Abstract
The domesticated almond [Prunus dulcis (L.) Batsch] and peach [P. persica (Mill.) D. A. Webb] originated on opposite sides of Asia and were independently domesticated ∼5000 yr ago. While interfertile, they possess alternate mating systems and differ in a number of morphological and physiological traits. Here, we evaluated patterns of genome-wide diversity in both almond and peach to better understand the impacts of mating system, adaptation, and domestication on the evolution of these taxa. Almond has around seven times the genetic diversity of peach, and high genome-wide values support their status as separate species. We estimated a divergence time of ∼8 MYA (million years ago), coinciding with an active period of uplift in the northeast Tibetan Plateau and subsequent Asian climate change. We see no evidence of a bottleneck during domestication of either species, but identify a number of regions showing signatures of selection during domestication and a significant overlap in candidate regions between peach and almond. While we expected gene expression in fruit to overlap with candidate selected regions, instead we find enrichment for loci highly differentiated between the species, consistent with recent fossil evidence suggesting fruit divergence long preceded domestication. Taken together, this study tells us how closely related tree species evolve and are domesticated, the impact of these events on their genomes, and the utility of genomic information for long-lived species. Further exploration of this data will contribute to the genetic knowledge of these species and provide information regarding targets of selection for breeding application, and further the understanding of evolution in these species.
Keywords: Prunus persica, peach, Prunus dulcis, almond, domestication, mating system
Prunus is a large genus in the family Rosaceae with ∼200 species, including multiple domesticated crops such as almond, apricot, cherry, peach, and plum (Rehder 1940; Potter 2011). Peach [P. persica (Mill.) D. A. Webb] and almond [P. dulcis (L.) Batsch] are two of the three most economically important domesticates in Prunus globally, and share a number of similarities, including perenniality, precocity, and genome size and organization (Baird et al. 1994; Arús et al. 2012). However, the two species also have striking differences. While peaches are harvested for their indehiscent fleshy mesocarp, almonds are harvested for their seed, encased in a stony endocarp and a leathery, dehiscent mesocarp and exocarp (see Supplemental Material, File S1 and Figure S1). And while almond, like most Prunus species, exhibits S-RNase-based gametophytic self-incompatibility, peach is self-compatible (Hedrick et al. 1917; Wellington et al. 1929). Almond and peach also differ for other traits, such as life span (Gradziel 2011), chilling requirements (Alonso et al. 2005; Dozier et al. 1990; Scorza and Okie 1991), and adventitious root generation (Kester and Sartori 1966).
Domestication of almond and peach occurred independently ∼5000 yr ago in the Fertile Crescent and China (Zohary et al. 2012), respectively, followed by global dissemination beginning before 1300 BCE (Hedrick et al. 1917; Edwards 1975; Gradziel 2011; Zheng et al. 2014). The few obvious domestication traits in almond are reduced toxicity, thinner endocarp, and increased seed size, while domestication in peach is characterized by diverse fruit morphology (size, color, texture, shape, etc.) and self-compatibility. Other traits not typically associated with domestication, such as precocity, adventitious rooting, graft compatibility, or tree architecture, may also have been selected during domestication or subsequent breeding (reviewed in Miller and Gross 2011; Spiegel-Roy 1986). Efforts to identify the wild progenitors of either almond or peach by examining species relationships within subgenus Amygdalus have produced inconsistent species trees and numerous polytomies (Mowrey et al. 1990; Browicz and Zohary 1996; Ladizinsky 1999; Aradhya et al. 2004; Bassi and Monet 2008; Zeinalabedini et al. 2010; Verde et al. 2013). Given uncertainty in the wild progenitors and the difficulties associated with long generation times, QTL-mapping approaches to investigate peach or almond domestication are thus impractical. In contrast, comparatively fast and inexpensive sequencing makes population genetic approaches (cf. Ross-Ibarra et al. 2007) an attractive option, enabling the identification of domestication loci and study of the genome-wide impacts of changes in mating system.
Both domestication and mating system have been shown to shape genomic patterns of diversity in annual species (Glémin et al. 2006; Doebley et al. 2006; Hazzouri et al. 2013; Slotte et al. 2013), but the impacts of these forces on tree species remain poorly documented (McKey et al. 2010; Miller and Gross 2011; Gaut et al. 2015; but see Hamrick et al. 1992 for relevant analyses of allozyme diversity data). Mating system differences between closely related species pairs has been shown to significantly affect many aspects of genome evolution in Arabidopsis, Capsella, and Collinsia, including lower nucleotide diversity, higher linkage disequilibrium (LD), and reduced effective population size (Ne) (Hazzouri et al. 2013; Slotte et al. 2013; Wright et al. 2013). Demographic bottlenecks associated with domestication may also reduce diversity genome-wide, and selection during domestication will reduce diversity even further at specific loci (Glémin et al. 2006; Doebley et al. 2006). While studies in perennials, particularly tree fruit crops, suggest they have lost little genetic diversity due to domestication (reviewed in Miller and Gross 2011), recent analysis of resequenced peach genomes are consistent with lower genetic diversity and higher LD across the genome compared to related wild species (Verde et al. 2013; Cao et al. 2014). No such genome-wide analysis of diversity in almonds currently exists, however, and little is known about how differences in mating system affect changes in diversity during domestication.
Here, we leverage both new and published genome sequences to present an evolutionary genomic analysis of the effects of domestication and mating system on diversity in both almond and peach. Understanding the impact of mating system will expand the basic knowledge of genome evolution in a perennial species pair with contrasting mating systems, and identification of candidate domestication loci will provide an opportunity to assess convergence during domestication and compare tree domestication to that of annual crops.
Materials and Methods
Samples
We used 13 almond and 13 peach genomes for all analyses, which included both public and newly resequenced data (Table 1 and Table S1). In addition, we used one peach-almond F1 hybrid and one peach with Nonpareil almond in its pedigree as checks for admixture analysis. For this study, we resequenced nine almonds, one peach, an almond-peach F1 hybrid, and the plum P. cerasifera as an outgroup (Table 1 and Table S1). Fresh leaves and dormant cuttings collected from multiple sources were either desiccated with silica or stored at 4° prior to DNA isolation. We isolated DNA following a modified CTAB method (Doyle 1987).
