SUMMARY
Retromer is a multi-protein complex that recycles transmembrane cargo from endosomes to the trans-Golgi network and the plasma membrane. Defects in retromer impair various cellular processes, and underlie some forms of Alzheimer’s disease and Parkinson’s disease. Although retromer was discovered over 15 years ago, the mechanisms for cargo recognition and recruitment to endosomes have remained elusive. Here we present an X-ray crystallographic analysis of a four-component complex comprising the VPS26 and VPS35 subunits of retromer, the sorting nexin SNX3, and a recycling signal from the divalent cation transporter DMT1-II. This analysis identifies a binding site for canonical recycling signals at the interface between VPS26 and SNX3. In addition, the structure highlights a network of cooperative interactions among the VPS subunits, SNX3 and cargo that couple signal-recognition to membrane recruitment.
Keywords: retromer, sorting nexins, endosomes, retrograde transport, endocytic recycling, cargo recognition, sorting signals, protein coats, membrane recruitment, membrane tubules
Graphical Abstract

INTRODUCTION
Retromer is a multi-protein complex that associates with the cytosolic face of endosomes, where it functions to recycle receptors, transporters, adhesion molecules, and other proteins to the trans-Golgi network (TGN) and the plasma membrane through sorting into tubular-vesicular carriers (Arighi et al., 2004; Seaman, 2004; Seaman et al., 1998; Steinberg et al., 2013; Temkin et al., 2011). Retromer dysfunction impairs many cellular processes, and underlies the pathogenesis of various neurodegenerative disorders including Alzheimer’s disease and Parkinson’s disease (Mecozzi et al., 2014; Small et al., 2005; Zimprich et al., 2011). The retromer complex comprises a VPS26-VPS29-VPS35 heterotrimer (herein referred to as “retromer”) that has been implicated in cargo recognition, and various combinations of sorting nexin (SNX) proteins that contribute to membrane recruitment and formation of recycling tubules (Carlton et al., 2004; Haft et al., 2000; Rojas et al., 2007; Seaman et al., 1998; Steinberg et al., 2013; Strochlic et al., 2007; Temkin et al., 2011; Wassmer et al., 2007).
Previous studies showed that the VPS26-VPS29-VPS35 trimer is an elongated structure in which VPS26 and VPS29 bind to the N- and C-terminal portions of VPS35 (VPS35N and VPS35C), respectively (Hierro et al., 2007). VPS26 has a bilobed β-sandwich, arrestin-like fold (Collins et al., 2008; Shi et al., 2006), but the structural details of its interaction with VPS35N are not known. VPS29, on the other hand, has a phosphoesterase fold (Collins et al., 2005; Wang et al., 2005) that serves as a scaffold for the α-helical solenoid structure of VPS35C (Hierro et al., 2007). Sorting nexins constitute a large family of proteins characterized by having a phospholipid-binding PX domain (Teasdale and Collins, 2012). Based on the absence or presence of additional domains, the SNX family has been subdivided into several subfamilies, three of which include members that interact with retromer: (1) the SNX-PX subfamily member SNX3, which consists of only a PX domain (Harrison et al., 2014; Strochlic et al., 2007); (2) SNX-BAR subfamily members such as the yeast Vps5-Vps17 and mammalian SNX1/2-SNX5/6 heterodimers, which have an additional BAR domain (Rojas et al., 2007; Wassmer et al., 2007); (3) the SNX-FERM subfamily member SNX27, comprising additional PDZ and FERM domains (Steinberg et al., 2013; Temkin et al., 2011).
Despite the widely held view that the VPS26-VPS29-VPS35 retromer trimer is responsible for cargo recognition, there is currently no structural evidence in support of this notion. In fact, recent X-ray crystallographic analyses of SNX27 have shown that a subset of retromer cargos with NPXY motifs and PDZ-ligands associate with the FERM domain and the PDZ domains of SNX27 (Clairfeuille et al., 2016; Gallon et al., 2014; Ghai et al., 2013). Nevertheless, another extensively-studied subset of retromer cargos, including the cation-independent mannose 6-phosphate receptor (CI-MPR) (Cereghino et al., 1995; Rojas et al., 2007; Seaman, 2007), sortilin (Canuel et al., 2008; Seaman, 2007), DMT1-II (Tabuchi et al., 2010), Wntless (Harterink et al., 2011; Zhang et al., 2011) and pIgR (Verges et al., 2004), lack NPXY and PDZ-ligand motifs, but instead share a canonical ØX(L/M) motif (where Ø is an aromatic amino acid) that mediates retromer-dependent sorting (Seaman, 2007; Tabuchi et al., 2010). How these cargos are recognized by retromer remains unknown.
In this article, we report the elucidation of the structural mechanism of cargo recognition and membrane recruitment of the retromer trimer bound to SNX3, using a combination of X-ray crystallography, small-angle X-ray scattering (SAXS), biochemistry and cellular analyses. In addition to completing the structural characterization of the whole retromer complex at the atomic level, we identify a binding site for canonical ØX(L/M) recycling signals at the interface between VPS26 and SNX3. The structure shows that SNX3 binds via an N-terminal extension and the PX domain to another site at the interface of VPS26 and VPS35. The SNX3 PX domain in turn binds phosphatidylinositol 3-phosphate (PtdIns3P) through a different surface, thus enabling retromer recruitment to the membrane. Remarkably, exposure of the binding site for ØX(L/M) signals requires a conformational change in VPS26 that is induced by the concomitant interaction with SNX3 and cargo, revealing that cargo recognition is coupled to membrane recruitment. The shared and cooperative nature of these interactions explains why previous attempts to characterize interactions of recycling signals with single subunits or partial complexes had limited success, and suggests a general mechanism for assembly of retromer coats on membrane tubules.
RESULTS
Overall Structures of VPS35N and VPS26-VPS35N-SNX3
Our initial X-ray crystallographic analyses focused on the unknown N-terminal portion of VPS35 (VPS35N). The crystal structure of residues 14–470, corresponding to ~60% of VPS35, was determined by single-wavelength anomalous diffraction using selenium as the anomalous scatterer, and was refined to 3.0 Å resolution (Figure S1A,B; Table S1). The structure displays an α/α-solenoid fold formed by 20 α helices (α1 to α20). The shape of the solenoid is slightly curved, except for the first three helices that are tilted ~45° relative to the solenoid axis. When combined with our previously determined structure of the C-terminal portion of VPS35 (VPS35C) (Hierro et al., 2007), we visualize VPS35 as having a total of 33 helices (Figure S1C). Comparison of the five copies of VPS35N in the asymmetric unit reveals larger Cα-RMS deviations together with higher temperature factors and weaker electron density for the last four helices of VPS35N (α17 to α20) (Figure S1D). These observations are consistent with electron microscopy data showing that VPS35 has an elongated structure with some bending capability around the midsection (Hierro et al., 2007).
We next sought to determine how VPS26 (VPS26A isoform) binds to VPS35, but attempts to co-crystallize a VPS26-VPS35N complex failed. Inclusion of SNX3 (Strochlic et al., 2007) in the crystallization trials, however, yielded crystals that diffracted to 2.7 Å. The structure of this complex was determined by molecular replacement (see Method Details) and the final model had a free R-factor of 0.25 and excellent stereochemistry (Figures 1 and S1E,F; Table S1).
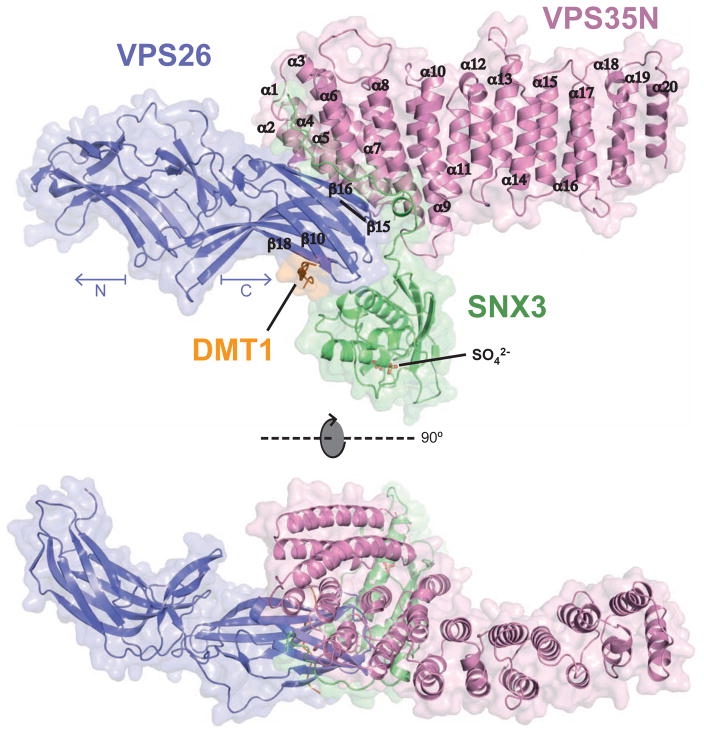
Figure 1. Overall structure of the VPS26-VPS35N-SNX3-DMT1-II complex.
The crystal structure is shown in two orthogonal views represented by a ribbon diagram with transparent surface. In the top view, the 20 α-helices (α1–20) that make up the solenoid of VPS35N and four β-strands from the C-terminal (CT) domain of VPS26 are labeled. Two sulphate ions (SO42−) found in the crystal structure, in stick representation, indicate the PtdIns3P-binding pocket on SNX3. See also Figure S1, Table S1 and Movie S1.
The overall complex has a T-shaped architecture, with VPS26 and VPS35N corresponding to both sides of the horizontal bar and SNX3 to the vertical bar (Figure 1 and Movie S1). The C-terminal lobe of VPS26 (VPS26C), previously shown to contact VPS35 (Collins et al., 2008; Shi et al., 2006), interacts through strands β15, β16 and the connecting loop, with helices α4, α5, α6 and α8 on the convex side of the VPS35 α-solenoid (Figure 1). SNX3 binds simultaneously through its N-terminal tail and PX domain to VPS26C and VPS35N. Notably, the PtdIns3P-binding pocket on the SNX3 PX domain occurs on the opposite side of the VPS26-VPS35-interaction surface (Figure 1), consistent with the role of SNX3 in retromer recruitment to endosomal membranes (Strochlic et al., 2007).
Association of VPS26 with VPS35
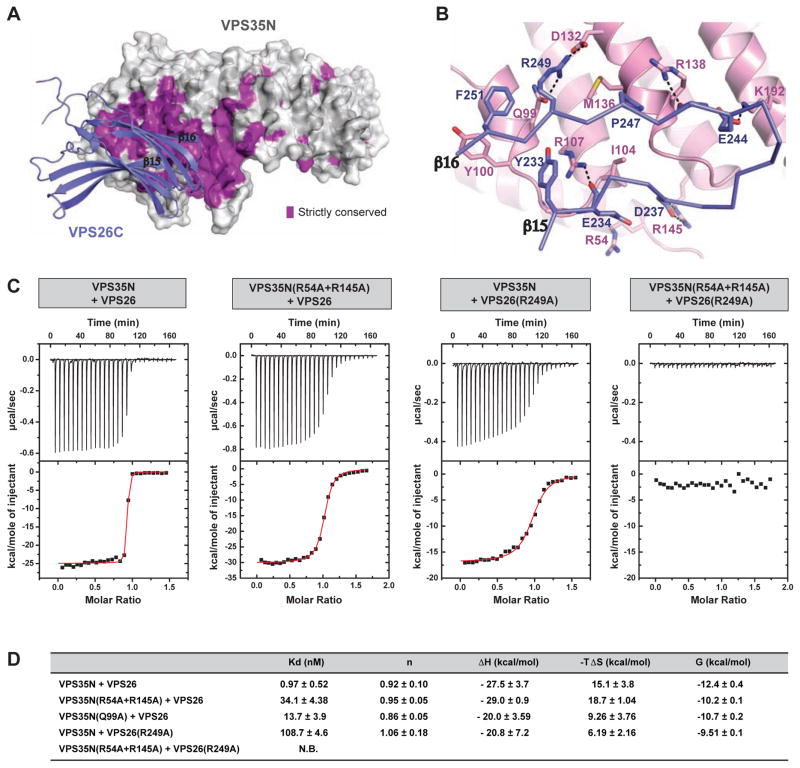
Conservation analysis and structure-based mutational studies established that the VPS26A and VPS26B mammalian paralogs bind to VPS35 through a flexible loop at the edge of the C-domain with the additional contribution of neighboring residues (Collins et al., 2008; Shi et al., 2006). The corresponding binding site on VPS35 includes a highly conserved 106PRLYL110 sequence (Gokool et al., 2007; Restrepo et al., 2007; Zhao et al., 2007) for which the two VPS26 paralogs compete (Collins et al., 2008). The structure of VPS26-VPS35N presented here enables rationalization of the previous mutational analyses and the contribution of specific residues to binding. The formation of the VPS26-VPS35N subcomplex buries approximately 829 Å2 of surface area. Contact regions concentrate most of the conserved residues (Figure 2A and Figures S2A,B), and include both polar and apolar interactions. Binding of VPS26 to VPS35 is through a central hydrophobic core dominated by P247 of VPS26, I104 and M136 of VPS35, and an extended H-bond network (Figure 2B). Accordingly, it has been shown that the double mutation P245S/R247S in VPS26B (analogous to P247S/R249S in VPS26A) prevented its incorporation into retromer in vitro and in vivo (Collins et al., 2008). To confirm the broad binding surface, we extended these analyses by substituting more peripheral contact residues (R54 and R145 of VPS35, and R249 of VPS26) with alanine, and found that these mutations also abolished the interaction (Figure 2C,D). Other mutations such as I235D/M236N in VPS26A or the equivalent I233D/M234N in VPS26B, which affect the VPS35 interaction in two-hybrid and pull-down assays (Collins et al., 2008; Shi et al., 2006), appear to contribute to structural destabilization of the VPS26 C-domain’s hydrophobic core rather than alter any direct contact. Furthermore, the crystal structures of VPS35N alone and in complex with VPS26A show that the 106PRLYL110 sequence in VPS35 is at a buried position in α-helix 5, acting as a major structural scaffold for stabilization of the surrounding helical solenoid. The only side chain from this motif that sticks out to the surface corresponds to R107, which interacts with E234 and Y233 of VPS26A. Thus, it is likely that mutations of R107 directly affect the interaction with VPS26, while other mutations within the 106PRLYL110 motif have a destabilizing effect that abolishes the binding.
