Abstract
We studied the antimicrobial susceptibility of 322 Streptococcus pyogenes throat isolates from French children and their serotype and genomic diversity. A total of 22.4% were erythromycin resistant, and 69.4, 4.2, and 26.4% of these isolates harbored ermB, ermA, and mefA, respectively. Increasing resistance in France is mainly associated with a few emm type 28 clones.
Streptococcus pyogenes pharyngitis is one of the most common bacterial upper respiratory tract infections in children. Group A streptococci (GAS) are uniformly susceptible to penicillin. Macrolides are recommended alternatives for penicillin-hypersensitive patients. However, resistance to erythromycin has become widespread among GAS (4, 6, 10, 18, 19; L. Mihaila-Amrouche, J. Loubinoux, and A. Bouvet, Abstr. 13th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. S6, 2003.). Here, we studied the prevalence of macrolide resistance among recent throat isolates of GAS collected from children throughout France. We also determined their mechanisms of resistance and their clonality by means of emm genotyping and pulsed-field gel electrophoresis (PFGE).
A total of 322 consecutive S. pyogenes isolates were collected between 2002 and 2003 throughout France. They were isolated from throat cultures of children 2 to 16 years of age (mean, 6 years) with pharyngitis. The isolates were identified as S. pyogenes by colony morphology, Gram staining, beta-hemolysis on blood agar, and the presence of group A antigen as determined by an agglutination test (Oxoid, Basingstoke, United Kingdom).
Antimicrobial susceptibility was tested as recommended previously (15). Erythromycin-resistant strains were identified by the disk diffusion method on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood (Bio-Rad, Marnes la Coquette, France) using 15-IU erythromycin disks (Bio-Rad) (15). Bacitracin susceptibility was studied by the disk diffusion method with 10-IU disks (Bio-Rad).
The MICs of amoxicillin, penicillin G, erythromycin, azithromycin, clarithromycin, telithromycin, clindamycin, and streptogramin B were determined for all isolates with erythromycin inhibition zone diameters of less than 21 mm (14). Telithromycin interpretative criteria for susceptibility and resistance were ≤0.5 and ≥4 μg/ml, respectively (NCCLS approved breakpoints are not available) (7).
The mefA, ermB, and ermA genes were detected by PCR amplification as previously described (3).
Biotypes were determined as previously described (5).
T serotypes were determined on trypsinated bacteria by slide agglutination with anti-T sera obtained from the Institute of Sera and Vaccines, Prague, Czech Republic (9).
emm-specific PCR products were obtained with the primers MF2 and MR1 described by Podbielski et al. (17). The 5′ end of the emm genes was sequenced as described by Beall et al. (2) (see the Centers for Disease Control and Prevention website [http://www.cdc.gov/ncidod/biotech/strep/protocols.htm]). DNA sequences were subjected to homology searches (http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm).
PFGE with SmaI was applied to all 72 erythromycin-resistant isolates, as previously described (4). The DNA of 19 isolates, all carrying mefA, could not be restricted by SmaI despite repeated attempts. We thus used SfiI for these isolates. Cluster analysis (unweighted pair group method with arithmetic mean) with whole-band analyzer software (Biogene, Vilber-Lourmat, Marne la Vallée, France) was used to calculate similarity or dissimilarity among GAS isolates. Clonally related PFGE patterns were defined by a similarity coefficient higher than 80% (usually corresponding to a difference of no more than four bands in our study) (20).
All isolates were susceptible to penicillin G and amoxicillin. Of the 322 isolates, 72 (22.4%) were resistant to erythromycin by the disk diffusion method. The MICs of the different antimicrobial agents are given in Table 1. Thirty-five isolates (10.9%) were resistant to bacitracin (disk diffusion zone diameters of 0 mm), and all these isolates were resistant to erythromycin.
TABLE 1.
