Abstract
Nanopores are powerful single-molecule sensors with nanometer scale dimensions suitable for detection, quantification, and characterization of nucleic acids and proteins. Beyond sequencing applications, both biological and solid-state nanopores hold great promise as tools for studying the biophysical properties of RNA. In this review, we highlight selected landmark nanopore studies with regards to RNA sequencing, microRNA detection, RNA/ligand interactions, and RNA structural/conformational analysis.
1. INTRODUCTION TO NANOPORES
Nanopores have garnered much attention for their ability to act as single molecule DNA sequencers when paired with a proper ratcheting enzyme and ionic current monitoring electronics. Despite their biological importance, much less work has been reported on the study of RNA molecules. Much like DNA, the direct sequencing of single RNA molecules is a topic of great importance. In addition, the secondary and tertiary structures formed by relatively short RNA fragments are also topics of equal biological importance. Nanopores are unique tools that have been shown to be extremely sensitive to variations in sequence as well as structure. In this chapter, we discuss the current state of RNA sequencing as well as structural interrogation using both solid-state and biological nanopores.
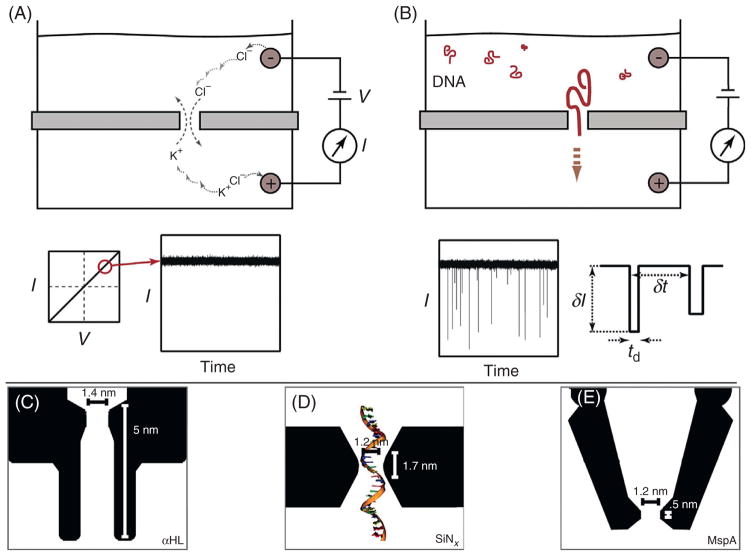
The principle of nanopore sensing is based on fast measurements of ion fluctuations through a nanoscale orifice. A cross-sectional view of a nanopore in an insulating membrane (not to scale) is shown in Fig. 1A. A concentrated electrolyte solution (>100 mM) is placed on both sides of the membrane such that the only electrical contact between the chambers is at the nanopore. An electrochemical bias applied across the membrane causes ion transport through the nanopore at rates that are determined by the nanopore dimensions, nanopore surface charge, applied voltage, ion mobility, and solution viscosity. In general for a thin pore of symmetric geometry, the current–voltage response is linear, as shown in Fig. 1A (bottom). Applying a constant DC bias results in a steady-state current that serves as a baseline measurement of the nanopore volume.
Figure 1.
Schematic of the nanopore ionic current detection of biomolecules,1 as well as illustrations of the most commonly used nanopores.2 (A) A partition containing a nanoscale aperture (nanopore) separates two chambers containing electrolyte solution. When a voltage bias is applied across the partition, a steady ionic current through the nanopore is detected (images not to scale). (B) When DNA is added to the grounded (cis) chamber, it is electrophoretically driven through the nanopore to the positively charged (trans) chamber. The passage of each individual DNA molecule results in a transient decrease in the ionic current through the nanopore. (C–E) Schematics of α-hemolysin (αHL), silicon nitride (SiN), and Mycobacterium smegmatis porin A (MspA) nanopores (left to right) are shown with dimension labels (SiN pores have variable geometry). Images obtained with permission from Venta et al.2
Transient entry and exit of biological macromolecules from the nanopore results in discrete disruptions in the steady-state baseline signal, as shown in Fig. 1B for the case of DNA molecules. Since nucleic acids are negatively charged the voltage applied to the analyte chamber (cis chamber) is more negative than the voltage applied to the trans chamber in order to electrophoretically drive the molecules through the nanopore. The transport kinetics of nucleic acids through nanopores is highly dependent on the nanopore geometry and other experimental conditions. While this topic has been studied exhaustively both theoretically and experimentally, it is beyond the scope of this chapter and the reader is referred to other reviews for a discussion on this topic.1,3,4
2. EARLY EXPERIMENTS TOWARD RNA SEQUENCING
2.1 Differentiation of RNA Homopolymers
It is of historical significance that the first experiment to report electrophoretically driven transport of biomolecules across a nanopore were done with homouridine RNA fragments (polyU).5 In these pioneering experiments the investigators formed a lipid bilayer across a 100 μm diameter orifice in a polytetrafluoroethylene (PTFE) partition; the bilayer and partition are used to separate two chambers containing a buffered electrolyte solution. When αHL nanopores are added to one of the chambers (cis), one porin will spontaneously insert itself into the lipid bilayer. In the presence of a trans-membrane potential, αHL insertion is observed as a discrete increase in ion current across the pore. Using αHL as a biomolecular sensor, researchers were able to detect the presence of RNA homopolymers (as well as DNA heteropolymers) as individual molecules translocated the nanopore. Kasianowicz et al. determined that the average time necessary for an RNA homopolymer to translocate the pore scales linearly with polymer length (ie, between 150 and 415 nt), thus demonstrating that nanopores can be used to measure the length of linear RNA fragments. The authors also hypothesized that this technique could be used to sequence DNA and RNA. While the former has been fully realized,6,7 experimenters have been making steady progress toward the latter.
