Abstract
Lariat RNAs formed as by-products of splicing are quickly degraded by the RNA debranching enzyme 1 (DBR1), leading to their turnover. Null dbr1 mutants in both animals and plants are embryo lethal, but the mechanism underlying the lethality remains unclear. Here we characterized a weak mutant allele of DBR1 in Arabidopsis, dbr1-2, and showed that a global increase in lariat RNAs was unexpectedly accompanied by a genome-wide reduction in miRNA accumulation. The dbr1-2 mutation had no effects on expression of miRNA biogenesis genes or primary miRNAs (pri-miRNAs), but the association of pri-miRNAs with the DCL1/HYL1 dicing complex was impaired. Lariat RNAs were associated with the DCL1/HYL1 dicing complex in vivo and competitively inhibited the binding of HYL1 with pri-miRNA. Consistent with the impacts of lariat RNAs on miRNA biogenesis, over-expression of lariat RNAs reduced miRNA accumulation. Lariat RNAs localized in nuclear bodies, and partially co-localize with HYL1, and both DCL1 and HYL1 were mis-localized in dbr1-2. Together with our findings that nearly four hundred lariat RNAs exist in wild type plants and that these lariat RNAs also associate with the DCL1/HYL1 dicing complex in vivo, we thus propose that lariat RNAs, as decoys, inhibit miRNA processing, suggesting a hitherto unknown layer of regulation in miRNA biogenesis.
Author Summary
It is known that lariat RNAs formed during pre-mRNA splicing are debranched by DBR1 (RNA debranching enzyme 1). Loss of function of DBR1 causes embryo lethality in both animals and plants. In animals, some debranched lariat RNAs could be further processed into mirtron miRNAs, a class of nonconventional miRNAs that bypass the microprocessor for their biogenesis. However, no mirtron has been functionally validated in plants, and how the accumulation of lariat RNA in dbr1 results in embryo lethality remains unclear. Here, we show that DBR1 is necessary for the regulation of genome-wide miRNA biogenesis in plants. By investigating the correlation between lariat RNA accumulation and miRNA processing, we showed that the DBR1-mediated lariat RNA debranching process provides a safeguard role for the binding of the dicing complex with miRNA precursors. As both the DBR1-mediated lariat RNA debranching process and miRNA biogenesis are common features in higher eukaryotes, the finding that lariat RNAs sequester the dicing complex in plants may have a broad implications for the non-coding RNA field.
Introduction
In higher eukaryotes, splicing of mRNA precursors (pre-mRNA), a critical step for gene expression, comprises two catalytic steps [1]. In the first step, the 5’ splice site is cleaved and concurrently the 5’ end of the intron is joined to the branch nucleotide by forming a phosphodiester bond. This results in the production of a 5’ exon and a 3’exon-containing lariat precursor. In the second step, the lariat precursor is cleaved at the 3’ splice site and the two exons are ligated to produce the mRNA. The excised intronic lariat RNA is linearized for degradation by an RNA debranching enzyme (DBR1) [2].
DBR1 was originally identified from budding yeast (Saccharomyces cerevisiae) in a study aimed at identifying host cellular factors involved in transposition of the Ty1 retrotransposon [3]. The dbr1Δ mutant of S. cerevisiae has reduced Ty1 transposition frequency and also shows increased accumulation of lariat RNAs. DBR1 is not essential for cell viability in S. cerevisiae. However, the dbr1Δ mutant in fission yeast (Saccharomyces pombe) exhibits severe growth defects [2]. Moreover, both the dbr1 mutant in Arabidopsis [4] and mice [5] are embryo lethal. DBR1 is a single copy gene in eukaryotes [6], and homologues of DBR1 isolated from plants or animals can complement the S. pombe dbr1 mutant [7], indicating that DBR1 function is conserved.
MicroRNAs (miRNAs) are a class of 21-24nt small RNAs generated from endogenous stem-loop transcripts. miRNA biogenesis begins with the processing of primary miRNAs (pri-miRNAs), which contain a hairpin structure that is cleaved twice by the dicing complex, which is mainly composed of the nuclear RNase III enzyme Dicer-Like 1 (DCL1) [8] and an RNA binding protein HYL1 [9], yielding mature miRNAs. DCL1 and HYL1, together with pri-miRNAs, are co-localized in nuclear dicing bodies [10,11], and HYL1 facilitates the binding of DCL1 to enhance the efficiency and accuracy of miRNA processing [12,13].
Previous findings showed that DBR1 is required for mirtron miRNA biogenesis in animals [14, 15], where it is independent of cleavage by the microprocessor. The existence of mirtron miRNAs in embryos might contribute the embryo lethality of the null dbr1 mutant in animals [14]. However, no mirtron miRNA has been functionally validated in plants [16]. In addition, earlier studies showed that DBR1 is required for intronic snoRNAs biogenesis in yeast [17, 18], but plant snoRNAs are originated from either an intronic or a non-intronic context [19, 20], indicating that the involvement of DBR1 in snoRNA biogenesis is not sufficient to explain the important role of DBR1 during embryo development. Here, we report that lariat RNAs inhibit global miRNA biogenesis in Arabidopsis, possibly explaining the detrimental effects of lariat over-accumulation. We provide evidence that the over-accumulation of lariat RNAs caused genome-wide reduction of miRNAs in Arabidopsis, and show that lariat RNAs associated with the DCL1/HYL1 dicing complex to compete with pri-miRNAs. Moreover, we demonstrated that hundreds of lariat RNAs exist in wild type plants, suggesting that these lariat RNAs might be biologically relevant. Given the observations of lariat RNAs in embryonic stem cells [21] and evidence for lariat RNAs acting as decoys of RNA binding proteins [22], we thus propose that lariat RNAs function as decoys of the dicing complex to maintain a proper processing of pri-miRNAs.
Results
DBR1 is required for miRNA accumulation in Arabidopsis
In an ethyl methanesulfonate mutagenesis screen aimed at isolating mutants compromised in miRNA biogenesis using dcl1-14 as a parental line [23], we isolated a mutant with pleiotropic developmental phenotypes, which included curly and serrated leaves, increased branching, short stature, and reduced fertility (Fig 1A). Whole genome sequencing identified a G-to-A mutation in the coding region of DBR1, which encodes an RNA debranching enzyme (DBR1) (S1A and S1B Fig). This mutation caused the conversion of a glycine, located within the LRL motif required for substrate binding (S1A and S1B Fig) [24], a region highly conserved in DBR1 in S. pombe, animals, and plants, to an arginine (S1A and S1B Fig). When DBR1 genomic fragments driven by the native promoter and fused to GFP, RFP, or Flag, respectively, were introduced into this mutant, the phenotypes were completely rescued (Fig 1A). We identified a T-DNA insertion allele in DBR1 only in a heterozygous state and crossed it to the homozygous mutant isolated from our genetic screen. Approximately half (83/178) of the F1 plants exhibited mutant phenotypes (S1C Fig). Therefore, the mutant is an allele of DBR1. Both a previously isolated loss-of-function allele of DBR1 [4] (here renamed dbr1-1) and the T-DNA insertion line (here named dbr1-3) are embryo lethal. The abundance of DBR1 was unaffected in our weak mutant allele (here named dbr1-2) (S1D Fig).
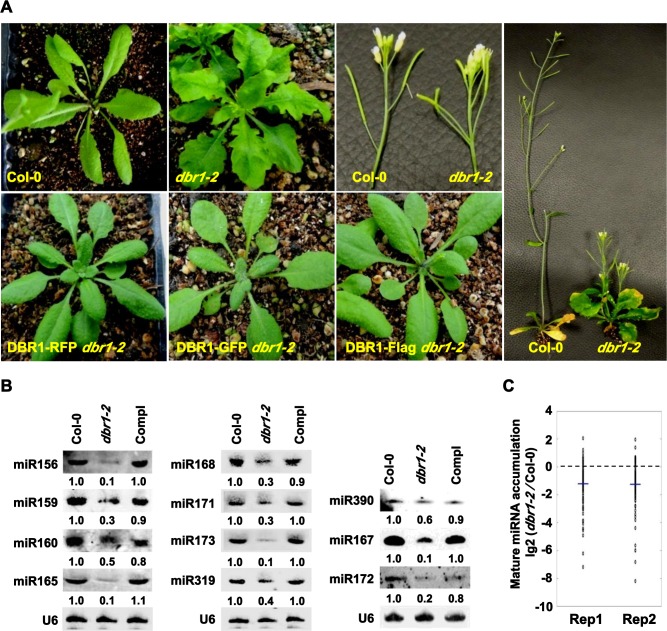
Fig 1. DBR1 is required for miRNA accumulation.
(A) Morphological phenotypes of Col-0 and dbr1-2 plants. DBR1-GFP dbr1-2, DBR1-RFP dbr1-2, and DBR1-Flag dbr1-2 represent rescued lines of dbr1-2 with different protein fusion constructs driven by the native DBR1 promoter. (B) miRNA northern blot analysis in Col-0, dbr1-2, and a dbr1-2 transgenic line containing the pDBR1::DBR1-RFP transgene (Compl). The numbers indicate the relative abundance of miRNAs among the three genotypes and represent the mean of three repeats (P < 0.05). U6 was used as a loading control. (C) Deep sequencing analysis of miRNAs in Col-0 and dbr1-2. Small RNA libraries were generated from inflorescences of two biological replicates. The normalized abundances of miRNAs were calculated as reads per ten million (RPTM), and log2-transformed ratios of dbr1-2/Col-0 were plotted. Each circle represents one miRNA. Thick lines indicate mean values. Rep1 and Rep2 denote replicate 1 and replicate 2, respectively.