Figure 2. Interacting surfaces between VPS26 and VPS35.
(A) Distribution of strictly conserved surface residues (violet) on VPS35N at the VPS26 contact site. (B) Relevant contacts of the VPS26-VPS35 interface. (C) Validation of the VPS26-VPS35 complex formation using ITC and site-directed mutagenesis. Baseline-corrected instrument response (upper) and integrated isotherms with the best fit curve to the data in red (lower) from ITC experiments measuring binding of VPS26 to VPS35N. (D) Thermodynamic binding parameters from ITC measurements. All ITC values are given as mean ± SD from at least three independent measurements. N.B., no appreciable binding. See also Figure S2
The flexible loop (residues 238–246) between strands β15 and β16 of VPS26A, shown to be critical for VPS35 binding (Shi et al., 2006), is structured in the VPS26-VPS35N-SNX3 complex with a partial segment of the loop (residues 243–246) contributing to β16 extension. Unexpectedly, only E244 within this region establishes H-bonds with VPS35 while other residues such V241 and K242 contact a conserved short α-helix (α1) of SNX3 (Figure 4B). These contacts are consistent with the finding that the triple mutant R240S/G241A/E242S at the equivalent position in the β15-β16 loop of VPS26B was able to bind VPS35 but could not be recruited to endosomal membranes (Collins et al., 2008), thus providing a structural explanation for the distinct effects of mutations in this loop.
Figure 4. Close-up of the SNX3 interfaces for retromer recruitment to membranes.
(A) SNX3 PX domain oriented to show the two sulphate ions at the phosphoinositide-binding pocket and superimposed with the Grd19p PX domain bound to C4-PtdIns(3)P. (B) Contacts between the N-terminal tail of SNX3 and the VPS26-VPS35 subcomplex. (C) Contacts between β1 of SNX3 and the α8-α9 connecting loop of VPS35. (D) Close-up view of the SNX3 P133 insertion between strands β10 and β18 of VPS26. See also Figure S4.
Structure of Retromer in Solution
Previous models based on the available crystal structure of VPS35C (residues 476–780) bound to VPS29, bioinformatic predictions, single-particle electron microscopy (EM) and small-angle X-ray scattering (SAXS) analyses revealed an overall elongated structure of retromer, with VPS26 and VPS29 bound to opposite ends of VPS35 (Hierro et al., 2007), and a tendency to form U-shaped dimers at high-protein concentration (Norwood et al., 2011). The structure of VPS35N (residues 12–469) bound to VPS26 presented here provides the missing piece of the retromer puzzle to understand more accurately the solution structure at molecular resolution. We observed that the concentration-dependent dimerization of retromer could be suppressed by increasing the ionic strength of the buffer, thus allowing the analysis of monomeric and dimeric states (Figure 3A). We performed inline size-exclusion chromatography (SEC) coupled to SAXS experiments and ab initio reconstructions to generate representative models for each state (Figure 3B-F). The bead model thus obtained for the monomeric state was elongated and slightly curved with a large lobe at one end and a smaller lobe at the other. Rigid-body refinement on the bead model using the crystal structures of VPS26-VPS35N (this study) and VPS29-VPS35C (Hierro et al., 2007) resulted in a very good fit between the theoretical scattering profile of this ensemble and the experimental SAXS data (Figure 3C,E), thus defining the solution structure more sharply.
Figure 3. Structure of the retromer complex in solution.
(A) SEC-MALS profiles for retromer at two protein concentrations under low and high ionic strength conditions. The value for the fitted molecular mass is shown as lines across the elution peak for each species. The predicted molecular mass of monomeric retromer is 150 kDa. (B) Normalized pair distance distribution P(r) functions for the monomeric and dimeric species of retromer. (C,D) Experimental spectrum of the small angle scattering of the monomeric (blue, C) and dimeric (red, D) species of retromer, and the simulated fit (grey) obtained from the model. (E,F) The ab initio shape reconstruction of the retromer complex by DAMMIN using P1 symmetry for the monomer (E) and P2 for the dimer (F), showing the fit with the crystallographic structures of VPS26-VPS35N and VPS26-VPS35C (PDB codes: 5F0L and 2R17). (G) Retromer dimer bound to two SNX3 molecules sits parallel to the membrane plane. Positively-charged VPS26 N-lobe provides a complementary surface for membrane interaction. See also Figure S3 and Movie S2.
The same ab initio and rigid-body modeling approach to characterize the dimeric self-assembly architecture of retromer under more physiological ionic conditions (150 mM NaCl) consistently resulted in an inverted U-shaped architecture. Positional mapping of retromer subunits using different MBP-tagged versions of the complex or subunit deletions showed that the VPS29-edges of two retromers are in close proximity at the center of the U (Figures 3D,F and S3, and Movie S2). Indeed, this morphology corresponds closely with an earlier SAXS reconstruction (Norwood et al., 2011). Remarkably, the VPS26 subunits at the distal ends of the U-shaped model, lay in an orientation parallel to the membrane plane with complementary surface charge distribution and compatible with SNX3 binding (Figure 3G). Although the molecular details for the dimerization are subject to some ambiguity given the intrinsic resolution limits of SAXS, it is tempting to speculate that the self-dimerization tendency could contribute to retromer coat assembly where very high local concentrations are achieved.
Molecular Determinants of Retromer Recruitment to Membranes by SNX3
SNX3 belongs to a sub-family of sorting nexins that comprise just a PX domain. A feature of most PX domains is their binding preference for PtdIns3P. This phospholipid is enriched at endosomal membranes and thus able to recruit PX-containing proteins to endosomes. The PX domain of the yeast ortholog of SNX3, Grd19p, consists of three β strands followed by three α helices and binds to PtdIns3P through a conserved pocket where two arginines and one lysine engage the 3-phosphate, 4/5-hydroxyl and 1-phosphate groups, respectively, while the inositol ring is stacked on a tyrosine side chain (Zhou et al., 2003). Our crystal structure of human SNX3 bound to VPS26-VPS35N revealed the presence of two sulfate ions within the PtdIns3P-binding pocket (Figure 1). Superposition of the SNX3 PX domain and Grd19p bound to diC4PtdIns3P shows that one sulfate ion occupies the same position as the 3-phosphate group while the second sulfate ion occupies the position of the 4/5-hydroxyl groups, mimicking the hydrogen bonding interactions that recognize a PtdIns3P ligand (Figure 4A). Interestingly, current data suggest that the association of PX domains with the membrane involves not only a direct contact with PtdIns3P but also residues within two mobile loops (L1 and L3) and the α1 helix that interact with the lipid bilayer (Jia et al., 2014; Stampoulis et al., 2012).
The crystal structure of VPS26-VPS35N-SNX3 shows that the interactions with the membrane and retromer occur on opposite sides of the SNX3 PX domain, consistent with SNX3 being a structural scaffold primed for retromer recruitment. The interaction between SNX3 and retromer can be described by three separate interfaces (Figure 4B–D and Figure S4A), one through the N-terminal tail and the other two involving the PX domain of SNX3. The N-terminal tail (residues 3–28) of SNX3 adopts a meandering conformation along the VPS26-VPS35 interface, alternating contacts with both subunits (Figure 4B). The nature of these contacts reveals a striking dichotomy between specific side-chain interactions proximal to the SNX3 PX domain, such as Y22 of SNX3 making H-bonds with N191 and Q249 within a groove between helices α8 and α10 of VPS35, and less specific interactions closer to the SNX3 N terminus (Figure 4B), consistent with the lower conservation of this region (Figure S4A). The second interface with VPS35 involves E30 and D32 of SNX3, which establish H-bonds with H202, S203 and R204 of VPS35 (Figure 4C). In line with these findings, E30 and D32 from human and yeast SNX3 are important for interaction with retromer in vivo and in vitro (Harrison et al., 2014). Finally, the third interface with VPS26 is dominated by P133 of SNX3, which sticks into a hydrophobic cavity at the tip of the C-terminal β-sandwich comprising I170, I202 and Y286 of VPS26 (Figure 4D). Taken together, these observations indicate that SNX3 integrates multiple binding sites within a single PX domain, enabling the recruitment of retromer to endosomal membranes. While the binding of SNX3 to PtdIns3P appears to follow a canonical mechanism, the interaction with retromer involves both flexible extensions and rigid segments of the PX domain in a multi-interface association with the VPS26 and VPS35 subunits.
A Mechanism for Cargo Recognition
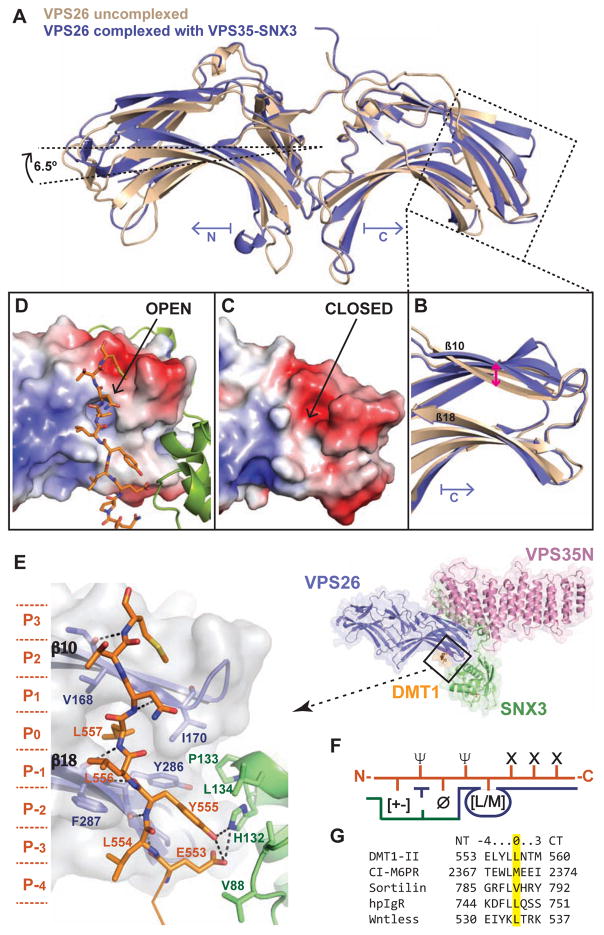
The overall structure of VPS26A in the complex exhibits several conformational changes as compared to uncomplexed forms of the protein (Shi et al., 2006). The N and C lobes, for instance, display a conformational twist relative to one another, with a 6.5° rotation around the middle axis (Figure 5A). A similar twist is observed when VPS26 is bound to the PDZ domain of SNX27 despite the use of a different interaction surface (Gallon et al., 2014). Yet, the most notable conformational change in VPS26 is the outward movement of strand β10, which generates a hydrophobic pocket between strands β10 and β18 in the context of the complex (Figure 5B–D).
Figure 5. Structural plasticity of VPS26 for cargo recognition.
(A) Superimposition of the crystal structures of VPS26A, in the free (PDB: 2FAU, brown) and VPS26-VPS35N-SNX3-DMT1-II complexed form (current work, PDB: 5F0L, blue). (B) Close-up view of the C-terminal domain. Pink arrow indicates changes in strand β10 from basal to active state. (C,D) Same view as in (B) showing the electrostatic surface potential (ranging from blue 5 kTe−1 to red - 5 kTe−1) of basal VPS26A (C) and activated VPS26A bound to the recycling signal of DMT1-II (D). (E) Close-up view showing the recognition of the DMT1-II recycling motif by the VPS26-SNX3 subcomplex. (F) Cartoon representing the consensus VPS26-SNX3 cargo binding motif (X stands for any residue, Ø a bulky aromatic residue, Ψ a residue having a hydrophobic or long aliphatic hydrocarbon tail, and [+-] any charged residue). (G) Sequence alignment of representative retromer-binding motifs. See also Figure S5 and Movie S3.
