Abstract
Aim:
Chhotanagpuri breed of sheep reared for mutton in Jharkhand, India, having problem of low litter size and body weight. The response of genetic improvement for traits with low heritability through traditional selection method is time-consuming. Therefore, marker-assisted selection based on a polymorphism study of suitable candidate gene can response quickly. Thus, this study was aimed at identification of different allelic and genotypic frequencies of Booroola fecundity (FecB) gene and its association with multiple birth and postnatal growth in Chhotanagpuri sheep.
Materials and Methods:
DNA isolation and gene-specific amplification of FecB gene was performed from blood samples of from 92 Chhotanagpuri lambs maintained under similar feeding and management conditions. Custom nucleotide sequencing and single-strand conformational polymorphism analysis were performed to identify different genotypes with respect to the target gene. Statistical analysis was performed for determination of allelic and genotypic frequencies of FecB gene polymorphisms and its association with multiple birth and postnatal growth of lambs from birth to 52 weeks age.
Results:
“AA,” “AB,” and “BB” genotypes were found at locus-1 as it is polymorphic for FecB gene while locus-2 was found to be monomorphic for FecB gene. Higher frequency of “A” allele at locus-1 was found in single born lambs, whereas “B” allele was predominant among multiple born lambs. The lambs having “BB” genotype weighed significantly (p≤0.01) heavier than those of “AB” and “AA” genotype at 52 weeks of age.
Conclusion:
“BB” genotype has emerged as favored genotype for multiple births and better growth indicator. Therefore, homozygous lambs for “B” allele should be selected and utilized in breeding program for better growth rate.
Keywords: body growth, Booroola fecundity gene, Chhotanagpuri sheep, multiple births, single-strand conformational polymorphism
Introduction
Sheep farming is of major economic importance, especially for small and marginal farmers because it requires minimum resource [1]. Inevitably, rearing of Chhotanagpuri sheep in its native tract mainly for mutton purpose is of vast significance in Jharkhand where rural and landless farmers are in the majority due to shortage of arable land. However, sheep production suffers from major constrain as the majority of sheep breeds in India is having low litter size except the Garole, Kendrapara, and NARI-Suvarna sheep breed (http://www.cswri.res.in/breed_profiles.asp).
The prolificacy trait is quantitative in nature and controlled by multiple genes [2]. Improvement of reproductive traits has conventionally been regulated using quantitative genetic methods. Hence, increase in litter size by selection within a breed will be a time-consuming process as the reproductive traits are having low heritability. If the major genes associated with reproduction are identified, they can be introduced in breeding through marker assisted selection and it can infuse superior genotypes rapidly in the breeding population [3,4]. The Booroola fecundity gene (FecB) is an autosomal gene, which enhances ovulation rate through codominant effect and litter size by partial dominance [5,6]. The FecB locus is situated in the region of ovine chromosome 6, which is syntenic to human chromosome 4 [7]. High prolificacy in Booroola sheep is associated with non-conservative mutation (q249r) in a conserved intracellular kinase signaling domain of the bone morphogenetic protein receptor-1B (BMPR-1B) expressed in ovary and granulosa cells [8,9]. The BMPR-1B, also known as activin-like kinase-6, is a multifunctional protein of transforming growth factor-β superfamily which regulates growth and differentiation in many cell types. In recent years, it has also been shown to be associated with embryogenesis, hematopoiesis, immunological responses, reproductive endocrinology, development of reproductive organs, litter size, and body biometrics [10-13].
Hence, this study has been undertaken to identify different allelic and genotypic frequencies of FecB gene and its association with multiple birth and postnatal growth in Chhotanagpuri sheep.
Materials and Methods
Ethical approval
All the animal experiments were conducted after approval of committee for the purpose of control and supervision of experiments on animals.
Experimental design
This investigation was conducted on 92 Chhotanagpuri lambs maintained under Mega sheep seed project, at Instructional Small Ruminant Farm of Ranchi Veterinary College, Birsa Agricultural University, Kanke, Jharkhand, India. The lambs were maintained under natural feeding system during pre-weaning period (0-3 months of age) with their dams. However, adult sheep were maintained under semi-intensive system of management with 7-8 h of grazing daily. To study the growth rate, absolute body weights of Chhotanagpuri sheep were recorded at birth, 4th, 8th, 12th, 24th, 36th, 48th, and 52nd week of age. Birth weights of lambs were recorded within 2 h of birth. Body weights were recorded in the morning before offering feed.