Table 1. P. dulcis, P. persica, and outgroup species used in analyses.
| Species | n | Average Depth | Reference |
|---|---|---|---|
| P. dulcis | 4 | 7.76 | Koepke et al. (2013) |
| P. dulcis | 9 | 19.34 | This study |
| P. persica | 10 | 19.13 | Verde et al. (2013) |
| P. persica | 2 | 13.78 | Ahmad et al. (2011) |
| P. persica | 1 | 37.36 | This study |
| P. cerasifera | 1 | 35.02 | This study |
Libraries for eight of the almond samples were prepared at UC Davis. We quantified the sample DNA with Quanti-iT Picogreen dsDNA assay (Invitrogen, Life Technologies) and then fragmented 1 μg with a Bioruptor (Diagenode) for 11 cycles of 30 sec ON and 30 sec OFF per cycle. The resulting DNA fragment ends were then repaired with NEBNext End Repair (New England BioLabs) followed by the addition of deoxyadenosine triphosphate to the 3’ ends with Klenow Fragment (New England BioLabs). We then ligated barcoded Illumina TrueSeq adapters (Affymetrix) to the A-tailed fragments with Quick Ligase (New England BioLabs). Between each enzymatic step, we washed the DNA with Sera-Mag SpeedBeads (GE Life Sciences, Pittsburgh). The resulting libraries were quantified with a Qubit (Life Technologies) and sized using a BioAnalyzer DNA 12,000 chip (Agilent Technologies). Libraries were sent to UC Berkeley (Berkeley, Qb3) for quantification by qPCR, multiplexing, and sequencing for 100 bp paired-end reads in a single HiSeq 2000 (Illumina) lane. DNA from the remaining samples (Table 1 and Table S1) was submitted to BGI (Shenzen, China) for library preparation and sequenced using 100 bp paired-end reads at their facility in Hong Kong. Sequence data are available from SRA (http://www.ncbi.nlm.nih.gov/sra) and the associated run numbers are given in Table S1.
Analysis
Quality control and mapping:
All FASTQ files were trimmed of remnant adapter sequences using Scythe (github.com/vsbuffalo/scythe) and then further trimmed for base quality with Sickle (github.com/najoshi/sickle) using default parameters for both. Trimmed reads were then aligned to the P. persica v1.0 reference (www.rosaceae.org) using BWA-MEM (Li 2013) with a minimum seed length of 10 and internal seed length of 2.85. After filtering for a minimum mapping quality of 30 and base quality of 20, sequence depth averaged 15.8× (4.7× to 34.6×) in almond and 19.7× (11.2× to 35.4× in peach; Figure S2, Table S1).
Diversity and candidate gene identification:
We estimated inbreeding coefficients using ngsF in the ngsTools suite (Fumagalli et al. 2014), and then calculated genotype likelihoods in ANGSD (Korneliussen et al. 2014) incorporating our inbreeding estimates. We calculated several population genetics statistics, including pairwise nucleotide diversity ( Nei and Li 1979), Tajima’s D (D; Tajima 1989), Fay and Wu’s H (H; Fay and Wu 2000), and Zeng’s E (E; Zeng et al. 2006) using the thetaStat subprogram in ANGSD. Diversity values were estimated in overlapping 1000 bp windows with 50 bp steps, removing windows with < 150 bp of sequence after filtering. Additionally, we calculated a normalized value by dividing per window by mean in each species. To identify candidate genes possibly selected during domestication, we filtered for genes in the lowest 5% empirical quantile of each diversity statistic. We further analyzed candidate loci for gene ontology (GO) using P. persica protein gene identifiers in the singular enrichment analysis tool and Fisher’s exact test using default statistical options at the AgriGO website (http://bioinfo.cau.edu.cn/agriGO/).
Population comparisons:
We treated peach samples and almond samples as two populations to evaluate population structure. We performed a principal component analysis (PCA) with ngsPopGen (Fumagalli et al. 2014), and used NGSadmix (Skotte et al. 2013) to perform an admixture analysis and assign proportions of almond and peach population to individuals using K = 2 through K = 6 as the number of potential subpopulations. Finally, we examined population differentiation by estimating genome-wide and in sliding windows (1000 bp windows with a 50 bp step) after removing windows with of sequence.
Estimating historical changes in Ne:
To model the history of these species and infer the historical changes in effective population size that may have occurred prior to or during domestication, we analyzed peach and almond samples using the Multiple Sequentially Markovian Coalescent (MSMC) method (Schiffels and Durbin 2014). This approach uses the observed pattern of mutations in multiple individuals to infer the time to the most recent common ancestor between pairs of sampled alleles, and provides maximum-likelihood estimation of population size as a function of time. Using the msmc software (github.com/stschiff/msmc) and msmc-tools (github.com/stschiff/msmc-tools), we applied this method to 10 individuals from our study (five peach and five almond samples; peach: PP02, PP03, PP04, PP05, and PP13; almond: PD03, PD04, PD05, PD06, and PD07) in two separate analyses. For each individual, we first identified SNPs for each chromosome using samtools mpileup (v. 1.3.1) with a minimum mapping and base quality cut off of 20. We filtered sites for depth < 15 using VCFtools (v. 0.1.13), and removed indels using bcftools (v. 1.3.1). To estimate population size changes during the recent past (since domestication), we ran the full MSMC model for peach and almond separately using the combined set of five samples for each run. To estimate changes in over a longer time period (2 MYA), we applied the PSMC’ model (see Schiffels and Durbin 2014) to each sample individually. To convert the mutation-scaled coalescent times and population sizes obtained from these analyses, we divided by a mutation rate of mutations per nucleotide per generation, and assumed a generation time of 10 yr for both peach and almond. The models and inference algorithms for PSMC’ and MSMC are available from github.com/stschiff/msmc, and our code for analyzing peach and almond samples is available from https://github.com/houghjosh/peach.
