Abstract
Resistance to antifungal agents is a recurring and growing problem among patients with systemic fungal infections. UV-induced Aspergillus nidulans mutants resistant to terbinafine have been identified, and we report here the characterization of one such gene. A sib-selected, 6.6-kb genomic DNA fragment encodes a salicylate 1-monooxygenase (salA), and a fatty acid synthase subunit (fasC) confers terbinafine resistance upon transformation of a sensitive strain. Subfragments carrying salA but not fasC confer terbinafine resistance. salA is present as a single-copy gene on chromosome VI and encodes a protein of 473 amino acids that is homologous to salicylate 1-monooxygenase, a well-characterized naphthalene-degrading enzyme in bacteria. salA transcript accumulation analysis showed terbinafine-dependent induction in the wild type and the UV-induced mutant Terb7, as well as overexpression in a strain containing the salA subgenomic DNA fragment, probably due to the multicopy effect caused by the transformation event. Additional naphthalene degradation enzyme-coding genes are present in fungal genomes, suggesting that resistance could follow degradation of the naphthalene ring contained in terbinafine.
The incidence of fungal infections has increased in the last 20 years, primarily because of the increase in the number of immunocompromised patients due to AIDS, malnutrition, the indiscriminate use of antibiotics, chemotherapy, and artificial immunosuppressive treatment in organ transplants (17). In addition, the widespread use of antifungal agents has led to clinical resistance. Thus, insights into molecular and genetic mechanisms involved in resistance are needed to define treatment strategies and to design new antifungal drugs.
Terbinafine is a generic antifungal agent used to treat superficial mycoses such as dermatophyte onychomycosis, dermatomycoses, tinea, and piedra (1, 6, 9, 20). Terbinafine interferes with ergosterol biosynthesis by inhibiting a membrane-bound squalene epoxidase (31). Inhibition of squalene epoxidase results in ergosterol deficiency with the accumulation of squalene, which may be responsible for the observed “in vitro” fungicidal activity (11, 26).
Besides the fact that fungi resistant to terbinafine have been consistently isolated, little is known about the molecular mechanisms associated with resistance (14, 22, 30, 38). In one case, a modified squalene epoxidase with reduced affinity for terbinafine conferred terbinafine resistance to Nectria haematococca mutants (15) and, in another, a Candida albicans strain was resistant to terbinafine due to activation of the multidrug efflux transporter CDR2 (35). Also, a C. albicans mutant carrying CDR1 deletion resulted in azoles and terbinafine hypersusceptibility (36).
We report here the involvement of naphthalene degradation as a possible mechanism of terbinafine resistance in Aspergillus nidulans. Transformation experiments demonstrate that terbinafine resistance is mediated through the overexpression of salA, a gene that encodes a salicylate 1-monooxygenase. In Pseudomonas putida, salicylate 1-monooxygenase catalyzes the formation of catechol from salicylate, a required intermediate of naphthalene degradation (2). Thus, terbinafine, which has a naphthalene nucleus in its chemical structure, may be a substrate for a salicylate-like part of an aromatic compound degradation pathway in A. nidulans.
MATERIALS AND METHODS
Strains, media, and culture conditions.
A. nidulans was cultivated at 37°C in complete medium (CM) or minimal medium (MM) (27). The following strains were used: the wild type, FGSCA26 (biA1 veA1), the terbinafine-resistant strains Terb7 (pabaA1 tebA7 veA1) (30) and Terb7A (acrA1 tebA7 veA1), the transformation recipient strains GR5 (pyroA4 pyrG89 wA3 veA1) and RPA26 (biA1 ΔargB::trpC801 trpC801 veA1). The mutant alleles were as follows: wA3, white conidia; tebA7 and acrA1, terbinafine and acriflavin resistance, respectively, and the auxotrophic markers; ΔargB::trpC801, biA1, pabaA1, pyrG89, and pyroA4 for arginine, biotin, p-aminobenzoic acid, uracil, and pyridoxine, respectively. Terbinafine [(E)-N-(6,6-dimethyl-2-hepten-4-yn-yl)-N-methyl-1-naphthalene methanamine] was from Sandoz AG, dissolved in dimethyl sulfoxide, and added to solid or liquid medium. An agar dilution assay was used for terbinafine susceptibility testing. A 100-μl suspension of the conidia of each strain (107 conidia per ml) was inoculated into solid-CM dishes with various concentrations of terbinafine, followed by incubation at 37°C for 3 days. The MIC corresponds to the lowest concentration of the drug at which there is no macroscopic growth. All assays were carried out in triplicate (8). Escherichia coli strain DH5α (12) or XL1-Blue (Stratagene) was used for the propagation of plasmid DNA, which was cultivated at 37°C in Luria-Bertani broth amended with 50 μg of ampicillin/ml.