In vitro activities of eight antimicrobials against erythromycin-resistant S. pyogenes isolates according to known mechanisms of resistance
| Group (n) | Antimicrobial agent | MIC (μg/ml)a
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| All isolates (72) | Erythromycin | 1->128 | >128 | >128 |
| Azithromycin | 4->128 | >128 | >128 | |
| Clindamycin | 0.064->128 | >128 | >128 | |
| Streptogramin B | 0.5->128 | 8 | 128 | |
| Clarithromycin | 0.5->128 | >128 | >128 | |
| Telithromycin | 0.064-128 | 2 | 4 | |
| Penicillin G | ≤0.008 | ≤0.008 | ≤0.008 | |
| Amoxicillin | ≤0.008-0.016 | 0.016 | 0.016 | |
| ermB (50) | Erythromycin | 1->128 | >128 | >128 |
| Azithromycin | 4->128 | >128 | >128 | |
| Clindamycin | 128->128 | >128 | >128 | |
| Streptogramin B | 2->128 | 8 | 128 | |
| Clarithromycin | 0.5->128 | >128 | >128 | |
| Telithromycin | 0.064-128 | 4 | 8 | |
| Penicillin G | ≤0.008 | ≤0.008 | ≤0.008 | |
| Amoxicillin | ≤0.008-0.016 | 0.016 | 0.016 | |
| ermA (3) | Erythromycin | 1->128 | ND | ND |
| Azithromycin | 4->128 | ND | ND | |
| Clindamycin | 0.125->128 | ND | ND | |
| Streptogramin B | 1-32 | ND | ND | |
| Clarithromycin | 0.5->128 | ND | ND | |
| Telithromycin | 0.064-0.25 | ND | ND | |
| Penicillin G | ≤0.008 | ND | ND | |
| Amoxicillin | ≤0.008 | ND | ND | |
| mefA (19) | Erythromycin | 8-16 | 8 | 16 |
| Azithromycin | 8-16 | 16 | 16 | |
| Clindamycin | 0.064-0.125 | 0.064 | 0.125 | |
| Streptogramin B | 0.5-2 | 1 | 2 | |
| Clarithromycin | 2-8 | 4 | 8 | |
| Telithromycin | 0.25-0.5 | 0.5 | 0.5 | |
| Penicillin G | ≤0.008 | ≤0.008 | ≤0.008 | |
| Amoxicillin | ≤0.008-0.016 | ≤0.008 | 0.016 | |
50% and 90%, MICs at which 50 and 90% of isolates are inhibited, respectively. ND, not determined.
Among the 72 erythromycin-resistant isolates, 69.4, 4.2, and 26.4% harbored the ermB, ermA, and mefA genes, respectively. Table 1 shows the MICs for erythromycin-resistant isolates according to the underlying mechanism. Isolates carrying ermB were resistant to 14-, 15- and 16-membered-ring macrolides and showed cross-resistance to clindamycin and streptogramin B; 47% of ermB-positive isolates were resistant to telithromycin. All but one of the 35 bacitracin-erythromycin-resistant isolates carried the ermB gene. Isolates carrying mefA were resistant to erythromycin, clarithromycin, and azithromycin but susceptible to clindamycin and telithromycin. Isolates carrying ermA (n = 3) were susceptible to telithromycin.
Clindamycin and telithromycin remained active against 29 and 32%, respectively, of the 72 erythromycin-resistant isolates.
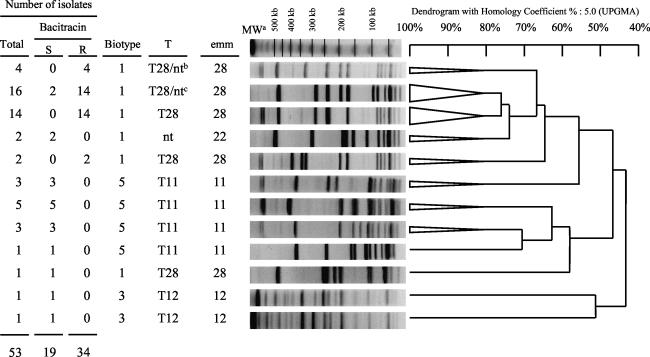
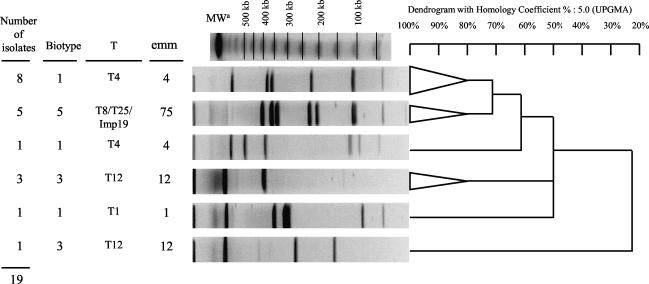
Molecular analysis of SmaI-digested DNA was possible for 53 of the 72 erythromycin-resistant isolates. The remaining 19 isolates could not be typed because SmaI did not restrict their DNA. These 53 GAS isolates were genetically diverse, as they yielded 35 PFGE patterns. However, 57% of isolates belonged to two clones (Fig. 1). The bacitracin-resistant isolates carrying ermB belonged to four clones, of which 82% belonged to the main two clones. The remaining 19 mef isolates yielded nine PFGE patterns after digestion with SfiI, but 68% of these isolates belonged to two clones (Fig. 2).
FIG. 1.