Gene expression:
We downloaded 10 SRA RNA-seq runs from four peach and almond tissues (Table S2). All runs were from either general transcriptome sequencing (Jo et al. 2015) or controls of differential expression experiments (Wang et al. 2013; Mousavi et al. 2014; Sanhueza et al. 2015). We then converted the runs into their paired FASTQ files using SRA-toolkit (v. 2.3.4) and quantified expression for each run separately against the peach transcriptome (v. 1.0) using kallisto (Bray et al. 2016). For each sequencing run, kallisto outputs the transcripts per million (TPM), a within library proportional measurement, for each gene. Each gene was then annotated with its candidate or noncandidate status based on Tajima’s D, Zeng’s E, or Fay and Wu’s H for both almond and peach. We also calculated the number of tissues in which each gene was expressed and the mean expression level in each tissue (across runs in which the gene was expressed).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
Diversity
Genome-wide nucleotide diversity ( Figure S5 and Figure S6) in almond is nearly sevenfold higher than in peach (0.0186 and 0.0027, respectively), and these differences were more pronounced in nongenic regions of the genome (Table 2 and Table S4). Though differences in diversity between peach and almond have been known from analyses using multiple marker systems (Mowrey et al. 1990; Byrne 1990; Martínez-Gómez et al. 2003; Aradhya et al. 2004), this study is the first comparison of whole genome sequences using multiple diverse individuals from both species. Previous genome scans of peach found low levels of genetic diversity compared to the closely related wild species, P. kansuensis, P. mira, and P. davidiana (Verde et al. 2013; Cao et al. 2014). Of these, only P. davidiana is outcrossing, and Verde et al. (2013) found it had the greatest nucleotide diversity of the species they examined, ∼threefold higher than domesticated peach. Despite its domesticated status, almond retains more genetic diversity than any of the peach species studied thus far, suggesting that mating system explains more of the differences in diversity among species than domestication. Finally, we observed considerable variation in diversity statistics among chromosomes in both species, including up to twofold differences in nucleotide diversity in peach (Table S4), perhaps suggesting the relatively recent effects of selection during domestication.
Table 2. Genome-wide, genic, and nongenic diversity statistics and neutrality test values.
| Species | Sites | D | H | E | |
|---|---|---|---|---|---|
| Almond | Genome | 18.37 | −1.15 | −0.12 | −0.22 |
| Genic | 10.57 | −1.49 | −0.03 | −0.35 | |
| Nongenic | 25.67 | −0.83 | −0.20 | −0.10 | |
| Peach | Genome | 2.70 | −0.49 | −0.56 | 0.14 |
| Genic | 1.67 | −0.51 | −0.50 | 0.10 | |
| Nongenic | 3.61 | −0.47 | −0.62 | 0.17 |
D, Tajima’s D; H, Fay and Wu’s H; E, Zeng’s E.
Mean values of Tajima’s D were negative for both almond and peach (Table 2), suggesting that a genome-wide excess of rare variants likely consistent with a history of population expansion. Strongly negative values of Tajima’s D have recently been reported in Populus tremuloides, a species also inferred to have undergone postglacial population expansion in the Quaternary Wang et al. (2016). While the wild progenitors of almond and peach are not definitively known, the current range of wild almond species is much larger than that of wild peach taxa, perhaps reflecting either contrasting initial population sizes or differential expansion of ancestral progenitors during interglacial periods following the Last Glacial Maximum (20 kbp; LGM).
Historical changes in Ne
To investigate the demographic factors that may have contributed to the strong allele frequency skews that we observed in both peach and almond (Table 2), we conducted a whole-genome analysis of coalescent rates between haplotypes through time using MSMC (Schiffels and Durbin 2014). The results from this analysis provide the first detailed comparisons of demography in both peach and almond, and enabled us to obtain estimates of population size changes from ∼2 MYA up to ≈1000 yr ago (i.e., the last 100 generations; Figure S8). We found no evidence for a domestication-associated population bottleneck in either peach or almond (Figure S8A). Instead, our results suggest that almond experienced a population expansion following a bottleneck ≈20,000 yr ago, consistent with our observations of a strongly negative Tajima’s D and perhaps due to rapid human-mediated dispersal from east Asia (Delplancke et al. 2012). In peach, our results suggest a gradual decline in beginning ≈2 MYA (Figure S8B), and extending to 5000 yr ago, after which the effective population size remains very low. Although our results do not support a bottleneck in peach, the gradual decline in starting in the distant past (≈2 MYA; Figure S8B) is consistent with the low overall diversity we observe (Table 2), and may reflect a shift to a higher selfing rate (Charlesworth 2003).
Overall, our analyses suggest that, although population bottlenecks or extreme population expansions have occurred during domestication in many crop species (Meyer et al. 2012; Beissinger et al. 2016), neither peach nor almond appear to have experienced such events. In this respect, our results mirror those from other domesticated woody perennial crop species, including grape and apple, which are also reported to lack domestication bottlenecks but maintain much of their ancestral genetic diversity (Myles et al. 2011; Gross et al. 2014). This difference between annual and perennial domesticated crops may be due to the unique life cycle features of perennials, including a long generation time, overlapping generations, and a typically outcrossing mating system, as well as a more recent period of domestication (Gaut et al. 2015). That we also found a large reduction in and neutral diversity in peach despite no evidence for a population bottleneck also highlights the possibility that, within woody perennials, mating system differences may play an important role in determining the propensity of these species to have domestication-associated bottlenecks.
Inbreeding
We estimated the average inbreeding coefficient (F) for almond and peach to be 0.002 (0.000–0.027) and 0.197 (0.000–0.737), respectively (Table S3 and Figure S3). Although two self-compatible almond varieties are included in this study, none of our almond samples are derived from self-fertilization, supporting the low estimated inbreeding values. Peaches in general are self-compatible (with the exception of male-sterile varieties), and three of the peach varieties sampled (PP06, PP08, and PP15) have inbreeding values consistent with self-pollination in the preceding generation (F = 0.74, 0.53, and 0.56, respectively). Consistent with its known history as the result of open-pollination (Hedrick et al. 1917), the Georgia Belle peach variety sampled was estimated to have
While the estimated inbreeding value for almond is not unexpected given that it is self-incompatible, the average for peach is lower than previously estimated selfing rates (s) of ( from Fogle and Dermen 1969; Fogle 1977; Miller et al. 1989; Akagi et al. 2016). While the widely cited Miller et al. (1989) estimate was based on a single isozyme marker and is thus unable to separate self-fertilization with outcrossing to close relatives, the Akagi et al. (2016) estimate based on 5180 SNP markers is also high ( from ). Our estimates are much closer to those from Aranzana et al. (2002), who estimated () from 35 microsatellites. In addition to differences in marker systems, these discrepancies are likely due at least in part to sampling, with estimates from outcrossed pedigrees (Aranzana et al. 2002) lower than those from landraces (Akagi et al. 2016). Broad examination of pedigree records, however, suggests our estimate of inbreeding is likely reasonable, as more than 67% of the 600 peaches in Okie (1998) were the result of outcrossing (Aranzana et al. 2002), including several of the varieties sampled here (Hedrick et al. 1917).