Molecular nucleic acid manipulation techniques and sequencing.
DNA manipulation and cloning procedures were performed as described either by Sambrook et al. (33) or by the supplier of enzymes and nucleic acid reagents. Shotgun subcloning, random clone sampling (Fig. 1C), and nucleotide sequence were determined by the dideoxynucleotide chain termination method (34) using BigDye terminator cycle sequencing (Perkin-Elmer) in an ABI Prism 377 DNA sequencer. DNA sequence data files were assembled by using the Phred, Phrap, and Consed packages (7, 10).
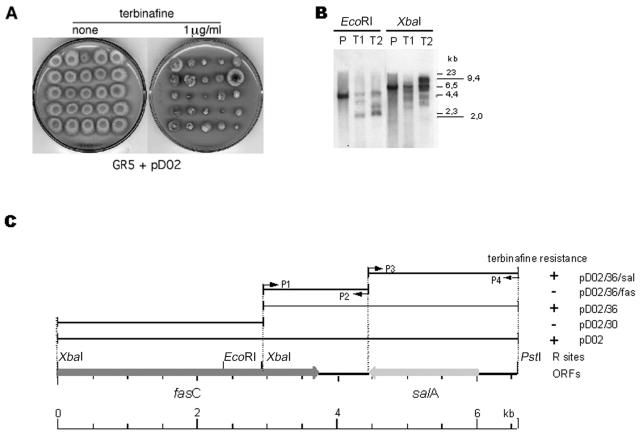
FIG. 1.
The salA gene confers terbinafine resistance in A. nidulans. (A) Identification of a subgenomic DNA fragment (pD02) that confers terbinafine resistance. Strain GR5 was transformed with plasmid pD02, which was shown in a sib selection experiment to induce terbinafine resistance. Colonies are independent transformants randomly selected after transformation with pD02 and grown on medium with or without 1 μg of terbinafine/ml. (B) Southern blot analysis of the genomic DNA of A. nidulans. Genomic DNA from GR5 (lanes P) and two pD02 transformant strains (lanes T1 and T2) were digested with EcoRI or XbaI, size fractionated through a 1.0% agarose gel, and blotted as described in Materials and Methods. The probe used was the salA fragment obtained from pD02/36/sal contained in pGEM-T Easy vector. Size markers (in kilobases) are indicated at right of the blot. (C) Salicylate 1-monooxygenase confers terbinafine resistance. Plasmid pD02 was subcloned, and the genomic DNA sequence of the region was determined. Two ORFs, fasC and salA, were identified. DNA-mediated transformation-dependent resistance analyses showed that only fragments that contain salA are terbinafine resistant. Primers P1 to P4 were used to construct the subclones pD02/36/sal and pD02/36/fas.
The original terbinafine-resistant plasmid was subcloned into smaller fragments by cleaving pD02 fragment with XbaI and PstI, resulting in 3.3-kb XbaI-PstI and 3.1-kb XbaI fragments. Further subcloning, isolating the fasC and salA open reading frames (ORFs), was pursued by PCR amplification (21) of the indicated regions on plasmid pD02/36 (shown in Fig. 1) with specific primers (see below). PCR product were cloned and propagated in E. coli with the pGEM-T Easy vector (Promega).
The following oligonucleotides were used: pUCM13 forward, 5′-GTAAAACGACGGCCAGT-3′; pUCM13 reverse, 5′-CAGGAAACAGCTATGAC-3′; P1 (fas1-5), 5′-GTGTTTTAGCATTCTCGCC-3′; P2 (Anfatty16), 5′-AGGATCACGACATTCACTTG-3′; P3 (ansalA4), 5′-CTCCAAGTCGCCCCCAATC-3′ and P4 (sal1-5), 5′-CACGGGACGGGAACACCATC-3′
Library construction.
