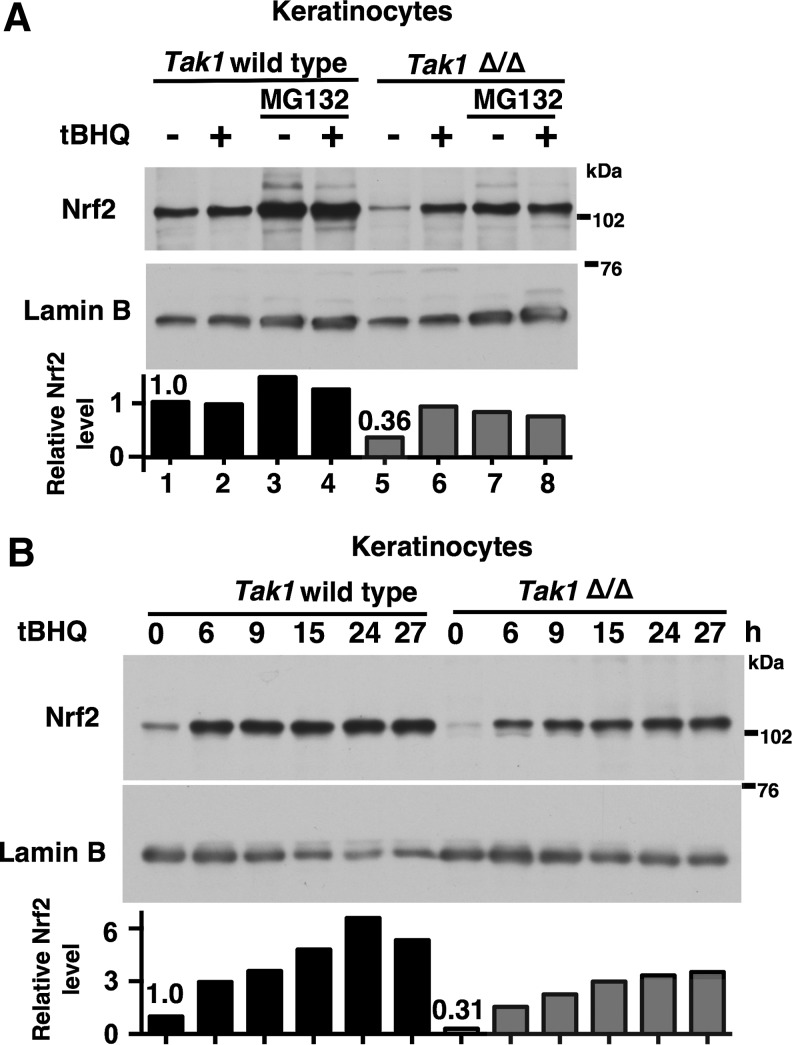
We next examined the relationship between the canonical oxidative stress-induced Nrf2 stabilization and TAK1-dependent Nrf2 regulation. We asked whether oxidative stress-induced Nrf2 stabilization still occurs without functional TAK1. We used wild-type and Tak1-deficient keratinocytes as model cells, which expressed a detectable level of Nrf2 without oxidative stress conditions as shown in Figure 1A. tBHQ, which is one of the most extensively studied electrophilic compounds and is known to covalently modify Keap1 at Cys151 (1, 49), was used as a model treatment. The steady-state level of Nrf2 was lower in Tak1-deficient cells compared with wild-type cells without tBHQ treatment, and Nrf2 was highly upregulated with tBHQ treatment in Tak1-deficient cells (Fig. 6A). In both wild-type and Tak1-deficient cells, Nrf2 was increased by proteasome inhibition. We also examined the time course of Nrf2 accumulation on tBHQ treatment in wild-type and Tak1-deficient keratinocytes (Fig. 6B). Although elevation of Nrf2 was weaker in Tak1-deficient cells, Nrf2 was increased in both wild-type and Tak1-deficient keratinocytes. These suggest that the redox regulation of Nrf2 by tBHQ treatment is not affected by ablation of TAK1, and that TAK1 is selectively involved in the steady-state basal level of Nrf2.
FIG. 6.