Population structure
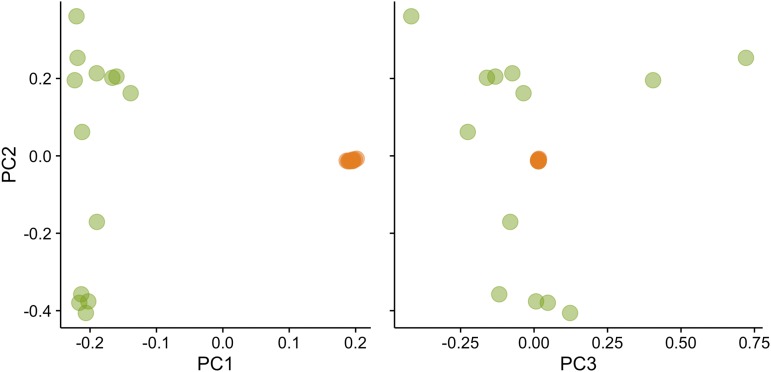
Genome-wide, our data are consistent with previous estimates (Aradhya et al. 2004) in finding strong genetic differentiation between almond and peach (weighted Figure S7 and Table S4). Like PCA also clearly distinguished almond from peach samples, primarily along PC1 (Figure 1). However, while PC2 and PC3 provided no further separation of peach samples, they do allow further separation of almond samples (Figure 1).
Figure 1.
Principle component (PC) analysis of almond (green) and peach (orange).
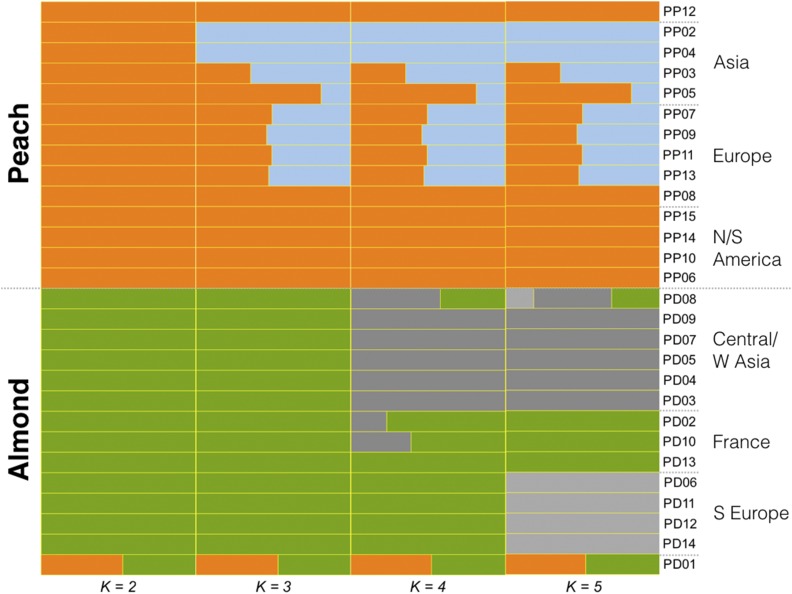
Admixture analysis clearly assigns individuals to either almond or peach populations at K = 2 (green and orange, respectively), including the correct identification of PD01 as an almond-peach F1 hybrid (Figure 2). Peach sample PP12, in contrast, should show ∼12.5% almond based on its pedigree (Fresnedo-Ramírez et al. 2013) but in this analysis does not differ from other New World peaches in its assigned proportions. The fact that PP12 shows fewer total variants than PP13 (“Georgia Belle”; Fresnedo-Ramírez et al. 2013) is also inconsistent with recent almond ancestry, suggesting possible errors in the recorded pedigree.
Figure 2.
Admixture proportion of almond (PD) and peach (PP) for K = 2 through K = 5. With the exception of the purported hybrids, PD01 and PP12, sample origins generally correspond with an east (top) to west (bottom) orientation for each type (Table S1).
Increasing the number of clusters (K), we find evidence for population substructure in both peach and almond (Figure 2 and Figure S4) distinguished by geographic origin or breeding status. In the admixture plot (Figure 2), within both almond and peach groups, samples at the top have more eastern origins (Central Asia or China, respectively), whereas those toward the bottom have more western origins (Spain or New World, respectively). Almond samples from China, Pakistan, Iran, and Turkey (PD09, PD07, PD05, PD04, and PD03) group together at both K = 4 and K = 5. At K = 5, a Mediterranean group of Italian and Spanish samples (PD06, PD11, PD12, and PD14) is identified, perhaps reflecting gene flow from North Africa into Spain and Italy (Delplancke et al. 2013). At K = 6, PD01 forms a unique cluster and several other almonds shift assignments, suggesting an overestimation of the number of subgroups (Figure S4). Similar overall patterns of structure in peach and almond were found in previous studies (Li et al. 2013; Micheletti et al. 2015; Shen et al. 2015; Delplancke et al. 2013) as well, suggesting the use of local varieties as founders, limited exchange between Asian and European breeding programs, or recent utilization of diverse genetic resources is not reflected in the sampling. The foundations of most modern almond breeding programs began within the past century, due in part to the challenges of understanding self-incompatibility, whereas the self-compatible peach has had more widespread efforts directed toward its development for millenia (though western breeding increased or intensified only within the past 10–20 generations).