Considering that the dbr1-2 mutant resembles mutants that are defective in miRNA accumulation, we examined miRNA accumulation in dbr1-2 by northern blot analysis. Compared to those in Col-0, the levels of tested miRNAs were reduced in dbr1-2 (Fig 1B). pDBR1::DBR1-RFP (Compl) fully restored the levels of these miRNAs (Fig 1B). To determine whether DBR1 is required for global miRNA accumulation, we compared mature miRNA levels in dbr1-2 and Col-0 by small RNA deep sequencing analysis. Results from two replicates confirmed a genome-wide reduction of miRNA levels in dbr1-2 (Fig 1C and S1 Table). Compared to many canonical miRNAs reduced in hyl1 [25, 26], our northern blot assay (Fig 1B) and deep sequencing analysis (S1 Table) showed that most HYL1-dependent miRNAs from 32 canonical miRNA families, such as miR156, miR159, and miR160, were also obviously reduced in dbr1-2 (S1 Table), indicating that DBR1 and HYL1 have overlapping functions in miRNA biogenesis. Due to the potential feedback regulation between lariat RNA debranching and pre-mRNA splicing, we wondered whether the effects of DBR1 on miRNA biogenesis was dependent on the possible function of DBR1 on splicing. Moreover, recent studies showed that many MIR genes contain introns and those MIR genes with introns usually stimulate biogenesis of miRNAs originating from such intron-containing precursors [27, 28]. By analyzing the genomic structure of 54 MIR genes with reduced miRNAs in dbr1-2, we showed that only 8 MIR genes contain introns (S1 Table), and that among more than 15 intronic miRNAs in Arabidopsis, only 4 intronic miRNAs were reduced in dbr1-2 (S1 Table), suggesting that the involvement of DBR1 in miRNA biogenesis might be unrelated to the possible function of DBR1 in pre-mRNA splicing, and thus that the effects of DBR1 on miRNA biogenesis are independent of the properties of MIR genes. We thus concluded that DBR1 is required for miRNA accumulation in plants.
Pri-miRNAs over-accumulate in the dbr1-2 mutant
To exclude the possibility that the reduced miRNA levels observed in dbr1-2 were due to altered expression of the miRNA pathway components, we performed quantitative RT-PCR (qRT-PCR) to determine transcript levels of genes that have been shown to act in miRNA biogenesis [8, 9, 29, 30]. As shown in S2A Fig, the expression levels of these genes were comparable to those in Col-0; Western blot analysis further confirmed that the protein levels of those key components in the miRNA pathway in dbr1-2 were comparable to those in Col-0 (S2B Fig). These results indicate that the dbr1-2 mutation had no effects on the expression of the miRNA pathway components.
The reduced miRNA accumulation in dbr1-2 could be caused by reduced pri-miRNA levels. To test whether it was the case, we examined the levels of pri-miRNAs in Col-0 and dbr1-2 plants. qRT-PCR analyses showed that the levels of the nine pri-miRNAs tested were increased by 2- to 4-fold in dbr1-2 (Fig 2A), a fold-change similar to that found in hyl1 (Fig 2A). The observation of increased levels of pri-miRNA prompted us to examine whether DBR1 plays a role in the transcription of miRNA genes (MIR). Pol II is responsible for MIR transcription in animals and plants [31–33]. Therefore, we first examined Pol II occupancy at promoters of MIR genes by chromatin immunoprecipitation (ChIP), using an antibody against the second largest subunit of Pol II (NRPB2). Compared to the no antibody control (S2C Fig), Pol II occupancy at MIR was comparable to that in dbr1-2 (S2D Fig). To further exclude the possibility that the over-accumulation of pri-miRNAs in dbr1-2 was due to increased transcription of MIR, we examined the effect of dbr1-2 on the expression of a GUS reporter gene driven by either the MIR172a promoter (MIR172a::GUS) or the MIR390b promoter (MIR390b::GUS). The transgenic lines MIR172a::GUS and MIR390b::GUS were separately crossed with dbr1-2 and progeny plants homozygous for both transgene and dbr1-2 were analyzed. Both GUS activity (S2E Fig) and the levels of GUS transcripts (S2F Fig) in dbr1-2 were comparable to those in Col-0, while the levels of pri-miR172a and pri-miR390b were increased in the dbr1-2 lines (S2F Fig). Taken together, these results indicate that DBR1 is not required for MIR transcription.
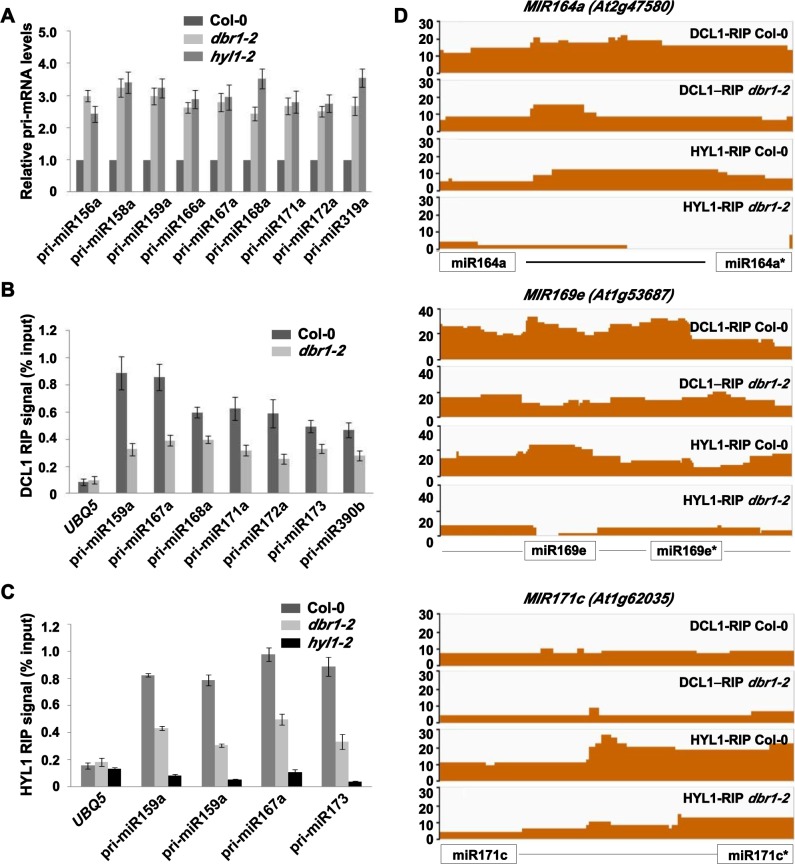
Fig 2. Pri-miRNA binding to DCL1 and HYL1 is reduced in dbr1-2.
(A) Detection of pri-miRNAs in Col-0, dbr1-2 and hyl1-2 by qRT-PCR. UBQ5 was the loading control. Standard deviations were calculated from three biological replicates. (B and C) Association between pri-miRNAs and the DCL1/HYL1 complex by RIP analysis. RNA was immunoprecipitated from inflorescences or seedlings of Col-0 and dbr1-2 using DCL1 or HYL1 antibodies, respectively. The amount of pri-miRNAs was determined by qRT-PCR and normalized to the input. hyl1-2 was used as the negative control for HYL1 antibody. UBQ5 was used as a negative control. Error bars show SE calculated from three biological replicates. (D) The occupancy of DCL1 and HYL1 at MIR164a, MIR169e, and MIR171c. The coverage regions are shown as normalized peaks. The x axis indicates the relative position of miRNA and miRNA* location. The y axis indicates normalized peaks from the genomic region. Reads counts were normalized to tag per 10 million (TP10M) to adjust for sequencing depth differences of the two RIP-seq libraries.
The binding of pri-miRNA with the dicing complex is impaired in dbr1-2
That pri-miRNAs over-accumulated and the levels of mature miRNAs were reduced in dbr1-2 suggested that miRNA processing might be impaired. We then investigated whether the association of pri-miRNAs with the dicing complex was compromised in dbr1-2 using RNA immunoprecipitation (RIP) experiments. We performed RIP assays, using DCL1 antibody, with Col-0 and dbr1-2 inflorescences, and using HYL1 antibody with Col-0, dbr1-2, and hyl1-2 seedlings. Pri-miRNAs were detected by qRT-PCR from immunoprecipitated RNAs. The amounts of the pri-miRNAs bound by both DCL1 (Fig 2B) and HYL1 (Fig 2C) were significantly reduced in the dbr1-2 background. To determine whether DBR1 is required for genome-wide pri-miRNA binding with the dicing complex, we compared the levels of DCL1- or HYL1-immunoprecipitated miRNA precursors (pri-miRNA) from Col-0 and dbr1-2 by RNA sequencing analysis. We found that most reads were predominantly mapped to the whole region of MIR genes. However because the boundaries of most pri-miRNAs are not clear, it was not feasible to calculate the abundance of pri-miRNAs, and thus we only counted reads uniquely mapped to pre-miRNA regions. We found that pre-miRNAs were overall enriched in both DCL1- and HYL1-immunoprecipitated samples in Col-0 (Fig 2D and S2 Table). Furthermore, enrichment of most pre-miRNAs with detectable RIP-seq abundances (more than 5 RPKM in either the dbr1-2 or Col-0 libraries), in both DCL1- and HYL1-immunoprecipitated samples, were reduced in dbr1-2 (Fig 2D and S2 Table). These results indicate that the association of pri-miRNAs with the dicing complex was disrupted in dbr1-2, which leads to reduced miRNA processing.