Total DNA from the Terb7A was extracted by the method of Reader and Broda (29) from a 12-h-old liquid culture shaken at 37°C after the inoculation of 107 conidia per ml. Partially Sau3AI-digested DNA fragments were ligated to the pRG4 shuttle vector (19), which confers uracil auxotropy when integrated into the genome of A. nidulans strain GR5, and transformed into E. coli XL1-Blue by electroporation. A total of 17,856 clones with an average insert size of 6.5 kb were recovered, and 7,680 of them were transformed into strain GR5 (∼1.6-fold genome coverage).
Transformation and sib selection.
Transformation experiments were performed as previously described (40), except that the protoplasts were made with Glucanex (Novo Nordisk) rather than with Novozym. A sib selection screen with modifications (33) was used in order to rescue a recombinant plasmid bearing the gene that confers terbinafine resistance. The approach is based on the concept of dividing a large genomic library into a manageable number of pools, each consisting of 960 clones (10 96-well plates). These pools are then tested for the ability to confer terbinafine resistance in 1 μg of terbinafine/ml. After a pool is identified, it is subdivided into successively smaller and smaller pools, until a unique recombinant plasmid is isolated. Once an individual plasmid was identified, the cloned insert was sequenced and characterized.
Southern and Northern blot hybridizations.
Southern blot analysis was performed by standard techniques (33). For Northern blots, total RNA isolation of 1 to 2 g of lyophilized ground mycelium was obtained by using the TRIzol method (4) and isolated according to manufacturer recommendations (Gibco-BRL). Northern blot analysis was performed as described elsewhere (42). Briefly, 10 μg of total RNA was separated on 1.1% agarose containing 3% formaldehyde and blotted onto nylon membranes (Hybond N+; Amersham Pharmacia Biotech). [32P]CTP-labeled probes were used to hybridize the membrane with modified Church buffer (5, 42) at 65°C for 16 to 20 h. Blots were washed and exposed to film and developed according to the method of Sambrook et al. (33).
The GenBank accession numbers for salA and fasC are AF316427 and AY120937, respectively.
RESULTS
Isolation of a gene that alters A. nidulans susceptibility to terbinafine.
To better understand terbinafine resistance, we screened for recombinant plasmids able to turn sensitive strains resistant that were made from genomic DNA of strain TerbA7, whose resistance to terbinafine had been genetically defined previously (30). We isolated a recombinant plasmid, pD02, which was able to change GR5 strain from being sensitive to being resistant to terbinafine. To confirm that the complementing activity did not arise from spontaneous reversions, the plasmid was retransformed into GR5.
Figure 1A shows that transformation of plasmid pD02 into GR5 results in multiple-level terbinafine-resistant strains. Figure 1C shows the subcloning, a physical map of the 6.6-kb chromosomal region contained in pD02, and mapping of the terbinafine resistance to a single ORF. The genomic DNA sequence reveals two ORFs: fasC, encoding the beta subunit of the A. nidulans fatty acid synthase (3), and salA, a salicylate 1-monooxygenase-like protein known to be associated with naphthalene degradation in Pseudomonas stutzeri AN10 (2). The subclones pD02/36 and pD02/30 were transformed into A. nidulans, and only pD02/36 was able to confer terbinafine resistance (Fig. 1C). Two additional genomic subregions were created by using PCR-amplified fragments and maintained in pGEM-T. Only the salA-containing fragment was capable to confer terbinafine resistance (Fig. 1C).
Genomic characterization and deletion of the salA locus.
The salA ORF is contained by 1,521 bp encoding a 473-amino-acid residue protein (GenBank accession number AF316427) interrupted by one putative intron (positions 578 and 680) deduced from consensus sequence of fungi. The predicted peptide encodes a salicylate 1-monooxygenase, an enzyme involved in naphthalene degradation. Table 1 shows that all fully sequenced fungal genomes to date contain at least one predicted protein product with high degree of homology to A. nidulans salA.
TABLE 1.