Since the first experiment, various research groups have made great strides in advancing the field of nanopore sensing by making favorable mutations to the αHL nanopore, as well introducing a number of new nanopore systems, both protein based and synthetic. The three types of nanopores we will discuss in this chapter can be seen in Fig. 1C–E, as adapted from Venta et al.,2 with αHL and MspA nanopores being the biological examples and SiN nanopores being the synthetic example. We will not cover device fabrication in this chapter, but many reviews exist on this topic.1,8
2.2 Observation of polyU and polyC Segments Within Diblock Copolymers
A sequence specificity study by Akeson et al. demonstrated that the same αHL nanopores are able to distinguish between RNA bases based on the unique current blockade level each RNA base produces during translocation.9 More specifically, their experiments revealed that RNA homopolymers of polyU, polyC, and polyA could be distinguished based upon the distinct current blockade of each molecule. The authors also showed that translocation of an RNA diblock copolymer containing repeats of A and C bases (ie, A(30)C(70)) produces bilevel events that are in good agreement with the current levels seen by polyA and polyC homopolymers. Interestingly, the authors noted that 90% of events produced blockades that correspond to 3′ entry of the polymer. The nature of this preferential orientation was later expanded upon by Butler et al. who observed that both C50 homopolymers as well as A25C50 coblock polymers favor entry from their polyC 3′ ends.10
2.3 DNA Sequencing Using an MspA Pore as a Reader and DNA Polymerase as a Molecular Ratchet
Over the next few years the promise of nanopores as a viable next-generation sequencing platform spurred development forward. The two major obstacles toward nanopore-based DNA sequencing were verification of true single base specificity and control over DNA transport during translocation. Through site-directed mutagenesis researchers were able to show that mutant αHL (E11N/K147N/M113Y)11,12 as well as mutant MspA nanopores13 are capable of individual DNA base recognition. Soon after, the Akeson and Gundlach groups slowed down DNA translocation to measurable rates (ie, milliseconds per nucleotide) by enzymatically ratcheting DNA base by base through a nanopore.6,7 Further information on the progress toward nanopore-based DNA sequencing can be found in several reviews.1,3,4
2.4 Discriminating Single Bases Using Immobilized RNA Strands
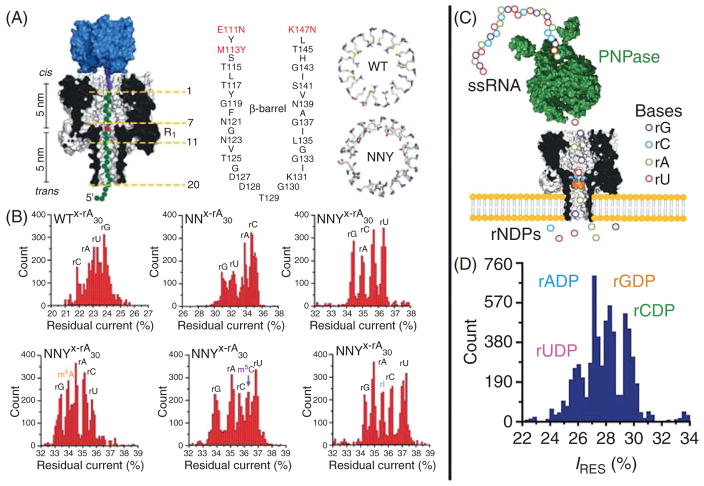
Ayub and Bayley later demonstrated that the αHL pore is sensitive to single base substitutions in RNA fragments.14 In this study the authors compared the wild-type (WT) αHL to the two αHL mutants that had previously shown sensitivity to base recognition. Here the authors used the same method that was previously employed to show that αHL and MspA nanopores are able to resolve all four DNA bases.12,13 For this experiment four oligonucleotides were synthesized, each with a different ribobase at the ninth nucleotide position (counted from the 3′ end) within an otherwise polyU sequence, which was chosen to eliminate formation of secondary structures. Each oligo was produced with a biotinylated 3′ end that was then conjugated to a streptavidin molecule. The ninth nucleotide position was specifically chosen as the site of variation because once a streptavidin–RNA complex is drawn into the pore (Fig. 2A), the ninth nucleotide will sit precisely at the central constriction of the αHL nanopore. Previous experiments have shown that the WT αHL pore contains four constrictions or recognition sites, but the central constriction (referred to as R1) appears to be the most sensitive of the four sites. As such the authors measured the ionic current through the pore when each of the four ribonucleobases was immobilized at the R1 site. While the WT and mutant NN pore displayed some level of base specificity, they were unable to completely resolve each base. In contrast, the NNY mutant produced substantial separation in the current levels observed for each base. The authors were also able to produce similar results for the N6-methyladenosine (m6A), 5-methylcytosine (m5C), and inosine (I) post-transcriptional modifications. By repeating the previous experiment with these modified bases at the ninth nucleotide position, they observed that each base has a current blockade level that is distinct from each of the four canonical RNA bases (Fig. 2B).
Figure 2.
The road toward direct RNA nanopore sequencing. (A) An illustration of the streptavidin immobilized RNA strand used by Ayub et al.14 The four proposed constrictions are labeled, and diagrams of the WT and NNY mutant αHL pores are shown. (B) Histograms of the residual ionic current while an RNA is immobilized within the nanopore are displayed. The top row shows the superior base discrimination of the NNY mutant when compared to WT and NN pores. The bottom row demonstrates the additional epigenetic sensing ability of the NNY mutant. (C) A diagram of the proposed exonuclease-assisted sequencing method.15 (D) Current histogram comparing the residual currents caused by binding of rADP, rGDP, rCDP, and rUDP to the molecular adapter inside the αHL β-barrel. Images obtained with permission from Ayub et al.14,15
2.5 Progress Toward Enzymatic Control of RNA Transport
While φ29 polymerase has been shown to be a suitable enzyme for controlling DNA motion by ratcheting DNA through nanopores, an enzyme for the ratcheting of RNA through nanopores is yet to be presented. Before the demonstration of nanopore sequencing via φ29 polymerase, researchers proposed to use an exonuclease-based method for nanopore sequencing. In this method, a DNA/exonuclease complex is electrophoretically captured by a nanopore where the exonuclease will sequentially cleave the DNA bases one at a time (Fig. 2C). As the bases are cleaved, the applied electric field pulls the bases to the pore where they encounter a molecular adapter that temporarily traps the base, and identifies it according to its unique current blockade.16 This method was revisited for the purpose of RNA sequencing and researchers showed that, as with DNA, this method can be used to accurately discriminate between individual RNA bases (Fig. 2D).15 Unfortunately, as with the exonuclease-based DNA sequencing method, significant problems persist with this method pertaining to the efficiency of capturing cleaved nucleotides.17 Once nucleotides are cleaved by the exonuclease, the electrophoretic force biases their motion toward the pore, but Brownian forces hamper this directionality. As a result of this diffusive effect, some bases will diffuse into the bulk solution and not be read by the nanopore, or be captured after diffusing out of the pore. This results in bases being read out of sequence, as well as insertion and deletion errors. Several improvements to this method have been proposed to circumvent the diffusion problem,18 but to date this remains an open problem.
2.6 Summary
The experiments described earlier represent an important step toward nanopore-based RNA sequencing. A single-molecule method for direct RNA sequencing that is able to resolve RNA modifications would be a valuable tool for medical diagnoses as well as academic studies of the regulatory roles played by these modifications. However, it remains to be seen whether these modifications can be resolved from one another in the context of a heavily modified RNA sequence, if these developments can be translated into a nanopore device with a single recognition site capable of sequencing or if there is an enzyme that is capable of long–read length ratcheting of RNA molecules. Though major roadblocks for this method persist, realization of such a device would certainly change the landscape of transcriptomics.
3. POINT-OF-CARE APPLICATIONS
Though nanopores have received a great deal of attention for their promise as tools for sequencing nucleic acids, they have also shown to be extremely effective platforms for potential single-molecule point-of-care (POC) systems. The nanopore’s extreme sensitivity to small differences in the charge and size of passing molecules makes them excellent systems for the detection of nucleic acid probes, RNA–ligand interactions, antibiotic complexes, etc. In addition the single-molecule nature of these systems allows them to perform rapid analysis without the need for error prone amplification steps or time consuming and costly labeling procedures.
3.1 MicroRNA Detection and Quantification
3.1.1 A Solid-State Nanopore System for Rapid Detection of Target microRNA Expression
One promising application for nanopores is the rapid detection of microRNA (miRNA) molecules. Several methods have been developed for the detection and quantification of miRNA molecules and many more are currently in development.19 The most conventional methods involve probe labeling using radioisotopes and/or some form of PCR amplification. Though these methods are effective, the use of radioactive labeling is hazardous and often times inconvenient, and amplification can be error prone and time consuming.
The Drndić Lab has demonstrated a promising technique for the analysis of miRNAs using solid-state nanopores,20 which takes advantage of the sensitivity of the nanopore platform to Angstrom differences in molecule size. The authors showed that by fabricating 3-nm diameter nanopores in thinned solid-state membranes (ie, 6-nm effective thickness) they can distinguish between dsDNA, dsRNA, and transfer RNA (tRNA) molecules, and previous papers have demonstrated the ability of solid-state nanopores to distinguish between single-stranded and double-stranded nucleic acids.21 This is accomplished by monitoring the ionic current through the pore for spontaneous reductions, each of which are representative of the diameter of the translocating molecule. By comparing the current blockades produced by translocation of an unknown sample with those produced by the translocation of known single-stranded and double-stranded samples, it becomes possible to identify the unknown sample based on its diameter. This same feature of nanopores is exploited to identify target miRNA sequences within total RNA extracts.