Characterization of lariat RNAs in Arabidopsis
Because DBR1 strongly binds to lariat RNAs [34], we hypothesized that DBR1 might directly act in the binding of pri-miRNAs with the dicing complex. To test this hypothesis, we performed a RIP assay using DBR1 antibodies, but found that DBR1 was not associated with pri-miRNA in vivo (S3A Fig). However, DBR1 is obviously associated with lariat RNAs (S3A Fig), which is consistent with previous studies [34]. In addition, a Co-IP assay showed no association between DBR1 and the miRNA biogenesis machinery (S3B Fig). These results therefore make it unlikely that DBR1 itself is a component of the dicing complex during miRNA processing.
Loss-of-function of DBR1 causes global accumulation of lariat RNAs in yeast [2]. To investigate the relationship between the accumulation of lariat RNAs and miRNA biogenesis, we tested genome-wide how many lariat RNAs accumulate in dbr1-2. As RNase R specifically degrades linear RNAs, while keeping the loop portion of a lariat RNA intact [35], our strategy was to globally compare total RNAs in dbr1-2 and Col-0, with or without RNase R-treatments. Therefore, we performed RNA-seq after constructing libraries of ribosomal RNA (rRNA)-depleted RNAs, with or without RNase R-treatments (S3C Fig). By selecting uniquely mapped intronic reads, 1560 intronic RNAs were identified as potential lariat RNAs in dbr1-2 (Fig 3A and S3 Table). After RNase R treatment, most of them (1534) were still detectable (Fig 3A and S3 Table), indicating that these lariat RNAs exist as stable circular forms. Unexpectedly, approximately 23% (360/1534) of these lariat RNAs also exhibited significant expression (> = 5 RPKM) in wild type plants (S3 Table), suggesting that these lariat RNAs naturally escaped debranching, a phenomenon recently shown in human cells [21].
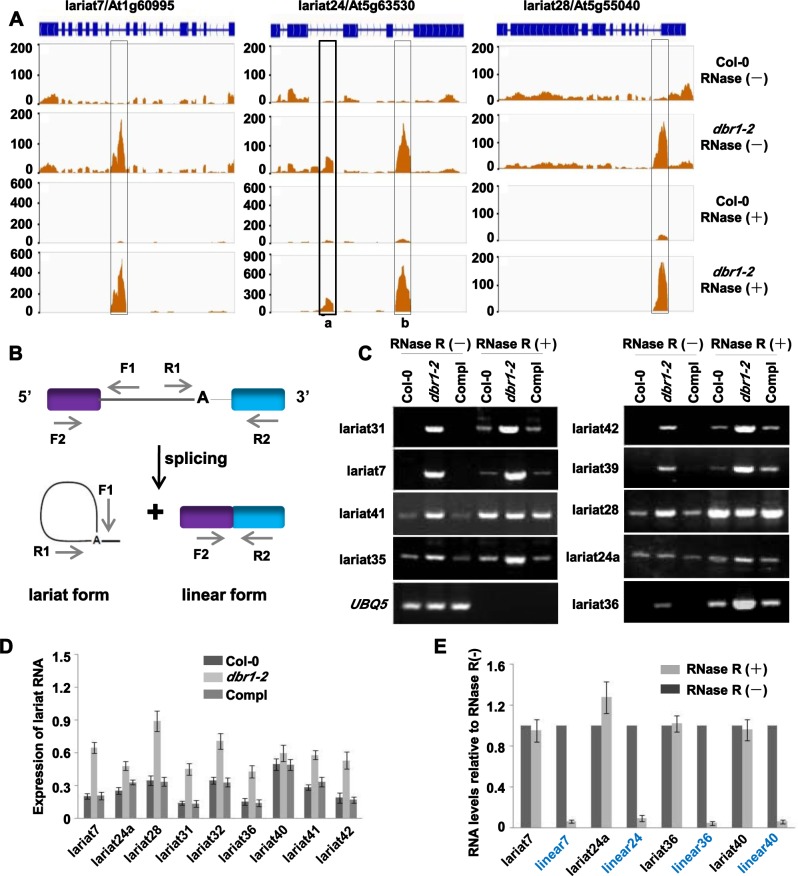
Fig 3. Characterization of lariat RNAs in plants.
(A) Three examples of identified lariat RNAs. Normalized peaks of lariat RNAs are highlighted by black rectangles. The exon is boxed in blue, and the intron is a line. The X-axis indicates the chromosomal location. The Y-axis indicates normalized peaks from the genomic region. Reads counts were normalized to tag per 10 million (TP10M) to adjust for differences in sequencing depth of the two RNA-seq libraries. (B) Schematic of divergent primers for lariat RNAs and convergent primers for linear mRNAs. The purple and blue boxes indicate exons; A represents the branch point. (C) Validation of lariat RNAs by RT-PCR with the divergent PCR primer pairs shown in (B) in Col-0, dbr1-2, and Compl (pDBR1::DBR1-RFP dbr1-2) with or without RNase R treatment. UBQ5 was used as the loading control. (D) Validation of lariat RNAs by qRT-PCR using total RNA of Col-0, dbr1-2, and Compl (pDBR1::DBR1-RFP dbr1-2). The amount of lariat RNAs was normalized to UBQ5. Error bars show SE calculated from three biological replicates. (E) qRT-PCR showing resistance of lariat RNAs to RNase R digestion. Linear mRNAs (blue font indicated) are positive controls for RNase R treatments. The amount of RNAs after RNase R treatment was normalized to the RNase R-untreated sample. Error bars show SE calculated from three biological replicates.
To validate lariat RNAs, we performed RT-PCR using sets of divergent primers (Fig 3B). As shown in Fig 3C and 3D, most tested lariat RNAs obviously over-accumulated in dbr1-2. Some lariat RNAs, such as lariat24a, lariat28, lariat35, and lariat41, were easily detected in Col-0 (Fig 3C), further confirming that these lariat RNAs naturally escaped the debranching activity of DBR1. Further qRT-PCR analyses of RNase R-untreated samples confirmed the results of RT-PCR (Fig 3D). To test whether the lariat RNAs were circular, we performed RT-PCR using RNase R-treated RNAs as templates, and showed that all tested lariat RNAs were circular (Fig 3C). To determine whether RNase R treatments were complete, we performed qRT-PCR analyses using both RNase R-untreated and RNase R-treated samples, and showed that lariat RNAs were stably detectable in both samples, while the corresponding linear mRNAs were almost eliminated by RNase R treatments (Fig 3E). Sanger sequencing further confirmed that lariat RNAs are circular (S3D Fig). Taken together, these results suggest that both lariat RNAs over-accumulated in dbr1-2, and lariat RNAs naturally present in wild type plants, might play certain roles in biological processes.
Lariat RNAs affect the binding of the DCL1/HYL1 complex with pri-miRNAs
To investigate whether lariat RNAs accumulated in dbr1-2 were correlated with reduced binding of the dicing complex to pri-miRNAs, we first investigated whether these lariat RNAs were associated with the DCL1/HYL1 complex in vivo. Lariat RNAs were present in both DCL1- and HYL1-immunoprecipitates in dbr1-2 (Fig 4A and 4B). Notably, compared to that of negative control, UBQ5, and other lariat RNAs hardly detected in Col-0 (lariat7, lariat31, lariat36), several tested lariat RNAs naturally present in Col-0, such as lariat24a, lariat28, lariat32, and lariat40, were also bound by both DCL1 and HYL1 (Fig 4A and 4B), indicating that lariat RNAs that naturally escaped debranching could be bound by the DCL1/HYL1 dicing complex.
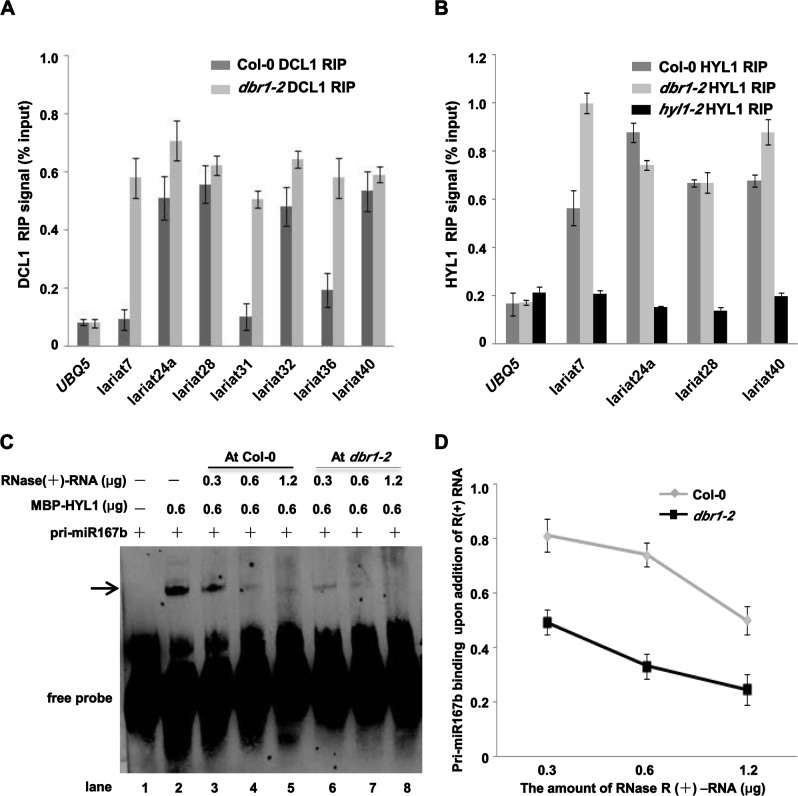
Fig 4. Lariat RNAs competitively inhibit DCL1/HYL1 binding to pri-miRNAs.