All of our analyses of differentiation provide unequivocal evidence distinguishing almonds from peaches, strongly supporting their status as distinct species. Previous molecular analyses have estimated a broad range of divergence times between these species, from 2.5 MYA (Vieira et al. 2008) to more than 47 MYA (Chin et al. 2014). One compelling idea for the origin of peach and almond is that climatic changes after Himalayan orogeny and Tibetan Plateau uplift led to isolation of an hypothesized ancestral species resulting in allopatric divergence of peach from almond (Chin et al. 2014). Consistent with this possibility, our estimates of and nucleotide diversity give a divergence time of ≈8 MY under a simple model of divergence in isolation (cf. Holsinger and Weir 2009), assuming a mutation rate of per nucleotide and generation time of ≈10 yr. This corresponds to a period of climatic change following significant geologic activity and uplift specifically in the northeastern section of the Tibetan Plateau (Fang et al. 2007; Molnar et al. 2010).
Candidate loci
We next scanned the genomes of both almond and peach for potential candidate genes targeted by selection during domestication. In the lowest 5% quantile of Zeng’s E, we found 1334 and 1315 genes in peach and almond, respectively. Of these, peach and almond share 104, nearly double that expected by chance (permutation p-value ) and suggesting convergence in the process of domestication. In almond, candidate genes showed enrichment for GO categories related to protein amino acid phosphorylation, ATP biosynthetic processes, regulation of ADP ribosylation factor (ARF) protein signal transduction, membrane and nucleus cellular components, ATP binding, ATPase and protein tyrosine kinase activities, and zinc ion binding; candidate genes in peach showed enrichment for the GO category related to cellular catabolic processes. We also identified the 1314 genes showing the greatest differentiation between species (top 5% quantile of ), but while these genes were enriched for a number of GO categories (Table S5) no clear patterns emerged.
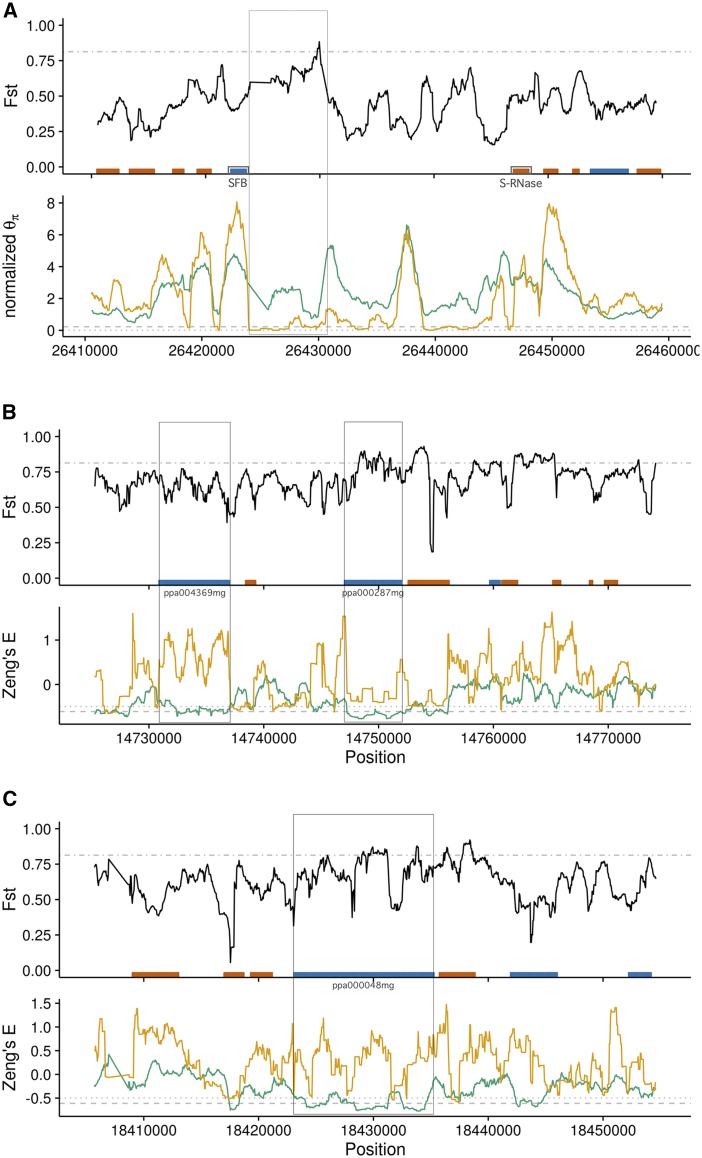
We first investigated the S-locus in order to examine a genomic region known to differ between almond and peach both in sequence and function (Tao et al. 2007; Hanada et al. 2014). The S-locus controls gametophytic self-incompatibility in Prunus (reviewed in Wu et al. 2013). The S-locus haplotype block consists of two genes, S-RNase and the S-haplotype-specific F-box (SFB), which function in the pistil and pollen, respectively. In our data, the intergenic region 3′ to both the S-RNase and SFB loci shows elevated differentiation with one extremely high peak and low nucleotide diversity in peach (Figure 3A), observations consistent with recent work showing peach having only five known S-haplotypes, two of which have identical SFB alleles (Tao et al. 2007; Hanada et al. 2014).
Figure 3.
Select 50 kb windows of the genome with high divergence () and either low normalized (A) or Zeng’s E (B and C) of almond (green) and peach (orange). Genes annotated in the peach reference genome are represented in the plot by boxes colored by their location on the plus strand (blue) or minus strand (red). In the plots, the gray lines indicate the upper 5% threshold, whereas in the and Zeng’s E panels the gray lines indicate the lower 5% thresholds of almond (dashed) and peach (dotted). Regions of interest, as described in the text, are boxed across adjacent panels and genes labeled. (A) S-locus divergence and diversity with S-locus genes, SFB (blue), and S-RNase (red), located on opposite sides of the central gap. Diversity in peach is drastically reduced immediately 3′ to SFB but only somewhat reduced 3′ to S-RNase, as might be expected for a linked locus. (B) and (C) Loci of interest on chromosome 3.
Windows in the lowest 5% quantile of the summary statistics investigated were generally enriched for genic regions of the genome in both taxa, but the signal in peach was weak and enrichment was not consistent across all statistics evaluated (Table S6). Nonetheless, a number of individual regions genome-wide showed strong signatures of selection. We examined 50 kb regions with contiguous windows in the bottom 5% quantile to focus our investigations of candidate genes. We focused on regions in both species for which there were overlapping regions of high and low or Zeng’s E, as these were significant for both peach and almond (permutation p-values Table 3).