Our first crystal structure of the VPS26-VPS35N-SNX3 complex revealed an unexpected electron density perpendicular to β10 and β18 of VPS26, which corresponded to a foreign C-terminal sequence (QPEMGLV) from a symmetrically-related VPS26 molecule resulting from vector construction (Figure S5A,B). Remarkably, this sequence fits the ØX(L/M) consensus motif for cargo selection by retromer (Seaman, 2007; Tabuchi et al., 2010) and strongly resembles the recycling signal (551QPELYLL557) of the divalent metal transporter 1 isoform II (DMT1-II), a known retromer cargo that cycles between endosomes and the plasma membrane (Tabuchi et al., 2010). Indeed, subsequent crystal structures obtained with an extended DMT1-II recycling signal (residues 545–568) and incorporation of specific selenomethionine markers unambiguously confirmed the binding mode (Figure 5E and Figures S5C–E). The central part of the interaction corresponds to L557 of DMT1-II, which is completely buried within the hydrophobic pocket between strands β10 and β18 of VPS26. Accordingly, we consider L557 position 0 (P0) of the consensus motif. The signal adopts an extended conformation, with residues P−3, P−1, P0, P1 and P3 making main-chain H-bonds with strands β10 and β18 of VPS26 (Figure 5E and Movie S3). Additional side-chain interactions of the signal make significant contributions to the binding specificity. Y555 at P−2 is accommodated within a large hydrophobic pocket at the VPS26-SNX3 interface and, together with E553 at P−4, makes H-bonding interactions with H132 of SNX3. The interaction is further stabilized through the side chains of L556 at P−1 and Leu554 at P−3, both embracing F287 of VPS26 in a clamp-like manner (Figure 5E). Comparison with other known retromer-sorting sequences shows that residues at P−1 and P−3 have aliphatic hydrocarbon tails, consistent with their clamping function around F287 of VPS26 (Figure 5F,G). Further support for the DMT1-II binding mode comes from cellular studies in which mutations of P0 to any hydrophobic residue except methionine cause strong DMT1-II recycling defects (Tabuchi et al., 2010). Similarly, shortening of the aliphatic side-chain of P−2, but not mutation to Trp, significantly decreases DMT1-II recycling, whereas hydrophobic substitution of Leu to Ala at P−1 or P−3 does not affect DMT1-II recycling (Tabuchi et al., 2010). In summary, the recycling signal of DMT1-II establishes an extensive network of hydrogen bonds and hydrophobic interactions engaging both SNX3 and VPS26.
DMT1-II Binding is Concomitant With Interaction of SNX3 with Retromer
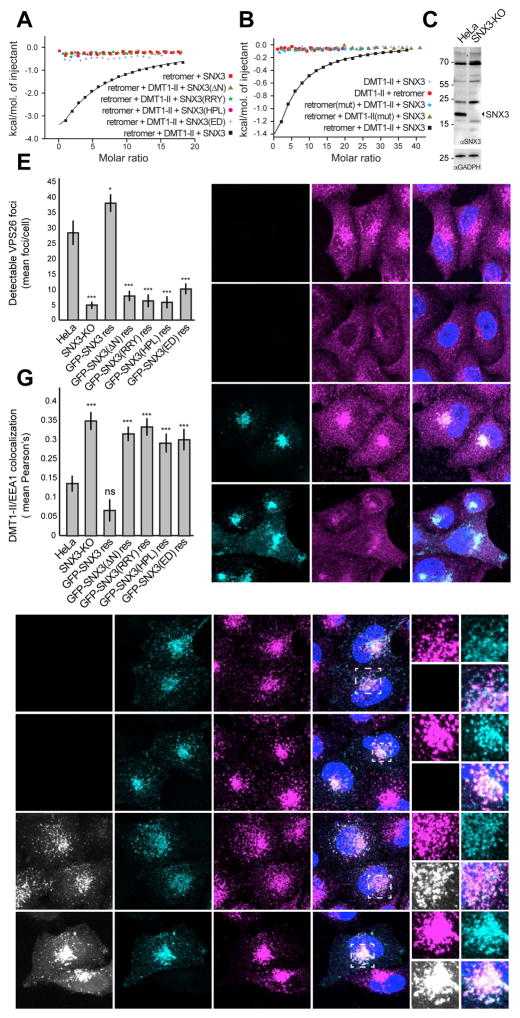
We used a combination of ITC and site-directed mutagenesis to assess the interaction of SNX3 with retromer and cargo. In isolation, SNX3 displayed a moderate affinity for retromer in the presence of cargo (Kd ~146 μM), but had no detectable affinity without cargo (Figure 6A). This interaction was completely abolished upon deletion of residues 1–25 [SNX3(Δ)] or targeted mutation to alanines of R9, R10 and Y22 [SNX3(RRY)] (Figure 4B) from the SNX3 N-terminal tail (Figures 6A and S6A,C), confirming the requirement of this flexible region for interaction of SNX3 with the VPS26-VPS35 interface. Similarly, mutation to alanines of the SNX3-PX domain residues H132, P133 and L134 at the interface of SNX3 with VPS26 and cargo [SNX3(HPL)] (Figures 4D and 5E), or residues E30 and D32 at the SNX3-VPS35 interface [SNX3(ED)] (Figure 4C), abrogated binding to retromer (Figures 6A and S6A,C). The DMT1-II recycling signal did not exhibit any detectable binding to isolated retromer or SNX3 but bound with Kd ~127 μM to retromer in the presence of SNX3 (Figures 6B and S6B,C), thus confirming that DMT1-II recognition involves binding to both retromer and SNX3. To further validate the cargo-binding site observed in the crystal structure, we introduced mutations on critical contact residues of the DMT1-II recycling signal (Y555A, L557A) or the VPS26 interface (V168N, F287A). As expected, these mutations completely abolished the interaction (Figures 6B and S6B,C), thus also validating the crystallographic interface between DMT1-II and VPS26. These findings support a mechanistic model in which the specific recognition of the DMT1-II recycling signal results from conformational changes that activate VPS26 together with additional contacts with SNX3 upon complex formation. The recruitment of retromer by SNX3 to PtdIns3P-enriched endosomes is thus concomitant with cargo selection.
Figure 6. SNX3 recruits retromer to membranes, promoting DMT1-II recycling.
(A) ITC isotherms of the binding of retromer to wild-type SNX3 and various SNX3 deletion/substitution mutants in the absence or presence of the DMT1-II recycling signal (residues 550–568). (B) ITC isotherms for the binding of peptides encompassing the normal DMT1-II recycling signal (residues 550–568) or a mutant of this sequence with Y555A and L557A substitutions (mut), to SNX3, retromer, or a combination of SNX3 with retromer or with retromer having VPS26 F287A and V168N substitutions in VPS26 (mut). (C) Immunoblot analysis of wild-type (WT) and SNX3-KO HeLa cells using antibodies to SNX3 and GADPH (loading control). The positions of molecular mass markers (in kDa) are indicated. (D) Immunofluorescence microscopy of endogenous VPS26 in WT, SNX3-KO, or SNX3-KO HeLa cells rescued (res) with GFP-SNX3 or GFP-SNX3(ΔN). Bars: 10 μm. (E) Quantification of the recruitment of endogenous VPS26 to membranes by different GFP-SNX3 constructs expressed in SNX3-KO cells. Datasets are from the stable cells lines shown in panel D, as well as stable cell lines expressing the GFP-SNX3(RRY), GFP-SNX3(HPL) or GFP-SNX3(ED) mutants shown in Figure S6F. Bars represent the mean ± SEM (n=22–34 cells; * p<0.05, *** p<0.005 by Student’s t-test). (F) Immunofluorescence microscopy of endogenous EEA1 and ectopically-expressed DMT1-II in the same cell lines from panel D. Bars: 10 μm. Magnified views of the boxed areas are shown on the right. Bars: 2 μm. Images in D and F are maximum intensity projections of Z-stacks. Nuclei were stained with DAPI (blue). (G) Quantification of DMT1-II–EEA1 colocalization in the cell lines shown in panel F and Figure S7. Bars represent the mean ± SEM of the Pearson’s correlation coefficient from three independent experiments (n=10–19; ns: not significant, *** p<0.005 by Student’s t-test). See also Figures S6 and S7.
To evaluate the functional relevance of the structure, we generated a SNX3 CRISPR knock-out (KO) HeLa cell line (Figures 6C and S6D), and tested the effects of rescuing this cell line with GFP-tagged forms of the various SNX3 variants described above. In agreement with previous findings (Harterink et al., 2011), SNX3-KO cells exhibited decreased association of retromer with membranes relative to control HeLa cells, as assessed by immunofluorescence microscopy of endogenous VPS26 (Figures 6D and S6E). This phenotype could be rescued by stable expression of GFP-SNX3, but not GFP-SNX3(Δ), GFP-SNX3(RRY), GFP-SNX3(HPL) or GFP-SNX3(ED) mutants (Figures 6D,E and S6F). Failure of retromer to associate with membranes would be expected to impair sorting of cargo from endosomes into recycling transport intermediates. Indeed, we found that DMT1-II displayed increased colocalization with the early-endosomal marker EEA1 in SNX3-KO cells relative to control HeLa cells, indicative of a defect in cargo export from endosomes (Figures 6F,G and S7A). This phenotype could also be corrected by stable expression of GFP-SNX3 but not the different GFP-SNX3 mutants described above (Figures 6F,G and S7B). Taken together, these findings demonstrated that concomitant interactions of SNX3 with VPS26, VPS35 and DMT1-II are required for recruitment of retromer to membranes and sorting of DMT1-II cargo out of endosomes and into recycling transport intermediates.
DISCUSSION
The retromer complex plays a critical role in endosomal recycling pathways, but the molecular mechanisms by which it is recruited to membranes and selects cargo proteins into transport carriers have remained elusive. Here we provide a structural framework for understanding how multivalent interactions involving retromer, SNX3, a recycling signal from the divalent cation transporter DMT1-II, and the PtdIns3P membrane lipid cooperate in a mechanism that couples membrane recruitment and cargo recognition. The crystal structure of the VPS26-VPS35N-SNX3-DMT1-II complex presented here shows that the recycling signal of DMT1-II is recognized by coincident interaction with SNX3 and retromer. This interaction involves a conformational change in VPS26 that exposes key residues of the signal-binding pocket, while complementary binding of SNX3 to PtdIns3P promotes membrane recruitment. In vitro binding and in vivo cellular studies support the functional relevance of this structure.
The resolution of the crystal structures of VPS26A and VPS26B revealed an arrestin-like fold (Collins et al., 2008; Shi et al., 2006). The strong structural homology of VPS26 with the arrestins hinted at an analogous role as an adaptor protein within the retromer complex. However, the sequence similarity with arrestins is low, and none of the VPS26 paralogs share the surface residues that are involved in arrestin binding to GPCRs, clathrin, adaptor proteins, phospholipids, or other signaling molecules (Collins et al., 2008; Shi et al., 2006). The structure of VPS26-VPS35N-SNX3-DMT1-II complex presented here reveals that VPS26 indeed functions as a cargo adaptor, but through a completely different mechanism. The C-terminal lobe of VPS26 undergoes a closed-to-open conformational change upon coincident interaction with SNX3 and the recycling signal of DMT1-II. In addition, VPS26-SNX3 coupling generates an additional binding surface in the VPS26-SNX3 interface that contributes to DMT1-II interaction, thus raising the possibility that other PX domains from distinct SNX proteins could display surface variations that contribute differently to cargo selection.
The use of short linear motifs with moderate-to-low affinity is a common feature of dynamic processes to make these interactions transient and reversible. Furthermore, the use of multiple low affinity interactions can provide high avidity and specificity, while maintaining the reversibility necessary to orchestrate dynamic assemblies. In this regard, the avidity of retromer for cargo might be increased by the dimerization of transmembrane receptors, the packaging of the receptors in a small area by coincident interaction with cargo, SNX proteins and phosphoinositides, the use of repetitive motifs on accessory proteins such FAM21 for binding multiple retromers (Jia et al., 2012), and the affinity-modulation by post-translational modifications, as recently demonstrated for some PDZ binding motifs to promote SNX27 association (Clairfeuille et al., 2016).
Retromer cargos include single-pass as well as multi-pass transmembrane proteins. It remains to be determined if the inter-domain loops of VPS26 establish additional interactions with the helical core of multi-pass transmembrane receptors, similar to recent descriptions of the rhodopsin-arrestin 1 complex based on its crystal structure (Kang et al., 2015). In this regard, the yeast DMT1-II homolog Ftr1p, which contains seven transmembrane domains, only requires the cytoplasmic C-terminal tail for effective endosomal sorting (Strochlic et al., 2007). It is interesting to note that the ability of arrestins to fully engage with the receptor core involves a 20° inter-domain twist together with the repositioning of three central loops, particularly the insertion of the finger loop within the receptor core (Kang et al., 2015; Kim et al., 2013; Shukla et al., 2013; Shukla et al., 2014). In the case of the human VPS26, the equivalent finger loop is much shorter, precluding a similar insertion mechanism, or at most contributing with a weaker interaction. Moreover, the inter-domain twist observed in VPS26A between the basal and cargo-bound states is only 6.5°. This limited flexibility may have only minor effects on local engagement with multi-pass transmembrane proteins, but in the context of the full-length retromer it could result in large vertical displacements of VPS29 bound at the opposite end of the complex, in turn influencing other intermolecular contacts.