Collection of blood samples and genomic DNA isolation
The jugular blood samples of 92 individuals (5 ml each) from Chhotanagpuri lambs were collected in vacutainer tubes containing ethylenediaminetetraacetic acid (EDTA) as anticoagulant. Cold chain was maintained throughout the sampling process. Genomic DNA was isolated from white blood cells using standard phenol-chloroform-Isoamyl alcohol method as per the standard protocol described by Sambrook et al. [14]. The extracted DNA samples were assessed for quantity and purity using NanoDrop Spectrophotometer (ND-1000 NanoDrop Spectrophotometer, Thermo Scientific, USA) while visual confirmation of the DNA integrity was also assessed by running on 0.6% agarose gel.
Amplification of FecB gene
The 190 bp fragment of the FecB gene was amplified from the genomic DNA of Chhotanagpuri lambs by polymerase chain reaction (PCR) using forward (5’- CCAGAGGACAATAGCAAAGCAAA-3’) and reverse primer (5’- CAAGATGTTTTCATGCCTCATCAACAGGTC-3’) reported elsewhere [9]. The amplification was performed in a thermocycler (GeneAmp® PCR system 9700, Applied Biosystems, USA) using PCR reaction mixture containing 1.5 µl 10× PCR buffer, 0.5 µl magnesium chloride (25 mM), 0.5 µl (10 mM) dNTPs, 0.5 µl (20 ng/µl) of each primer, 1U of Taq polymerase (SIGMA, USA), and 1.5 µl diluted genomic DNA (50 µg/µl). The final volume was adjusted to 25 µl by adding nuclease-free water. The thermocycling steps consisted of initial denaturation at 94°C for 3 min; while the cycling parameter consisted of denaturing step of 30s at 94°C, an annealing step of 30s at 58°C and an extension step of 30s at 72°C for 33 cycles; while the final extension step of 7 min was performed at 72°C. A non-template control reaction was simultaneously run to eliminate reagent contamination. The amplified products were resolved on 2% agarose gel in Tris-acetate EDTA buffer (×1). The agarose gel was stained with ethidium bromide and documented under UV light in a gel documentation system (UMAX PowerLook 2100XL-USB with MagicScan, Alpha Innotech Corporation, USA).
Single-strand conformational polymorphism (SSCP) of PCR amplified FecB gene fragment.
The SSCP analysis of FecB gene was carried out by diluting 4 µl amplified PCR product to 10 µl of SSCP gel loading dye (0.05% bromophenol blue, 0.05% xylene cyanol, 95% formamide, 20 mM EDTA). After denaturation at 95°C for 5 min, the samples were snap chilled immediately on ice for 10 min. Then, the contents were separated using 12% vertical acrylamide: bis-acrylamide gel electrophoresis at 8 V/cm for 16 h under refrigerated condition (29:1 acrylamide to bisacrylamide). The SSCP patterns of DNA fragments were visualized by silver staining of the polyacrylamide gel after electrophoresis [15].
Purification of PCR products and custom nucleotide sequencing
The PCR products were purified using PureLink® PCR purification kit (Thermo Scientific, USA) to remove primer dimers and other PCR ingredients before custom sequencing following manufacturer’s instructions. Custom sequencing of the purified PCR products from four selected animals was performed in both the directions using DNA sequencing service provided by Xcelris, Hyderabad following BigDye terminator v3.1 Cycle Sequencing Method (Applied Biosystems, USA).
In-silico analysis of FecB nucleotide sequence
The sequence chromatogram was annotated with BioEdit Sequence Alignment Editor Software version 7.0.5 (Isis Therapeutics, USA). The annotated sequences were identified based on being the closest match to the sequences submitted in the nonredundant GenBank nucleotide database (http://www.ncbi.nlm.nih.gov) on BLASTN searches [16].