Electrophilic activation of Nrf2 is intact in Tak1-deficient cells. (A) Wild-type and Tak1-deficient (Tak1 Δ/Δ) keratinocytes were pretreated with 10 μM MG132 or vehicle (DMSO) for 5 h and, subsequently, stimulated with 100 μM tBHQ for 20 h. Nuclear proteins were analyzed by immunoblotting. Lamin B is shown as a loading control. The levels of Nrf2 were quantified, and the relative values to those of lamin B are shown (bottom). The data are representative of two similar experiments. (B) Time course of Nrf2 accumulation after 100 μM tBHQ stimulation. Lamin B is shown as a loading control. The levels of Nrf2 were quantified, and the relative values to those of lamin B are shown (bottom). The data are representative of three similar experiments. tBHQ, tert-butylhydroquinone.
Electrophilic activation of Nrf2 prevents ROS in Tak1-deficient intestinal epithelium
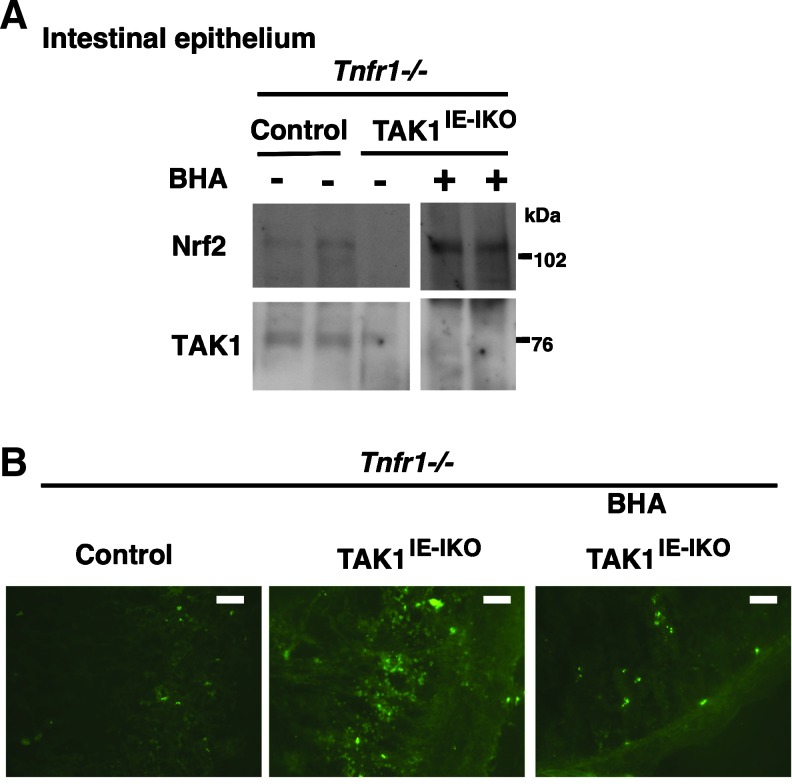
We previously reported that intestinal epithelial-specific inducible Tak1 deletion (Tak1flox/flox Villin-CreERT2, Tak1IE-IKO) mice develop inflammatory bowel disease-like tissue injury on Tak1 gene deletion, which is associated with a profound increase of ROS in the small intestine (24). Severe tissue injury can be blocked by additional gene deletion of TNF receptor 1, which is through blockade of TNF-dependent inflammation. However, increased ROS are still observed in Tak1IE-IKO in the background of TNF receptor 1 deletion (Tnfr1−/−) (24). We further demonstrated that treatment of butylated hydroxyanisole (BHA), which is metabolized to tBHQ in the liver, could prevent the increase of ROS (24). However, the involvement of Nrf2 was not explored in the in vivo setting.
Our current study revealed that TAK1 regulation of Nrf2 is independent of tBHQ-induced Nrf2 activation. This raises the possibility that BHA treatment may upregulate Nrf2 in the Tak1-deficient intestinal epithelium. Control (Tnfr1−/−) and Tak1IE-IKO Tnfr1−/− mice were fed with BHA-containing food starting from 1 week before the induction of gene deletion, and Tak1 gene deletion was induced by injection of a chemical activator of the CreERT2 system, tamoxifen. Proteins from control and Tak1-deficient intestinal epithelium were analyzed by immunoblotting (Fig. 7A). Nrf2 protein was highly upregulated by BHA treatment in Tak1IE-IKO Tnfr1−/− intestinal epithelium, which was correlated with the reduction of ROS (Fig. 7B). These results demonstrate that electrophilic chemicals can activate Nrf2 in Tak1-deficient cells and reduce ROS in the in vivo setting.