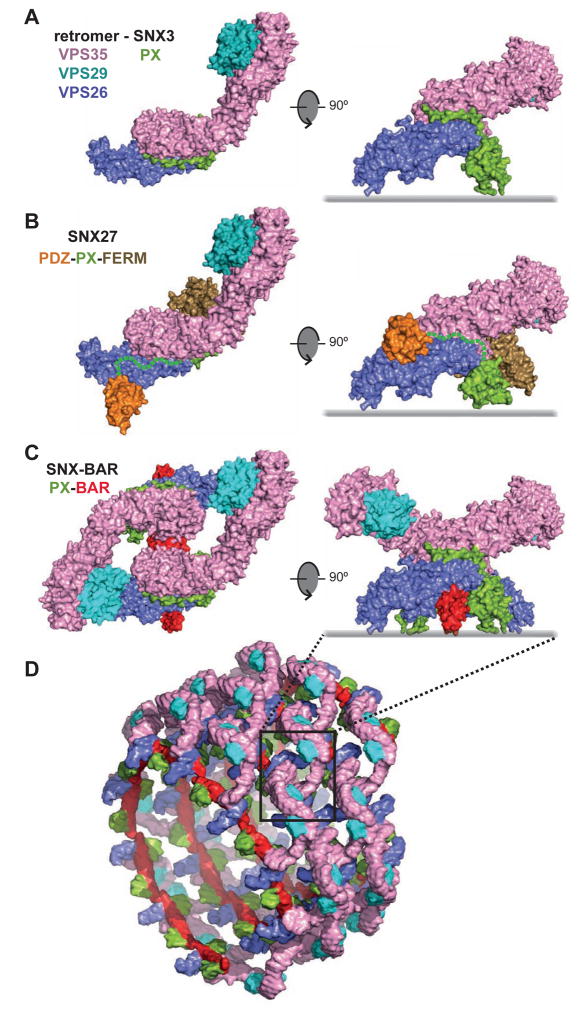
Based on the VPS26-VPS35N structure presented here, the previously solved VPS29-VPS35C structure (Hierro et al., 2007), and the low-resolution SAXS data of the VPS26-VPS29-VPS35 retromer core (Figures 3F,G and S3), we can picture the entire retromer architecture bound to a SNX-PX family member such as SNX3 (Figure 7A). It remains to be determined whether this architecture is conserved for the SNX-BAR (i.e., SNX1/2-SNX5/6) and SNX-FERM (i.e., SNX27) subfamilies (Figure S4C). In this regard, the residues of the SNX3 PX domain involved in the interaction with VPS26-VPS35 are conserved in the SNX-BAR PX domains, arguing for a similar binding mode (Figure S4B). This conservation is not so evident in the VPS27 PX domain; yet, the binding sites for the PDZ and PX domains on VPS26 are ~80 Å apart, a distance compatible with the 33-residue linker that connects both domains (Figure 7B). This arrangement would place the FERM domain of VPS27 at the C-terminal end of the PX domain, parallel to the membrane (Ghai et al., 2015), and on the concave side of VPS35. In the case of SNX-BAR proteins, the only PX-BAR tandem structure solved to date corresponds to SNX9 (Pylypenko et al., 2007). Assuming a similar inter-domain arrangement in SNX-BAR-retromer, superposition of the PX domains would place one retromer complex on each of the distal parts of the curved BAR dimer in a trans orientation. In this configuration, the C-terminal lobe of VPS26 sits over the tips of the BAR arms while the rest of retromer protrudes as extended wings (Figure 7C). Based on the tip-loop contacts between SNX-BAR assemblies (van Weering et al., 2012; van Weering et al., 2010), the observed oligomeric lattices of the N-BAR domain of endophilin (Mim et al., 2012), and the tendency of retromer to form dimers through the VPS29 side ends (Figure 3F), we posit a speculative model of how retromer dimers and PX-BAR dimers might be combined together in a helical arrangement held by tip-loop contacts between BAR domains (Figure 7D and Movie S4). Assuming that PX-BAR homo-dimers and hetero-dimers are capable of assembling into helical arrangements, the model presented here would be equally valid for hetero-dimers. Although several factors can contribute to a less regularly arranged lattice during the elongation of the tube, such as the flexibility of the VPS35 subunit, fluctuations in the orientation of the BAR domains, the presence of extra domains among different SNXs, and the packing of cargos with distinct sizes, this intuitive model suggests an architecture that involves a double zipper helical assembly of SNX-BAR-retromer where lateral and longitudinal contacts may contribute to tubule morphology.
Figure 7. Proposed architecture of different SNX-retromer assemblies.
(A) Proposed model of the SNX3-retromer complex. The entire retromer (VPS26-VPS29-VPS35) structure was generated by fitting the crystal structures of VPS26-VPS35N (current work) and VPS29-VPS35C (Hierro et al., 2007) within experimental SAXS data (Figure 3E) and superimposed on the crystal structure of SNX3 (green) bound to VPS26-VPS35N. (B) Proposed model of the SNX27-retromer complex. The retromer (VPS26-VPS29-VPS35) structure was superimposed on the crystal structure of the SNX27 PDZ domain (orange) bound to VPS26 (Gallon et al., 2014). Residues for the linker segment between the PDZ and PX domains of SNX27 are indicated with a green dashed line. The PX domain (green) of SNX27 (PDB: 4HAS) was superimposed on the PX domain of SNX3 and linked to the crystal structure of the FERM-like domain of SNX17 (Ghai et al., 2013). (C) Proposed model of a SNX-BAR-retromer complex. Two VPS26-VPS29-VPS35 retromer complexes were superimposed on the SNX9 PX-BAR dimer structure (Pylypenko et al., 2007) using the PX-domain of SNX3 as reference. (D) Cartoon showing a speculative helical coat organization formed by the combination of retromer and PX-BAR dimers. See also Movie S4.
In conclusion, the crystal structure of VPS26-VPS35N-SNX3-DMT1-II presented here not only uncovers the atomic details for the interaction of a consensus retromer-binding motif, but also suggests a mechanism that couples membrane recruitment with cargo selection. An exciting ‘Cryptex code’ thus emerges from our observations, where combinatorial retromer-SNXs interactions can reshape the cargo binding surface to favor multivalent contacts in cargo selection. These findings should stimulate further research to decipher the specific assemblies behind distinct endosomal export pathways and their role in protein homeostasis and disease.
STAR★Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests should be addressed to Aitor Hierro at ahierro@cicbiogune.es
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Corning), supplemented with 2 mM L-glutamine (Corning), 100 IU/ml penicillin (Corning), 100 μg/ml streptomycin (Corning) and 10 % v/v heat-inactivated fetal bovine serum (FBS, Corning). Cells were cultured at 37 °C with 5 % CO2 and 95 % humidity. For stable transformants, complete medium was supplemented with 0.5 mg/ml G418.
METHOD DETAILS
Recombinant DNA Procedures
The DNA sequence encoding the N-terminal part of human VPS35 (VPS35N) (residues 14–470) was cloned into the vector pGST-Parallel2 (Sheffield et al., 1999) with a cleavable N-terminal Glutathione S-transferase (GST) tag. DNA encoding full-length human VPS26A was cloned into pET28-Sumo3 vector (EMBL, Heidelberg) to express protein with a N-terminal cleavable 6xHis-Sumo3 tag. DNAs encoding fusion constructs of VPS26A with human DMT1 (DMT1-II isoform) were cloned using pET28-Sumo3-VPS26 as template. The following plasmids were cloned: pET28-Sumo3-VPS261–317-DMT1545–568 and pET28-Sumo3-VPS261–321-DMT1549–560 with an additional His tag SHHHHH at the C terminus. DNAs encoding full-length human SNX3 and SNX3ΔN (residues 26–162) were cloned into pHisMBP-Parallel2 (Sheffield et al., 1999) to express these proteins with a N-terminal cleavable 6xHis-maltose binding protein (MBP) tag. The plasmids pET28-Sumo3-MBP-VPS26 and pMR101A-MBP-VPS29 were cloned in order to express VPS26 and VPS29 with an N-terminal non-cleavable MBP tag followed by a small linker of three serines. Site-directed mutations in VPS35-, VPS26-, SNX3- and DMT1-coding sequences were introduced using mutagenic primers and the Phusion polymerase (Thermo). All constructs were verified by DNA sequencing. For the expression of VPS26A, VPS29 and VPS35, the following plasmids were used: pmr101A-VPS26 (Shi et al., 2006) that expresses VPS26 with an extra MG at the N terminus and GLVPRGSHHHHH at the C terminus, pMR101A-VPS29 (Hierro et al., 2007), and pGST-Parallel2-VPS35 (Hierro et al., 2007).
A GFP-SNX3 plasmid (Harterink et al., 2011) (kindly donated by Prof. Peter J. Cullen, University of Bristol, UK) was used as a template to amplify GFP-SNX3ΔN using the primer pair (GCCGA GGAAT TCCTC GAGAT CGATG TGAGC AACCC GCAAA CG and GATCC GGTGG ATCCT CAGGC ATGTC), which was subsequently sub-cloned into GFP-SNX3 (EcoRI/BamHI). For generation of point mutants, gBlock gene fragments (IDT) were ordered containing the desired mutations, which were cloned by Gibson assembly (Gibson et al., 2009) into GFP-SNX3 (PCR primers: CAGAA CGAAC GTTGT CTTCA CATG, CCACG GTCTC CGCGA ATTCG TTCAG G) as above. pMT423 (3xHA-DMT1) (Tabuchi et al., 2002) used for localization of DMT1-II in HeLa cells was kindly donated by Dr. Mitsuaki Tabuchi (Kagawa University, Japan).
Protein Expression and Purification
Native proteins were expressed in E. coli BL21(DE3) grown in Luria-Bertani (LB) broth at 37 °C, and protein expression was induced at an OD600 of 0.8 by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). Cells were harvested after 16 hours of growth at 18 °C. Seleno-L-methionine (SeMet) derivative proteins were expressed in E. coli B834(DE3) grown in LB medium at 37 °C to an OD600 of 1.0. Cells were harvested, resuspended in SeMet medium base plus nutrient mix (Molecular Dimensions) and starved of methionine for 1 hour. 0.2 mM SeMet (Acros Organics) and 0.5 mM IPTG were added to the medium. Cells were harvested after 16 hours of growth at 18 °C. All following purification steps were performed at 4 °C. The concentration of all purified proteins was calculated using the theoretical extinction coefficient.
VPS35N, VPS35N labeled with SeMet and VPS35N mutants were purified using the following protocol. The cell pellet was resuspended in buffer A [50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM dithiothreitol (DTT)] supplemented with 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM benzamidine, disrupted by sonication, and the lysate was cleared by centrifugation at 50000 g for 45 min. The supernatant was incubated for 2 hours in batch with glutathione-Sepharose beads (GE Healthcare) followed by extensive washing of the beads with buffer A in a gravity column. Protein was released from the beads by overnight cleavage of the N-terminal GST-tag with tobacco etch virus (TEV) protease in buffer A. The cleaved protein was further purified by ion-exchange chromatography (HiTrapQ, GE Healthcare) using a gradient of 150–1000 mM NaCl, followed by size-exclusion chromatography (Superdex 200 16/60, GE Healthcare) in 50 mM Tris-HCl pH 7.5, 750 mM NaCl and 10 mM β-mercaptoethanol (BME).
For the purification of SNX3 and SNX3 mutants, the cell pellet was lysed by sonication in buffer B (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM DTT, 10 mM imidazol) supplemented with 0.1 mM PMSF and 1 mM benzamidine. After centrifugation at 50000 g for 45 min, the soluble fraction was incubated for 2 hours in batch with Ni2+-nitrilotriacetate (NTA) agarose resin (Qiagen). After extensive washing of the beads with buffer B, the protein was eluted with buffer B and 250 mM imidazol. TEV protease was added to the eluted sample to remove the N-terminal HisMBP-tag and linker. The mixture was dialyzed overnight against 50 mM Tris-HCl pH 6.7, 100 mM NaCl and 1 mM DTT. Following ion-exchange chromatography (HiTrapSP, GE Healthcare) using a gradient of 15–1000 mM NaCl, SNX3 was further purified by size-exclusion chromatography (Superdex 75 16/60, GE Healthcare) in 50 mM Tris-HCl pH 7.5, 150 mM NaCl and 1 mM DTT.
VPS26 and VPS26 mutants were expressed with a 6xHis-Sumo3 tag. The lysis and Ni-NTA affinity chromatography were performed as described for SNX3. The N-terminal 6xHis-Sumo3-tag was cleaved with Sentrin-specific protease 2 (SENP2) by overnight dialysis against 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM DTT and 10 mM imidazol. A second Ni2+-NTA chromatography was carried out to remove cleaved 6xHis-Sumo3 and uncleaved protein. VPS26 was subsequently purified by ion-exchange chromatography (HitrapQ, GE Healthcare) using a gradient of 15–1000 mM NaCl followed by size-exclusion chromatography (Superdex 200 16/60, GE Healthcare) in buffer C [25 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP)].