Salicylate monooxygenase salA genes in filamentous fungi
| Analysis type and ORF or locus | Organism | BLASTp or TBLASTn analysisa
|
||||||
|---|---|---|---|---|---|---|---|---|
| P | Bit score | Identities
|
Positives
|
No. of gaps (%) | ||||
| aa or bp | % | aa or bp | % | |||||
| BLASTp | ||||||||
| AN3382.1 | Aspergillus nidulans | 1E+00 | 878 | 435/473 | 91 | 435/473 | 91 | 0 |
| MG10012.3 | Magnaporthe grisea | 1E−70 | 263 | 159/439 | 36 | 245/439 | 55 | 6 |
| NCU07598.1 | Neurospora crassa | 1E−54 | 209 | 136/438 | 31 | 214/438 | 48 | 12 |
| FG03657.1 | Fusarium graminearum | 2E−52 | 202 | 131/407 | 32 | 206/407 | 50 | 9 |
| nahG | Pseudomonas putida | 2E−27 | 122 | 114/441 | 25 | 197/441 | 44 | 17 |
| TBLASTn | ||||||||
| 1.190 (sfd 16) | Ustilago maydis | 2E−25 | 113 | 118/437 | 27 | 183/437 | 41 | 4 |
| 1.143 (sfd 7) | Coprinus cinereus | 3E−08 | 57 | 57/218 | 26 | 89/218 | 40 | 20 |
BLASTp, the A. nidulans salA amino acid sequence was compared with the predicted ORFs of fully sequenced genomes; TBLASTn, the A. nidulans salA amino acid sequence was compared with the translated genomic nucleotide sequence. BLASTp identities are expressed as amino acids (aa); TBLASTn identities are expressed as base pairs. Identity is the extent to which two amino acid sequences are invariant, expressed as the number identical amino acids/total amino acids in the protein. Positive is the extent to which protein sequences are related, expressed as the number of similar amino acids/total amino acids in the protein.
Restriction enzyme Southern genomic mapping indicates that salA is present as a single copy in the genome (Fig. 1B, lanes P). Hybridization of a radioactively labeled salA DNA fragment with blotted pulsed-field electrophoresis separated chromosomes (data not shown) and the minimal tiling cosmid collection (28) identify a single cosmid (data not shown), suggesting that salA is physically located on chromosome VI. The profile of Southern blot hybridization was consistent with salA being a single-copy gene suitable for targeted gene disruption experiments. The inactivation of gene salA in strain RPA26 did not change the sensitivity of the fungus to terbinafine, i.e., the relative terbinafine resistance of wild-type, RPA26, and ΔsalA mutant strains, expressed as the terbinafine MIC, was 0.2 μg/ml.
Terbinafine-dependent salA transcript accumulation.
The mutation in Terb7, which confers semidominant terbinafine resistance, has been mapped to chromosome IV (30). Thus, the pD02 DNA insert, which maps to chromosome VI and encodes SalA, mediates terbinafine resistance, apparently independently of the tebA7 mutation.
Figure 2 shows terbinafine-dependent, salA transcript accumulation in a wild-type strain, a pD02-transformant and Terb7 (Fig. 1A and C). Vegetative mycelia were incubated in liquid shake-flask cultures with the indicated amounts of terbinafine and total RNA extracted, separated by gel electrophoresis, blotted onto nylon membranes, and hybridized with a radioactively labeled salA probe.
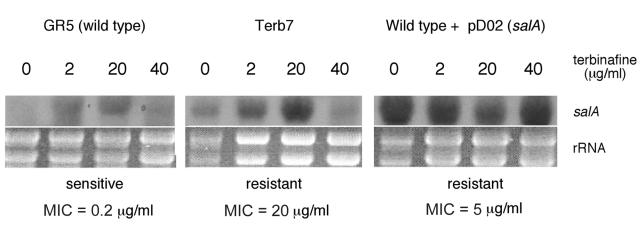
FIG. 2.
Terbinafine-dependent and overexpressed salA transcript accumulation in wild-type and mutant strains. salA transcript accumulation analysis in response to various concentrations of terbinafine in wild type, Terb7, and a pD02 transformant was carried out.