FIG. 7.
Electrophilic activation of Nrf2 reduces ROS in Tak1-deficient intestinal epithelium. (A, B) Control (Tak1flox/flox Tnfr1−/−) and TAK1IE-IKO Tnfr1−/− mice were fed with BHA-containing food from 1 week before injection of tamoxifen for 3 consecutive days, and the small intestine was harvested at day 3 (B) or 7 (A). Protein extracts from the intestinal epithelium were analyzed with immunoblotting (A). Each lane represents an individual animal. The immunoblot shown is the same exposure film, and unnecessary lanes were removed. Fresh frozen sections without any fixation were stained by CM-H2DCFDA (B). Scale bars, 50 μm. ROS, reactive oxygen species. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Discussion
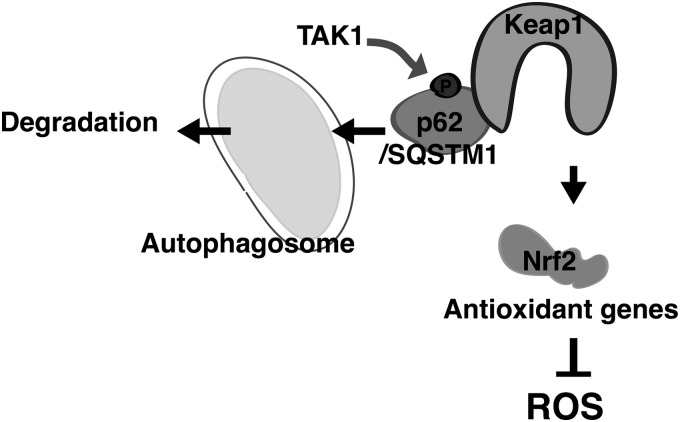
Nrf2 protein level is highly inducible dependent on the redox status in cells, in which Keap1 is electrophilically modulated and Cul3-directed ubiquitylation of Nrf2 is inhibited. Our study has revealed that TAK1 regulates Nrf2 through the mechanism that is different from the canonical redox regulation of Nrf2 (Fig. 8). Unlike the redox-dependent regulation, activation of TAK1 facilitates the interaction between Keap1 and p62/SQSTM1 through upregulation of serine 351 phosphorylation of p62/SQSTM1. p62/SQSTM1 is known to recruit Keap1 in inclusion bodies, and the p62/SQSTM1-Keap1 complex is subsequently degraded by autophagy (4).
FIG. 8.
Model. TAK1 upregulates the binding of Keap1 with p62/SQSTM1 through mediating S351 phosphorylation of p62/SQSTM1, which facilitates Keap1 degradation and upregulates Nrf2. This mechanism provides antioxidant protection under steady-state conditions in the intestinal epithelium.
p62/SQSTM1 was initially found to be a regulatory molecule of innate immune signaling complexes, including TRAF6 (46), and its physiological importance was revealed by a critical finding that mutations within the ubiquitin-binding region of p62/SQSTM1 gene are associated with Paget disease, which is characterized by abnormal bone turnover (14). p62/SQSTM1 serves as a cargo-like protein bringing ubiquitylated proteins into inclusion bodies, which leads to autophagic clearance of protein aggregates (29). An interaction between Keap1 and p62/SQSTM1 is induced under pathogenic conditions in autophagy-deficient cells such as hepatocellular carcinoma cells (16, 32, 33). Phosphorylation at serine 351 of p62/SQSTM1 was determined as a modification to increase the binding affinity between p62/SQSTM1 and Keap1 under several pathological conditions (16). Elevated S351 phosphorylation of p62/SQSTM1 promotes Keap1 degradation and releases Nrf2. As a result, highly upregulated Nrf2 promotes tumor growth by transcriptional activation of a battery of anti-oxidant and anti-cell death genes [reviewed in (50)].