Table 3. Permutation probability for the overlap of neutrality test or selected candidate genes with high selected candidate genes.
| Species | Tajima’s D | Fay and Wu’s H | Zeng’s E | |
|---|---|---|---|---|
| Almond | 0 | 0 | 0 | 0 |
| Peach | 0.5854 | 0.3336 | 0.0342 | 0 |
While many intergenic and putative regulatory regions also showed interesting patterns in diversity statistics, we examined two regions of chromosome 3 with moderate to high and divergent values of Zeng’s E between peach and almond, specifically low values of Zeng’s E in almond (Figure 3, B and C). The first of these regions (Figure 3B), contains the uncharacterized genes ppa004369mg (position 3:14730867..14736998; Uniprot identifier M5WRK6_PRUPE) and ppa00287mg (position 3:14747030..14752018; Uniprot identifier M5WX95_PRUPE), which have similarity to γ-aminobutyrate (GABA) transaminases in Malus domesticus and Myosin-1 in Gossypium arboreum, respectively. GABA is involved in signaling and nuclear regulation of cell wall modification and cell death through repression and activation, respectively, while GABA transaminases degrade GABA in the mitochondria and are reported to have a role in pollen–pistil interactions. Myosins are cellular motor proteins that act in concert with actin filaments for intracellular transport and cellular structure. The second region of interest on chromosome 3 (Figure 3C) contains the uncharacterized gene ppa000048mg (position 3: 18423078..18435264, Uniprot identifier M5XGZ7_PRUPE). This gene is in the GO category of protein N-linked glycosylation, and though it has high protein BLAST similarity among many species, few were annotated. Further investigation of additional regions with limited homology to characterized genes or functional information may be warranted given the poor characterization of genes in these species.
Given the importance of fruit morphology in peach, we hypothesized that selection during domestication and subsequent breeding may have targeted genes primarily expressed in fruit tissue. To test this hypothesis, we compared gene expression in four tissues (peach fruit and leaf, and almond ovary and anther) to candidate gene status. Candidates were overrepresented among genes expressed in all tissues, and we saw no evidence of enrichment for tissue-specific expression in any of the four tissues ( test showed significant underenrichment in most cases; Table S7). Even among genes showing tissue-specific expression, we found no difference in expression between domestication candidates and noncandidates. We did, however, find that genes showing strong differentiation between almond and peach (highest 5% tail of ) showed higher levels of expression in both leaves and fruit. While we have no clear a priori hypothesis predicting differences in leaf-specific expression, higher fruit-specific expression among is certainly of note given the striking differences in fruit morphology between the species.
Contrary to our predictions, we find no evidence that domestication candidates are enriched for genes showing unusual patterns or levels of expression. Recent results, however, suggest that larger fruits may have much predated domestication. Seeds of a 2.6 MY-old fossil peach, P. kunmingensis, were recently reported to be nearly identical to modern peaches (Su et al. 2015), and the observed correlation between seed size and fruit size in peach (Zheng et al. 2014) suggests that fruit size was likely larger as well. Our finding that fruit-specific genes showing the strongest differentiation between species are more highly expressed is, thus, at least consistent with the possibility of selection for differences in fruit morphology between peach and almond predating domestication.
Conclusions
One of the primary questions regarding the domestication of perennial crops, particularly tree crops, is its genetic basis (Miller and Gross 2011). Here, we have examined two closely related domesticated tree species with alternate mating systems in an attempt to tease apart the genomic signatures of domestication and mating system, and better understand these processes in perennial species. In addition to presenting evidence consistent with mating system effects in determining overall patterns of genetic diversity, our results identify numerous genes and genomic regions showing evidence of selection, and provide evidence of convergence in the domestication of almond and peach, and that fruit was not preferentially targeted during domestication but likely selected much earlier during species divergence. Finally, the high-coverage sequence we provide for a number of important cultivars may be useful to breeders and geneticists in identifying the causal basis of quantitative trait loci or developing marker sets for marker-assisted selection or genomic prediction.
Supplementary Material
Acknowledgments
We thank Anne Lorant for almond sequencing library preparations and Emily Josephs, Michelle Stitzer, and two anonymous reviewers for their helpful comments and suggestions on the manuscript. Support for D.V. was provided by the McDonald Endowment for University of California (UC), Davis Plant Sciences Graduate Student Research Assistantship and the Almond Board of California (ABC; grant HORT16-Aradhya/Ledbetter). Resequencing was funded by the ABC (grant HORT16-Aradhya/Ledbetter) and a Henry A. Jastro Research Fellowship. Resequencing funded by the Henry A. Jastro Research Fellowship used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by National Institutes of Health S10 Instrumentation grants S10RR029668 and S10RR027303. J.R.-I. and J.H. were supported by funding from National Science Foundation Plant Genome project IOS-1238014.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.032672/-/DC1.