The presence of terbinafine promotes accumulation salA transcript, which appears to be a transcriptionally regulated event in the wild-type (sensitive) and Terb7 (resistant) strains but not in a pD02 transformant (resistant). Figure 2 shows that the salA transcript accumulates when terbinafine was added to the culture medium at 2 and 20 μg/ml in a wild-type strain and Terb7; however, the expression of salA is terbinafine independent in a pD02 transformant at the concentrations assayed (2 to 40 μg/ml). Interestingly, salA transcript does not accumulate when terbinafine is 40 μg/ml, except for the pD02 transformant (terbinafine independent) (Fig. 2).
Other fungal genes associated with naphthalene degradation in filamentous fungi.
One possible explanation for the observations described in Fig. 2 relies on the notion that the mutation in strain Terb7 targets a regulatory gene located on chromosome IV, which affects the expression of salA, located on chromosome VI. In pD02 transformants overexpression of the salicylate 1-monooxygenase transcript causes terbinafine resistance and is apparently independent of tebA7. Terbinafine contains a naphthalene ring, a possible substrate for the A. nidulans SalA. Naphthalene degradation as is typically observed in bacteria involves a series of enzymes and has not yet been described in fungi.
Table 2 gives the homology scores of the closest fungal homolog to microorganism genes involved in naphthalene degradation. Genes that are present in A. nidulans and other filamentous fungi are as follows: 2-hydroxychromene-2-carboxylate isomerase, P = 4e−40 in A. nidulans only; salicylaldehyde dehydrogenase; P = 6e−92, 1e−91, 2e−96, and 8e−94; salicylate 1-monooxygenase, P = 2e−63, 1e−32, 6e−42, and 8e−53; and 2-hydroxymuconic semialdehyde dehydrogenase, P = 9e−94, 4e−98, 2e−91, and 3e−90 in A. nidulans, Magnaporthe grisea, Neurospora crassa, and Fusarium graminearum, respectively. Genes that could not be found through homology matching fungal genome database entries include 1,2-dihydroxy-naphthalene dioxygenase, 2-hydroxychromene-2-carboxylate isomerase, and 4-hydroxy-2-oxovalerate aldolase.
TABLE 2.
Naphthalene degradation genes in filamentous fungi
| Homology search source
|
Homology target genome
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Function | Organism | Protein |
Aspergillus nidulans
|
Magnaporthe grisea
|
Neurospora crassa
|
Fusarium graminearum
|
|||||
| ORF | Bit score | P | Bit score | P | Bit score | P | Bit score | P | |||
| Naphthalene 1,2-dioxygenase | Pseudomonas sp. strain ND6 | AAP44248 | AN8998 | 122 | 9E−29 | 111 | 3E−25 | 110 | 3E−25 | 119 | 2E−27 |
| Tetrahydroxynaphthalene reductase | Neurospora crassa | EAA29497 | AN1886 | 82 | 1E−16 | 81 | 3E−16 | 92 | 2E−19 | 84 | 3E−17 |
| 1,2-Dihydroxy-naphthalene dioxygenase | No hit | No hit | No hit | No hit | |||||||
| 2-Hydroxychromene-2-carboxylate isomerase | Rhodopseudomonas palustris | ZP_00010435 | AN6936 | 159 | 4E−40 | No hit | No hit | No hit | |||
| Pseudomonas syringae | NP_793938 | No hit | 68 | 2E−12 | 96 | 5E−21 | 99 | 1E−21 | |||
| Salicylaldehyde dehydrogenase | Sphingomonas sp. | AAD45415 | AN1689 | 333 | 6E-92 | 332 | 1E−91 | 327 | 2E−96 | 340 | 8E−94 |
| Pseudomonas sp. strain ND6 | AAP44246 | AN4050 | 330 | 4E−91 | 314 | 4E−86 | 313 | 8E−86 | 307 | 4E−84 | |
| Salicylate 1-monooxygenase | Pseudomonas stutzeri | AAD02146 | AN7382 | 238 | 2E−63 | 136 | 1E−32 | 167 | 6E−42 | 203 | 8E−53 |
| Mesorhizobium loti | BAB53776 | AN3569 | 140 | 5E−34 | 154 | 3E−38 | 128 | 2E−30 | 169 | 2E−42 | |
| Catechol oxygenase | Pseudomonas putida | AAA66204 | AN4532 | 130 | 6E−31 | 133 | 7E−32 | 141 | 2E−34 | 129 | 1E−90 |
| 2-Hydroxymuconic semialdehyde dehydrogenase | Pseudomonas putida | S107722 | AN9034 | 339 | 9E−94 | 353 | 4E−98 | 331 | 2E−91 | 328 | 3E−90 |
| 4-Hydroxy-2-oxovalerate aldolase | Novosphingobium aromaticivorans | AAD04028 | AN0080 | 140 | 4E−34 | No hit | No hit | No hit | |||
DISCUSSION