Activation of mTOR pathway (cell growth signal) and Toll-like receptor (TLR) signaling-induced TBK1 (TANK-Binding Kinase 1) have been implicated in phosphorylation of p62/SQSTM1 and facilitate the interaction between p62/SQSTM1 and Keap1 (16, 19, 32, 35). Here, we show that TAK1 also mediates S351 phosphorylation of p62/SQSTM1. TAK1 is a critical signaling intermediate in TNF, IL-1, and TLR signaling pathways (36). TLR signaling is branched at the receptor proximal signaling complex, where one branch activates TBK1 and the other branched pathway activates TAK1 (26). Unlike TBK1, TAK1 participates not only in TLR but also in nucleotide-binding oligomerization domain (NOD)-like receptor TNF-, IL-1-, and stressor-induced pro-inflammatory signaling pathways (12, 17, 28, 36, 51, 54). TAK1 regulation of p62/SQSTM1-Keap1 and Nrf2 may be a previously uncharacterized link between inflammation signaling and redox homeostasis.
Importantly, this regulation determines the steady-state level of Nrf2 but does not affect the redox-induced activation of Nrf2. Inflammatory signaling is basally active, to some extent, in some tissues such as the intestine, which is constantly exposed to environmental challenge. In such tissues, TAK1 constitutively upregulates Nrf2, which may provide basal protection from oxidative damage.
Ablation of TAK1 causes accumulation of ROS and tissue damage in the intestinal epithelium and the epidermis (24, 42). We show that enforced activation of Nrf2 in Tak1-deficient intestinal epithelium reduces ROS, which suggests that disruption of TAK1 regulation of Nrf2 is the cause of increased ROS. However, gene deletion of Nrf2 in mouse models does not cause any abnormalities in tissues, including the intestine (5). Thus, it is clear that reduced Nrf2 in Tak1-deficient intestinal epithelium is not solely the cause of ROS accumulation. Accordingly, TAK1 is likely to modulate multiple biological processes, which include non-Nrf2 pathways that are associated with redox regulation. We previously reported that ablation of a transcription factor, cJun, is partially involved in accumulation of ROS in Tak1-deficient keratinocytes (41). Tak1 deletion seems to impair Nrf2, cJun, and potentially other unidentified processes, resulting in ROS accumulation. Nonetheless, activation of Nrf2 can reduce ROS in the Tak1-deficient tissue, demonstrating effectiveness of Nrf2 activation in reduction of unattended ROS accumulation regardless of the causes of ROS.
In summary, our results demonstrate that TAK1 is a previously uncharacterized regulator of Nrf2, and this regulation is likely through TAK1-induced Keap1 downregulation. TAK1 seems to limit the steady-state Keap1 protein level through facilitating phosphorylation of p62/SQSTM1. Our in vivo results further suggest that TAK1 regulation of Keap1 is important for normal tissue homeostasis.
Materials and Methods
Tissue culture and mouse models
Human embryonic kidney (HEK) 293 cells and MEFs were cultured in Dulbecco's modified Eagle's medium that was supplemented with 10% bovine growth serum (Hyclone GE Healthcare Life Sciences, Pittsburgh, PA) and 50 I.U./ml penicillin-streptomycin at 37°C in 5% CO2. In some experiments, HEK293 cells stably expressing exogenous Nrf2 as described later were used, and they were cultured in the same condition as parent HEK293 cells. Keratinocytes (immortalized) were originally isolated from epidermal-specific Tak1-deficient and wild-type mice (40), and they were cultured in Ca2+-free Eagle's minimal essential medium (BioWhittaker, Allendale, NJ) that was supplemented with 4% Chelex-treated bovine growth serum, 10 ng/ml of human epidermal growth factor (Invitrogen, Carlsbad, CA), 0.05 mM calcium chloride, and 50 I.U./ml penicillin-streptomycin at 33°C in 8% CO2. Some cells were treated with tBHQ, MG132 (Sigma, St. Louis, MO), and 5z-7oxozeaenol (38). Mice carrying a floxed Map3k7 allele (Tak1fl/fl) (47) were backcrossed to C57BL/6 mice for at least seven generations. Tnfr1-deficient (Tnfr1−/−) (43) (Jackson lab, Bar Harbor, ME) and intestinal epithelium-specific inducible deleter (villin.CreERT2) (11) mice with a C57BL/6 background were used. We generated inducible intestinal epithelium-specific Tak1-deficient (villin.CreERT2 Tak1flox/floxl, TAK1IE-IKO) mice on a Tnfr1−/− background. To induce gene deletion, 6–12 week-old mice were given intraperitoneal injections of tamoxifen (1 mg per mouse, approximately 20 g body weight, per day) for 3 consecutive days. The first day of tamoxifen injection is referred to here as day 1. Some mice were fed with food containing 0.7% butylated hydroxyanisole (BHA; Sigma) from 1 week before the tamoxifen treatment, and mice were continuously given BHA food for the entire period of the experiments. All animal experiments were conducted with the approval of the North Carolina State University Institutional Animal Care and Use Committee. All efforts were made to minimize animal suffering.