Statistical analysis
The genotypic frequency, as well as the frequency of different alleles of FecB gene, was computed after sequence alignment. Chi-square test was used for statistical analysis in GraphPad Software (GraphPad Software, Inc., USA). The statistical significance of various fixed effect was studied by F-test. Whenever, the effect was significant, the differences were tested for significance by Duncan’s multiple range test as modified by Kramer [17] at 5% level of significance using the inverse of coefficient matrix.
Results
PCR amplification of FecB gene fragment
The 260/280 nm absorbance ratio for all the DNA samples ranged from 1.7 to 1.9 indicating acceptable quality of genomic DNA which was confirmed by visualization in agarose gel. PCR amplified product showed a prominent band of 190 bp size shown in Figure-1.
Figure-1.
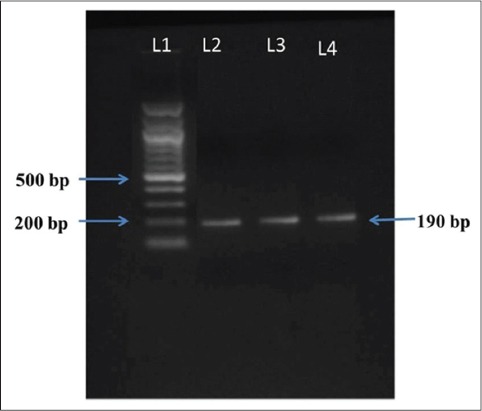
Polymerase chain reaction amplified product of Booroola fecundity (FecB) gene (190 bp) along with 100 bp DNA ladder; L1: DNA ladder; L2, L3, and L4: FecB gene product.
SSCP analysis of PCR amplified FecB gene product
SSCP revealed a polymorphic pattern with three genotypes, depicted as AA, AB, and BB, based on different band pattern observed. Three bands were present in the case of AA and BB genotype, whereas four bands were present in AB genotype (Figure-2).
Figure-2.
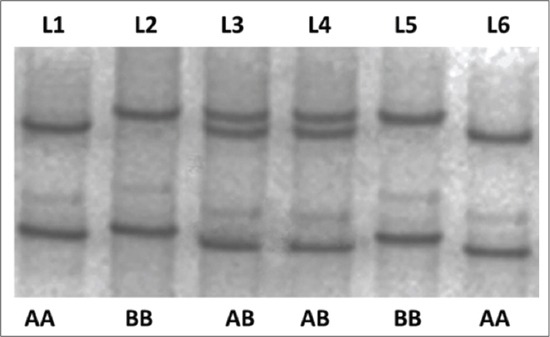
Different genotypic pattern (AA, AB, BB) of Booroola fecundity gene in single-strand conformational polymorphism analysis. L1: AA; L2: BB; L3: BBCD; L4: AB; L5: BB; L6: AA.
Nucleotide sequence analysis of PCR-SSCP variants
Manual annotation of the sequence chromatograms by BioEdit Sequence Alignment Editor Software version 7.0.5 revealed gene sequences of the partially amplified FecB gene fragment of Chhotanagpuri sheep and submitted to NCBI GenBank (Accession number KX896751). Custom nucleotide sequencing revealed single A/G polymorphism at 746 position (locus-1) of the coding region of FecB (BMPR-IB) gene whereas locus-2 was found to be monomorphic at 750 position as compare to reference sequences (AF312016, GU979816).
Genotype and gene frequency analysis
The genotypic and allelic frequencies in single and multiple born lambs were given in Table-1. The frequency of “A” allele at locus-1 was found to be higher in the case of single born lambs, whereas the frequency of “B” allele was found to be higher in the case of multiple born lambs.
Table 1.