Failure in treating fungal infections with terbinafine has been reported for patients infected with Trichophyton rubrum (22) and C. albicans (32). In C. albicans, multidrug efflux transporters have been implicated in resistance (35, 36). Thus, in order to obtain data that might contribute to a better understanding of antifungal resistance mechanisms, mutants resistant to terbinafine were isolated and characterized by classic genetic analysis (30). We constructed a genomic library of one such mutant, Terb7A, and through sib selection we identified clone pD02 (Fig. 1), which encodes two previously unreported genes: salA, which codes for a prokaryotic salicylate 1-monooxygenase (2), and the C-terminal portion of the fasC gene, which codes for the β subunit of the enzyme fatty acid synthase.
Subcloning of pD02 and creation of plasmids carrying the fasC (pD02/36fas)- or salA (pD02/36/sal)-encoding fragments of pD02 (see Fig. 1C) revealed that only fragments containing salA confer terbinafine resistance upon transformation into a sensitive A. nidulans strain. Bacterial salicylate 1-monooxygenase catalyzes the formation of catechol from salicylate, an intermediate from naphthalene degradation (2). Thus, terbinafine, which has a naphthalene nucleus in its chemical structure (23-25, 39), could be the substrate for a similar degradation pathway in fungi, generating salicylate or an analogous compound as substrate for the enzyme salicylate 1-monooxygenase in A. nidulans.
Most of the information about naphthalene metabolism has been obtained from bacteria such as P. putida (41), with which salicylate 1-monooxygenase is an extracellular flavoprotein that catalyzes conversion of salicylate to catechol by introduction of a hydroxyl group with concomitant removal of a carboxyl group (16). In addition, naphthalene degradation is a regulated process and involves nahR, a positive regulator of transcription induced by salicylate (37).
In fungi, aromatic compounds are initially converted into orthodiphenolic intermediates, e.g., catechol and protocatechuate, hydrolyzed between two hydroxyl groups and catabolized via the β-ketoadipate pathway, producing succinate and acetyl-coenzyme A (13).
Even though the genomic DNA library used in transformation experiments contained the tebA7 allele, which is genetically mapped to chromosome IV, we found one terbinafine-resistance-conferring DNA fragment, which contains the salA gene, physically mapped to chromosome VI. The physical location of the salA DNA fragment was verified by three means: (i) by DNA-DNA hybridization to pulsed-field-gel-electrophoresis-separated chromosomes, (ii) hybridization to a single chromosome VI cosmid of the A. nidulans physical map (28), and (iii) the determination that salA is on chromosome VI of the fully sequenced genome A. nidulans database at the Whitehead Institute (Cambridge, Mass.). Thus, we hypothesize that an apparent multicopy effect of the salA gene observed by Southern analysis of the pD02-salA transformant strains (Fig. 1B) may confer resistance to terbinafine in a manner similar to the mutation in the tebA7 gene, supporting the suggestion of Rocha et al. (30) that terbinafine resistance should involve multiple genes that appear to act within a genetically regulated circuit.
Northern blot experiments show (Fig. 2) increased salA transcript accumulation by terbinafine concentrations in wild-type and Terb7, indicating that salA transcript accumulation is affected by the presence of terbinafine and likely to be regulated by tebA7. Furthermore, in pD02 transformants, salA is expressed at high levels, independent of the presence of terbinafine, suggesting that salicylate or an intermediary degradation product is catalyzed more effectively in Terb7 and pD02 transformant than in the wild type. Finally, when salA is less abundant, levels of intracellular salicylate or an analogous compound may be more elevated. In P. aeruginosa membrane protein, synthesis associated with multiple drug resistance is suppressed by the presence of salicylate in the cell, rendering cells sensitive to different drugs (18).