Representative SmaI PFGE patterns of the 53 erm-positive erythromycin-resistant S. pyogenes isolates according to biotype, T serotype, and emm type. The triangles are collapsed branches gathering isolates with 80% similarity of banding patterns. MWa, molecular weight marker (lambda ladder; Bio-Rad); T28/ntb, one isolate nontypeable for T antigen. T28/ntc, three isolates nontypeable for T antigen; UPGMA, unweighted pair group method with arithmetic mean.
FIG. 2.
Representative SfiI PFGE patterns of the 19 mefA erythromycin-resistant S. pyogenes isolates according to biotype, T serotype, and emm type. The triangles are collapsed branches gathering isolates with 80% similarity of banding patterns. MWa, molecular weight marker (lambda ladder; Bio-Rad); UPGMA, unweighted pair group method with arithmetic mean.
Various emm types were observed among the 72 erythromycin-resistant S. pyogenes isolates (Table 2). However, most of them were of the genotype emm28. There was only one emm type 1 resistant isolate. The emm type 28 isolates which contain either the ermB or the ermA gene exhibited two highly related PFGE patterns (Fig. 1).
TABLE 2.
T serotypes and emm types of the 72 erythromycin-resistant S. pyogenes isolates with various resistance mechanisms
| T serotype and emm typea | Total no. of isolates | No. of isolates with resistance gene
|
||
|---|---|---|---|---|
| ermB | ermA | mefA | ||
| emm22 (T nontypeable) | 2 | 2 | ||
| emm28 (T nontypeable) | 4 | 4 | ||
| T1 emm1 | 1 | 1 | ||
| T11 emm11 | 12 | 11 | 1 | |
| T12 emm12 | 6 | 2 | 4 | |
| T28 emm28 | 33 | 31 | 2 | |
| T4 emm4 | 9 | 9 | ||
| T8/T25/lmp19 emm75 | 5 | 5 | ||
The prevalence of erythromycin resistance among GAS isolates from French children was only 6.2% two years ago (3). Our study shows a threefold increase in the prevalence of erythromycin resistance among GAS in France compared to the results of a study carried out in 2000 (6.2% in 2000 and 22.4% in 2003; P < 0.001) (3).
The gene ermB was the main resistance mechanism in our isolates. In contrast, the M phenotype and/or the mefA gene predominates in other European countries such as Finland (11), Spain (16), and Italy (1) and also in the United States (13) and Canada (12). The increase in erythromycin resistance among GAS isolates has been linked to the spread of serotype T28M28 in Quebec (21) and T28 emm28 in Spain (16). In our study, 46% of the strains resistant to macrolides were associated with the type T28 emm28, with 94% carrying ermB. The major type encountered among our isolates carrying mefA was T4 emm4 (47%). This type was also related to the increase in erythromycin resistance found in Finland (11).
Interestingly, in our study, all isolates whose DNA was not restricted by SmaI carried the mefA gene, in keeping with a previous report (8). The diversity of the erythromycin-resistant isolates was shown by the fact that 35 different profiles were obtained with SmaI and 9 were obtained with SfiI, although a small number of PFGE patterns accounted for most of the isolates. Indeed, two highly related SmaI PFGE patterns included 57% of the erm isolates, and two different SfiI PFGE patterns included 68% of the mefA isolates. Thus, our results suggest that the emergence of macrolide resistance among GAS in France is due to the dissemination of a limited number of clones as described in Europe and North America (11-13; Mihaila-Amrouche et al., Abstr. 13th Eur. Congr. Clin. Microbiol. Infect. Dis.). The increase of prevalence of macrolide resistance in GAS in our study may be due to both the epidemic spread of resistant clones and the dissemination of resistance determinants among isolates of different genetic background.
Contrary to the situation in the United States, Spain, and Italy, where clindamycin remains active against M phenotype strains, the main mechanism of erythromycin resistance in our isolates involved the erm genes, leading to high-level resistance to both macrolides and clindamycin (16% of our isolates were resistant to clindamycin). Moreover, 15% of our isolates were intermediately resistant or resistant to telithromycin. Current data indicate that macrolide resistance is clinically relevant in GAS pharyngitis (13). Indeed, erythromycin-resistant GAS eradication failed in 47% of children in Finland (19) and 40% of children in Italy (1).
Macrolide GAS resistance must therefore be closely monitored in each country. Furthermore, the use of rapid group A antigen test must be followed by sensitivity testing when the patient is allergic to β-lactam agents.
Acknowledgments
Financial support was provided in part by Pfizer (France).