Transfection, immunoprecipitation, and immunoblotting
Expression vectors for HA-tagged TAK1, TAB1 (39), Nrf2 (15), and T7-tagged Keap1 (13) were used and transiently transfected into HEK293 cells. The standard calcium phosphate transfection method was used. HEK293 cells stably expressing Nrf2 were generated by transfection of a mammalian expression vector pCMV-neo-Nrf2, which was generated by inserting human Nrf2 cDNA (Invitrogen) into pcDNA3.1 (Invitrogen). Transfected cells were selected by incubation with 100 μg/ml G418 (Millipore, Billerica, MA) for 2 weeks, and a pool of G418-resistant colonies was used.
Cell lysates were prepared at 48 h post-transient transfection using an extraction buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM DTT, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 20 μM aprotinin, and 0.5% Triton X-100. In some experiments for detecting endogenous Nrf2, nuclear extracts were prepared using Nuclear Extraction Kit (Active Motif, Carlsbad, CA). Cell extracts were immunoprecipitated with anti-TAK1 (39), anti-HA (16B12) (Biolegend, San Diego, CA), anti-T7 (Millipore), anti-p62/SQSMT1 (BD Transduction Laboratories, Franklin Lakes, NJ), or nonimmunized control IgG (Sigma) with Protein G Sepharose 4 fast flow (GE Healthcare) or SureBeads Magnetic beads (Bio-Rad, Hercules, CA). The beads were washed thrice with a wash buffer (20 mM HEPES pH 7.4, 10 mM MgCl2, 0.5 M NaCl) and once with a kinase buffer (10 mM HEPES pH 7.4, 1 mM DTT, 5 mM MgCl2).
Immunoprecipitates and whole cell lysates were resolved on SDS-PAGE and transferred to Hypond-P membranes (GE Healthcare). The membranes were immunoblotted with anti-TAK1, anti-HA, anti-T7, anti-Keap1 (clone 144; Millipore), anti-Nrf2 (C2 and H-300; Santa Cruz, Santa Cruz, CA), anti-p62/SQSTM1, and anti-phospho serine 351 p62/SQSTM1 (Medical and Biological Laboratories, Woburn, MA); the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit, rat, or mouse IgG using the ECL Western blotting system (GE Healthcare).
For cycloheximide chase assay, cells were treated with 100 μg/ml cycloheximide (Sigma). ImageJ software (National Institutes of Health, Bethesda, MD) was used for quantification of the bands in unsaturated exposure films of immunoblotting. Nrf2 half-lives were calculated by GraphPad software (GraphPad, La Jolla, CA).
TAK1, KEAP1, and βTrCP knockdown
siRNAs TAK1 and nontargeting control siRNAs were obtained from Sigma (TAK1 siRNA, 5′-GAGAUCGACUACAAGGAGA-3′ and Nontargeting siRNA, 5′-UUCUCCGAACGUGUCACGU-3′), and Keap1 and βTrCP siRNAs were obtained from Dharmacon (Keap1 siRNA, 5′-GGACAAACCGCCUUAAUUC-3′ and βTrCP siRNA, 5′-CACAUAAACUCGUAUCUUA-3′). HEK293 cells were transfected siRNAs by a standard electroporation method using Gene Pulser Xcell™ Electroporation System (Bio-Rad).
Quantitative real-time PCR analysis
Total RNA was isolated from HEK293 cells using Trizol (Invitrogen) and transcribed into cDNA using MultiScribe reverse transcriptase (Life Technology). Expression levels of KEAP1 and βTrCP were determined by real-time PCR and normalized to mRNA levels of GAPDH. Primers used were as follows: KEAP1-forward, 5′-CTGGAGGATCATACCAAGCAGG-3′; KEAP1-reverse, 5′-GGATACCCTCAATGGACACCAC-3′. βTrCP-forward, 5′-CCAGACTCTGCTTAAACCAAGAA-3′; βTrCP -reverse, 5′-GGGCACAATCATACTGGAAGTG-3′.GAPDH-forward, 5′-GAAGGTCGCTGTGAACGGA-3′; GAPDH-reverse, 5′-GTTAGTGGGGTCTCGCTCCT-3′.