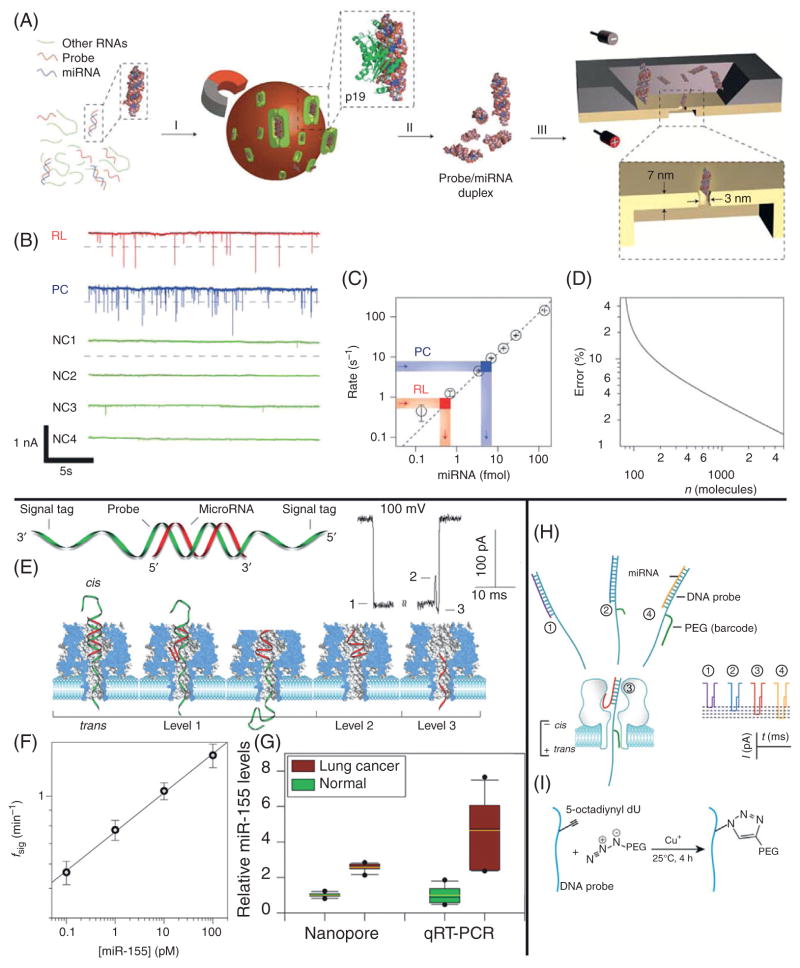
In this method, RNA extracted from tissue cells is incubated with probe RNA molecules whose sequence is complimentary to that of the target molecule and therefore binds to any target miRNA molecule present. This sample is then incubated with magnetic beads that are functionalized with the p19 protein from the Carnation Italian ringspot virus. This protein has been shown to selectively bind to dsRNA in a size-dependent, but sequence-independent manner, specifically binding to 21–23 bp dsRNA. Accordingly, the p19 protein will bind specifically to any probe/miRNA duplex molecules present in the sample. The sample is then washed to remove the remaining RNA molecules that are not bound to the magnetic beads and finally the desired probe/miRNA duplex is eluted from the protein (see Fig. 3A for a diagram of this entire process).
Figure 3.
Nanopore-based miRNA detection schemes. (A) A cartoon of the miRNA detection scheme used by the Drndić Lab.20 Total RNA extract is incubated with probe molecules complimentary to a target miRNA sequence. This mixture is then incubated with magnetic beads that are functionalized with the p19 protein, which selectively binds to 21–23 bp dsRNA molecules. A washing step then isolates the target miRNA/probe complex, which can be readily detected using a thin SiN nanopore. (B) A sample of the ionic current traces resulting from translocations of the isolated miRNA/probe complexes. RL is the trace resulting from enrichment of miR122a, PC is a positive control, and NC refers to various negative control experiments. (C) A calibration curve is constructed by plotting the event rates as a function of miRNA concentration. The event rates of the PC and RL experiments are shown as indicated and the miRNA concentration is inferred from the calibration curve. (D) The error in concentration as predicted from the nanopore experiments, as a function of number of translocation events. (E) A diagram of the unzipping method used by the Gu Lab.22 A probe/target miRNA complex is shown along with the translocation process of this molecule and its resulting ionic current trace. (F) A calibration curve constructed by plotting event rate versus miRNA concentration is shown. (G) The miRNA levels as detected by the nanopore, as well as qRT-PCR are compared. (H) A schematic of the revised Gu Lab strategy,23 employing four different polyethylene glycol (PEG) tags. (I) A diagram of the click chemistry tagging process. Images obtained with permission from Wanunu et al.20,22
Once the probe/miRNA duplex has been isolated, these ultrathin nanopore platforms can easily distinguish any dsRNA molecules present, thereby verifying the presence of the target miRNA. Example nanopore traces of this sample (RL), a positive control (PC), and negative controls (NC) are shown in Fig. 3B. In order to quantify the amount of target miRNA present in a sample, a calibration curve is constructed. The calibration curve is populated by measuring the rate of translocation events from control dsRNA samples with varying concentrations. From this calibration curve the authors concluded that there is a linear relationship between the rate of translocation events and the concentration of molecules in the sample. Finally, when an unknown sample is tested its event rate can easily be linked to a corresponding target miRNA concentration, as shown in Fig. 3C. The authors confirm that this method can quantify a 1 fM sample in ~4 min, making it a promising platform for the rapid detection of biologically relevant levels of miRNA.
3.1.2 Detecting Lung Cancer–Related miRNAs and Single Nucleotide Polymorphisms Using a Biological Nanopore System
The Gu group have presented a similar probe-based approach to quantification of miRNAs using an αHL nanopore.22 This method does not use a simple threshold technique for detecting probe bound molecules, as the αHL nanopore constriction is too narrow to allow the passage of dsRNA. Instead, this method relies upon the identification of what the authors call a “signature signal.” To produce this signature signal, the authors use a probe molecule that is similar to the one used by the Drndić Lab in that it is designed to target a specific miRNA sequence by Watson–Crick base pairing in its central region, but is extended from both its 3′ and 5′ ends with a polydC30 tag. The capture of this probe/miRNA complex produces a distinctive signal, which is shown in detail in Fig. 3E. As the complex enters the vestibule of the pore the extended end of the probe molecule threads through the β-barrel of the pore (Level 1), here the miRNA is sterically required to “unzip” from the probe molecule as the probe translocates, the current momentarily increases as the probe exits the β-barrel region (Level 2), then quickly reduces to a similar level as the miRNA enters the β-barrel (Level 3), and finally returns to the open pore level upon translocation of the miRNA. Counting the rate of these signature events, and using a similar calibration process as described by the Drndić Lab enables the authors to quantify miRNA concentrations as low as 100 fM (Fig. 3F).
Notably, this platform is capable of detecting single nucleotide mismatches between the miRNA and probe sequence, corresponding to miRNA single nucleotide polymorphisms (SNPs). A fully complementary probe molecule will bind more tightly to the target miRNA and thus take longer to unzip than the molecule containing an SNP. The authors demonstrated that by first calibrating the system with a fully complimentary prove/miRNA complex, they are able to detect three different lung cancer-related SNPs, as each SNP results in a distinct decrease in the unzipping time.
The authors used this technique to detect the presence of miR-155, an miRNA that has elevated levels in lung cancer patients. The results obtained from this method matched the diagnosis of those obtained from qRT-PCR, but with a higher degree of precision (Fig. 3G).
3.1.3 Multiplexing microRNA Detection Using Probe Specific Tags
The Gu Lab recently published a paper representing a significant advance in the development of miRNA detection technology.23 The authors demonstrate a method for overcoming the problem of simultaneous detection of several miRNA targets by conjugating PEG tags of varying lengths to the probe molecules through copper(I)-catalyzed click chemistry, with each PEG length corresponding to a different miRNA (Fig. 3H and I). This method relies on the ability of PEG to modulate the ionic current, and hence producing characteristic current levels depending upon the size of the translocating PEG molecule.24,25 As probe/miRNA complexes translocate the pore, each produces a “signature signal,” similar to that described earlier, but with the addition of a variable-length PEG tag discrimination of several miRNA targets is possible. This tagging system is used to simultaneously detect the concentrations of four different miRNAs down to 10 pM (though, the authors suggest a limit of 100 fM).