(A) Lariat RNAs associate with DCL1 using a RIP assay performed as in Fig 2B. Immunoprecipitated RNAs were analyzed by qRT-PCR with divergent primers to detect the indicated lariat RNAs. UBQ5 was used as the loading and negative control. Error bars show SE calculated from three biological replicates. (B) Lariat RNAs associate with HYL1 using a RIP assay performed as in Fig 2C. qRT-PCR was performed according to (A). hyl1-2 was used as the negative control. UBQ5 was used as the loading and negative control. Error bars show SE calculated from three biological replicates. (C) R-EMSA to determine HYL1 binding to pri-miR167b in the presence of circular RNAs from Col-0 or dbr1-2 plants. Recombinant MBP-HYL1-D1D2 (MBP-HYL1) was incubated with a 5’_biotin_labeled pri-miR167b probe after addition of different amounts of circular RNAs isolated from Col-0 or dbr1-2 inflorescences, respectively. The arrow indicates the HYL1-pri-miR167b complex. (D) Hybridization intensities were quantified and normalized to the controls (lane 2 in C), and are shown in the line graph. Bars represent the average normalized intensity of three biological replicates.
Due to the observations that lariat RNAs are circular and that most circular RNAs accumulated in dbr1-2 could be lariat-derived, we then investigated whether the association of the DCL1/HYL1 complex with lariat RNAs affected binding of the DCL1/HYL1 complex to pri-miRNAs. We performed RNA electrophoretic mobility shift assays (R-EMSA) with Col-0 and dbr1-2, to compare the binding capacity of recombinant HYL1 (S4A Fig) with biotin-labeled pri-miR167b in the presence or absence of lariat RNAs. Consistent with a previous study [36], HYL1 specifically bound pri-miR167b as indicated by the arrow (S4B Fig), while other unrelated recombinant proteins MBP and GST (S4A Fig) did not bind pri-miR167b (S4B Fig). Notably, DBR1 recombinant protein showed no binding with pri-miR167b (S4B Fig), further supporting that DBR1 itself is not directly required for the binding of pri-miRNA with the dicing complex. To investigate whether lariat RNAs regulate HYL1 binding with pri-miRNA, we performed competition assays. Notably, due to the limitations of synthesizing lariat RNA in vitro in our conditions, and because lariat RNAs should be the most abundant population of circular RNAs in dbr1 mutants, we used RNase R-digested RNAs (R(+)-RNA) to perform the competition assays. Increasing amounts (0.3 to 1.2 μg) of cold R(+)-RNA were added to the binding reaction mixture containing HYL1 and labeled pri-miR167b. As shown in Fig 4C and 4D, the signal corresponding to the HYL1-pri-miR167b complex was decreased proportionally to the amount of cold circular RNA added from dbr1-2. Unexpectedly, the addition of cold circular RNA from Col-0 in the binding reaction also produced moderate competition effects (Fig 4C and 4D), while linear single-stranded RNA of GAPDH had minor competition effects on HYL1 binding (S4C Fig).
Considering that the function of DBR1 is highly conserved in eukaryotes, we speculated that lariat RNAs from other species might play similar roles in binding the dicing complex. To test this idea, we examined the effects of circular RNAs from a fission yeast dbr1Δ strain on the binding capacity of HYL1 with pri-miR167b. Compared to the binding in the control (S4D Fig), circular RNAs from the yeast dbr1 mutant greatly attenuated the binding of HYL1 (S4D and S4E Fig). Taken together, these results suggest that circular RNAs, most likely lariat RNAs, could play a regulatory role in binding of the dicing complex to pri-miRNAs.
Over-expression of lariat RNAs causes reduced miRNA accumulation
Since it seems that lariat RNAs compete for HYL1/DCL1 binding to pri-miRNAs, thus reducing their processing and then the accumulation of miRNAs, we hypothesized that any method that changed lariat RNAs levels would have an impact on miRNA accumulation. To test this, we generated over-expression (OE) lines of lariat RNA and assessed miRNA accumulation. Although how some lariat RNAs escape debranching in wild type plants remains unknown, we hypothesized that over-expressing the corresponding genomic DNA should lead to increased levels of intron-derived lariat RNAs. Here, we selected lariat41, which is detectable in wild type plants (Fig 3C and S3 Table), to investigate whether over-expression of lariat RNA would affect miRNA accumulation. We generated more than 10 independent transgenic plants over-expressing the genomic DNA of At5g37720 (lariat41-OE) or the CDS of At5g37720 (local41-OE) (Fig 5A). Only lariat41-OE transgenic plants exhibited pleiotropic phenotypes (Fig 5B), which were reminiscent of mutants deficient in miRNA accumulation. Both RT-PCR and western blot analysis showed comparable mRNA and protein levels of At5g37720 in local41-OE and lariat41-OE plants (Fig 5C). As expected, lariat41 was significantly increased in lariat41-OE lines but not in local41-OE lines (Fig 5C), while lariat28, an unrelated lariat RNA, was equal among the three genotypes (Fig 5D). Importantly, we showed that levels of miR159 and miR167 were reduced in lariat41-OE but not in local41-OE plants (Fig 5E), indicating that increased lariat41 levels were anti-correlated with accumulation of miRNAs. To further support this anti-correlation, we transiently expressed lariat42 in tobacco leaves (S5 Fig) with a split YFP separated by lariat42-originated intron sequences (S5A Fig). The YFP signal was detected, indicating that the lariat42-originated intron was properly spliced (S5B Fig). RT-PCR analysis showed that lariat42 accumulated in the infiltrated leaves but not in control leaves (S5C Fig). Northern blot analysis showed that miR167 was reduced in leaves over-expressing lariat42 (lariat42-OE). Taken together, these results indicate that lariat RNA accumulation is negatively correlated with miRNA levels.
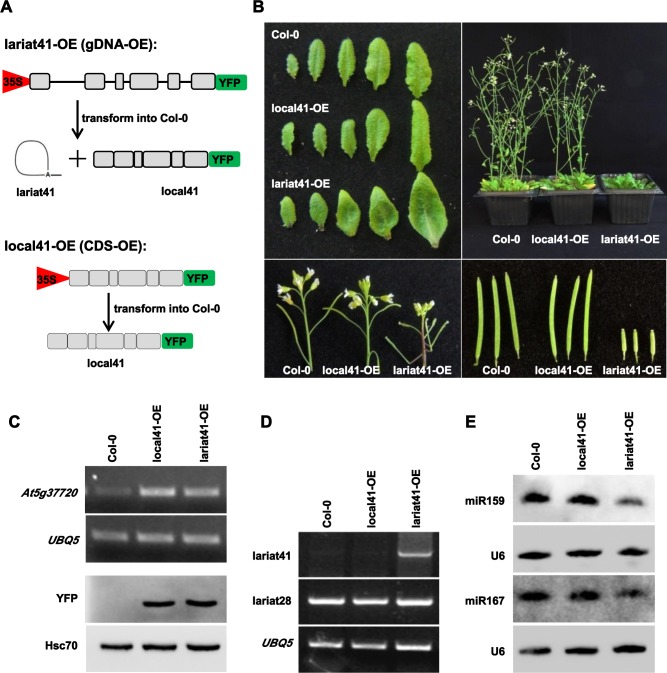
Fig 5. Over-expression of lariat41 caused reduced miRNA accumulation.
(A) Strategy to over-express lariat41 in Arabidopsis. Full length genomic sequence (indicated as lariat41-OE/gDNA-OE) or full length coding sequence (indicated as local41-OE/CDS-OE) was driven by the 35S promoter and fused by an YFP tag, and introduced into plants. Grey boxes indicate exons, and lines indicate the intron. (B) Morphological phenotypes of transgenic plants over-expressing lariat 41 (lariat41-OE) and the corresponding gene of lariat41 (local41-OE). Curly leaves, late flowering, altered phyllotaxy, and reduced fertility were shown in lariat41-OE. More than 10 independent transgenic lines were obtained for each. (C) RT-PCR and western blot analysis to detect expression of the corresponding gene of lariat41. Total RNA from inflorescences of a representative T3 transgenic line for each was used for cDNA synthesis, and At5g37720 was amplified to indicate the mRNA level of the corresponding gene, UBQ5 as the loading control. In similar, YFP was detected using total protein from inflorescences by western blot, Hsc70 as the loading control. Two additional biological replicates were performed, and similar results were obtained. (D) RT-PCR to detect expression of lariats. Total RNA from inflorescences of a representative T3 transgenic line for each was used for cDNA synthesis, and lariat41 was amplified, lariat28 was used as the negative control, and UBQ5 as the loading control. Two additional biological replicates were performed, and similar results were obtained. (E) miR159 and miR167 northern blot analysis in Col-0, local41-OE, and lariat41-OE. U6 was used as a loading control. Another biological replicate was performed, and similar results were obtained.