The genotypic (“AA,” “AB,” and “BB” at locus 1) and allelic (“A” and “B”) frequencies of lambs born in singlet and multiple birth condition on the basis of SSCP variants of FecB gene.
| Genotypes | Genotype frequency | Allele | Allelic frequency |
|---|---|---|---|
| Singlet condition | |||
| Locus-1 | |||
| AA | 0.424 | A | 0.587 |
| AB | 0.326 | B | 0.413 |
| BB | 0.250 | ||
| Multiple birth | |||
| Locus-1 | |||
| AA | 0.048 | A | 0.22 |
| AB | 0.343 | B | 0.78 |
| BB | 0.608 |
SSCP: Single-strand conformational polymorphism, FecB: Booroola fecundity
Impact of FecB gene polymorphism on growth of lambs
Body weights of lambs were studied from birth to 12th week of age at an interval of 4 weeks and thereafter at 24, 36, 48, and 52 weeks of age. Least square analysis of variance was performed to see the effect of FecB genotypes (“AA,” “AB”, and “BB” at locus-1)) on body weight of lambs at birth and different stages of growth (Table-2).
Table 2.
Comparative analysis of body weights (least-square means±SE) with different genotypes (“AA,” “AB,” and “BB”) at locus 1 of FecB gene in Chhotanagpuri lambs from birth to 52 week of age.
| Effects | Number of observed | Body weights (kg) at different stages of growth | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Birth | 4-week | 8-week | 12-week | 24-week | 36-week | 48-week | 52-week | ||
| Overall (µ) | (92) | 1.76±0.03 | 4.04±0.11 | 5.64±0.18 | 6.64±0.23 | 8.42±0.26 | 9.79±0.28 | 10.92±0.27 | 11.83±0.32 |
| Genotypes at Locus 1 | |||||||||
| AA | (39) | 1.72±0.04 | 4.10±0.16 | 5.78±0.26ab | 6.75±0.33ab | 8.52±0.38ab | 9.61±0.39ab | 10.80±0.38ab | 11.72±0.45b |
| AB | (30) | 1.78±0.04 | 3.90±0.16 | 5.33±0.26b | 6.18±0.33b | 7.90±0.38b | 9.26±0.39b | 10.37±0.38b | 11.22±0.45b |
| BB | (23) | 1.77±0.06 | 4.12±0.17 | 5.80±0.27a | 6.98±0.34a | 8.85±0.40a | 10.48±0.42a | 11.60±0.40a | 12.54±0.48a |
Values bearing same superscript in a column for each effect separately did not differ significantly (p≤0.01). SE=Standard error, FecB=Booroola fecundity
The lambs having “BB” genotype weighed significantly (p≤0.01) heavier than lambs of “AB” genotype from 8 to 52 weeks of age. However, the difference between “AA” and “BB” genotype was found to be non-significant (p≥0.01). Similarly, the difference between “AB” and “AA” genotypes was also found to be non-significant (p≥0.01) up to 48 weeks of age (Table-2). However, finally at 52 weeks of age, the weight of lambs having “BB” genotype weighed significantly (p≤0.01) higher than lambs of both “AA” and “AB” genotype.
Discussion
Chhotanagpuri sheep as the sole recognized sheep breed of Jharkhand carries ample significance but suffers from low litter size. The FecB and related fecundity associated genes such as FecB, FecX1, FecXH, FecXB, and FecXG genes are well recognized to be associated with fertility, growth, and developmental parameters [18-25]. In this study, we have analyzed the frequencies of different genotypes and alleles in different loci of partially amplified 190 bp FecB gene fragment and its association with postnatal growth rate up to 52 weeks of age in Chhotanagpuri sheep. Three SSCP variants of FecB gene at locus-1 (“AA,” “AB” and “BB” genotypes) were observed. Genotypic frequencies at locus-1 among multiple birth lambs were 0.048, 0.343, and 0.608 for “AA,” “AB,” and “BB” genotypes, respectively, which revealed that “BB” genotypes are favored genotype for multiple births. This is in accordance with the finding of Guan et al. [12] where mean litter size of Chinese Merino sheep with genotypes “BB” and “AB” was found to be significantly higher than that of genotype “AA.” Genotypic frequencies at locus-1 revealed that the overall frequency of “A” allele is more (0.587) as compared to “B” allele (0.413). Among multiple born lambs, the frequencies of “B” allele (0.78) was found to be predominant than its counterparts “A” allele (0.22). The present findings are in agreement with the finding of Jia et al. [26], who found three types of genotype of FecB gene in small tailed Han sheep and poll Dorset sheep. Similarly, Guan et al. [12] have also reported three different Booroola genotypes in Chinese Merino sheep. Polley et al. [27] reported FecB gene polymorphism in Garole sheep and obtained two types of allele of that gene as observed in this study. In a related study by Sun et al. [28], on microsatellite markers have revealed different polymorphisms in FecB gene having a significant effect on litter size in Hu sheep. Similar studies on related fecundity genes have also documented several polymorphisms, and many of them were found to be associated with fertility parameters in sheep as well as goats. Genome-wide association studies by Demars et al. [29] have identified two novel mutations in the BMP15 or FecX gene associated with increased litter size and ovulation rate in Grivette and Olkuska ewes. In a related study by Feng et al. [30] have revealed GDF9 gene or FecG polymorphism is having a positive significant association with high litter size in Jining Gray goats.