Though these experiments represent significant progress toward a mature nanopore miRNA detector, a number of problems must still be addressed. The Gu Lab has shown simultaneous detection of four different miRNAs, but an ideal miRNA detection platform would be able to detect hundreds or thousands of miRNA in parallel. This need dictates that any mature system should incorporate a large array of nanopores, though the strategy proposed by the Gu Lab can simplify this engineering task by decreasing the size of the array by at least a factor of four. The optical detection scheme developed by the Wallace and Bayley labs (but not covered in this chapter) may be able to address this issue.26 Additionally, other proposed miRNA detection schemes have demonstrated attomolar detection limits.27 While such a low detection limit may be unnecessary, demonstration of a similar sensitivity would prove useful for detection of rare miRNAs or early stage diagnoses. Many techniques have been developed to push nanopore detection limits,28–30 but none have been demonstrated in conjunction with miRNA quantification to date.
3.2 RNA/Ligand Interactions and Nanopore-Based Drug Screening
3.2.1 Antibiotic Drug Screening Through Detection of RNA/Drug Binding
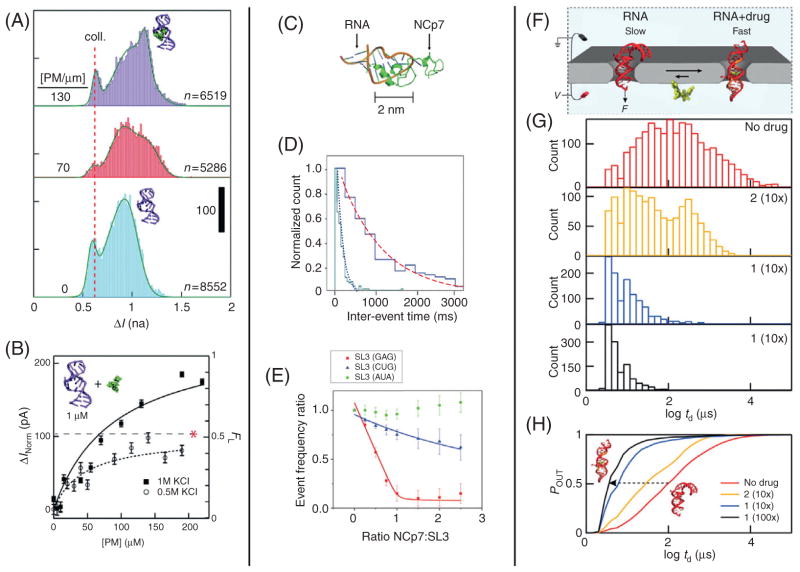
Another potential POC application for the nanopore platform is as a monitor for RNA/ligand interactions. The Drndić Lab have presented a proof-of-principle experiment showing that the previously described thin solid-state nanopore system can be used as a platform for high-throughput drug screening assays, particularly for RNA-mediated diseases.31 To perform this first demonstration, the authors tested the ability of nanopores to detect the binding of several aminoglycoside antibiotics. Aminoglycosides target the prokaryotic ribosomal RNA decoding site or A site and are of clinical importance in treating bacterial infections. For these nanopore experiments, a truncated RNA construct containing the A site was used as the RNA target. Previous experiments confirm that this truncated RNA construct mimics the function of the intact A site and it interacts with ligands and antibiotics in a manner that correlates with normal subunit function.32–34 Neomycin, paromomycin, and kanamycin are three such antibiotic agents that have been shown to bind to this truncated construct and are clinically effective in disrupting protein synthesis. To first characterize the sensitivity of the nanopore system to drug-binding events, bare RNA was first translocated through a 3-nm diameter SiN pore (N = 8552); this process was then repeated in the presence of 70 μM paromomycin (N = 5286) and 130 μM paromomycin (N = 6519) (Fig. 4A). Histograms were then constructed for the mean value of the current blockade from each event. A persistent low blockade peak can be observed at a value of ~0.6 nA for all samples, which the authors attributed to failed attempts by the molecules to translocate the pore, or “collisions.” Each sample also displayed a population near ~0.9 nA, representing bare RNA translocations. While the control sample containing no paromomycin fits very well to a double-Gaussian distribution, a third peak seems to develop with increasing levels of paromomycin near ~1.15 nA, which is most likely due to binding of paromomycin to the A-site molecule. As the complexed molecule passes through the pore, it blocks more of the ionic current than that of the A-site molecule alone, mainly due to an increase in the excluded volume of ions.
Figure 4.
Detection and quantification of RNA/ligand interactions. (A) Current blockade (ΔI) histograms at various ratios of paromomycin to RNA as indicated. Bottom histogram is fit to a double Gaussian and the top two are fit to triple Gaussians. A third peak emerges at large ΔI attributed to the RNA/paromomycin complex. (B) Normalized current blockade (ΔInorm) is plotted versus paromomycin concentration. Right axis shows FL values (see main text) and data is fit to Eq. (1). The dashed line and star indicate the saturated values at 0.5 M KCl. (C) Diagram of the RNA/NCp7 complex used by the Movileanu group.35 (D) Histograms of the interevent times for RNA only [dotted (purple in the web version) curve] and with added NCp7 [dashed (red in the web version) curve]. (E) Event frequencies normalized to RNA; only event rate are plotted against the ratios of NCp7/RNA constructs. (F) A cartoon diagram of the hepatitis C virus (HCV) conformational analysis system used by the Wanunu Lab.36 (G) Histograms showing the log of the dwell time (td) for various drug concentrations as indicated. (H) Integrated normalized dwell-time distributions are plotted for various drug concentrations. Images obtained with permissions from Wanunu et al.,31 Niedzwiecki et al.,35 and Shasha et al.36
To quantify the binding affinity of paromomycin to the A site and construct a binding isotherm, it is necessary to know the fraction of drug-bound RNA molecules. Assuming a simple 1:1 binding model (ie, Drug + RNA ⇌ Drug:RNA) and equal capture rates for the RNA and RNA/drug complex, a simple relationship for the dissociation rates can be inferred. For such a system, the dissociation constant is Kd = [Drug][RNA]/[Drug:RNA]. With the dissociation constant we can calculate the fraction of RNA molecules that form a complex with the drug as a function of drug concentration (FC).
| (1) |
As can be seen in Fig. 4A, the bare RNA and RNA/drug complex have largely overlapping current blockade distributions; as a result it is not possible to simply count the fraction of bound molecules. Although, the authors find that they can effectively quantify the fraction of bound molecules by monitoring the mean of the current blockade distribution and plotting its deviation from the mean current blockade of the bare RNA molecule (ΔInorm) as a function of drug concentration. To estimate the fraction of molecules bound, first ΔInorm is measured with drug concentration in excess to RNA concentration to obtain the current blockade for a completely saturated system (ΔImax). The fraction of molecules bound is then found by normalizing ΔInorm to this saturated value (FL = ΔInorm/ΔImax). By fitting the ΔInorm versus drug concentration data to Eq. (1), as displayed in Fig. 4B, it is possible to extract Kd for paromomycin. Using this analytical method, the authors estimated a Kd value of 90 ± 34 μM in the presence of 1 M KCl and 44 ± 17 μM in the presence of 0.5 M KCl. The increase in affinity of aminoglycosides (such as paromomycin) to the A-site molecule in the presence of reduced salt concentrations is a well-documented effect resulting from the decreased screening of electrostatic interactions.34 This same method was also used to extract Kd values for two additional A-site targeting antibiotic molecules, neomycin and kanamycin. A Kd value of 14.4 ± 3.3 μM is reported for neomycin at 1 M KCl, while the observed Kd value of kanamycin was too low to be observed at this salt concentration. These measurements are consistent with previous observations made using an independent FRET-based assay, showing that at 0.5 M NaCl, neomycin has a much higher affinity to the A-site construct than paromomycin, which has a much higher affinity than kanamycin.37
In the system presented here, a statistically significant amount of data (~500 events) is collected in under 1 min using ~1 pmol of RNA, demonstrating that nanopores may be used as an effective tool for rapid drug screening assays.