The inalterable terbinafine sensitivity exhibited by the null salA allele supports the idea that resistance is due to an altered accumulation of salA transcript and the consequent increase in salicylate 1-monooxygenase and degradation of the naphthalene ring of terbinafine in the pD02 transformant compared to the wild-type strain.
Acknowledgments
This study was supported by grants from FAPESP, CNPq, CAPES, and FAEPA.
We thank R. A. P. Ferreira, M. Mazucatto, A. C. Crescenço, and L. N. Oliveira for technical support.
REFERENCES
- 1.Aly, R., R. Forney, and C. Bayles. 2001. Treatments for common superficial fungal infections. Dermatol. Nurs. 13:91-101. [PubMed] [Google Scholar]
- 2.Bosch, R., E. Garcia-Valdes, and E. R. Moore. 2000. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245:65-74. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D. W., T. H. Adams, and N. P. Keller. 1996. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 93:14873-14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomczynski, P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532-537. [PubMed] [Google Scholar]
- 5.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Doncker, P., and A. K. Gupta. 1999. Itraconazole and terbinafine in perspective: from petri dish to patient. Postgrad. Med. Spec. No. 1999:6-11. [PubMed] [Google Scholar]
- 7.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accurate assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 8.Fachin, A. L., C. M. Maffei, and N. M. Martinez-Rossi. 1996. In vitro susceptibility of Trichophyton rubrum isolates to griseofulvin and tioconazole. Induction and isolation of a resistant mutant to both antimycotic drugs. Mutant of Trichophyton rubrum resistant to griseofulvin and tioconazole. Mycopathologia 135:141-143. [DOI] [PubMed] [Google Scholar]
- 9.Goodfield, M. J., and E. G. Evans. 1999. Treatment of superficial white onychomycosis with topical terbinafine cream. Br. J. Dermatol. 141:604-605. [DOI] [PubMed] [Google Scholar]
- 10.Green, P. 2000. Documentation for Phrap. Washington University, Bozeman. [Online.] http://bozeman.mbt.washington.edu/phrap.docs/phred.html.
- 11.Gupta, A. K., and N. H. Shear. 1997. Terbinafine: an update. J. Am. Acad. Dermatol. 37:979-988. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hondmamann, D. H. A., and J. Visser. 1994. Carbon metabolism, p. 61-139. In S. D. Martinelli and J. R. Kinghorn (ed.), Aspergillus: 50 years, vol. Elsevier, Amsterdam, The Netherlands.
- 14.Klobucnikova, V., P. Kohut, R. Leber, S. Fuchsbichler, N. Schweighofer, F. Turnowsky, and I. Hapala. 2003. Terbinafine resistance in a pleiotropic yeast mutant is caused by a single point mutation in the ERG1 gene. Biochem. Biophys. Res. Commun. 309:666-671. [DOI] [PubMed] [Google Scholar]
- 15.Lasseron-De Falandre, A., D. Debieu, J. Bach, C. Malosse, and P. Leroux. 1999. Mechanisms of resistance to fenpropimorph and terbinafine, two sterol biosynthesis inhibitors, in Nectria hematococca, a phytopathogenic fungus. Pestic. Biochem. Physiol. 64:167-184. [Google Scholar]
- 16.Lee, J., J. Oh, K. R. Min, and Y. Kim. 1996. Nucleotide sequence of salicylate hydroxylase gene and its 5′-flanking region of Pseudomonas putida KF715. Biochem. Biophys. Res. Commun. 218:544-548. [DOI] [PubMed] [Google Scholar]
- 17.Loeffler, J., and D. A. Stevens. 2003. Antifungal drug resistance. Clin. Infect. Dis. 36(Suppl. 1):S31-S41. [DOI] [PubMed] [Google Scholar]
- 18.Masuda, N., E. Sakagawa, and S. Ohya. 1995. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May, G. S., R. B. Waring, S. A. Osmani, N. R. Morris, and S. H. Denison. 1989. The coming of age of molecular biology in Aspergillus nidulans. EMBO-Alko Workshop on Molecular Biology of Filamentous Fungi, Helsinki, Finland.