For the purification of the VPS26-VPS35N complex, the cell pellets of overexpressed VPS35N and VPS26 (from pmr101A-VPS26) labeled with SeMet were mixed. Lysis and glutathione-Sepharose purification were carried out with the same protocol as for VPS35N, with the difference that the cells were disrupted by high-pressure homogenization (20 kpsi) and the TEV proteolysis was done for 4 hours. The VPS26-VPS35N complex was further purified with Ni2+-NTA beads in buffer D (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM DTT, 20 mM imidazol). Elution was performed with buffer D and 200 mM imidazol. After overnight dialysis in buffer A, the complex was further purified by ion-exchange chromatography (HitrapQ, GE Healthcare) using a gradient of 150–1000 mM NaCl followed by size-exclusion chromatography (Superdex 200 16/60, GE Healthcare) in buffer A.
VPS26-VPS35N-DMT1-II complexes were purified as described for VPS26-VPS35N with the difference that dialysis was carried out in the presence of SENP2 protease. For the purification of VPS26-VPS35N-DMT1549–560 and VPS26-VPS35N-DMT1549–560(L557M) complexes, VPS26-DMT1 was labeled with SeMet.
Full-length retromer complex (VPS26-VPS29-VPS35) was purified by mixing the cell pellet of coexpressed VPS29 and GST-VPS35 with the cell pellet of His-Sumo3-VPS26. The purification was carried out as for the VPS26-VPS35N-DMT1-II complex but differed in the buffer composition of lysis and GST beads purification (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT), Ni2+-NTA agarose purification (50 mM Tris-HCl pH 8.5, 100 mM NaCl, 1 mM DTT, 20 mM imidazol) and size-exclusion chromatography (buffer C).
MBP tagged full-length retromer complex [MBP-VPS26]-VPS29-VPS35 was purified by mixing the cell pellet of coexpressed VPS29 and GST-VPS35 with the cell pellet of His-MBP-VPS26. The purification was carried out as for the full retromer complex but skipping the SENP2 proteolysis step. Similarly, full-length retromer complex with an MBP tag in VPS29, VPS26-[MBP-VPS29]-VPS35, was purified by mixing the cell pellets of His-Sumo3-VPS26, MBP-VPS29 and GST-VPS35. The purification was carried out as described for the full retromer complex.
Protein Crystallization
A detailed description of the constructs crystallized in this work is shown in Table S1. All five crystal forms were obtained by hanging-drop vapor diffusion at 18 °C by mixing 1 μl protein solution and 1 μl precipitant solution. VPS35N crystallization drops were set after concentrating the gel-filtration purified protein to 4.7 mg/ml using as precipitant 1.65 M ammonium sulfate, 2% (w/v) polyethylene glycol (PEG) 1000 and 0.1 M HEPES pH 7.6. Rod-shaped crystals appeared after three to five days. Individual crystals were cryoprotected by immersion in a precipitant solution supplemented with 20% (w/v) sucrose and 5% (v/v) glycerol. VPS26-VPS35N-SNX3 crystallization was achieved by mixing VPS26-VPS35N purified complex (45 μM) with a three-fold molar excess of SNX3 (135 μM) in Tris 25 mM, 250 mM NaCl, 1 mM DTT and 5% (v/v) glycerol. Oval-shaped crystals grew after 5–10 days in crystallization solutions containing 0.75–0.9 M ammonium sulfate, 0.1 M MES pH 6.0 and 0–15% (v/v) glycerol. Streak seeding was required in order to grow diffraction-quality crystals. Prior to flash freezing in liquid nitrogen, the crystals were transferred for 1–5 minutes into a reservoir solution containing 25% ethylene glycol for cryoprotection. VPS26-VPS35N-SNX3-DMT1-II complexes were crystallized and cryoprotected using the same protocol as for VPS26-VPS35N-SNX3.
Data Collection and Structure Determination
Diffraction data were collected in the following synchrotrons: VPS35N and VPS26-VPS35N-SNX3-DMT1-II data sets at SOLEIL beamline Proxima 1 (Paris, France); VPS26-VPS35N-SNX3 data sets at ALBA beamline XALOC (Barcelona, Spain); VPS26-VPS35N-SNX3-DMT1-II SeMet labeled datasets at Diamond Light Source (Didcot, UK) beamlines I03 and I02.
For structure determination, the CCP4 software suite (Winn et al., 2011) (SHELX, Parrot, Buccaneer, DM, Refmac5, Phaser, QtPISA) and Phenix (Adams et al., 2010) were used. VPS35N diffraction data were integrated and scaled using XDS (Kabsch, 2010). The space group was determined to be C2 with five molecules in the asymmetric unit. The structure was solved using the SAD approach in SHELX, with one SeMet dataset at 3.1 Å. Phases were improved by density modification using Parrot, and an initial model was built with Buccaneer. The model was further improved by phase extension using DM to the native data at 3.0 Å and by iterative cycles of refinement and manual building using Phenix, Refmac5 and Coot (Emsley et al., 2010). Initial NCS restraints were gradually removed in the final cycles of the refinement to allow some structural variation. In the final model, residues 382–390 and 445–455 located in connecting loops and residue 470 at the C-terminus could not be modeled because of poor electron density in these regions.
Diffraction data from VPS26-VPS35N-SNX3 were processed with XDS. The crystal belonged to space group C2 and contained one copy of the complex in the asymmetric unit. The structure was solved by molecular replacement with Phaser using the coordinates of human VPS26A (PDB: 2FAU), human SNX3 (PDB: 2YPS) and our previously solved structure of VPS35N as search models. Owing to the anisotropic diffraction, the dataset was subjected to ellipsoidal truncation and anisotropic scaling with the UCLA Diffraction Anisotropy Server. Refining the model against the anisotropy corrected data significantly improved the quality of the resulting electron-density maps. The final structure was obtained through iterative cycles of manual building and refinement using Phenix, Refmac5 and Coot. The electron density maps clearly showed an extra density at the VPS26 surface that could be traced as the C-terminal part of VPS26 from a symmetrically related molecule (Figure S5B). VPS26-VPS35N-SNX3 was crystallized with VPS26 labeled with SeMet. The position of the selenium anomalous scatters confirmed the correctness of the atomic model. In the final model, residues 1–7 and 301–320 of VPS26, 470 of VPS35, and 1–3; 159–162 of SNX3 could not be modeled because of poor electron density in these regions. The structures of VPS26-VPS35N-SNX3-DMT1-II, native or SeMet-labeled, complexes were solved by molecular replacement using Phaser and the initial structure of VPS26-VPS35N-SNX3 as search model. Refinement of the structures was done as described before. The anomalous signal from two different SeMet-labeled VPS26-DMT1-II fused constructs was used to confirm the identity and orientation of the recycling signal (Figure S5D,E). Crystallographic data collection and model statistics are summarized in Table S1. Model validation was carried out using the Molprobity tool in Phenix. The Ramachandran statistics calculated by Molprobity are: 97.1%/2.6%/0.3%, 97.9%/2.1%/0%, 96.6%/3.3%/0.1%, 96.9%/3.1%/0%, 97.0%/2.9%/0.1% (favored/allowed/outliers) for VPS35N, VPS35N-VPS26-SNX3, VPS35N-VPS26-SNX3-DMT1, VPS35N-VPS26-SNX3-DMT1 (SeMet labeled) and VPS35N-VPS26-SNX3-DMT1L557M (SeMet labeled), respectively. The surface buried in the complex interface was calculated using QtPISA. Graphics presented in this manuscript were generated using the program PyMOL (http://www.pymol.org/) and UCSF Chimera package (Pettersen et al., 2004).
Size-exclusion Chromatography Coupled to Multiangle Light Scattering
The oligomerization state of full-length retromer complex was determined by size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS). 45 μl sample (1–10 mg/ml) were autoinjected onto a Shodex KW403-4F column at 0.16 ml/min with an Agilent 1200 Series HPLC at 25 °C. Two different buffers were assayed with 25 mM HEPES pH 7.5 and 0.5 mM TCEP that differ in the salt concentration, 150 mM NaCl or 300 mM NaCl. The column output was inline with a DAWN HELEOS II MALS detector (Wyatt Technology) followed by an Optilab T-rEX differential refractometer (Wyatt Technology). Light scattering and refractive index data were collected and analyzed with ASTRA 6 software (Wyatt Technology). Bovine serum albumin was used as the calibration standard. Molecular masses were calculated across individual eluted peaks with a dn/dc value set to 0.185 ml/g.
Small-angle X-ray Scattering
Synchrotron small-angle X-ray scattering (SAXS) data of full-length retromer complex (VPS26-VPS29-VPS35), VPS35-VPS29 and MBP-tagged retromer complexes were collected on beamline B21 at Diamond Light Source (Didcot, United Kingdom) with an inline HPLC system. Scattering was recorded on a Pilatus 2M detector over an angular range qmin = 0.015 Å−1 to qmax = 0.3 Å−1. X-ray scattering patterns at high (300 mM NaCl) and low (150 mM NaCl) ionic strength were recorded after 45 μl injection protein samples at 7–11 mg/ml in Shodex column KW403-4F equilibrated in 25 mM HEPES pH 7.5, 300 mM NaCl and 0.5 mM TCEP with a flow-rate of 0.16 ml/min at 20 °C. Initial data processing (background subtraction, radius of gyration Rg, maximum distance Dmax and distance distribution function calculation) was performed using ScÅtter (Version 3.0 by Robert P. Rambo, Diamond Light Source, UK). The subsequent data processing was performed with the ATSAS package (DAMMIN, DAMAVER, DAMFILT, CORAL, CRYSOL) (Petoukhov et al., 2012). For each dataset, twenty independent ab initio models of the scattering particles were obtained with DAMMIN. These models were averaged and filtered using DAMAVER, and DAMFILT, respectively, to find the most representative compact map filled with 278 dummy-atoms, for the monomer and 544 for the dimmer with at P2 symmetry. A full retromer model was generated fitting the structures of VPS26-VPS35N and VPS29-VPS35C (PDB: 2R17) with the envelope using CHIMERA (Pettersen et al., 2004). The probable conformation of missing loops in the crystal structures were found with CORAL and modeled with dummy atoms. The fitting of the theoretical scattering curves between the model and the experimental data was obtained using CRYSOL with a discrepancy factor χ2 of 1.4 for the monomer and χ2 of 1.7 for the dimmer. The program MONSA was used to locate MBP tags relatively to the full retromer complex. MONSA is multiphase bead modeling that allows the simultaneous fitting of multiple SAXS curves. The untagged constructs of the full retromer complex were represented by three phases (VPS35-VPS29-VPS26 for phase 1, VPS35-VPS29 for Phase 2, and VPS26 for phase 3), the MBP-tagged construct were represented by three phases ([MBP-VPS26]-VPS29-VPS35 or VPS26-[MBP-VPS29]-VPS35 for phase 1, VPS35-VPS26-VPS29 for phase 2, and MBP for phase 3). Simulated annealing was used to search, starting from a random phase distribution, which simultaneously fitted the multiple SAXS curves from untagged and MBP-tagged species, to minimize overall discrepancy. For each phase combination, twenty independent ab initio models of the scattering particles were obtained and in a similar process of DAMMIN models the most representative was selected. The resulting bead model was converted to a map envelope and visualized using CHIMERA (Pettersen et al., 2004).
Isothermal Titration Calorimetry Assays
Isothermal titration calorimetry (ITC) experiments were carried out on a VP-ITC titration microcalorimeter (MicroCal/GE Healthcare) at 25 °C. All the proteins and peptides used for ITC experiments were dialyzed overnight at 4 °C against 50 mM HEPES 7.5, 300 mM NaCl and 0.5 mM TCEP and degassed for 5 minutes in a ThermoVac sample degasser before titration. The titration sequence consisted of an initial 2 μl injection to prevent artifacts arising from filling of the syringe (not used in data fitting), followed by 20 or 30 s injections of 10 or 15 μl aliquots with a spacing of 360 s between injections. Similar injections of protein or peptides in buffer were performed to determine the heat of dilution used to correct the experimental data. The resulting titration data were integrated and fitted to a one-site model using the Origin ITC software package supplied by MicroCal. The binding constant (Ka, Kd=1/Ka), the molar binding stoichiometry (n) and binding enthalpy (ΔH) were extracted directly from the fit. The free energy (ΔG) and entropy (ΔS) of binding was calculated from ΔG = −RTlnKa = ΔH-TΔS, where R is the gas constant and T is the absolute temperature. For the ITC analysis of the VPS26-VPS35N complex formation, 70–89 μM VPS26 (wt or R249A) solution was titrated into 9–10 μM VPS35N solution (wt, or Q99A, or R249A, or R54A+R145A). The interaction of SNX3 with retromer in the presence of the DMT1-II peptide was analyzed by titrating 730–950 μM SNX3 or SNX3 mutants into 10 μM full-length retromer complex and 150 μM peptide DMT1550–568 (AQPELYLLNTMDADSLVSR). The analysis of the binding of the DMT1-II recycling signal with retromer was carried out with the peptides DMT1550–568 and DMT1(mut)550–568 (AQPELALANTMDADSLVSR) that contains the mutations Y555A and L557A. 10 μM retromer, or 150 μM SNX3, or 10 μM retromer + 150 μM SNX3, or 10 μM retromer(mut) (harboring VPS26 mutations F287A and V168N) + 150 μM SNX3 in the calorimetric cell was titrated by successive injections of 1821–2000 μM DMT1550–568, or DMT1(mut)550–568. Data are the mean of a minimum of three replicate titrations for each experiment.