However, Chu et al. [31] reported that the FecB gene had no significant effect on prolificacy of Jining Gray goats whereas Ahlawat et al. [32] have shown that FecB, BMP15, and GDF9 genes have no effect on prolificacy of seven Indian goat breeds including the highly prolific Black Bengal goats. Findings of Ghaffari et al. [33] in Shal sheep and Borni et al. [34] in North African Barbarine ewes suggested that the genetic factor controlling twinning in these sheep breeds is not related to FecB mutation. Shafieiyan et al. [35] have also reported no mutations of FecB and FecGH genes in high prolific Iranian Lory sheep. The difference in opinion may be due to environmental factors, the difference in breeds or genetic materials used.
This study revealed a significant effect of genotypes at locus-1 on body weights of lambs from 8th to 52nd week of age, though its effect was not significant for birth weight and weight at 4th week of age. At all stages of growth from 8 to 52-week of age, lambs with “BB” genotype weighed significantly more than heterozygous (“AB”) lambs, but the difference with those with “AA” genotype was not significant except at 52 weeks of age. The superiority of “BB” genotype over “AB” for body weights from 8 to 52-week of age ranged between 8.82% (8 weeks) and 13.17% (36 weeks). However, its superiority over “AA” genotype ranged between 0.35% (8 weeks) and 9.05% (36 weeks). This is in accordance with findings of Guan et al. [12] where the body weights of “BB/BA” Chinese Merino lambs at 90 days after birth were found to be significantly higher than “AA” lambs. It is also evident that “BB” genotype has emerged as superior to “AA” and “AB” genotypes with the progression of age.
Conclusion
Lambs of BB genotype were found to be favorable for multiple birth and B allele was found to have better growth potential. Thus, the lambs for “BB” genotype may be selected and utilized for breed improvement under breeding program.
Authors’ Contributions
TO, LBS, and DKS have designed the research plan TO have performed the wet lab analysis MG, SSK, and RK have analyzed the data. TO, MG, and DKS have drafted the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
The authors fully acknowledge the financial and infrastructural support provided by the competent authorities of Birsa Agricultural University, Ranchi, Jharkhand, India.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Naqvi S.M.K, Sejian V, Karim S.A. Effect of feed flushing during summer season on growth, reproductive performance and blood metabolites in Malpura ewes under semi-arid tropical environment. Trop. Anim. Health Prod. 2013;45:143–148. doi: 10.1007/s11250-012-0185-2. [DOI] [PubMed] [Google Scholar]
- 2.Miao X, Luo Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction. 2013;145:587–596. doi: 10.1530/REP-12-0507. [DOI] [PubMed] [Google Scholar]
- 3.Dekkers J.C.M. Commercial application of marker and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004;82:313–328. doi: 10.2527/2004.8213_supplE313x. [DOI] [PubMed] [Google Scholar]
- 4.Williams J.L. The use of marker-assisted selection in animal breeding and biotechnology. Rev. Sci. Tech. 2005;24:379–391. [PubMed] [Google Scholar]
- 5.Piper L.R, Bindon B.M, Davis G.H. The single gene inheritance of the high litter size of the Booroola Merino. In: Land R.B, Robinson D.W, editors. Genetics of Reproduction in Sheep. London: Butterworths; 1985. pp. 115–125. [Google Scholar]