Lariat RNAs are recruited into nuclear bodies and partially co-localize with HYL1
Pre-mRNA splicing occurs in the nucleus and lariat RNAs are byproducts of splicing, so we reasoned that DBR1 might localize in the nucleus. Transient expression of DBR1-RFP in tobacco as well as transgenic Arabidopsis plants expressing DBR1-GFP showed that DBR1 was distributed in both the nucleus and cytoplasm (Fig 6A), which is consistent with the finding that DBR1 is a nucleocytoplasmic shuttling protein in humans [37]. The DBR1-2 mutant protein did not affect the distribution of DBR1 (S6A Fig).
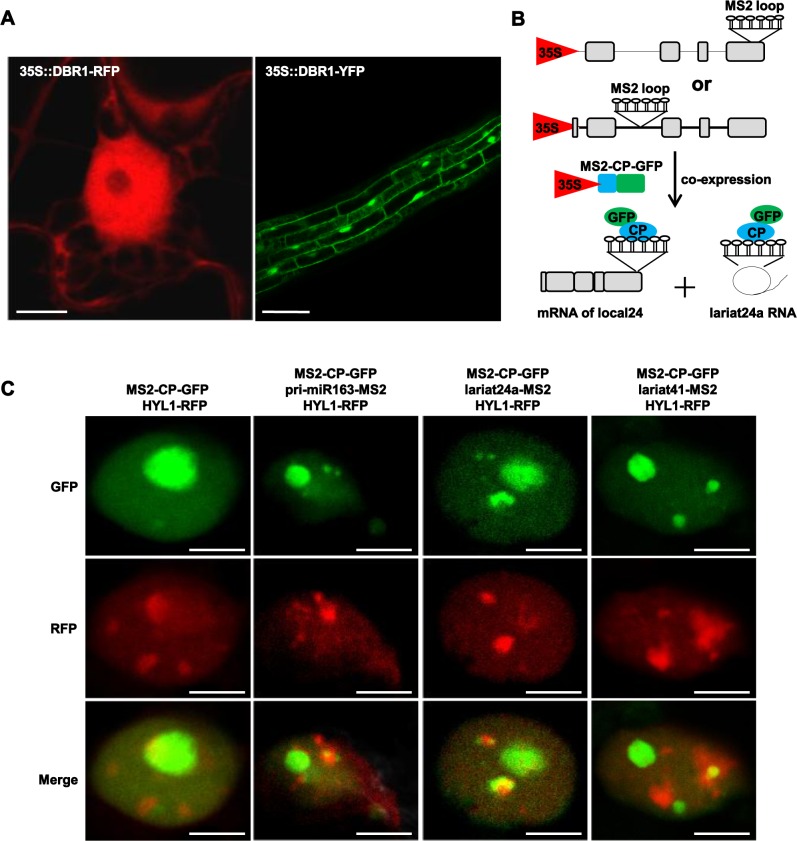
Fig 6. Subcellular localization of DBR1 and lariat RNA.
(A) Subcellular localization of Arabidopsis DBR1. 35S::DBR1-RFP was transiently expression in tobacco leaves and RFP signal was observed after 48 hr. Roots from 35S::DBR1-GFP transgenic plants were observed under the GFP channel. (B) Strategy to visualize lariat RNAs in live cells. The MS2 sequence (indicated as stem-loops) was inserted into a lariat24a-located intron. A co-expressed GFP-tagged MS2-CP protein was used to visualize lariat RNAs. Grey boxes indicate exons, and lines indicate the intron. (C) Genomic DNA of lariat24a and lariat41 was fused to 6XMS2 repeats and co-infiltrated into tobacco leaves with MS2-CP-GFP and HYL1-RFP. The genomic DNA of pri-miR163 was used as the positive control. Scale bars = 10 μm.
Because processing of pri-miRNAs occurs in the nucleus, we hypothesized that nuclear lariat RNAs might be co-localized with components of the DCL1/HYL1 complex. To test this idea, we modified the MS2 system [38] to visualize endogenous lariat RNAs in plants (Fig 6B and S6B Fig). Six copies of the binding site for the RNA-binding MS2 coat proteins (MS2-CP) were inserted into the lariat24a- or lariat41-generating intronic regions of At5g63530 (Fig 6B) and At5g37720 (S6B Fig), respectively. Expression of MS2-CP fused to GFP carrying an NLS signal (MS2-CP-GFP) enables the visualization of lariat24a and lariat41 tagged with MS2-binding sites. We co-transformed tobacco with different combinations of plasmids harboring MS2-CP-GFP, HYL1-RFP or lariat24a-MS2 and lariat41-MS2 under the control of the 35S promoter. To differentiate the subcellular localization of lariat RNA and the mRNA of the corresponding gene, we co-transformed a plasmid harboring the full length genomic DNA fused to 6X MS2 at the end of the last exon of the lariat24a corresponding gene, under control of the 35S promoter. As a positive control for dicing bodies, we co-transformed a plasmid harboring pri-miR163-MS2 under control of the 35S promoter. We analyzed the subcellular localization of MS2-CP-GFP by fluorescence microscopy. MS2-CP-GFP uniformly accumulated in one big dot in the nucleus (Fig 6C), while HYL1-RFP accumulated in small dots in addition to the big dot in the nucleus in some nuclei (Fig 6C). Notably, in many nuclei HYL1-RFP was also distributed more evenly without any dots (S6C Fig), a localization consistent with a previous study [39]. As expected, when the plasmid of pri-miR163-MS2 was introduced, we observed small bodies surrounding the big dot in some nuclei (Fig 6C), and these nuclear bodies were partially co-localized with HYL1-nuclear bodies (Fig 6C), consistent with previous localization studies [10, 11]. Intriguingly, lariat24a and lariat41 also accumulated in small nuclear bodies surrounding the big dot in some nuclei (Fig 6C and S6D and S6E Fig), and similar to pri-miR163, both lariat24a and lariat41 partially co-localized with HYL1-nuclear bodies (Fig 6C and S6D and S6E Fig). As a negative control, we showed that the mRNA of the lariat24a corresponding gene was uniformly distributed in all examined nuclei (S6C Fig). Collectively, our data indicate that lariat RNAs are partially recruited into nuclear bodies.
Localization of the dicing complex in the nucleus was impaired in dbr1-2
Several mutants deficient in miRNA biogenesis exhibit abnormal patterns of dicing bodies [40–42], which might reflect a deficiency in the function of the dicing complex. To examine whether DBR1 affects localization of the dicing complex, we examined the effects of dbr1-2 on the sub-nuclear localization of DCL1 and HYL1. Progeny homozygous for both transgenes (DCL1-YFP and YFP-HYL1) and dbr1-2 were obtained. In Col-0, more than 85% of 1168 root cells harbored three or fewer dicing bodies in the nucleus and less than 10% of root cells harbored more than three dicing bodies (Fig 7A). In contrast, dbr1-2 had significantly more DCL1-containing nuclear bodies (Fig 7A); more than 40% of 1027 root cells contained four or more dicing bodies (Fig 7A). A similar observation was made for HYL1-containing dicing bodies (Fig 7B). Taken together, these results suggest that DBR1 is required for the proper sub-nuclear localization of the dicing complex.
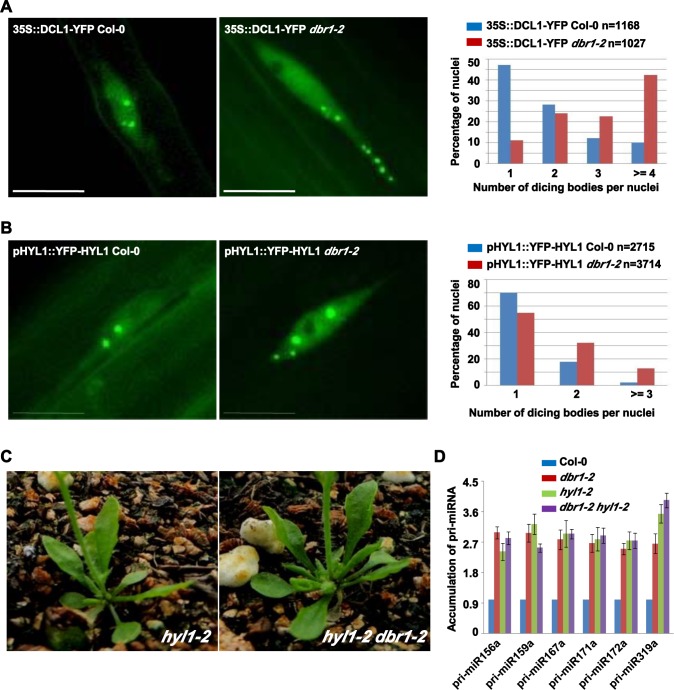
Fig 7. DCL1 and HYL1 are mis-localized in dbr1-2 mutants.
(A-B) Subnuclear localization of DCL1 (A) and HYL1 (B) in Col-0 and dbr1-2 root cells. Right panels show the percentage distribution of dicing bodies per cell. The X-axis represents the number of dicing bodies per cell, and the Y-axis represents the percentage of cells with the corresponding numbers. “n” represents the numbers of analyzed cells. (C) Morphological phenotypes of the hyl1-2 and the dbr1-2 hyl1-2 double mutant. Pictures were taken of 5-week-old plants. (D) Expression levels of indicated pri-miRNAs in Col-0, dbr1-2, hyl1-2, and dbr1-2 hyl1-2 plants by qRT-PCR. UBQ5 was used as an internal control and for normalization of the data. Standard deviations were calculated from three technical replicates. The results shown were reproduced with three biological replicates. Scale bars = 10 μm.