Genetic deletion of SNX3 using CRISPR/Cas9
The SNX3 gene in HeLa cells was mutated using the CRISPR/Cas9 system (Cong et al., 2013). Target sequences were designed using the CRISPR design tool (http://crispr.mit.edu/). 24-mer (bp) oligonucleotides including the targeting sequence (sense CACCGGGGTCCGTAGGCGTCATTC, antisense AAACGAATGACGCCTACGGACCCC) were synthesized (Eurofins), annealed and introduced into plasmid px330 (Cong et al., 2013) (Addgene). HeLa cells were transfected with the plasmid in a 24-well plate and re-seeded at low confluency after 72 hours to allow single colony formation. After ~10 days, 100 colonies were picked and seeded into 24-well plates. After a further 4–5 days, each clone was split into 2 wells of a 24-well plate. From one of the wells, the cells were lysed using Laemmli sample buffer (Laemmli, 1970) and subjected to immunoblot analysis using antibody to SNX3 to screen for SNX3-KO cells. Of the 52 clones screened, one was found to be deficient for expression of SNX3. The KO was confirmed further by immunoblot analysis on the isolated clone, using GADPH and clathrin heavy chain as loading controls.
Generation of Stable Rescue Cell Lines
SNX3-KO cells were transfected with plasmids encoding GFP-SNX3, GFP-SNX3(ΔN), GFP-SNX3(HPL), GFP-SNX3(RRY) or GFP-SNX3(ED) in a 24-well plate. Cells were re-seeded at medium confluency after 72 hours to allow single colony formation and selected in medium containing 0.5 mg/ml G418 (Geneticin, Invitrogen). After ~10 days and multiple rounds of splitting and re-plating in G418 containing medium cells were split onto a 100mm plate. Cells were lifted from the plate and 40000–100000 cells FACS sorted for GFP-positive expression, thus preventing clonal expression specific artifacts. Stable expression was verified by microscopic analysis.
Confocal Laser-scanning Microscopy
Cells were cultured on glass coverslips (Daigger), fixed with 4% paraformaldehyde, and permeabilized either with 0.2% Triton X-100 or 0.2% saponin (Sigma) in PBS supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2. Antibodies and Alexa-conjugated secondary antibodies (Life Technologies) diluted in 1% BSA-containing PBS supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2 were used to label proteins for localization. GFP fluorescence was observed either directly or using GFP-booster (Chromotek, 1:200), which was added at the same time as the secondary antibodies. Glass slides with a drop of Fluoromount-G 9 (with DAPI) (EMS) were used to mount the coverslips that were then observed on a Zeiss LSM710 confocal microscope (Zeiss, Germany). For comparative datasets, Z-stacks were taken of each observed cell and converted to a maximum intensity projection using ImageJ (http://rsb.info.gov/ij/). Objective and pixels per micron were kept consistent between comparable datasets.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Microscopic Data
To quantify the number of VPS26-positive foci in an unbiased way, Imaris spot detection was used. Three datasets (repeats) were quantified. Per dataset a threshold was set for the presence of VPS26-positive foci based on control cells from that dataset (dataset 1: 4.74x104, dataset 2: 4.03x104, dataset 3: 2.2x104). Spot size was consistently set to 0.5 μm, and background subtraction was turned off to prevent false positives. Homoscedastic t-test statistical analysis was performed in Python [2.7.10] (www.python.org) using the SciPy package.
To quantify colocalization of EEA1 with DMT1-II, image analysis was performed with ImageJ and the PSC colocalization plug-in with three repeat experiments with multiple cells per experiment (French et al., 2008). The degree of correlation is given as the Pearson’s rank correlation. A threshold level of 50 was set, under which pixel values were considered noise and not included in the statistical analysis. The Pearson’s rank correlation for each cell was compiled and the mean of each population calculated. Homoscedastic t-test statistical analysis was performed in Python [2.7.10] (www.python.org) using the SciPy package.
DATA AND SOFTWARE AVAILABILITY
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession numbers 5F0K, 5F0J, 5F0L, 5F0M and 5F0P (see Table S1).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SNX3 (IB, 1:500; IF, 1:100) | Abcam | ab56078 |
| GADPH (IB, 1:200) | Santa Cruz | sc-20357 |
| VPS35 (IB, 1:1000) | Haft et al., 2000 | N/A |
| VPS26 (IB, 1:2000, IF, 1:750) | Haft et al., 2000 | N/A |
| Clathrin heavy chain (IB, 1:10000) | BD Biosciences | 610499 |
| GFP (IB, 1:1000) | MACS Miltenyi Biotec | 130-091-833 |
| HA epitope (IF, 1:1000) | Thermo Scientific | OPA1-10980 |
| EEA1 (IF, 1:1000) | BD Biosciences | 610457 |
| GFP booster nanobody conjugate (IF, 1:200) | Chromotek | gba488 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Selenomethionine medium base plus nutrient mix | Molecular Dimensions | Cat# MD12-501 |
| L(+) - Selenomethionine | Acros Organics | Cat# 259960025 |
| Peptide DMT1550–568 (AQPELYLLNTMDADSLVSR) | Genscript | N/A |
| Peptide DMT1(mut)550–568 (AQPELALANTMDADSLVSR) | Genscript | N/A |
| Critical Commercial Assays | ||
| Glutathione Sepharose 4B | GE Healthcare | Cat# 17-0756-05 |
| Ni-NTA Agarose | Qiagen | Cat# 30230 |
| HiTrap Q HP 5ml column | GE Healthcare | Cat# 17-1154-01 |
| HiTrap SP HP 5ml column | GE Healthcare | Cat# 17-1152-01 |
| HiLoad 16/60 Superdex 75 column | GE Healthcare | Cat# 17-1068-01 |
| HiLoad 16/60 Superdex 200 column | GE Healthcare | Cat# 17-1069-01 |
| KW403-4F column | Shodex | Cat# F6989202 |
| Deposited Data | ||
| VPS35C+VPS29 | Hierro et al., 2007 | PDB: 2R17 |
| VPS26A | Shi et al., 2006 | PDB: 2FAU |
| SNX3 | unpublished | PDB: 2YPS |
| VPS35N | This study | PDB: 5F0K |
| VPS26-VPS35-SNX3 | This study | PDB: 5F0J |
| VPS26-VPS35-SNX3-DMT1 | This study | PDB: 5F0L |
| VPS26-VPS35-SNX3-DMT1 (SeMet) | This study | PDB: 5F0M |
| VPS26-VPS35-SNX3-DMT1 (L557M) (SeMet) | This study | PDB: 5F0P |
| Experimental Models: Cell Lines | ||
| HeLa | N/A | N/A |
| HeLa SNX3-KO | This study | N/A |
| HeLa SNX3-KO GFP-SNX3 rescue | This study | N/A |
| HeLa SNX3-KO GFP-SNX3(ΔN) rescue | This study | N/A |
| HeLa SNX3-KO GFP-SNX3(HPL) rescue | This study | N/A |
| HeLa SNX3-KO GFP-SNX3(RRY) rescue | This study | N/A |
| HeLa SNX3-KO GFP-SNX3(ED) rescue | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Escherichia coli BL21(DE3) | Invitrogen | Cat# C600003 |
| Escherichia coli B834(DE3) | Novagen | Cat# 69041 |
| Recombinant DNA | ||
| pET28-Sumo3 | EMBL, Heidelberg | N/A |
| pGST-Parallel2 | Sheffield et al., 1999 | N/A |
| pHisMBP-Parallel2 | Sheffield et al., 1999 | N/A |
| pmr101A-VPS26tail | Shi et al., 2006 | N/A |
| pET28-Sumo3-VPS26 | This study | N/A |
| pET28-Sumo3-VPS261–317- DMT1545–568 | This study | N/A |
| pET28-Sumo3-VPS261–321- DMT1549–560 | This study | N/A |
| pET28-Sumo3-VPS261–321- DMT1549–560 (L557M) | This study | N/A |
| pET28-Sumo3-VPS26 (R249A) | This study | N/A |
| pET28-Sumo3-VPS26 (F287A+V168N) | This study | N/A |
| pET28-Sumo3-MBP-VPS26 | This study | N/A |
| pMR101A-VPS29 | Hierro et al., 2007 | N/A |
| pMR101A-MBP-VPS29 | This study | N/A |
| pGST-Parallel2-VPS35 | Hierro et al., 2007 | N/A |
| pGST-Parallel2-VPS35-NT | This study | N/A |
| pGST-Parallel2-VPS35-NT (R54A+R145A) | This study | N/A |
| pGST-Parallel2-VPS35-NT (Q99A) | This study | N/A |
| pHisMBP-Parallel2-SNX3 | This study | N/A |
| pHisMBP-Parallel2-SNX3(ΔNT) | This study | N/A |
| pHisMBP-Parallel2-SNX3(ED) | This study | N/A |
| pHisMBP-Parallel2-SNX3(HPL) | This study | N/A |
| pHisMBP-Parallel2-SNX3(RRY) | This study | N/A |
| GFP-SNX3 | Harterink et al., 2011 | N/A |
| GFP-SNX3(ΔN) | This study | N/A |
| GFP-SNX3(RRY) | This study | N/A |
| GFP-SNX3(HPL) | This study | N/A |
| GFP-SNX3(ED) | This study | N/A |
| pMT423 (3xHA-DMT1) | Tabuchi et al., 2002 | N/A |
| Sequence-Based Reagents | ||
| Software and Algorithms | ||
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de |
| CCP4 | Winn et al., 2011 | http://www.ccp4.ac.uk |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| ATSAS | Petoukhov et al., 2012 | https://www.embl-hamburg.de/biosaxs/atsas-online/ |
| UCSF CHIMERA | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/uc |
| PYMOL | Molecular Graphics System, Version 1.8 Schrödinger, LLC | https://www.pymol.org/ |
| UCLA Diffraction Anisotropy Server | UCLA | https://services.mbi.ucla.edu/anisoscale/ |
| Clustal Omega Server | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/esp |
| ESPript 3.0, Easy Sequencing in PostScript | SBGrid | http://espript.ibcp.fr/ESPript/ESPript/ |
| STRIDE | Technische Universität München | http://webclu.bio.wzw.tum.de/stride/ |
| PROMALS3D | University of Texas Southwestern Medical Center | http://prodata.swmed.edu/promals3d/promals3d.php |
| ImageJ | NIH | http://rsb.info.gov/ij/ |
| ImageJ PSC colocalization plugin | French et al., 2008 | |
| Python | Python Software Foundation | www.python.org [2.7.10] |
| Other | ||
Supplementary Material
Table S1. Crystallographic data collection and refinement statistics. (Related to Figure 1 and S1).
Movie S1. Crystal structure of the VPS26-VPS35N-SNX3-DMT1-II complex. (Related to Figure 1).
Movie S2. Dimeric structure of retromer by small-angle X-ray scattering. (Related to Figure 3 and S3).
(A) Representative electron density map (2Fo-Fc) calculated with phases derived from the final refined model and contoured at 1.5σ, showing the refined VPS35N structure in a stick model. (B) Packing of the C2 crystal structure of VPS35N. (C) Extended VPS35 model incorporating the VPS35N(aa14–470) and the VPS35C(aa476–780)-VPS29 crystal structures. (D) The VPS35N model with the B factor in colors ranging from blue (low fluctuations) to red (high fluctuations). (E) Electron density map (2Fo-Fc) calculated with phases derived from the final refined model and contoured at 1.5σ in the vicinity of the VPS26-VPS35N-SNX3 interface, showing the refined structure in a stick model. (F) Crystal lattice packing of the VPS26-VPS35N-SNX3 complex.