Communicating editor: J. Wendel
Literature Cited
- Ahmad R., Parfitt D. E., Fass J., Ogundiwin E., Dhingra A., et al. , 2011. Whole genome sequencing of peach (Prunus persica L.) for SNP identification and selection. BMC Genomics 12: 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T., Hanada T., Yaegaki H., Gradziel T. M., Tao R., 2016. Genome-wide view of genetic diversity reveals paths of selection and cultivar differentiation in peach domestication. DNA Res. 23: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J., Ansón J., Espiau M., R. Socias i Company , 2005. Determination of endodormancy break in almond flower buds by a correlation model using the average temperature of different day intervals and its application to the estimation of chill and heat requirements and blooming date. J. Am. Soc. Hortic. Sci. 130: 308–318. [Google Scholar]
- Aradhya M. K., Weeks C., Simon C. J., 2004. Molecular characterization of variability and relationships among seven cultivated and selected wild species of Prunus L. using amplified fragment length polymorphism. Sci. Hortic. (Amsterdam) 103: 131–144. [Google Scholar]
- Aranzana M., Garcia-Mas J., Carbo J., Arús P., 2002. Development and variability analysis of microsatellite markers in peach. Plant Breed. 121: 87–92. [Google Scholar]
- Arús P., Verde I., Sosinski B., Zhebentyayeva T., Abbott A. G., 2012. The peach genome. Tree Genet. Genomes 8: 531–547. [Google Scholar]
- Baird W. V., Estager A. S., Wells J. K., 1994. Estimating nuclear DNA content in peach and related diploid species using laser flow cytometry and DNA hybridization. J. Am. Soc. Hortic. Sci. 119: 1312–1316. [Google Scholar]
- Bassi D., Monet R., 2008. Botany and taxonomy, pp. 1–36 in The Peach: Botany, Production and Uses, chap 1., edited by Layne D. R., Bassi D. CABI, Wallingford, UK. [Google Scholar]
- Beissinger T. M., Wang L., Crosby K., Durvasula A., Hufford M. B., et al. , 2016. Recent demography drives changes in linked selection across the maize genome. Nature Plants 2: 16084. [DOI] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L., 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Browicz K., Zohary D., 1996. The genus Amygdalus L. (Rosaceae): species relationships, distribution and evolution under domestication. Genet. Resour. Crop Evol. 43: 229–247. [Google Scholar]
- Byrne D., 1990. Isozyme variability in four diploid stone fruits compared with other woody perennial plants. J. Hered. 81: 68–71. [Google Scholar]
- Cao K., Zheng Z., Wang L., Liu X., Zhu G., et al. , 2014. Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 15: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2003. Effects of inbreeding on the genetic diversity of populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S.-W., Shaw J., Haberle R., Wen J., Potter D., 2014. Diversification of almonds, peaches, plums and cherries–molecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol. 76: 34–48. [DOI] [PubMed] [Google Scholar]
- Delplancke M., Alvarez N., Espíndola A., Joly H., Benoit L., et al. , 2012. Gene flow among wild and domesticated almond species: insights from chloroplast and nuclear markers. Evol. Appl. 5: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delplancke M., Alvarez N., Benoit L., Espíndola A., Joly H. I., et al. , 2013. Evolutionary history of almond tree domestication in the mediterranean basin. Mol. Ecol. 22: 1092–1104. [DOI] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D., 2006. The molecular genetics of crop domestication. Cell 127: 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doyle J. J., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Dozier W., Powell A., Caylor A., McDaniel N., Carden E., et al. , 1990. Hydrogen cyanamide induces budbreak of peaches and nectarines following inadequate chilling. HortScience 25: 1573–1575. [Google Scholar]
- Edwards, S., 1975 The almond industry of Mexico. Master’s Thesis, Oregon State University, Corvallis. [Google Scholar]
- Fang X., Zhang W., Meng Q., Gao J., Wang X., et al. , 2007. High-resolution magnetostratigraphy of the Neogene Huaitoutala section in the eastern Qaidam Basin on the NE Tibetan Plateau, Qinghai Province, China and its implication on tectonic uplift of the NE Tibetan Plateau. Earth Planet. Sci. Lett. 258: 293–306. [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle H., 1977. Self-pollination and its implications in peach improvement. Fruit Var. J. 31: 74–75. [Google Scholar]
- Fogle H. W., Dermen H., 1969. Genetic and chimeral constitution of three leaf variegations in the peach. J. Hered. 60: 323–328. [Google Scholar]
- Fresnedo-Ramírez J., Martínez-García P. J., Parfitt D. E., Crisosto C. H., Gradziel T. M., 2013. Heterogeneity in the entire genome for three genotypes of peach [Prunus persica (L.) Batsch] as distinguished from sequence analysis of genomic variants. BMC Genomics 14: 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Vieira F. G., Linderoth T., Nielsen R., 2014. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics 30: 1486–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Díez C. M., Morrell P. L., 2015. Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 31: 709–719. [DOI] [PubMed] [Google Scholar]
- Glémin S., Bazin E., Charlesworth D., 2006. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. Biol. Sci. 273: 3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradziel T. M., 2011. Origin and dissemination of almond. Hortic. Rev. (Am. Soc. Hortic. Sci.) 38: 23–81. [Google Scholar]
- Gross B. L., Henk A. D., Richards C. M., Fazio G., Volk G. M., 2014. Genetic diversity in Malus × domestica (Rosaceae) through time in response to domestication. Am. J. Bot. 101: 1770–1779. [DOI] [PubMed] [Google Scholar]
- Hamrick J. L., Godt M. J. W., Sherman-Broyles S. L., 1992. Factors influencing levels of genetic diversity in woody plant species, pp. 95–124 in Population Genetics of Forest Trees. Springer, New York. [Google Scholar]
- Hanada T., Watari A., Kibe T., Yamane H., Wünsch Blanco A., et al. , 2014. Two novel self-compatible S haplotypes in peach (Prunus persica). J. Jpn. Soc. Hortic. Sci. 83: 203–213. [Google Scholar]
- Hazzouri K. M., Escobar J. S., Ness R. W., Killian Newman L., Randle A. M., et al. , 2013. Comparative population genomics in Collinsia sister species reveals evidence for reduced effective population size, relaxed selection, and evolution of biased gene conversion with an ongoing mating system shift. Evolution 67: 1263–1278. [DOI] [PubMed] [Google Scholar]
- Hedrick U. P., Howe G. H., Taylor O. M., Tubergen C. B., 1917. The Peaches of New York. JB Lyon Company, Albany, NY. [Google Scholar]
- Holsinger K. E., Weir B. S., 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y., Chu H., Cho J. K., Choi H., Lian S., et al. , 2015. De novo transcriptome assembly of two different peach cultivars grown in Korea. Genom. Data 6: 260–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester D. E., Sartori E., 1966. Rooting of cuttings in populations of peach (Prunus persica l.), almond (Prunus amygdalus batsch) and their F1 hybrids. Proc. Am. Soc. Hortic. Sci. 88: 219–223. [Google Scholar]
- Koepke T., Schaeffer S., Harper A., Dicenta F., Edwards M., et al. , 2013. Comparative genomics analysis in Prunoideae to identify biologically relevant polymorphisms. Plant Biotechnol. J. 11: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T. S., Albrechtsen A., Nielsen R., 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladizinsky G., 1999. On the origin of almond. Genet. Resour. Crop Evol. 46: 143–147. [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997v1 [q-bio.GN].