- 6.Montgomery G.W, McNatty K.P, Davis G.H. Physiology and molecular genetics of mutations that increases ovulation rate in sheep. Endocr. Rev. 1992;13(2):309–327. doi: 10.1210/edrv-13-2-309. [DOI] [PubMed] [Google Scholar]
- 7.Montgomery G.W, Crawford A.M, Penty J.M, Dodds K.G, Ede A.J, Henry H.M, Pierson C.A, Lord E.A, Galloway S.M, Schmack A.E, Sise J.A, Swarbrick P.A, Hanrahan V, Buchanan F.C, Hill D.F. The ovine Booroola fecundity gene (FecB) is linked to markers from a region of human chromosome 4q. Nat. Genet. 1993;4:410–414. doi: 10.1038/ng0893-410. [DOI] [PubMed] [Google Scholar]
- 8.Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, Cribiu E, Thimonier J, Teyssier J, Bodin L, Cognie Y, Chitour N, Elsen J.M. Mutation in the bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola ewes. Proc. Natl. Acad. Sci. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson T, Yang X.W, Jennifer L.J, Ian K.R, Joanne M.L, Eric A.L, Ken G.D, Grant A.W, John C.M, Anne R.O, Kenneth P.M, Grant W.M. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 10.Letterio J.J, Roberts A.B. Regulation of immune responses by TGF-beta. Annu Rev Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 11.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 12.Guan F, Liu S.R, Shi G.Q, Yang L.G. Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Anim. Reprod. Sci. 2007;99:44–52. doi: 10.1016/j.anireprosci.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 13.Hua G, Yang L. A review of research progress of FecB gene in Chinese breeds of sheep. Anim. Reprod. Sci. 2009;116:1–9. doi: 10.1016/j.anireprosci.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E.F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. NY: Cold Spring Harbour, Cold Laboratory Press; 1989. [Google Scholar]
- 15.Bassam B.J, Gustavo C.A, Gresshoff P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F, Madden T.L, Schäffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer C.Y. Extension of multiple range tests to group correlated adjust means. Biometrics. 1957;13:13. [Google Scholar]
- 18.Davis G.H. Fecundity genes in sheep. Anim. Reprod. Sci. 2004;82-83:247–253. doi: 10.1016/j.anireprosci.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Davis G.H, Balakrishnan L, Ross I.K, Wilson T, Galloway S.M, Lumsden B.M, Hanrahan J.P, Mullen M, Mao X.Z, Wang G.L, Zhao Z.S, Zeng Y.Q, Robinson J.J, Mavrogenis A.P, Papachristoforou C, Peter C, Baumung R, Cardyn P, Boujenane I, Cockett N.E, Eythorsdottir E, Arranz J.J, Notter D.R. Investigation of the Booroola (FecB) and Inverdale (FecXI) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim. Reprod. Sci. 2006;92(1-2):87–96. doi: 10.1016/j.anireprosci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Vacca G.M, Dhaouadi A, Rekik M, Carcangiu V, Pazzola M, Dettori M.L. Prolificacy genotypes at BMPR 1B, BMP15 and GDF9 genes in North African sheep breeds. Small Rumin. Res. 2010;88:67–71. [Google Scholar]
- 21.Silva B.D, Castro E.A, Souza C.J, Paiva S.R, Sartori R, Franco M.M, Azevedo H.C, Silva T.A, Vieira A.M, Neves J.P, Melo E.O. A new polymorphism in the growth and differentiation factor 9 (GDF9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim. Genet. 2011;42:89–92. doi: 10.1111/j.1365-2052.2010.02078.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y.Q, Li Y.X, Zhang N, Wang Z.B, Bai J.Y. Polymorphism of exon 2 of BMP15 gene and its relationship with litter size of two Chinese goats. Asian Aust. J. Anim. Sci. 2011;24(7):905–911. [Google Scholar]