The alignments were generated by using the Clustal Omega server at EMBL-EBI and plotted with ESPript. (A) Secondary structure elements corresponding to human VPS35 are shown above the alignment. α-helices are represented by cylinders; invariant residues are colored red on a pink background, and conserved residues are colored blue. The residue numbering shown above the alignment corresponds to human VPS35. VPS35 residues that interact with VPS26 or SNX3 are indicated under the alignment; the types of interactions are specified in the text box. VPS35 residues that reduce the affinity of VPS35 for VPS26 when mutated are indicated. The residues mutated in this work are marked with yellow stars (R54A+R145A and Q99A) and those of previous work are marked with black stars (R107A and L108P) (Norwood et al., 2011). The accession numbers of the aligned sequences are the following: Homo sapiens (NP_060676), Gallus gallus (NP_001005842), Danio rerio (NP_001020688), Drosophila melanogaster (NP_611651), Caenorhabditis elegans, (CCD70154), Saccharomyces cerevisiae (NP_012381), and Arabidopsis thaliana (BAF93445). (B) Sequence alignment of human VPS26 and orthologs. The alignment was produced as described above. β-strands are represented as arrows and 310 helices as cylinders labeled with η; invariant residues are colored red on a blue background and conserved residues are colored blue. The residue numbering shown above the alignment corresponds to human VPS26. Secondary structure was assigned using Stride and β-strands were numbered as reported before (Shi et al., 2006). Residues involved in inter-subunit contacts are indicated under the alignment; the types of interactions are specified in the text box. Residues of VPS26 that reduce the affinity for VPS35 when mutated are indicated. Residues mutated in this work are marked with yellow stars (R249A). Residues mutated in other studies are marked with black stars (I235S+M236D, G238P, Δ238–246 GG, 238–246 polyS) (Shi et al., 2006); VPS26B mutations/VPS26A equivalent residues I233D/I235D, I233D+M234N/I235D+M236N, P245S+R247S/P247S+R249S, L197S+R199E/L199S+R201E, R240S+G241A+E242S/K242S+G243A+E242S (Collins et al., 2008). Residues of VPS26 that reduce the affinity for DMT1-II when mutated are indicated with yellow stars (V168N and F287A). The accession numbers of the aligned sequences are: Homo sapiens (NP_004887), Gallus gallus (XP_421577), Danio rerio (NP_957201), Drosophila melanogaster (AAF45679), Caenorhabditis elegans (CCQ25652), Saccharomyces cerevisiae (EDV12710), and Arabidopsis thaliana (NP_200165).
(A) Spatial location of the VPS26 subunit and the N-terminus of the VPS26 and VPS29 polypeptides within the SAXS derived model. (B) Normalized P(r) distance distribution function of the full retromer complex VPS26-VPS29-VPS35, the VPS29-VPS35 subcomplex, and the N-terminal MBP-tagged constructs [MBP-VPS26]-VPS29-VPS35 and VPS26-[MBP-VPS29]-VPS35. (C-E) Multiphase envelopes calculated by MONSA in surface representation indicating the calculated phase for (C) the VPS26 (slate) subunit, (D) MBP(red)-tagged VPS26, and (E) MBP(red)-tagged VPS29.
(A) Sequence alignment of human SNX3 and orthologs. The alignment was produced as described for Figure S2. α-helices and 310 helices (η) are represented by cylinders, and β-strands as arrows; invariant residues are colored red on a green background and conserved residues are colored blue. Residues involved in inter-subunit contacts are indicated under the alignment; the types of interactions are specified in the text box. Residues of SNX3 mutated in this study are indicated as yellows stars (R9A+R10A+Y22A, E30A+D32A and H132A+P133A+L134A) and residues of SNX3 that reduce the affinity for retromer when mutated in other studies are indicated as black stars (yeast mutations/equivalent human mutations: E37A+E39A+H41A/E30A+D32A+S34A) (Harrison et al., 2014). The residue numbering shown above the alignments corresponds to human SNX3. Green frames highlight the relevant residues for the SNX3-VPS26 interaction. The accession numbers of the aligned sequences are the following: Homo sapiens (NP_003786), Gallus gallus (NP_001006408), Danio rerio (NP_001032183), Drosophila melanogaster (AAF54838), Caenorhabditis elegans, (CAA22253) and Saccharomyces cerevisiae (NP_015002) (B) Sequence alignment of human SNX3 with other human sorting nexins. The alignment was generated with the multiple sequence and structure alignment server PROMALS3D, using the following PDB entries: SNX3 (this work), SNX1 (2I4K), SNX5 (3HPC) and SNX27 (4HAS). The accession numbers of the aligned sequences are the following: SNX1 (NP_003090), SNX2 (AAC17181), SNX5 (NP_055241), SNX6 (AAD27829) and SNX27 (XP_005245566). (C) Domain architecture of sorting nexins important for retromer function in mammals. The following domains are depicted: PX (phox), BAR (Bin-Amphiphysin-Rvs), FERM (band4.1-ezrin-radixin-moesin), and PDZ (postsynaptic density 95-discs large-zonula occludens).
(A) Crystal packing of the VPS26-VPS35N-SNX3 complex with a zoomed-in view of the asymmetric unit complex and a symmetry-related VPS26 molecule (VPS26-sym). The C-terminal end of VPS26, SAEQPM (residues 321–327) followed by the extra sequence GLVPRG derived from cloning is highlighted in red. No electron density was observed for residues 303–320 of VPS26, indicating flexibility. We assume that the observed C terminus of VPS26 corresponds to a symmetry-related VPS26 molecule according to the observed distance (17 Å red dashed line). (B–E) The sequence of the VPS26 with the C-terminal cloning tag and three different VPS26-DMT1-II fusion constructs used in this work are indicated on the upper part of each figure. In blue is the sequence of VPS26, in red the sequence derived from the cloning, in orange the sequence of DMT1-II, in bold the residues that could be modeled in the crystal structures, and in magenta the methionines that were labeled with selenium. The following electron density maps of VPS26 or DMT1-II are superimposed on the corresponding crystal structures: in (B) and (C), omit difference electron density map (Fo-Fc) in green contoured at 2.0 σ and in (B) anomalous map of Se in magenta contoured at 4 σ; in (D), electron density map (2Fo-Fc) in blue contoured at 1.5 σ and the anomalous map of Se in magenta contoured at 4 σ; and in (E), electron density map (2Fo-Fc) in blue contoured at 1.5 σ and the anomalous map of Se in magenta contoured at 6 σ. The color code for the atoms in is as follows: slate blue (carbon of VPS26), pink (carbon of VPS35), green (carbon of SNX3), orange (carbon of DMT1), dark red (carbon of extra residues from cloning), red (oxygen), dark blue (nitrogen), and yellow (sulphur or selenium).
(A) ITC thermograms for the titration of retromer (VPS26-VPS29-VPS35), in the presence or absence of DMT1550–568 peptide, with WT SNX3 or SNX3 mutants. (B) ITC thermograms for the titration of DMT1550–568 peptide or the mutated peptide DMT1(mut) 550–568 (with the mutations Y555A and L557A) with retromer, SNX3, or the preformed complex of retromer with SNX3 or retromer(mut) (trimeric core having VPS26 F287A and V168N substitutions). (C) Thermodynamic binding parameters from ITC measurements in (A) and (B). (D) Immunofluorescence microscopy of endogenous SNX3 in HeLa cells and in a SNX3 CRISPR-KO cell line, using antibody to SNX3. Bar: 5 μm. (E) Immunofluorescence microscopy of endogenous VPS26 in WT and SNX3 CRISPR-KO HeLa cell lines. Bar: 10 μm. Magnifications of the boxed regions are shown at right. (F) Recovery of endogenous VPS26 recruitment to membranes in stable cell lines rescued with different SNX3 GFP constructs. Bars: 10 μm. Magnifications of the boxed regions are shown at right. Notice recruitment of VPS26 comparable to HeLa WT cells in recovery with full-length GFP-SNX3, but not SNX3 with truncation of the whole N-terminus (ΔN), or with point mutations (RRY, HPL and ED) that prevent interaction with VPS26, VPS35 and/or the DMT1 recycling signal.
(A) Immunofluorescence microcopy colocalization of ectopically expressed DMT1-II-HA with the endogenous early-endosomal marker EEA1 in WT and SNX3-KO HeLa cells. (B) Colocalization of DMT1-II-HA and EEA1 in the same SNX3-GFP rescue cell lines described in the legend to Figure S6. Bars: 10 μm. Magnifications of the boxed regions are shown at right.
Movie S3. Recycling motif of DMT1-II bound to VPS26. (Related to Figure 5).
Movie S4. Speculative model for the SNX-BAR-retromer coat on a tubular membrane. (Related to Figure 7).
Acknowledgments
We thank Alberto Marina (CIC bioGUNE) for technical assistance. This work was supported by the Carlos III Health Institute grant PI11/00121, the Basque Government grant PI2011-26, the Spanish Ministry of Economy and Competitiveness Grant BFU2014-59759-R (to A.H.), and the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH (ZIA HD001607) (to J.S.B.). This study made use of the Diamond Light Source (Oxfordshire, UK), synchrotron SOLEIL (Gif-sur Yvette, France), the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and ALBA synchrotron beamline BL13-XALOC, funded in part by the European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement N°283570). We thank all the staff from these facilities, and in particular to Andrew Thomson from SOLEIL, for assistance with X-ray data collection and processing, and Robert Rambo from Diamond for assistance with SAXS data collection. We also thank Peter Cullen, Carol R. Haft and Mitsuaki Tabuchi for kind gifts of reagents, and Philip McCoy (NHLBI, NIH) for cell sorting.
Footnotes
AUTHOR CONTRIBUTIONS
M.L. performed construct design, cloning, purification, crystallization, structure solution, model building, structural analysis, ITC, and SAXS data collection and processing; D.C.G. performed cellular studies; A.V. performed cloning, purification and crystallization experiments; A.L.R. performed crystallization, structure solution, model building, structural analysis, and SAXS data collection and processing. A.H and J.S.B designed the research, analyzed the data and wrote the manuscript with help from M.L. and D.C.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. The Journal of cell biology. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuel M, Lefrancois S, Zeng J, Morales CR. AP-1 and retromer play opposite roles in the trafficking of sortilin between the Golgi apparatus and the lysosomes. Biochemical and biophysical research communications. 2008;366:724–730. doi: 10.1016/j.bbrc.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Current biology : CB. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Molecular biology of the cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairfeuille T, Mas C, Chan AS, Yang Z, Tello-Lafoz M, Chandra M, Widagdo J, Kerr MC, Paul B, Merida I, et al. A molecular code for endosomal recycling of phosphorylated cargos by the SNX27-retromer complex. Nature structural & molecular biology. 2016 doi: 10.1038/nsmb.3290. [DOI] [PubMed] [Google Scholar]
- Collins BM, Norwood SJ, Kerr MC, Mahony D, Seaman MN, Teasdale RD, Owen DJ. Structure of Vps26B and mapping of its interaction with the retromer protein complex. Traffic. 2008;9:366–379. doi: 10.1111/j.1600-0854.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Collins BM, Skinner CF, Watson PJ, Seaman MN, Owen DJ. Vps29 has a phosphoesterase fold that acts as a protein interaction scaffold for retromer assembly. Nature structural & molecular biology. 2005;12:594–602. doi: 10.1038/nsmb954. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nature protocols. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- Gallon M, Clairfeuille T, Steinberg F, Mas C, Ghai R, Sessions RB, Teasdale RD, Collins BM, Cullen PJ. A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3604–3613. doi: 10.1073/pnas.1410552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Bugarcic A, Liu H, Norwood SJ, Skeldal S, Coulson EJ, Li SS, Teasdale RD, Collins BM. Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E643–652. doi: 10.1073/pnas.1216229110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Tello-Lafoz M, Norwood SJ, Yang Z, Clairfeuille T, Teasdale RD, Merida I, Collins BM. Phosphoinositide binding by the SNX27 FERM domain regulates its localization at the immune synapse of activated T-cells. Journal of cell science. 2015;128:553–565. doi: 10.1242/jcs.158204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gokool S, Tattersall D, Reddy JV, Seaman MN. Identification of a conserved motif required for Vps35p/Vps26p interaction and assembly of the retromer complex. The Biochemical journal. 2007;408:287–295. doi: 10.1042/BJ20070555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Molecular biology of the cell. 2000;11:4105–4116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MS, Hung CS, Liu TT, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:267–272. doi: 10.1073/pnas.1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, van Weering JR, van Heesbeen RG, Middelkoop TC, Basler K, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nature cell biology. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Billadeau DD, Rosen MK. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Molecular biology of the cell. 2012;23:2352–2361. doi: 10.1091/mbc.E11-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Ghai R, Collins BM, Mark AE. The recognition of membrane-bound PtdIns3P by PX domains. Proteins. 2014;82:2332–2342. doi: 10.1002/prot.24593. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta crystallographica Section D, Biological crystallography. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–146. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mecozzi VJ, Berman DE, Simoes S, Vetanovetz C, Awal MR, Patel VM, Schneider RT, Petsko GA, Ringe D, Small SA. Pharmacological chaperones stabilize retromer to limit APP processing. Nature chemical biology. 2014;10:443–449. doi: 10.1038/nchembio.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Structural basis of membrane bending by the N-BAR protein endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood SJ, Shaw DJ, Cowieson NP, Owen DJ, Teasdale RD, Collins BM. Assembly and solution structure of the core retromer protein complex. Traffic. 2011;12:56–71. doi: 10.1111/j.1600-0854.2010.01124.x. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. New developments in the program package for small-angle scattering data analysis. Journal of applied crystallography. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. The EMBO journal. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo R, Zhao X, Peter H, Zhang BY, Arvan P, Nothwehr SF. Structural features of vps35p involved in interaction with other subunits of the retromer complex. Traffic. 2007;8:1841–1853. doi: 10.1111/J.1600-0854.2007.00659.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R, Kametaka S, Haft CR, Bonifacino JS. Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Molecular and cellular biology. 2007;27:1112–1124. doi: 10.1128/MCB.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. The Journal of cell biology. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. Journal of cell science. 2007;120:2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. The Journal of cell biology. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein expression and purification. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shi H, Rojas R, Bonifacino JS, Hurley JH. The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain. Nature structural & molecular biology. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497:137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK, Westfield GH, Xiao K, Reis RI, Huang LY, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Annals of neurology. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- Stampoulis P, Ueda T, Matsumoto M, Terasawa H, Miyano K, Sumimoto H, Shimada I. Atypical membrane-embedded phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2)-binding site on p47(phox) Phox homology (PX) domain revealed by NMR. The Journal of biological chemistry. 2012;287:17848–17859. doi: 10.1074/jbc.M111.332874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, Tavare JM, Cullen PJ. A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nature cell biology. 2013;15:461–471. doi: 10.1038/ncb2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. The Journal of cell biology. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Tanaka N, Nishida-Kitayama J, Ohno H, Kishi F. Alternative splicing regulates the subcellular localization of divalent metal transporter 1 isoforms. Molecular biology of the cell. 2002;13:4371–4387. doi: 10.1091/mbc.E02-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Yanatori I, Kawai Y, Kishi F. Retromer-mediated direct sorting is required for proper endosomal recycling of the mammalian iron transporter DMT1. Journal of cell science. 2010;123:756–766. doi: 10.1242/jcs.060574. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. The Biochemical journal. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jager S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nature cell biology. 2011;13:715–721. doi: 10.1038/ncb2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JR, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, Carlsson SR, Hurley JH, Cullen PJ. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. The EMBO journal. 2012;31:4466–4480. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JR, Verkade P, Cullen PJ. SNX-BAR proteins in phosphoinositide-mediated, tubular-based endosomal sorting. Seminars in cell & developmental biology. 2010;21:371–380. doi: 10.1016/j.semcdb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges M, Luton F, Gruber C, Tiemann F, Reinders LG, Huang L, Burlingame AL, Haft CR, Mostov KE. The mammalian retromer regulates transcytosis of the polymeric immunoglobulin receptor. Nature cell biology. 2004;6:763–769. doi: 10.1038/ncb1153. [DOI] [PubMed] [Google Scholar]
- Wang D, Guo M, Liang Z, Fan J, Zhu Z, Zang J, Zhu Z, Li X, Teng M, Niu L, et al. Crystal structure of human vacuolar protein sorting protein 29 reveals a phosphodiesterase/nuclease-like fold and two protein-protein interaction sites. The Journal of biological chemistry. 2005;280:22962–22967. doi: 10.1074/jbc.M500464200. [DOI] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. Journal of cell science. 2007;120:45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta crystallographica Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wu Y, Belenkaya TY, Lin X. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell research. 2011;21:1677–1690. doi: 10.1038/cr.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Nothwehr S, Lara-Lemus R, Zhang BY, Peter H, Arvan P. Dominant-negative behavior of mammalian Vps35 in yeast requires a conserved PRLYL motif involved in retromer assembly. Traffic. 2007;8:1829–1840. doi: 10.1111/j.1600-0854.2007.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CZ, Li de La Sierra-Gallay I, Quevillon-Cheruel S, Collinet B, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, Graille M, et al. Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. The Journal of biological chemistry. 2003;278:50371–50376. doi: 10.1074/jbc.M304392200. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. American journal of human genetics. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Crystallographic data collection and refinement statistics. (Related to Figure 1 and S1).