To further dissect the genetic relationship between lariat RNA debranching and pri-miRNA processing, we crossed dbr1-2 to hyl1-2. The double mutant was morphologically similar to hyl1-2 (Fig 7C), and expression levels of pri-miRNAs (Fig 7D) in hyl1-2 dbr1-2 were comparable to that in hyl1-2, suggesting that DBR1 and HYL1 act in miRNA biogenesis in the same genetic pathway. Taken together, these observations indicate the debranching process of lariat RNAs is tightly coupled with pri-miRNA processing, and thus that non-debranched lariat RNAs are conveniently accessible for the dicing complex to balance the binding of pri-miRNAs.
Discussion
Although a few plant miRNAs originate from intronic regions, it is becoming increasingly apparent that miRNA biogenesis has to be coordinately regulated by pre-mRNA splicing, since many mutants of genes involved in splicing exhibit defects in miRNA biogenesis [29, 43–45]. However, the mechanism by which pre-mRNA splicing affects miRNA biogenesis remains largely unexplored. Here we demonstrated that lariat RNAs, as so-called by-products of pre-mRNA splicing, play a negative role in regulation of miRNA homeostasis. Lariat RNAs act as a molecular sponge to sequester the dicing complex, and thus maintain a steady level of mature miRNAs. Therefore, our data provide new insights into how by-products of pre-mRNA splicing, other than the previously thought spliceosome itself, play a role in miRNA biogenesis.
Our results suggest that the debranching process of lariat RNAs contributes to the proper processing of pri-miRNA by balancing the binding of pri-miRNA to the DCL1/HYL1 complex. As we proposed, in wild type plants (Fig 8, upper panel), DBR1 is able to degrade most lariat RNAs, but some lariat RNAs naturally escape debranching, as also shown in human embryonic stem cells [21]. In Arabidopsis, we showed that these non-debranched lariat-derived circular RNAs act as a molecular sponge of the dicing complex, in which lariats play a potential competitive role to avoid excessive pri-miRNA processing by attracting the DCL1/HYL1 complex. In contrast, in dbr1-2 (Fig 8, lower panel), the over-accumulation of lariat RNAs disrupts the binding of the DCL1/HYL1 complex to pri-miRNA, and thus causes less miRNA production. In conclusion, our results uncover a new layer of miRNA biogenesis regulation by other RNA molecules, and establish the link between the splicing process and the generation of small RNAs that had not been conceived previously.
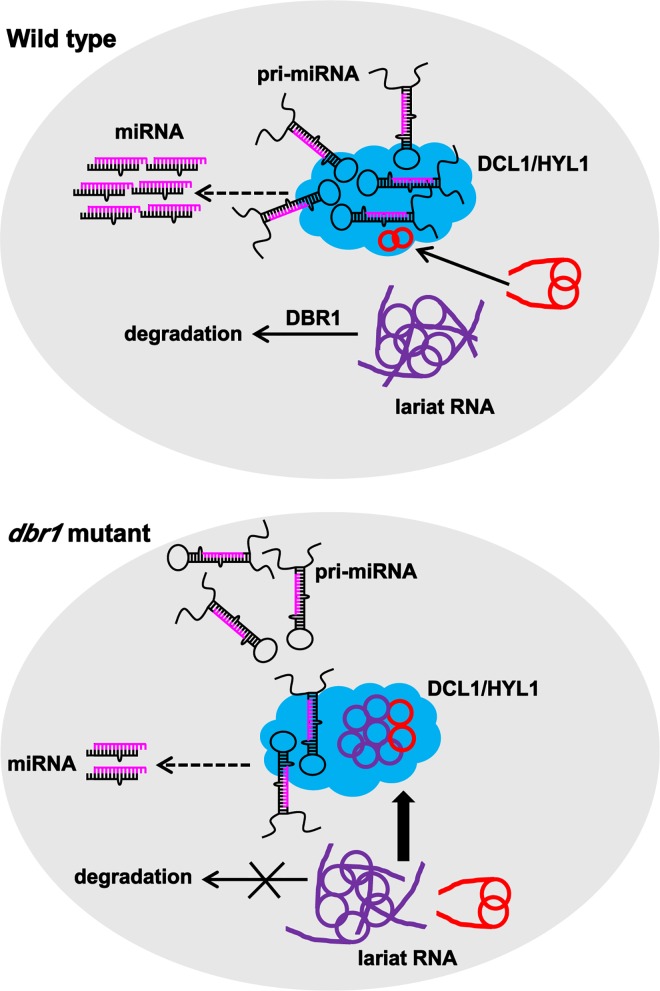
Fig 8. A Model for lariat RNA inhibits miRNA biogenesis in plants.
Pri-miRNAs are specifically recruited to the DCL1/HYL1 complex (blue indicated) for processing. In wild type plants, most lariat RNAs (indicated in purple) are quickly debranched by DBR1, only a small portion of lariat RNAs (indicated in red) naturally escapes debranching, also associates with the DCL1/HYL1 complex. In contrast, in dbr1-2, over-accumulated lariat RNAs might be recognized by the DCL1/HYL1 complex, and thus attenuate the association between the DCL1/HYL1 complex and pri-miRNAs, producing less miRNA.
It is well known that pri-miRNA binding to the dicing complex is an important step in miRNA biogenesis, but how the dicing complex differentiates pri-miRNAs from other structurally related RNAs is unclear. A previous study in plants showed that SINE RNAs, potentially with loop structures, compete with pri-miRNAs for HYL1 binding [46], and a study in human cells showed that the microprocessor recognizes the basal UG and apical UGU motifs of pri-miRNAs [47]. Interestingly, lariat RNAs contain the GU/UG motif from the 5’ splicing site next to the branch point, indicating that the GU/UG motif might be wrongly recognized by the DCL1/HYL1 complex in plants. In addition, recent studies in animals showed that DGCR8 binds other RNAs besides pri-miRNAs [48], and that Dicer globally binds to many loop RNAs [49], which thus sequesters Dicer to reduce miRNA accumulation [49], suggesting that binding of the miRNA biogenesis machinery could be regulated.
Indeed, together with the findings that lariat RNA acts as sources for both mirtron miRNA biogenesis in animals [14, 15] and siRNA biogenesis in yeast [50], our finding that lariat RNA inhibits global miRNA processing in plants implicates a widespread involvement of lariat RNA in small RNA biogenesis. In species such as yeast, which lack miRNAs, lariat RNAs are further processed into siRNA for gene silencing [50]. In contrast, in eukaryotes such as Drosophila and Arabidopsis, which are enriched in miRNAs, lariat RNA might have switched its role from siRNA biogenesis to miRNA biogenesis. Elucidation of additional genetic modifiers of lariat RNA binding and processing should provide more insights into how lariat RNA acts in small RNA biogenesis.
Materials and Methods
Plant materials, yeast strains, and antibodies
Arabidopsis thaliana Columbia (Col-0) is wild type. Seeds of dbr1-3 (SALK_047099), hyl1-2 (SALK_064863), and MIR390b::GUS (CS66477) were obtained from the Arabidopsis Biological Resources Center (ABRC, www.arabidopsis.org). 35S::DCL1-YFP seeds were generated in our laboratory. pHYL1::YFP-HYL1 seeds were obtained from Yuda Fang’s lab. Wild-type and dbr1 fission yeast strains were purchased from Bioneer (http://pombe.bioneer.co.kr). Anti-DCL1 (Agrisera, #AS122102), anti-HYL1 (Agrisera, #AS06136), anti-SE (Agrisera, #AS09532), anti-Hsc70 (Stressgen, SPA-018), anti-GFP (Covance, #MMS-118R), and anti-FLAG (Sigma, #F7425) antibodies were purchased. Anti-DBR1, anti-NRPB2, and anti-AGO1 antibodies were generated by our laboratory.