- Li X.-w., Meng X.-q., Jia H.-j., Yu M.-l., Ma R.-j., et al. , 2013. Peach genetic resources: diversity, population structure and linkage disequilibrium. BMC Genet. 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gómez P., Arulsekar S., Potter D., Gradziel T. M., 2003. An extended interspecific gene pool available to peach and almond breeding as characterized using simple sequence repeat (SSR) markers. Euphytica 131: 313–322. [Google Scholar]
- McKey D., Elias M., Pujol B., Duputié A., 2010. The evolutionary ecology of clonally propagated domesticated plants. New Phytol. 186: 318–332. [DOI] [PubMed] [Google Scholar]
- Meyer R. S., DuVal A. E., Jensen H. R., 2012. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196: 29–48. [DOI] [PubMed] [Google Scholar]
- Micheletti D., Dettori M. T., Micali S., Aramini V., Pacheco I., et al. , 2015. Whole-genome analysis of diversity and SNP-major gene association in peach germplasm. PLoS One 10: e0136803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. J., Gross B. L., 2011. From forest to field: perennial fruit crop domestication. Am. J. Bot. 98: 1389–1414. [DOI] [PubMed] [Google Scholar]
- Miller P. J., Parfitt D. E., Weinbaum S. A., 1989. Outcrossing in peach. HortScience 24: 359–360. [Google Scholar]
- Molnar P., Boos W. R., Battisti D. S., 2010. Orographic controls on climate and paleoclimate of Asia: thermal and mechanical roles for the Tibetan Plateau. Annu. Rev. Earth Planet. Sci. 38: 77. [Google Scholar]
- Mousavi S., Alisoltani A., Shiran B., Fallahi H., Ebrahimie E., et al. , 2014. De novo transcriptome assembly and comparative analysis of differentially expressed genes in Prunus dulcis Mill. in response to freezing stress. PLoS One 9: e104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey B. D., Werner D. J., Byrne D. H., 1990. Isozyme survey of various species of Prunus in the subgenus Amygdalus. Sci. Hortic. (Amsterdam) 44: 251–260. [Google Scholar]
- Myles S., Boyko A. R., Owens C. L., Brown P. J., Grassi F., et al. , 2011. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 108: 3530–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W.-H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okie W. R., 1998. Handbook of Peach and Nectarine Varieties. Performance in the Southeastern United States and Index of Names. Agriculture Handbook, Washington. [Google Scholar]
- Potter D., 2011. Prunus, pp. 129–145 in Wild Crop Relatives: Genomic and Breeding Resources. Springer, New York. [Google Scholar]
- Rehder A., 1940. Manual of Cultivated Trees and Shrubs. Macmillan Company, New York. [Google Scholar]
- Ross-Ibarra J., Morrell P. L., Gaut B. S., 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104: 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhueza D., Vizoso P., Balic I., Campos-Vargas R., Meneses C., 2015. Transcriptomic analysis of fruit stored under cold conditions using controlled atmosphere in Prunus persica cv. “Red Pearl.” Front. Plant Sci. 6: 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S., Durbin R., 2014. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorza R., Okie W. R., 1991. Peaches (Prunus). Acta Hortic. (290): 177–234. [Google Scholar]
- Shen Z., Ma R., Cai Z., Yu M., Zhang Z., 2015. Diversity, population structure, and evolution of local peach cultivars in china identified by simple sequence repeats. Genet. Mol. Res. 14: 101. [DOI] [PubMed] [Google Scholar]
- Skotte L., Korneliussen T. S., Albrechtsen A., 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195: 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F., et al. , 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. [DOI] [PubMed] [Google Scholar]
- Spiegel-Roy P., 1986. Domestication of fruit trees. Dev. Agric. Managed-For. Ecol. 16: 201–211. [Google Scholar]
- Su T., Wilf P., Huang Y., Zhang S., Zhou Z., 2015. Peaches preceded humans: fossil evidence from SW China. Sci. Rep. 5: 16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R., Watari A., Hanada T., Habu T., Yaegaki H., et al. , 2007. Self-compatible peach (Prunus persica) has mutant versions of the S haplotypes found in self-incompatible Prunus species. Plant Mol. Biol. 63: 109–123. [DOI] [PubMed] [Google Scholar]
- Verde I., Abbott A. G., Scalabrin S., Jung S., Shu S., et al. , 2013. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45: 487–494. [DOI] [PubMed] [Google Scholar]
- Vieira J., Fonseca N. A., Santos R. A., Habu T., Tao R., et al. , 2008. The number, age, sharing and relatedness of S-locus specificities in Prunus. Genet. Res. 90: 17–26. [DOI] [PubMed] [Google Scholar]
- Wang J., Street N. R., Scofield D. G., Ingvarsson P. K., 2016. Natural selection and recombination rate variation shape nucleotide polymorphism across the genomes of three related Populus species. Genetics 202: 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhao S., Gu C., Zhou Y., Zhou H., et al. , 2013. Deep RNA-seq uncovers the peach transcriptome landscape. Plant Mol. Biol. 83: 365–377. [DOI] [PubMed] [Google Scholar]
- Wellington R., Stout A. B., Einset O., Van Alstyne L. M., 1929. Pollination of fruit trees. Bulletin of the New York State Agricultural Experiment Station 577: 3–54. [Google Scholar]
- Wright S. I., Kalisz S., Slotte T., 2013. Evolutionary consequences of self-fertilization in plants. Proc. R. Soc. Lond. B Biol. Sci. 280: 20130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Gu C., Khan M. A., Wu J., Gao Y., et al. , 2013. Molecular determinants and mechanisms of gametophytic self-incompatibility in fruit trees of Rosaceae. Crit. Rev. Plant Sci. 32: 53–68. [Google Scholar]
- Zeinalabedini M., Khayam-Nekoui M., Grigorian V., Gradziel T., Martinez-Gomez P., 2010. The origin and dissemination of the cultivated almond as determined by nuclear and chloroplast SSR marker analysis. Sci. Hortic. (Amsterdam) 125: 593–601. [Google Scholar]
- Zeng K., Fu Y.-X., Shi S., Wu C.-I., 2006. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics 174: 1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Crawford G. W., Chen X., 2014. Archaeological evidence for peach (Prunus persica) cultivation and domestication in China. PLoS One 9: e106595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D., Hopf M., Weiss E., 2012. Domestication of Plants in the Old World. Oxford University Press, Oxford. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.