Movie S1. Crystal structure of the VPS26-VPS35N-SNX3-DMT1-II complex. (Related to Figure 1).
Movie S2. Dimeric structure of retromer by small-angle X-ray scattering. (Related to Figure 3 and S3).
(A) Representative electron density map (2Fo-Fc) calculated with phases derived from the final refined model and contoured at 1.5σ, showing the refined VPS35N structure in a stick model. (B) Packing of the C2 crystal structure of VPS35N. (C) Extended VPS35 model incorporating the VPS35N(aa14–470) and the VPS35C(aa476–780)-VPS29 crystal structures. (D) The VPS35N model with the B factor in colors ranging from blue (low fluctuations) to red (high fluctuations). (E) Electron density map (2Fo-Fc) calculated with phases derived from the final refined model and contoured at 1.5σ in the vicinity of the VPS26-VPS35N-SNX3 interface, showing the refined structure in a stick model. (F) Crystal lattice packing of the VPS26-VPS35N-SNX3 complex.
The alignments were generated by using the Clustal Omega server at EMBL-EBI and plotted with ESPript. (A) Secondary structure elements corresponding to human VPS35 are shown above the alignment. α-helices are represented by cylinders; invariant residues are colored red on a pink background, and conserved residues are colored blue. The residue numbering shown above the alignment corresponds to human VPS35. VPS35 residues that interact with VPS26 or SNX3 are indicated under the alignment; the types of interactions are specified in the text box. VPS35 residues that reduce the affinity of VPS35 for VPS26 when mutated are indicated. The residues mutated in this work are marked with yellow stars (R54A+R145A and Q99A) and those of previous work are marked with black stars (R107A and L108P) (Norwood et al., 2011). The accession numbers of the aligned sequences are the following: Homo sapiens (NP_060676), Gallus gallus (NP_001005842), Danio rerio (NP_001020688), Drosophila melanogaster (NP_611651), Caenorhabditis elegans, (CCD70154), Saccharomyces cerevisiae (NP_012381), and Arabidopsis thaliana (BAF93445). (B) Sequence alignment of human VPS26 and orthologs. The alignment was produced as described above. β-strands are represented as arrows and 310 helices as cylinders labeled with η; invariant residues are colored red on a blue background and conserved residues are colored blue. The residue numbering shown above the alignment corresponds to human VPS26. Secondary structure was assigned using Stride and β-strands were numbered as reported before (Shi et al., 2006). Residues involved in inter-subunit contacts are indicated under the alignment; the types of interactions are specified in the text box. Residues of VPS26 that reduce the affinity for VPS35 when mutated are indicated. Residues mutated in this work are marked with yellow stars (R249A). Residues mutated in other studies are marked with black stars (I235S+M236D, G238P, Δ238–246 GG, 238–246 polyS) (Shi et al., 2006); VPS26B mutations/VPS26A equivalent residues I233D/I235D, I233D+M234N/I235D+M236N, P245S+R247S/P247S+R249S, L197S+R199E/L199S+R201E, R240S+G241A+E242S/K242S+G243A+E242S (Collins et al., 2008). Residues of VPS26 that reduce the affinity for DMT1-II when mutated are indicated with yellow stars (V168N and F287A). The accession numbers of the aligned sequences are: Homo sapiens (NP_004887), Gallus gallus (XP_421577), Danio rerio (NP_957201), Drosophila melanogaster (AAF45679), Caenorhabditis elegans (CCQ25652), Saccharomyces cerevisiae (EDV12710), and Arabidopsis thaliana (NP_200165).
(A) Spatial location of the VPS26 subunit and the N-terminus of the VPS26 and VPS29 polypeptides within the SAXS derived model. (B) Normalized P(r) distance distribution function of the full retromer complex VPS26-VPS29-VPS35, the VPS29-VPS35 subcomplex, and the N-terminal MBP-tagged constructs [MBP-VPS26]-VPS29-VPS35 and VPS26-[MBP-VPS29]-VPS35. (C-E) Multiphase envelopes calculated by MONSA in surface representation indicating the calculated phase for (C) the VPS26 (slate) subunit, (D) MBP(red)-tagged VPS26, and (E) MBP(red)-tagged VPS29.
(A) Sequence alignment of human SNX3 and orthologs. The alignment was produced as described for Figure S2. α-helices and 310 helices (η) are represented by cylinders, and β-strands as arrows; invariant residues are colored red on a green background and conserved residues are colored blue. Residues involved in inter-subunit contacts are indicated under the alignment; the types of interactions are specified in the text box. Residues of SNX3 mutated in this study are indicated as yellows stars (R9A+R10A+Y22A, E30A+D32A and H132A+P133A+L134A) and residues of SNX3 that reduce the affinity for retromer when mutated in other studies are indicated as black stars (yeast mutations/equivalent human mutations: E37A+E39A+H41A/E30A+D32A+S34A) (Harrison et al., 2014). The residue numbering shown above the alignments corresponds to human SNX3. Green frames highlight the relevant residues for the SNX3-VPS26 interaction. The accession numbers of the aligned sequences are the following: Homo sapiens (NP_003786), Gallus gallus (NP_001006408), Danio rerio (NP_001032183), Drosophila melanogaster (AAF54838), Caenorhabditis elegans, (CAA22253) and Saccharomyces cerevisiae (NP_015002) (B) Sequence alignment of human SNX3 with other human sorting nexins. The alignment was generated with the multiple sequence and structure alignment server PROMALS3D, using the following PDB entries: SNX3 (this work), SNX1 (2I4K), SNX5 (3HPC) and SNX27 (4HAS). The accession numbers of the aligned sequences are the following: SNX1 (NP_003090), SNX2 (AAC17181), SNX5 (NP_055241), SNX6 (AAD27829) and SNX27 (XP_005245566). (C) Domain architecture of sorting nexins important for retromer function in mammals. The following domains are depicted: PX (phox), BAR (Bin-Amphiphysin-Rvs), FERM (band4.1-ezrin-radixin-moesin), and PDZ (postsynaptic density 95-discs large-zonula occludens).
(A) Crystal packing of the VPS26-VPS35N-SNX3 complex with a zoomed-in view of the asymmetric unit complex and a symmetry-related VPS26 molecule (VPS26-sym). The C-terminal end of VPS26, SAEQPM (residues 321–327) followed by the extra sequence GLVPRG derived from cloning is highlighted in red. No electron density was observed for residues 303–320 of VPS26, indicating flexibility. We assume that the observed C terminus of VPS26 corresponds to a symmetry-related VPS26 molecule according to the observed distance (17 Å red dashed line). (B–E) The sequence of the VPS26 with the C-terminal cloning tag and three different VPS26-DMT1-II fusion constructs used in this work are indicated on the upper part of each figure. In blue is the sequence of VPS26, in red the sequence derived from the cloning, in orange the sequence of DMT1-II, in bold the residues that could be modeled in the crystal structures, and in magenta the methionines that were labeled with selenium. The following electron density maps of VPS26 or DMT1-II are superimposed on the corresponding crystal structures: in (B) and (C), omit difference electron density map (Fo-Fc) in green contoured at 2.0 σ and in (B) anomalous map of Se in magenta contoured at 4 σ; in (D), electron density map (2Fo-Fc) in blue contoured at 1.5 σ and the anomalous map of Se in magenta contoured at 4 σ; and in (E), electron density map (2Fo-Fc) in blue contoured at 1.5 σ and the anomalous map of Se in magenta contoured at 6 σ. The color code for the atoms in is as follows: slate blue (carbon of VPS26), pink (carbon of VPS35), green (carbon of SNX3), orange (carbon of DMT1), dark red (carbon of extra residues from cloning), red (oxygen), dark blue (nitrogen), and yellow (sulphur or selenium).
(A) ITC thermograms for the titration of retromer (VPS26-VPS29-VPS35), in the presence or absence of DMT1550–568 peptide, with WT SNX3 or SNX3 mutants. (B) ITC thermograms for the titration of DMT1550–568 peptide or the mutated peptide DMT1(mut) 550–568 (with the mutations Y555A and L557A) with retromer, SNX3, or the preformed complex of retromer with SNX3 or retromer(mut) (trimeric core having VPS26 F287A and V168N substitutions). (C) Thermodynamic binding parameters from ITC measurements in (A) and (B). (D) Immunofluorescence microscopy of endogenous SNX3 in HeLa cells and in a SNX3 CRISPR-KO cell line, using antibody to SNX3. Bar: 5 μm. (E) Immunofluorescence microscopy of endogenous VPS26 in WT and SNX3 CRISPR-KO HeLa cell lines. Bar: 10 μm. Magnifications of the boxed regions are shown at right. (F) Recovery of endogenous VPS26 recruitment to membranes in stable cell lines rescued with different SNX3 GFP constructs. Bars: 10 μm. Magnifications of the boxed regions are shown at right. Notice recruitment of VPS26 comparable to HeLa WT cells in recovery with full-length GFP-SNX3, but not SNX3 with truncation of the whole N-terminus (ΔN), or with point mutations (RRY, HPL and ED) that prevent interaction with VPS26, VPS35 and/or the DMT1 recycling signal.
(A) Immunofluorescence microcopy colocalization of ectopically expressed DMT1-II-HA with the endogenous early-endosomal marker EEA1 in WT and SNX3-KO HeLa cells. (B) Colocalization of DMT1-II-HA and EEA1 in the same SNX3-GFP rescue cell lines described in the legend to Figure S6. Bars: 10 μm. Magnifications of the boxed regions are shown at right.
Movie S3. Recycling motif of DMT1-II bound to VPS26. (Related to Figure 5).
Movie S4. Speculative model for the SNX-BAR-retromer coat on a tubular membrane. (Related to Figure 7).