Plasmid construction
For construction of the DBR1-GFP, DBR1-RFP, DBR1-Flag, 35S::DBR1-RFP, and 35S::DBR1-YFP plasmids containing the endogenous promoter, DBR1 was amplified from Col-0 genomic DNA with the primer pair DBR1F1/R1, cloned into pENTR-D/TOPO, and then transferred into the plant expression destination vector pMDC107 to construct DBR1-GFP, into pMDC163 (GUS was replaced by mRFP) to construct DBR1-RFP, into pEarleyGate302 to construct DBR1-Flag, into pEarleyGate101 (HA-GFP was replaced by mRFP) to construct 35S::DBR1-RFP, and into pB7YWG2 to construct 35S::DBR1-YFP. To construct 35S::DBR1-Flag, the DBR1 CDS was obtained using the primer pair DBR1F2/R2 and cloned into the plant expression vector pCambia1306. To construct 35S::DBR1-2-YFP, the CDS sequences of DBR1 was amplified from dbr1-2 with primer pairs DBR1F2/R2 and cloned into pCambia2302. To construct DBR1-GST, the CDS sequences of DBR1 was amplified with primer pairs DBR1F3/R2 and cloned into pGEX2TK. To construct DCL1-YFP, HYL1-RFP, and HYL1-Flag, the CDS sequences of DCL1 and HYL1, respectively, were obtained using the primer pairs DCL1F1/R1 and HYL1F1/R1, cloned into pENTR-D/TOPO, and then transferred into the plant expression destination vectors pEarleyGate101, pEarleyGate101 (YFP-HA was replaced by mRFP), and pEarleyGate202, respectively. To construct SE-Flag, the CDS sequence of SE was obtained using the primer pair SEF1/R1, and cloned into the plant expression vector pCambia1306. To construct lariat41-OE, the full length genomic region of At5g37720 was amplified using primer pair lariat41F2/R2 and ligated to pENTR1A, and transferred into pB7YWG2. To construct local41-OE, the full length coding region of At5g37720 was amplified from Col-0 cDNA using primer pair lariat41F2/R2 and ligated to pENTR1A, and transferred into pB7YWG2. To construct lariat42-OE, the intron sequences of At5g43100 were amplified using primer pair lariat42F2/R2 and inserted between nYFP and cYFP, and transferred into pB7WG2. To transfer lariat24a and lariat41 into the MS2 system, we first constructed lariat24a-pENTR-D/TOPO and lariat41-pENTR1A by introducing full length genomic fragments of At5g63530 and At5g37720 into pENTR-D/TOPO and pENTR1A using the primer pairs gL24F/R and gL41F/R, respectively, and then the 6XMS2 loops were amplified with primers 6XMS2Loops_F1/R1 and 6XMS2Loops_F2/R2 from 35S::GW-6XMS2, then ligated into lariat24a-pENTR-D/TOPO by XmnI and lariat41-pENTR1A by SacI, respectively, then transferred into the plant expression vector pB7WG2. To construct the mRNA control of At5g63530 into the MS2 system (local24-MS2), the plasmid of lariat24a-pENTR-D/TOPO were introduced into 35S::GW-6XMS2 by LR reaction. To construct pri-miR163 into the MS2 system, we first constructed pri-miR163-pENTR-D/TOPO by introducing full length genomic fragments of MIR163 into pENTR-D/TOPO with the primer pair miR163-MS2-F/R, and then transferred into the plant expression vector 35S::GW-6XMS2. Primer sequences are listed in S4 Table.
Small RNA sequencing analysis
Illumina sequencing of small RNA libraries from inflorescences was performed by Genergy (Shanghai, China), and small RNA libraries were constructed according to Illumina's instructions using a TruSeq Small RNA Library Preparation kit. The small RNA sequencing profiles were analyzed as described [51]. Briefly, the sequencing reads were filtered to remove reads with more than 5 nucleotides with sequencing scores smaller than 20. Adaptor sequences were removed to extract small RNAs. Redundant sequences were eliminated, and unique small RNAs were counted. Unique small RNAs were mapped back to the A. thaliana reference genome using SOAP2 [52]; mature miRNAs and pre-miRNAs (downloaded from miRBase, http://www.mirbase.org/, v19) were found using BLASTN. Then, custom programs were used to calculate the normalized abundance (RPTM, Reads Per Ten Million sequencing tags) of mature miRNAs. miRNAs with at least 100 RPTM in either Col-0 or dbr1-2 were used to calculate log2 fold changes of their RPTM values. Deep sequencing datasets were deposited in the SRA database of National Center for Biotechnology Information with accession No. SRP062035.
miRNA northern blot
Total RNA was extracted using Trizol reagent from inflorescences. 5 μg small RNAs enriched by PEG8000 were separated by denaturing 15% (w/v) PAGE and transferred to a nylon membrane. 5’_biotin_labeled-oligo nucleotide sequences complementary to miRNA were synthesized as probes. Hybridization was performed using hybridization buffer (Ambion), and signals were detected using chemiluminescent nucleic acid detection module. U6 was used as a loading control. The probes are listed in S4 Table.
R-EMSA
R-EMSA was performed according to [36] with modifications. Biotin-labeled pri-miR167b was synthesized by in vitro transcription. Using an RNA EMSA Kit (Thermo Scientific), electrophoresis mobility shift assay experiments were performed in a 20 μl reaction system containing binding buffer, 5% (v/v) glycerol, 2 μg tRNA, 2 nM of biotin-labeled pri-miR67b transcripts, and purified MBP-HYL1_D1D2 (only two DsRBD domains). For the competition assay, total RNA treated by RNase R was added to the reaction at different concentrations. The reaction was incubated for 30 min, electrophoresed on a 6.5% (w/v) native PAGE gel, and transferred to a nylon membrane, then detected using a chemiluminescent nucleic acid detection module.
RIP (RNA IP)
Chromatin from inflorescences of Col-0 and dbr1-2 was immunoprecipitated with anti-DCL1 and anti-HYL1 antibodies. For the DBR1 RIP assay, chromatin from inflorescences of Col-0 was immunoprecipitated with anti-DBR1 antibodies. RNA recovered from the immunoprecipitates was used for cDNA synthesis with Oligo dT. The primer sets used for PCR are listed in the S4 Table. For RIP-seq analysis, recovered RNAs were used for library preparation with Illumina TruSeq Stranded Total RNA HT Sample Prep Kit (P/N15031048), and then subjected to deep sequencing with Illumina HiSeq 2000 at Genergy (Shanghai, China). The RIP-seq libraries were aligned to the genome with Cufflinks 2 [53]. The "bedtools genomecov" command of bedtools [51] was used to calculate genome coverage of RIP-seq libraries. Then, a custom program was used to calculate the RPKMs (Reads Per Kilo basepairs and per Million sequencing tags) of pre-miRNAs in miRBase (v21), using the genome coverage of RIP-seq libraries. Deep sequencing datasets were deposited in the SRA database of National Center for Biotechnology Information with accession No. SRP063916.
ChIP (Chromatin Immunoprecipitation)
ChIP was performed similar to RIP, the difference is that DNA not RNA is recovered. Pol II occupancy at miRNA loci was determined by ChIP using anti-RPB2 antibody and 2-week-old seedlings from Col-0 and dbr1-2, respectively. Immunoprecipitated DNA was quantified by qPCR relative to total input DNA. The results shown were consistent in three biological replicates. The primer sets used for the PCR are listed in S4 Table.
GUS Staining
Three week-old seedlings from transgenic plants in Col-0 and dbr1-2 backgrounds was fixed in 90% acetone for 2–3 h and then stained for 12 h in 50 mM sodium phosphate buffer, pH 7.0, containing 0.2% Triton X-100, 5 mM potassium ferrocyanide, 5mM potassium ferricyanide, and 1 mM X-Gluc, then washed in 70% ethanol three times. Images were taken with a Leica DFC295 stereoscope.
Pipeline for lariat RNA annotation by RNA-seq analysis
Total RNA isolated from Col-0 and dbr1-2 was first treated with a Ribo Zero kit (Epicenter) to obtain a ribosomal RNA-depleted RNA (ribo- RNA). Then ribo- RNA was incubated with or without RNase R (Epicentre) and then subjected to phenol:chloroform purification. Purified RNAs were further used for library preparation with Illumina TruSeq Stranded Total RNA Sample Prep Kit, and then subjected to deep sequencing with Illumina HiSeq 2000 at Genergy (Shanghai, China). Stranded RNA-seq reads were mapped to the Arabidopsis genome annotation (TAIR 10) with Cufflinks 2 [53]. CuffDiff in Cufflinks 2 was used to find deregulated genes. Genes with at least 5 RPKM in either dbr1-2 or Col-0 and q-values smaller than 0.05 were designated deregulated genes. The "bedtools genomecov" command of bedtools [53] was used to calculate the genome coverage of RNA-seq libraries. Then, a custom program was used to calculate the RPKMs (Reads Per Kilo basepairs and per Million sequencing tags) of introns of annotated genes in TAIR10, using the genome coverage of RNA-seq libraries. Introns that had at least 5 RPKM in dbr1-2 profiles and had multiple test corrected P-values of smaller than 0.05, calculated with edgeR [54], were designated as enriched introns. Deep sequencing datasets were deposited in the SRA database of National Center for Biotechnology Information with accession No. SRP062035.
Detection of lariat RNAs
Lariat RNAs across the branch site were detected by RT-PCR as described [35]. Total RNA with or without RNase R-treatment were used as templates. cDNA synthesis was carried out using SuperScript III (Invitrogen) with random hexamers. The reaction mixtures were incubated at 30°C for 10 min, at 42°C for 120 min, at 50°C for 30 min, at 60°C for 30 min, and at 99°C for 5 min. We then used divergent primer sets to detect lariat RNAs by PCR or qPCR. Primer sequences used are listed in S4 Table.
Recombinant protein expression and purification
The corresponding plasmids of DBR1-pGEX2TK and HYL1 (D1D2)-pMAL were introduced into the expression host BL21-CodonPlus (DE3). Cells was grown to an OD600 of 0.4–0.6, and expression of GST or MBP fused proteins were induced by addition of 0.1 mM IPTG and incubation at 16°C for overnight. Cells carrying the pGEX2TK or pMAL empty vectors were also grown and induced as controls. Cell pellets were resuspended in lysis buffer and purified according to the recommended protocols of Glutathione Sepharose4B resin (for GST) or the amylose resin (for MBP), respectively.
Transient expression in N. benthamiana and co-immunoprecipitation
35S::DCL1-YFP with 35S::DBR1-Flag, 35S::HYL1-Flag, and 35S::SE-Flag, respectively, were co-expressed in N. benthamiana. Leaves were ground in liquid nitrogen and homogenized in lysis buffer (50 mM Tris_HCl (pH 8.0), 150 mM NaCl, 0.2% Nonidet P-40, 2 mM DTT, 5% glycerol, proteinase inhibitor) and centrifuged for 15 min at 13200 rpm. The lysate was incubated with GFP-Trap agarose beads (Chromotek) for 2 h. The immune complexes were then washed with lysis buffer. Proteins retained on the beads were resolved on SDS-PAGE. Anti-GFP and anti-Flag antibodies were used to detect DCL1, and DBR1, HYL1 or SE, respectively, by using western blot analysis.