3.2.2 A Nanopore Assay for the Detection of an RNA-Binding HIV Biomarker
The Movileanu group has shown that nanopores can be used to monitor RNA–ligand interactions even when the size of the ligand does not permit transport of the RNA/ligand complex through the nanopore.35 In this work, SiN nanopores are used to detect binding of the human immunodeficiency virus 1 (HIV-1) nucleocapsid protein p7 (NCp7)38 to RNA aptamers. NCp7 is a protein that is involved in the packaging of the HIV-1 viral genome and in functions critical to viral replication. This protein contains two zinc finger domains that recognize the packaging domains of genomic RNA by preferentially binding to exposed guanosines in RNA stem loops (Fig. 4C). To test the nanopore’s ability to detect the binding of NCp7, translocation experiments were conducted with three 20-nucleotide RNA aptamers with various degrees of binding affinity. These aptamers were derived from a region of the retroviral ψ-packaging element of the HIV-1 genome known as stem-loop 3 (SL3). The aptamers derived from the SL3 region are known to bind the NCp7 protein with high affinity. The aptamers designed for this experiment form stem-loop structures, but each varies in the number of guanosine bases in the stem loop, thus varying its binding affinity to NCp7.
Two methods for calculating binding affinities are described by the authors: one using nanopores that are small enough to deny the passage of the RNA/protein complex, but large enough to translocate the bare RNA aptamer (diameter < 6 nm) and another method using large nanopores that are able to translocate the complex (diameter of 7–15 nm). First using small nanopores, translocation data was collected for the standard SL3 aptamer (GAG) at a concentration of 500 nM and then with the NCp7 protein added to the system at a concentration of 500 nM. Since the protein/RNA complex is too large to translocate the pore, a decrease in the translocation event rate was observed upon addition of NCp7 since only the fraction of molecules not bound by NCp7 produce translocation events. Creating a histogram of the interevent time and fitting the data to single exponential distributions (Fig. 4D) show that the interevent time increases from 149 ± 12 to 1030 ± 60 ms upon addition of NCp7, which confirms the binding of NCp7 to the GAG aptamer. This technique is then used to extract the binding affinities of NCp7 to each of the RNA aptamers by constructing titration curves of the interevent rates for each aptamer, which are presented in Fig. 4E. Though the binding affinities calculated using nanopores do not agree quantitatively with those found using FRET techniques,39 they do agree qualitatively with FRET results, as NCp7 was observed to bind more tightly to aptamers with greater guanosine concentrations in their stem-loop regions (ie, GAG > CUG > AUA).
The authors also provide evidence that this system may be used to test HIV-1 drug candidates that target the NCp7 protein. By introducing an excess of N-ethylmaleimide (a compound that is known to inhibit the binding of NCp7) to the system, they are able to restore the event frequency to a level that is comparable to the rate observed before the addition of NCp7.
These results show that using nanopores small enough to prohibit the translocation of the target complex, one can extract binding affinity information solely by measuring the interevent times. In addition, the authors utilized nanopores that are large enough to translocate the RNA/protein complex (diameter ~7 – 15 nm) and observed that the addition of NCp7 results in an increase in events that block significantly more current than that observed for the RNA molecule alone (data not shown here). This result is very similar to that described earlier by the Drndić Lab,31 except it does not find equal capture rates for the RNA and RNA/protein complex. As a result, a more complex equation is derived to fit the titration curve and the authors extracted a Kd value for NCp7 affinity to the GAG aptamer using the same experimental protocol as described by the Drndić Lab.
3.2.3 Screening for Conformational Changes Induced by RNA Drug Targets
The techniques presented by the Drndić and Movileanu labs represent two effective methods for detecting the binding affinities of ligands that add a considerable size to their target molecule. However, the Wanunu Lab has recently shown that it is still possible to detect RNA/ligand interactions, even when the binding molecule does not add considerable size, provided that there is some conformational change induced in the target molecule by the binding of the ligand.36 The internal ribosome entry site (IRES) motif of the hepatitis C virus (HCV) was employed to demonstrate the sensitivity of nanopores to conformational changes in a target biomolecule. The HCV IRES domain is a highly conserved motif with a structure that promotes high affinity interactions with the 40S subunit of its host ribosome.40 These interactions allow the HCV to hijack the host translational machinery and use it to translate the viral genome instead. A class of benzimidazole-based HCV inhibitors have been shown to disrupt HCV RNA translation.41 These inhibitors bind to the HCV IRES subdomain IIa and induce a conformational change that results in widening of the RNA interhelical angle.42 This conformational change promotes undocking of the IIa subdomain from the ribosome, thus preventing HCV translation. FRET has been an effective tool for the screening of potential inhibitors,43 as it can be used to report on conformational changes in the nanometer range, but it requires tedious and often expensive labeling steps.
The Wanunu Lab has shown that nanopores can be used to detect the conformational changes that are induced by these benzimidazole derivatives in a rapid and label-free method, eliminating some of the drawbacks of FRET-based methods. As seen in Fig. 4F, translocation of the HCV subdomain IIa RNA construct is sterically hindered when using a ~3 nm diameter nanopore, while passage of the construct after it adopts its drug-induced linear conformation is largely unhindered. To sense this conformational shift, nanopore experiments were conducted with the RNA construct in its natively bent conformation, as well in the presence of two benzimidazole derived drugs that bind with different affinities (1: Kd = 3.4 μM, 2: Kd = 64 μM, measured by an independent FRET assay42,44). Dwell-time histograms in the presence of different drug concentrations are shown in Fig. 4G. The peak in the dwell-time distribution for the RNA molecule alone appears to be around 100 μs. Upon addition of drug 2 (10:1 ratio of drug: RNA), there is a slight shift in the distribution, with a second population that develops near 10 μs. Addition of drug 1 (also in a 10:1 ratio) almost completely eliminates the longer timescale population and when the concentration is increased to 100:1 for drug 1 all events display timescales below 100 μs. To visualize this in a more quantitative fashion, the integrated normalized dwell-time distributions are plotted in Fig. 4H. The relative inflection of these curves match with what would be theoretically expected based on the binding affinity of the two drug molecules. Since drug 1 has a much lower dissociation constant than drug 2, it binds to the RNA construct more tightly, resulting in more RNA constructs in the linearized conformation that translocate the nanopore faster. Though this study does not contain enough data points to extract accurate measurements of the dissociation constants, it does show that solid-state nanopores of a proper geometry can be used for rapid, label-free drug-screening assays to detect conformational shifts in target molecules.
3.3 Summary
The collection of papers discussed here demonstrate the ability of appropriately sized nanopores to sense and quantify RNA/ligand interactions. This platform offers a variety of sensing modalities, as each parameter (ie, current blockade, interevent time, or dwell time) has been shown to shift in response to different ligand-induced changes in the target molecule (ie, internal structure occupancy,31 increased cross-sectional area,35 or conformational shifts36). In each of the cases presented the nanopore provides rapid sensing of RNA/ligand interactions without the need for application-specific labeling techniques, making it a valuable tool for drug-screening assays as well as studies of molecule interactions.
4. RNA STRUCTURAL ANALYSIS
Though Shasha et al. were the first to demonstrate the sensitivity of nanopores to ligand-induced conformational changes, there are several other examples of nanopore-based RNA structural analysis. Borrowing from studies of nanopore DNA unzipping,45,46 theoretical studies have shown that nanopores may be able to infer details about the sequence of an RNA molecule by sequentially unzipping and “ironing out” the secondary structure.47,48 In addition, it has already been shown that solid-state nanopores are able to discriminate between ssDNA and dsDNA,21 as well as dsDNA and dsRNA20 based on their differing helical diameters. Here we will highlight a few of these important nanopore-based RNA structural analysis papers.