- 23.Moradband F, Rahimi G, Gholizadeh M. Association of polymorphisms in fecundity genes of GDF9, BMP15 and BMP15-1B with litter size in Iranian Baluchi sheep. Asian Aust. J. Anim. Sci. 2011;24(9):1179–1183. [Google Scholar]
- 24.Pramod R.K, Sharma S.K, Singhi A, Pan S, Mitra A. Differential ovarian morphometry and follicular expression of BMP15, GDF9 and BMPR1B influence the prolificacy in goat. Reprod. Domest. Anim. 2013;48(5):803–809. doi: 10.1111/rda.12165. [DOI] [PubMed] [Google Scholar]
- 25.Ahlawat S, Sharma R, Maitra A. Screening of indigenous goats for prolificacy associated DNA markers of sheep. Gene. 2013;517:128–131. doi: 10.1016/j.gene.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Jia C.L, Li N, Zhao X.B, Zhu X.P, Jia Z.H. Association of single nucleotide polymorphisms in exon 6 region of BMPR1B gene with litter size traits in sheep. Asian Aust. J. Anim. Sci. 2005;18:1375–1378. [Google Scholar]
- 27.Polley S, De S, Brahma B, Mukherjee A, Vinesh P.V, Batabyal S, Arora J.S, Pan S, Samanta A.K, Datta T.K, Goswami S.L. Polymorphism of BMPR1B, BMP15 and GDF9 fecundity genes in prolific Garole sheep. Trop. Anim. Health Prod. 2010;42(5):985–993. doi: 10.1007/s11250-009-9518-1. [DOI] [PubMed] [Google Scholar]
- 28.Sun W, Chang H, Musa H.H, Chu M. Study on relationship between microsatellite polymorphism and producing ability on litter size trait of Hu sheep in China. Afr. J. Biotechnol. 2010;9(50):8704–8711. [Google Scholar]
- 29.Demars J, Fabre S, Sarry J, Rossetti R, Gilbert H, Persani L, Tosser-Klopp G, Mulsant P, Nowak Z, Drobik W, Martyniuk E, Bodin L. Genome-wide association studies identify two novel BMP15 mutations responsible for an atypical hyperprolificacy phenotype in sheep. PLoS Genet. 2013;9(4):e1003482. doi: 10.1371/journal.pgen.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng T, Geng C.X, Lang X.Z, Chu M.X, Cao G.L, Di R, Fang L, Chen H.Q, Liu X.L, Li N. Polymorphisms of caprine GDF9 gene and their association with litter size in Jining Grey goats. Mol. Biol. Rep. 2011;38(8):5189–5197. doi: 10.1007/s11033-010-0669-y. [DOI] [PubMed] [Google Scholar]
- 31.Chu M.X, Zhao X.H, Zhang Y.J, Jin M, Wang J.Y, Di R, Cao G.L, Feng T, Fang L, Ma Y.H. Polymorphisms of BMPR-IB gene and their relationship with litter size in goats. Mol. Biol. Rep. 2010;37(8):4033–4039. doi: 10.1007/s11033-010-0062-x. [DOI] [PubMed] [Google Scholar]
- 32.Ahlawat S, Sharma R, Roy M, Mandakmale S, Prakash V, Tantia M.S. Genotyping of novel SNPs in BMPR1B, BMP15, and GDF9 genes for association with prolificacy in seven Indian goat breeds. Anim. Biotechnol. 2016;27(3):199–207. doi: 10.1080/10495398.2016.1167706. [DOI] [PubMed] [Google Scholar]
- 33.Ghaffari M, Javaremi A.N, Rahimi G. Detection of polymorphism in BMPR-IB gene associated with twining in Shal sheep using PCR-RFLP method. Int. J. Agric. Biol. 2009;11(1):97–99. [Google Scholar]
- 34.Borni J, Sonia B, Naouer D.M. PCR-RFLP of BMPR1B gene in North African Barbarine sheep. Res. Opin. Anim. Vet. Sci. 2011;1(4):200–203. [Google Scholar]
- 35.Shafieiyan Z, Mohammadi G, Jolodarzadeh A, Amiri S. No mutations of FecB and FecGH in Iranian Lory sheep. Vet. Res. Forum. 2013;4(4):265–268. [PMC free article] [PubMed] [Google Scholar]