ROS staining
Ileums were embedded into optimum cutting temperature compounds and frozen immediately. Cryosections (8 μm) were incubated with the ROS staining dye (CM-H2DCFDA, Life Technologies, Carlsbad, CA) for 30 min at room temperature. Images were visualized using a fluorescent microscope (BX41; Olympus, Tokyo, Japan) that was controlled by the CellSens imaging software (Olympus). Random portions of the intestine were selected, and images were visualized and photographed using the same exposure times.
Abbreviations Used
- ARE
antioxidant responsive element
- BHA
butylated hydroxyanisole
- CNC/bZIP
Cap‘n’ Collar/basic leucine zipper
- GST
glutathione S-transferase
- HEK293
human embryonic kidney 293
- HO1
heme oxygenase-1
- IKK
IκB kinase
- Keap1
Kelch-like ECH-associated protein 1
- MAPK
mitogen-activated protein kinase
- MEF
mouse embryonic fibroblast
- NF-E2
nuclear factor erythroid 2
- NOD
nucleotide-binding oligomerization domain
- NQO1
NADPH quinone oxidoreductase-1
- Nrf2
nuclear factor erythroid 2 (NF-E2)-related factor 2
- ROS
reactive oxygen species
- SQSTM1
Sequestosome-1
- TAB1
TAK1-binding protein 1
- TAB2
binding protein 2
- TAK1
TGF-β-activated kinase 1
- tBHQ
tert-butylhydroquinone
- TBK1
TANK-Binding Kinase 1
- TLR
toll-like receptor
Acknowledgments
The authors thank Dr. Yue Xiong for Keap1 expression vector, Dr. Shizuo Akira for Tak1flox/flox mice, Dr. Robine for villin.CreERT2 mice, and North Carolina State University Biological Resources Facility for animal care. This work was supported by the National Institutes of Health Grants GM068812 and GM112986 (to J.N-T.), and GM088392 (to Y.T).
Contribution Statements
K.H., A.N.S., R.K-S, and J.N-T designed and conducted all the experiments. Y.T. and J.N-T. wrote the article.
Author Disclosure Statement
The authors declare no conflicts of interest exist.
References
- 1.Abiko Y, Miura T, Phuc BH, Shinkai Y, and Kumagai Y. Participation of covalent modification of Keap1 in the activation of Nrf2 by tert-butylbenzoquinone, an electrophilic metabolite of butylated hydroxyanisole. Toxicol Appl Pharmacol 255: 32–39, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, Lee HE, Kang D, and Rhee SG. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab 17: 73–84, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Baird L, Lleres D, Swift S, and Dinkova-Kostova AT. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc Natl Acad Sci U S A 110: 15259–15264, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A, and Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Chan K, Lu R, Chang JC, and Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93: 13943–13948, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 3765–3781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, and Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullinan SB, Gordan JD, Jin J, Harper JW, and Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, and Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A 99: 11908–11913, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggler AL, Luo Y, van Breemen RB, and Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem Res Toxicol 20: 1878–1884, 2007 [DOI] [PubMed] [Google Scholar]
- 11.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39: 186–193, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Franchi L, Munoz-Planillo R, and Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 13: 325–332, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa M. and Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol 25: 162–171, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hocking LJ, Lucas GJ, Daroszewska A, Mangion J, Olavesen M, Cundy T, Nicholson GC, Ward L, Bennett ST, Wuyts W, Van Hul W, and Ralston SH. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget's disease. Hum Mol Genet 11: 2735–2739, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Huang BW, Ray PD, Iwasaki K, and Tsuji Y. Transcriptional regulation of the human ferritin gene by coordinated regulation of Nrf2 and protein arginine methyltransferases PRMT1 and PRMT4. FASEB J 27: 3763–3774, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, and Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell 51: 618–631, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Inagaki M, Omori E, Kim JY, Komatsu Y, Scott G, Ray MK, Yamada G, Matsumoto K, Mishina Y, and Ninomiya-Tsuji J. TAK1-binding protein 1, TAB1, mediates osmotic stress-induced TAK1 activation but is dispensable for TAK1-mediated cytokine signaling. J Biol Chem 283: 33080–33086, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inokuchi S, Aoyama T, Miura K, Osterreicher CH, Kodama Y, Miyai K, Akira S, Brenner DA, and Seki E. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci U S A 107: 844–849, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishimura R, Tanaka K, and Komatsu M. Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett 588: 822–828, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Itoh K, Mimura J, and Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal 13: 1665–1678, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, and Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8: 379–391, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, and Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285: 22576–22591, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaramillo MC. and Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27: 2179–2191, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajino-Sakamoto R, Omori E, Nighot PK, Blikslager AT, Matsumoto K, and Ninomiya-Tsuji J. TGF-β-activated kinase 1 signaling maintains intestinal integrity by preventing accumulation of reactive oxygen species in the intestinal epithelium. J Immunol 185: 4729–4737, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, and Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci U S A 101: 2046–2051, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai T. and Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34: 637–650, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Kensler TW, Wakabayashi N, and Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Kim JY, Omori E, Matsumoto K, Nunez G, and Ninomiya-Tsuji J. TAK1 is a central mediator of NOD2 signaling in epidermal cells. J Biol Chem 283: 137–144, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkin V, McEwan DG, Novak I, and Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 34: 259–269, 2009 [DOI] [PubMed] [Google Scholar]
- 30.Kishimoto K, Matsumoto K, and Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J Biol Chem 275: 7359–7364, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi A, Kang M-I, Watai Y, Tong KI, Shibata T, Uchida K, and Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 26: 221–229, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, and Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213-U217, 2010 [DOI] [PubMed] [Google Scholar]
- 33.Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, and Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Johnson D, Calkins M, Wright L, Svendsen C, and Johnson J. Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol Sci 83: 313–328, 2005 [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto G, Shimogori T, Hattori N, and Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet 24: 4429–4442, 2015 [DOI] [PubMed] [Google Scholar]
- 36.Mihaly SR, Ninomiya-Tsuji J, and Morioka S. TAK1 control of cell death. Cell Death Differ 21: 1667–1676, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscat J. and Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137: 1001–1004, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ninomiya-Tsuji J, Kajino T, Ono K, Ohtomo T, Matsumoto M, Shiina M, Mihara M, Tsuchiya M, and Matsumoto K. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem 278: 18485–18490, 2003 [DOI] [PubMed] [Google Scholar]
- 39.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, and Matsumoto K. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398: 252–256, 1999 [DOI] [PubMed] [Google Scholar]
- 40.Omori E, Matsumoto K, Sanjo H, Sato S, Akira S, Smart RC, and Ninomiya-Tsuji J. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J Biol Chem 281: 19610–19617, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omori E, Matsumoto K, Zhu S, Smart RC, and Ninomiya-Tsuji J. Ablation of TAK1 upregulates reactive oxygen species and selectively kills tumor cells. Cancer Res 70: 8417–8425, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omori E, Morioka S, Matsumoto K, and Ninomiya-Tsuji J. TAK1 regulates reactive oxygen species and cell death in keratinocytes, which is essential for skin integrity. J Biol Chem 283: 26161–26168, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeffer K, Matsuyama T, Kundig TM, Wakeham A, Kishihara K, Shahinian A, Wiegmann K, Ohashi PS, Kronke M, and Mak TW. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 73: 457–467, 1993 [DOI] [PubMed] [Google Scholar]
- 44.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rada P, Rojo AI, Offergeld A, Feng GJ, Velasco-Martin JP, Gonzalez-Sancho JM, Valverde AM, Dale T, Regadera J, and Cuadrado A. WNT-3A regulates an Axin1/NRF2 complex that regulates antioxidant metabolism in hepatocytes. Antioxid Redox Signal 22: 555–571, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz L, Diaz-Meco MT, Nakano H, and Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J 19: 1576–1586, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, and Akira S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol 6: 1087–1095, 2005 [DOI] [PubMed] [Google Scholar]
- 48.Taguchi K, Motohashi H, and Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140, 2011 [DOI] [PubMed] [Google Scholar]
- 49.Takaya K, Suzuki T, Motohashi H, Onodera K, Satomi S, Kensler TW, and Yamamoto M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med 53: 817–827, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, and Tergaonkar V. ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol Cell 40: 75–86, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, and Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23: 33–42, 1998 [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, and Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28: 2758–2770, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, Gandhi P, Munson M, Miyamoto S, and Kelliher MA. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-κB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol Cell Biol 31: 2774–2786, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang DD. and Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23: 8137–8151, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, and Karin M. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 164: 896–910, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]