4.1 Observation of Helix-Coil Conformational Fluctuations
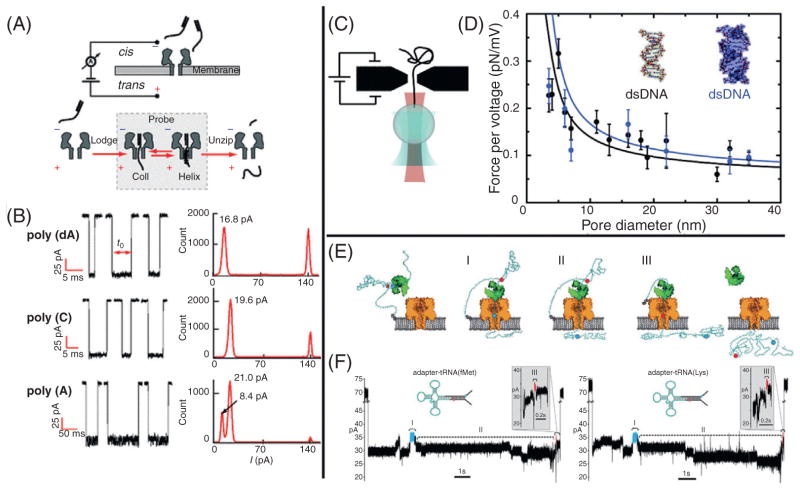
The first nanopore unzipping study to focus on RNA structures was conducted by Lin et al.,49 in which they aimed to observe the helix-coil transition of a polyA RNA molecule on the single-molecule level. Temperature-jump experiments have been able to measure the submicro-second conformational fluctuations between helix and coil conformations of polyA molecules at low-ionic strengths (50 mM salt).50 To observe this conformational shift and accurately measure its timescale using an αHL nanopore, it is beneficial to increase the time that the molecule resides in the pore (polyA has been observed to translocate αHL at a rate of ~3 μs/nt with an applied bias of 120 mV at room temperature).9 To increase the dwell time, the authors employed the DNA unzipping strategy as used in earlier nanopore studies.45,46 By designing duplex nucleic acid molecules with single stranded overhangs (similar to those later used by the Gu Lab for miRNA detection22,23), the time needed for a molecule to translocate is significantly prolonged. The researchers designed three partial duplex molecules, each of which contains an identical 10 bp duplex region, but differs in its overhang region: (1) dA50, 50 nt polydA overhang; (2) C25, 25 nt polyC overhang; and (3) A25, 25 nt polyA overhang. These constructs extend the dwell time of each molecule because the duplex region is too large to thread into the β-barrel and must completely unzip before the single-stranded segment can fully translocate the pore (Fig. 5A).
Figure 5.
RNA structural analysis using nanopores. (A) A schematic of the unzipping technique used to observe the helix-coil transition.49 (B) Characteristic sample events are shown for each of the molecules studied, with each accompanied by an all-point histogram of the current data. (C) A cartoon of the hybrid optical tweezers/solid-state nanopore setup used by the Cees Dekker Lab.51 (D) Force per voltage as a function of nanopore diameter is plotted for both dsDNA (black) and dsRNA [gray (blue in the web version)]. (E) A schematic for the tRNA unfolding and transport process used by Smith et al.52 (F) Characteristic ionic current traces observed during the capture, unfolding, and translocation of tRNAfMet and tRNALys. Images obtained with permission from Van Den Hout et al.51
Employing this unzipping strategy, the authors observed dwell times of ~5 ms for the dA50 and C25 constructs. Interestingly, the A25 construct displays dwell times that are nearly an order of magnitude longer than dA50 and C25. In addition to these longer timescales, large fluctuations can be observed in the ionic current when the A25 molecule occupies the nanopore cavity. To visualize these fluctuations, all-point current histograms are plotted from several representative translocation events of each molecule (Fig. 5B). As previously described, the authors observe that each of the polynucleotides exhibit a unique current blockade level, which can be seen in the all-point histograms (dA50, C25, and A25 display large peaks at 16.8, 19.6, and 21 pA, respectively). In addition to these characteristic peaks, a second peak can be seen in the all-point histogram for A25. In the publication, it is theorized that the ionic current fluctuations observed must be the result of the translocating molecule oscillating between helix and coil conformational states, with the lower peak observed at 8.4 pA being the result of the more dense helix structure. This hypothesis also explains the increase in dwell time for the A25 construct, since the helix form may slow down the translocation process due to increased interaction with the pore walls, as well as steric considerations.
To provide additional evidence that the ionic current fluctuations observed were in fact due to the helix-coil transition, the authors next constructed two additional RNA molecules: (1) C5A35 and (2) C15A25, where both possess the same 10 bp duplex region as the previously discussed molecules. Lin et al. hypothesized that the addition of a spacer larger than 12 nt should eliminate the two-state fluctuations in the ionic current, as the β-barrel region of the αHL nanopore is only ~5 nm long. This implies that the observed fluctuations should decrease as the length of the spacer increases as more of the helix-forming region is located outside the barrel region during the unzipping process. Indeed this theory was confirmed with all-point histograms showing slightly less two-state behavior for C5A35 when compared to A25, and C15A25 being virtually indistinguishable from C25 (data not shown).
Using a simple threshold analysis, the lifetimes of each of the two states was extracted from >2000 transitions. Fitting the lifetime data to exponential distributions, the mean lifetimes were found to be: τhelix = 1.0 ms and τcoil = 3.1 ms. Interestingly, these timescales are nearly three orders of magnitude slower than those observed by laser temperature-jump experiments.50 The authors theorize that this increase in lifetime is due to stabilizing effects that arise from the confinement of the molecule in the nanopore. Finally, by conducting a temperature-dependent study, enthalpic barriers and free energy changes are calculated for the helix-coil transition as well as pore/RNA interaction energies for the two states. Measurements such as these may be extremely beneficial in understanding the effects of confinement of nucleic acids and their conformational states.
4.2 A Hybrid Optical-Tweezer/Nanopore System for Studying Confinement Effects on RNA
Optical tweezers have been a valuable tool in the study of secondary and tertiary structures formed by nucleic acids.53 By optically trapping one end of a nucleic acid structure and attaching the other to some manipulator (micropipette, glass cover slip, etc.), one can stretch the captured molecule and monitor its response to the applied force (Fig. 5C). Alternatively, it is possible to optically trap both ends of the nucleic acid structure and measure changes in the positions of the optically trapped ends in response to some outside force (eg, proteins, ligands, etc.). These systems have been used to study RNA secondary and tertiary structures through force-induced melting,54 as well as real-time monitoring of RNA folding events.55 Optical tweezers have been combined with a number of different tools to provide an additional method for manipulating the molecule in question or to establish multiple, concurrent measurement systems. For a more in-depth account of RNA force measurements using optical tweezers, see the chapter: Applying Mechanical Force to Single RNA Molecules Reveals Structure and Function.