REFERENCES
- 1.Bassetti, M., G. Manno, A. Collida, A. Ferrando, G. Gatti, E. Guilty, M. Cruciani, and D. Bassetti. 2000. Erythromycin resistance in Streptococcus pyogenes in Italy. Emerg. Infect. Dis. 6:180-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen, E., R. Leclercq, F. Fitoussi, N. Brahimi, B. Malbruny, D. Deforche, and R. Cohen. 2002. Emergence of group A streptococcus strains with different mechanisms of macrolide resistance. Antimicrob. Agents Chemother. 46:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet, A., P. Geslin, P. Kriz-Kuzemenska, V. Blanc, C. Devine, and F. Grimont. 1994. Restricted association between biotypes and serotypes within group A streptococci. J. Clin. Microbiol. 32:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozdogan, B., P. C. Appelbaum, L. M. Kelly, D. B. Hoellman, A. Tambic-Andrasevic, L. Drukalska, W. Hryniewicz, H. Hupkova, M. R. Jacobs, J. Kolman, M. Konkoly-Thege, J. Miciuleviciene, M. Pana, L. Setchanova, J. Trupl, and P. Urbaskova. 2003. Activity of telithromycin compared with seven other agents against 1039 Streptococcus pyogenes pediatric isolates from ten centers in central and eastern Europe. Clin. Microbiol. Infect. 9:741-745. [DOI] [PubMed] [Google Scholar]
- 7.Canton, R., E. Loza, M. I. Morosini, and F. Baquero. 2002. Antimicrobial resistance amongst isolates of Streptococcus pyogenes and Staphylococcus aureus in the PROTEKT antimicrobial surveillance programme during 1999-2000. J. Antimicrob. Chemother. 50:9-24. [DOI] [PubMed] [Google Scholar]
- 8.De Azavedo, J. C., R. H. Yeung, D. J. Bast, C. L. Duncan, S. B. Borgia, and D. E. Low. 1999. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob. Agents Chemother. 43:2144-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, D. R., E. L. Kaplan, J. Sramek, R. Bicova, J. Havlicek, H. Havlickova, H. Motlova, and P. Kriz. 1996. Determination of T-protein agglutination patterns, p. 37-41. In D. R. Johnson (ed.), Laboratory diagnosis of group A streptococcal infections. World Health Organization, Geneva, Switzerland.
- 10.Kaplan, E. L. 1997. Recent evaluation of antimicrobial resistance in beta-hemolytic streptococci. Clin. Infect. Dis. 24:S89-S92. [DOI] [PubMed] [Google Scholar]
- 11.Kataja, J., P. Huovinen, A. Muotiala, J. Vuopio-Varkila, A. Efstratiou, G. Hallas, H. Seppala, et al. 1998. Clonal spread of group A streptococcus with the new type of erythromycin resistance. J. Infect. Dis. 177:786-789. [DOI] [PubMed] [Google Scholar]
- 12.Katz, K. C., A. J. McGeer, C. L. Duncan, A. Ashi-Sulaiman, B. M. Willey, A. Sarabia, J. McCann, S. Pong-Porter, Y. Rzayev, J. S. de Azavedo, and D. E. Low. 2003. Emergence of macrolide resistance in throat culture isolates of group A streptococci in Ontario, Canada, in 2001. Antimicrob. Agents Chemother. 47:2370-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 14.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. NCCLS, Wayne, Pa.
- 15.NCCLS. 2003. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A7. NCCLS, Wayne, Pa.
- 16.Perez-Trallero, E., C. Garcia, B. Orden, J. M. Marimon, and M. Montes. 2004. Dissemination of emm28 erythromycin-, clindamycin- and bacitracin-resistant Streptococcus pyogenes in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 8:8. [DOI] [PubMed] [Google Scholar]
- 17.Podbielski, A., B. Melzer, and R. Lutticken. 1991. Application of the polymerase chain reaction to study the M protein (-like) gene family in beta-hemolytic streptococci. Med. Microbiol. Immunol. (Berlin) 180:213-227. [DOI] [PubMed] [Google Scholar]
- 18.Schlegel, L., B. Merad, H. Rostane, V. Broc, and A. Bouvet. 2001. In vitro activity of midecamycin diacetate, a 16-membered macrolide, against Streptococcus pyogenes isolated in France, 1995-1999. Clin. Microbiol. Infect. 7:362-366. [DOI] [PubMed] [Google Scholar]
- 19.Seppala, H., A. Nissinen, H. Jarvinen, S. Huovinen, T. Henriksson, E. Herva, S. E. Holm, M. Jahkola, M. L. Katila, T. Klaukka, et al. 1992. Resistance to erythromycin in group A streptococci. N. Engl. J. Med. 326:292-297. [DOI] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss, K., J. De Azavedo, C. Restieri, L. A. Galarneau, M. Gourdeau, P. Harvey, J. F. Paradis, K. Salim, and D. E. Low. 2001. Phenotypic and genotypic characterization of macrolide-resistant group A Streptococcus strains in the province of Quebec, Canada. J. Antimicrob. Chemother. 47:345-348. [DOI] [PubMed] [Google Scholar]