- 20.McClellan, K. J., L. R. Wiseman, and A. Markham. 1999. Terbinafine: an update of its use in superficial mycoses. Drugs 58:179-202. [DOI] [PubMed] [Google Scholar]
- 21.McPherson, M. J., and B. D. Hames. 1995. PCR: a practical approach. Oxford University Press, New York, N.Y.
- 22.Mukherjee, P. K., S. D. Leidich, N. Isham, I. Leitner, N. S. Ryder, and M. A. Ghannoum. 2003. Clinical Trichophyton rubrum strain exhibiting primary resistance to terbinafine. Antimicrob. Agents Chemother. 47:82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussbaumer, P., G. Dorfstatter, I. Leitner, K. Mraz, H. Vyplel, and A. Stutz. 1993. Synthesis and structure-activity relationships of naphthalene-substituted derivatives of the allylamine antimycotic terbinafine. J. Med. Chem. 36:2810-2816. [DOI] [PubMed] [Google Scholar]
- 24.Nussbaumer, P., I. Leitner, K. Mraz, and A. Stutz. 1995. Synthesis and structure-activity relationships of side-chain-substituted analogs of the allylamine antimycotic terbinafine lacking the central amino function. J. Med. Chem. 38:1831-1836. [DOI] [PubMed] [Google Scholar]
- 25.Nussbaumer, P., G. Petranyi, and A. Stutz. 1991. Synthesis and structure-activity relationships of benzo[b]thienylallylamine antimycotics. J. Med. Chem. 34:65-73. [DOI] [PubMed] [Google Scholar]
- 26.Petranyi, G., J. G. Meingassner, and H. Mieth. 1987. Activity of terbinafine in experimental fungal infections of laboratory animals. Antimicrob. Agents Chemother. 31:1558-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. McDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 28.Prade, R. A., J. Griffith, K. Kochut, J. Arnold, and W. E. Timberlake. 1997. In vitro reconstruction of the Aspergillus (Emericella) nidulans genome. Proc. Natl. Acad. Sci. USA 94:14564-14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reader, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 30.Rocha, E. M., C. B. Almeida, and N. M. Martinez-Rossi. 2002. Identification of genes involved in terbinafine resistance in Aspergillus nidulans. Lett. Appl. Microbiol. 35:228-232. [DOI] [PubMed] [Google Scholar]
- 31.Ryder, N. S. 1992. Terbinafine: mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 126(Suppl. 39):2-7. [DOI] [PubMed] [Google Scholar]
- 32.Ryder, N. S., S. Wagner, I. Leitner, and B. Favre. 1998. In vitro activities of terbinafine against cutaneous isolates of Candida albicans and other pathogenic yeasts. Antimicrob. Agents Chemother. 42:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1987. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y..
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 36.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schell, M. A., and P. E. Wender. 1986. Identification of the nahR gene product and nucleotide sequences required for its activation of the sal operon. J. Bacteriol. 166:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuetzer-Muehlbauer, M., B. Willinger, G. Krapf, S. Enzinger, E. Presterl, and K. Kuchler. 2003. The Candida albicans Cdr2p ATP-binding cassette (ABC) transporter confers resistance to caspofungin. Mol. Microbiol. 48:225-235. [DOI] [PubMed] [Google Scholar]
- 39.Stutz, A., and G. Petranyi. 1984. Synthesis and antifungal activity of (E)-N-(6,6-dimethyl-2-hepten-4-ynyl)-N-methyl-1-naphthalenemethanamine (SF86-327) and related allylamine derivatives with enhanced oral activity. J. Med. Chem. 27:1539-1543. [DOI] [PubMed] [Google Scholar]
- 40.Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington, and R. W. Davies. 1983. Transformation by integration in Aspergillus nidulans. Gene 26:205-221. [DOI] [PubMed] [Google Scholar]
- 41.Uz, I., Y. P. Duan, and A. Ogram. 2000. Characterization of the naphthalene-degrading bacterium, Rhodococcus opacus M213. FEMS Microbiol. Lett. 185:231-238. [DOI] [PubMed] [Google Scholar]
- 42.Yu, J. H., R. A. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]