In 2010, the Cees Dekker Lab was able to combine the solid-state nanopore platform with optical tweezers to measure the effective force on a dsDNA molecule as it is electrophoretically driven through a SiN nano-pore.56 To combine these systems, DNA molecules were first functionalized with biotin at one end, then attached to a streptavidin-coated polystyrene bead, which is optically trapped by a tightly focused laser beam. When the trapped bead is brought close to a solid-state nanopore, the DNA is pulled into the pore by the emanating electric field. The transmembrane potential can then be varied while the optical tweezers platform is used to report the force on the DNA molecule. Collection of force versus voltage data allows the authors to extract the effective charge of the molecule while confined inside the nanopore. Subsequently, this method has been used to investigate the effects of DNA/protein interaction,57 localize individual proteins along a DNA strand,58 and investigate the electroosmotic force and hydrodynamic slip on the nanopore translocation process.59
Nynke Dekker Lab has used this system for the study of individual RNA molecules.51 Using this dual optical tweezers/nanopore system they compared the forces experienced by dsRNA and dsDNA molecules as they translocate a SiN nanopore as a function of the pore diameter (Fig. 5D). Their results indicate that the effective charge of the confined dsRNA is on the order of, but slightly less than the effective charge of dsDNA during the translocation process. Importantly, the authors note that their system is able to make accurate force measurements using nanopores as small as ~3 nm in diameter. This system is of potential interest as it may be adapted to monitor the folding of individual RNA molecules and report the effects of extreme confinement on RNA folding. In addition, this system could be used to detect the presence of individual RNA-binding proteins, monitor their position along a strand and observe conformation changes they induce on RNA molecules.
4.3 Discriminating Individual tRNA Molecules Through Sequential Unfolding
tRNA molecules are good examples of the complex structures that RNA molecules can form. The tRNAs that encode for each amino acid differ slightly from one another, and isodecoder as well as isoacceptor tRNAs add an additional layer of complexity. Their nucleotide sequences and the post-transcriptional modifications that they undergo lead to slight differences in their folded structures. Gerland et al. proposed that by threading structured polynucleotides through an αHL nanopore, it may be possible to sequentially break the intramolecular bonds that they form and “iron out” their secondary structures.46 They stated that with adequate control over the speed of this process, it should be possible to distinguish the identity of the translocating molecules.
Smith et al. have presented a system that is able to discriminate between two Escherichiacoli tRNAs51 by drawing upon the ideas put forth by Gerland et al. The system developed by Smith et al. aims to sequentially unfold and translocate individual tRNA molecules at speeds significantly slower than those observed by simple duplex unzipping experiments. To achieve this, the authors ligate the ends of a tRNA to a synthetic DNA adapter molecule. The 3′ end of the DNA adapter possesses a terminal TEG-linked (triethylene glycol) cholesterol, while the 5′ end possesses an ssDNA leader. The cholesterol on the 3′ end serves to concentrate the molecules at the lipid bilayer, and hence increasing the capture rate of tRNA molecules. This leaves the 5′ end free to be threaded into the nanopore, assisted by the free ssDNA leader. To slow the rate of translocation, the sample is incubated with φ29 DNA polymerase (DNAP), which has been previously used to processively ratchet DNA through a nanopore and read the sequence as each base moves through the pore.12,13 While this method cannot be used to ratchet the DNA/tRNA hybrid, as the DNAP cannot process the RNA, the authors are able to use an inactive φ29 DNAP as a “molecular brake” to slow the transport. The polymerase preferentially binds near the free 5′ ssDNA end of the DNA/tRNA hybrid, but does not process along the strand, as there are no free DNTPs in the solution. A schematic of what the authors hypothesize the transport process to look like is shown in Fig. 5E. When a voltage bias is applied, any φ29 DNAP adapted DNA/tRNA complexes located near the pore will be electrophoretically captured by the nanopore from their free 5′ ends. From here, the authors partition the translocation process into three steps: (I) translocation of the 5′ end of the adapter beginning with the ssDNA leader, (II) translocation of the tRNA portion, and (III) translocation of the 3′ end of the adapter. The sequence of the DNA adapter contains two abasic residues to assist in identification of these three steps (Fig. 5F). When an abasic residue passes through the pore it results in a significantly larger ionic current, serving as a marker for the strand position, and allowing the authors to identify the part of the signal corresponding to tRNA translocation (II). Characteristic current traces for tRNAfMet and tRNALys are shown in Fig. 5F.
Approximately 80 events were collected for each of the tRNAs and a custom support vector machine (SVM) algorithm was employed to use the ionic current parameters contained in each of the three regions in order to distinguish between the two tRNAs. First, each translocation event is analyzed and segregated into the three steps previously mentioned. The mean current and dwell time of each step is then extracted for each translocation event. The SVM plots the extracted mean current and dwell-time values and attempts to draw a line that best separates the points that correspond to the two different tRNAs. In this way, an unknown tRNA can be classified by measuring its dwell time and mean current values and determining on which side of the line this data point resides. The authors find that by analyzing the two adapter regions (I and III), the identity of a translocating tRNA molecule can be classified with accuracies of 60.0 ± 6.9 and 59.4 ± 7.0%, for the two regions respectively. By analyzing only the tRNA region (II) they are able to distinguish the identity of the translocating molecule with an accuracy of 87.2 ± 5.3%.
The inaccuracy of the SVM when applied to the adapter region and the impressive accuracy when applied to the tRNA region, serve as evidence that this method is able to distinguish between two different tRNAs based upon their structural properties. This method employs a sequential bond breaking and unfolding approach to distinguish between molecules, but cannot be used to outright sequence tRNA molecules. In its current incarnation, this technique may have problems scaling up to discriminate more than two tRNAs at a time, but the authors propose that by using an active enzymatic motor that is able to process RNA in single nucleotide steps, they may be able to adapt this method for direct sequencing of tRNA molecules. A nanopore-based system for direct tRNA sequencing would be an important tool for the study of tRNA biology, the effects of posttranslational modifications, and the links between tRNAs and human diseases, adding exciting potential applications for nanopores in RNA studies.
5. CONCLUSIONS
In conclusion, over the past 20 years there have been many studies that evaluate the applicability of nanopores for RNA detection and analysis. Due to their differing chemical structure from their parent DNA, RNA molecules adopt more complex tertiary structures that often resemble protein folding in terms of both structure and function. Both protein-based and solid-state nanopores have been used to study various aspects of RNA molecules, such as their globular (tertiary) structure, sequence, structural dynamics, and their interactions with small molecules and proteins. The further development of nanopore based measurement systems could provide many useful instruments for POC applications and RNA structural studies. Based on the success of single-molecule DNA sequencing using nanopores in recent years, it is logical that a major milestone in nanopore science will be achieved when long-read RNA sequencing is demonstrated. While RNA molecules are typically sequenced using bulk methods by conversion to complementary DNA (cDNA) molecules followed by DNA sequencing, bias errors in library preparation for RNA sequencing and the method’s lack of sensitivity toward important RNA modifications are both grand challenges that can be addressed by direct single-molecule RNA sequencing. In the coming years, it will be interesting to see what nanopore-based methods can do in order to facilitate direct RNA sequencing.
References
- 1.Wanunu M. Nanopores: a journey towards DNA sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venta K, Shemer G, Puster M, et al. Differentiation of short, single-stranded DNA homopolymers in solid-state nanopores. ACS Nano. 2013;7(5):4629–4636. doi: 10.1021/nn4014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carson S, Wanunu M. Challenges in DNA motion control and sequence readout using nanopore devices. Nanotechnology. 2015;26(7):74004. doi: 10.1088/0957-4484/26/7/074004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manrao EA, Derrington IM, Laszlo AH, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30(4):349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2(4):209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 9.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler TZ, Gundlach JH, Troll MA. Determination of RNA orientation during translocation through a biological nanopore. Biophys J. 2006;90(1):190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACSNano. 2009;3(9):2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 12.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. ProcNatl Acad Sci USA. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoSOne. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayub M, Bayley H. Individual RNA base recognition in immobilized oligonucleotides using a protein nanopore. Nano Lett. 2012;12(11):5637–5643. doi: 10.1021/nl3027873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayub M, Hardwick SW, Luisi BF, Bayley H. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013;13(12):6144–6150. doi: 10.1021/nl403469r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 17.Reiner JE, Balijepalli A, Robertson JWF, Drown BS, Burden DL, Kasianowicz JJ. The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine. J Chem Phys. 2012;137:214903. doi: 10.1063/1.4766363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brady KT, Reiner JE. Improving the prospects of cleavage-based nanopore sequencing engines. JChem Phys. 2015;143:074904. doi: 10.1063/1.4928647. [DOI] [PubMed] [Google Scholar]
- 19.Hunt EA, Broyles D, Head T, Deo SK. MicroRNA detection: current technology and research strategies. Annu RevAnal Chem. 2015;8(1):217–237. doi: 10.1146/annurev-anchem-071114-040343. [DOI] [PubMed] [Google Scholar]
- 20.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5(11):807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 21.Skinner GM, Van Den Hout M, Broekmans O, Dekker C, Dekker NH. Distinguishing single- and double-stranded nucleic acid molecules using solid-state nanopores. Nano Lett. 2009;9(8):2953–2960. doi: 10.1021/nl901370w. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zheng D, Tan Q, Wang MX, Gu L-Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6(10):668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Wang Y, Fricke BL, Gu L-Q. Programming nanopore ion flow for encoded multiplex microRNA detection. ACS Nano. 2014;8(4):3444–3450. doi: 10.1021/nn406339n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Tao C, Chien M, et al. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Sci Rep. 2012;2:684. doi: 10.1038/srep00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues CG, Machado DC, Chevtchenko SF, Krasilnikov OV. Mechanism of KCl enhancement in detection of nonionic polymers by nanopore sensors. BiophysJ. 2008;95(11):5186–5192. doi: 10.1529/biophysj.108.140814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Romero-Ruiz M, Castell OK, Bayley H, Wallace MI. High-throughput optical sensing of nucleic acids in a nanopore array. Nat Nanotechnol. 2015;10:986–991. doi: 10.1038/nnano.2015.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramnani P, Gao Y, Ozsoz M, Mulchandani A. Electronic detection of microRNA at attomolar level with high specificity. AnalChem. 2013;85(17):8061–8064. doi: 10.1021/ac4018346. [DOI] [PubMed] [Google Scholar]
- 28.Bayley H, Cronin B, Heron A, et al. Droplet interface bilayers. Mol Biosyst. 2008;4(12):1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer A, Holden MA, Pentelute BL, Collier RJ. Ultrasensitive detection of protein translocated through toxin pores in droplet-interface bilayers. Proc Natl Acad Sci USA. 2011;108(40):16577–16581. doi: 10.1073/pnas.1113074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced translocation of single DNA molecules through alpha-hemolysin nanopores by manipulation of internal charge. ProcNatl Acad Sci USA. 2008;105(50):19720–19725. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wanunu M, Bhattacharya S, Xie Y, Tor Y, Aksimentiev A, Drndic M. Nanopore analysis of individual RNA/antibiotic complexes. ACS Nano. 2011;5(12):9345–9353. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recht MI, Fourmy D, Blanchard SC, Dahlquist KD, Puglisi JD. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J Mol Biol. 1996;262(4):421–436. doi: 10.1006/jmbi.1996.0526. [DOI] [PubMed] [Google Scholar]
- 33.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichiacoli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274(5291):1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 34.Pilch DS, Kaul M, Barbieri CM. Ribosomal RNA recognition by aminoglycoside antibiotics. Anion Sens. 2005;70(1):179–204. doi: 10.1007/b100447. [DOI] [Google Scholar]
- 35.Niedzwiecki DJ, Iyer R, Borer PN, Movileanu L. Sampling a biomarker of the human immunodeficiency virus across a synthetic nanopore. ACSNano. 2013;7(4):3341–3350. doi: 10.1021/nn400125c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shasha C, Henley RY, Stoloff DH, Rynearson KD, Hermann T, Wanunu M. Nanopore-based conformational analysis of a viral RNA drug target. ACSNano. 2014;8(6):6425–6430. doi: 10.1021/nn501969r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie Y, Dix AV, Tor Y. FRET enabled real time detection of RNA-small molecule binding. JAm Chem Soc. 2009;131(48):17605–17614. doi: 10.1021/ja905767g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBOJ. 1992;11(8):3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paoletti AC, Shubsda MF, Hudson BS, Borer PN. Affinities of the nucleocapsid protein for variants of SL3 RNA in HIV-1. Biochemistry. 2002;41(51):15423–15428. doi: 10.1021/bi026307n. [DOI] [PubMed] [Google Scholar]
- 40.Spahn CM, Kieft JS, Grassucci RA, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291(5510):1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 41.Seth PP, Miyaji A, Jefferson EA, et al. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. JMed Chem. 2005;48(23):7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 42.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5(11):823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou S, Rynearson KD, Ding K, Brunn ND, Hermann T. Screening for inhibitors of the hepatitis C virus internal ribosome entry site RNA. Bioorg Med Chem. 2013;21(20):6139–6144. doi: 10.1016/j.bmc.2013.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rynearson KD, Charrette B, Gabriel C, et al. 2-Aminobenzoxazole ligands of the hepatitis C virus internal ribosome entry site. Bioorg Med Chem Lett. 2014;24(15):3521–3525. doi: 10.1016/j.bmcl.2014.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathé J, Visram H, Viasnoff V, Rabin Y, Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys J. 2004;87(5):3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys Rev Lett. 2003;90(23):238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 47.Gerland U, Bundschuh R, Hwa T. Translocation of structured polynucleotides through nanopores. Phys Biol. 2004;1(1–2):19–26. doi: 10.1088/1478-3967/1/1/002. [DOI] [PubMed] [Google Scholar]
- 48.McCauley M, Forties R, Gerland U, Bundschuh R. Anomalous scaling in nanopore translocation of structured heteropolymers. Phys Biol. 2009;6(3):036006. doi: 10.1088/1478-3975/6/3/036006. [DOI] [PubMed] [Google Scholar]
- 49.Lin J, Kolomeisky A, Meller A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys Rev Lett. 2010;104(15):158101. doi: 10.1103/PhysRevLett.104.158101. [DOI] [PubMed] [Google Scholar]
- 50.Dewey TG, Turner DH. Laser temperature-jump study of stacking in adenylic acid polymers. Biochemistry. 1979;18(26):5757–5762. doi: 10.1021/bi00593a002. [DOI] [PubMed] [Google Scholar]
- 51.Van Den Hout M, Vilfan ID, Hage S, Dekker NH. Direct force measurements on double-stranded RNA in solid-state nanopores. Nano Lett. 2010;10(2):701–707. doi: 10.1021/nl903925a. [DOI] [PubMed] [Google Scholar]
- 52.Smith AM, Abu-Shumays R, Akeson M, Bernick DL. Capture, unfolding, and detection of individual tRNA molecules using a nanopore device. Front Bioeng Biotechnol. 2015;3:91. doi: 10.3389/fbioe.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Recent advances in optical tweezers. Annu Rev Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 54.Wen J-D, Manosas M, Li PTX, et al. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys J. 2007;92(9):2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338(6105):397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keyser UF, Koeleman BN, van Dorp S, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2(7):473–477. [Google Scholar]
- 57.Hall AR, Van Dorp S, Lemay SG, Dekker C. Electrophoretic force on a protein-coated DNA molecule in a solid-state nanopore. Nano Lett. 2009;9(12):4441–4445. doi: 10.1021/nl9027318. [DOI] [PubMed] [Google Scholar]
- 58.Sischka A, Spiering A, Khaksar M, et al. Dynamic translocation of ligand-complexed DNA through solid-state nanopores with optical tweezers. JPhysCondensMatter. 2010;22(45):454121. doi: 10.1088/0953-8984/22/45/454121. [DOI] [PubMed] [Google Scholar]
- 59.Galla L, Meyer AJ, Spiering A, et al. Hydrodynamic slip on DNA observed by optical tweezers-controlled translocation experiments with solid-state and lipid-coated nano-pores. Nano Lett. 2014;14(7):4176–4182. doi: 10.1021/nl501909t. [DOI] [PubMed] [Google Scholar]