Data deposition
The data reported in this paper have been deposited in the SRA database of National Center for Biotechnology Information with accession No. SRP062035 for small RNA seq and RNA-seq, and accession No. SRP063916 for RIP-seq.
Supporting Information
(A) Partial amino acid sequence alignment of DBR1 from various species. DBR1 from Mouse (Mus musculus, NP_113580.2), Human (Homo sapiens, NP_057300.2), Fish (Danio rerio, NP_955947.1), Plant (Arabidopsis thaliana, NP_567881.1), Fly (Drosophila melanogaster, NP_996022.1), Worm (Caenorhabditis elegans, NP_491868.2), and fission yeast (Schizosaccharomyces pombe, NP_593470.2), were aligned using ClustalW. Identical residues are highlighted in yellow. Asterisk indicates the Gly that was mutated to Arg (G135R) in dbr1-2. (B) The protein structure of DBR1. The closed triangle indicates the T-DNA insertion site in dbr1-1 and dbr1-3. The arrow indicates the location of the mutation of glycine to arginine in dbr1-2. MPE indicates the metallophosphoesterase domain, LRL indicates the lariat recognition loop, and CTD indicates a novel C-terminal domain. (C) Allelic analysis of dbr1-2 and dbr1-3. A small field was shown for 3-week-old growing F1 progeny, and dbr1-2-like plants are highlighted by red circles. (D) Western blot to determine DBR1 levels in wild type (Col-0) and dbr1-2 using anti-DBR1 antibody. # indicates the endogenous DBR1 in Col-0 and dbr1-2, and * indicates DBR1-Flag and DBR1-GFP in 35S::DBR1-Flag or 35S::DBR1-GFP transgenic plants, respectively. Notably, the loading amount in Col-0 and dbr1-2 is half of that of two transgenic plants. Hsc70 served as a loading control.
(TIF)
(A) RT-qPCR analysis of mRNA levels of miRNA biogenesis genes. SE was calculated from three biological replicates. (B) Western blot to determine levels of DCL1, HYL1, SE, AGO1, and NRPB2 in Col-0 and dbr1-2. Hsc70 was the loading control. (C) and (D) ChIP performed with Col-0 (dark gray bars) or dbr1-2 (light gray bars) with anti-NRPB2 antibody. SE was calculated from three biological replicates. (E) Representative GUS staining images of MIR172a::GUS and MIR390b::GUS transgenic plants in Col-0 and dbr1-2 backgrounds. (F) RT-qPCR analysis of the transcript levels of GUS, pri-miR172a, and pri-miR390b in the indicated plants. SE was calculated from three biological replicates.
(TIF)
(A) RIP performed with dbr1-2 inflorescences using anti-DBR1 antibody (light gray bars) or no antibody (dark gray bars) as the negative control. RNA was immunoprecipitated from inflorescences of dbr1-2. The amount of pri-miRNAs was determined by RT-qPCR and normalized to the input. UBQ5 was used as a negative control. Lariat RNAs served as positive controls. Error bars show SE calculated from three biological replicates. (B) Co-immunoprecipitation between DCL1 and DBR1, SE, and HYL1. DCL1-YFP was co-expressed with DBR1-Flag, SE-Flag, or HYL1-Flag in N. benthamiana. HYL1-Flag and SE-Flag were used as positive controls. (C) Schematic flow of RNA-seq analysis. 5 μg total RNA were digested by Dnase I, and then RNA were removed ribosomal RNA using Ribo-Zero kit. The half amount of RNAs were treated by RNase R as R(+) RNA to remove linear RNAs, the other half amount of RNAs were used as R(-) RNA. Those RNA fragments with size larger than 100 nt were used for library construction and sequencing. (D) Chromas figures of partial sequences from lariat7 and lariat24a by Sanger sequencing. PCR products of lariat7 and lariat24a from Fig 3C were sequenced using the primer lariat7R and lariat24aR, respectively. The asterisk represents branch points “A” in the DNA sequence, here it was reversed to “T” after cDNA synthesis. The “GT” downstream of the branch point was indicated the 5’ splice site. The 5’ intron sequence means the partial intron sequences starting with the splice site “GT”. The 3’ intron sequence means the partial intron sequences ending with the branch point.
(TIF)
(A) Recombinant proteins of MBP, MBP-HYL1 (only the N-terminal harboring two dsRNA binding domains, D1D2 was fused to MBP), GST, GST-DBR1. (B) R-EMSA to determine if above recombinant proteins binding Biotin-labeled pri-miR167b. The arrow indicates the HYL1-pri-miR167b complex. (C) R-EMSA to determine if linear RNAs compete for HYL1 binding to pri-miR167b. Cold GAPDH single-stranded linear RNA transcribed by in vitro transcription using the T7 promoter was gradually increased in the reaction, and cold pri-miR167b with corresponding concentrations was used as the positive control. The arrow indicates the HYL1-pri-miR167b complex. (D) R-EMSA to determine if circular RNAs compete for HYL1 binding, using wild type or a dbr1 yeast strain. Experiments were performed as Fig 4C, except total RNAs from WT or dbr1 yeast cells. The arrow indicates the HYL1-pri-miR167b complex. (E) Hybridization intensities were quantified and normalized to the controls (lane 1 in D), and are shown in the line graph. Bars represent the average normalized intensity of three biological replicates.
(TIF)
(A) Strategy to over-express lariat42 in tobacco leaves. The intron sequence (indicated as lariat42-OE) was inserted into the split YFP and transiently expressed in tobacco leaves. (B) YFP signals were shown in tobacco leaves infiltrated by the above plasmid. Bright dots indicate YFP signals in nuclei. (C) RT-PCR analysis to detect the expression of lariat42. Total RNA from tobacco leaves infiltrated by the above plasmid or the blank control was used for cDNA synthesis, and lariat42 was amplified to indicate the level of lariat42, Actin7 as the loading control. (D) Northern blot analysis of miR167 in the control and lariat42-OE. U6 was used as a loading control.
(TIF)
(A) Subcellular localization of DBR1 and DBR1-2. 35S::DBR1-YFP and 35S::DBR1-2-YFP were transiently expressed in tobacco leaves and fluorescence signals were observed after 48 hr. (B) Strategy to visualize lariat RNAs in live cells. The MS2 sequence (indicated as stem-loops) was inserted into the lariat41-located intron. A co-expressed GFP-tagged MS2-CP protein was used to visualize lariat41. Grey boxes indicate exons, and lines indicate the intron. (C) HYL1-RFP alone or MS2-CP-GFP and local24-MS2 were transiently expressed in N. benthamiana leaves. Representative images were shown. (D-E) Three additional representative images of lariat24a (D) and two additional representative images of lariat41 (E) were shown. Scale bars = 10 μm.
(TIF)
More than 100 miRNAs with more than 100 RPTM were identified in small RNA sequencing libraries, and are listed with information of their ID, normalized reads (expression, RPTM), and log2 ratio of dbr1-2/Col-0, from two biological replicates. The first sheet lists 57 miRNAs that were significantly reduced in the replicate 1 of dbr1-2, the second sheet lists 56 miRNAs that were significantly reduced in the replicate 2 of dbr1-2, the third sheet lists 54 miRNAs that were commonly reduced in the two replicates of dbr1-2, the fourth sheet lists the information of genome structure of 54 commonly reduced miRNAs in dbr1-2, and the fifth sheet lists miRNAs co-regulated by both DBR1 and HYL1.
(XLSX)
There were 55 and 48 miRNA genes with log2 fold changes of smaller than -1 or larger than 1 identified as deregulated in binding to DCL1 and HYL1, respectively. Most of them, i.e. 47 in the DCL1- and 29 in the HYL1- RIP-seq profiles, had less occupancy of DCL1 and HYL1 in the dbr1-2 mutant. The genomic location, strand information, binding capacity (RPKM), and log2 ratios of dbr1-2/Col-0 from DCL1 and HYL1 RIP-seq were listed in the first and second sheets of the table.
(XLSX)
The genomic location, strand information, intron ID, and normalized abundance (RPKM) in different RNA-seq libraries, log fold change (logFC), log count per million sequencing reads (logCPM), P-value and False Discovery Rate (FDR) are listed. The intron ID is named according to the accession ID of the corresponding gene, followed by "I" and the intron sequential number. The first sheet lists all 1560 intron RNAs that have significantly different expression levels between Col-0 and dbr1-2 in RNase R (-) libraries, the second sheet lists the 1534 intron RNAs whose average abundance in dbr1-2_RNase R (+) libraries are ≥ 5 from 1560 introns, and the third sheet lists the 360 introns whose average abundance values in Col_RNase R (-) libraries are ≥ 5 RPKM from the 1534 introns on the second sheet.
(XLSX)
(DOC)
Acknowledgments
We thank the ABRC for sharing mutant and transgenic seeds, and appreciate Jorge Muschietti for providing plasmids for the MS2 system. We thank Profs. Sheila McCormick and Xuemei Chen for editing and helpful comments on the manuscript.
Data Availability
All RNA sequencing data files are available from the SRA database of National Center for Biotechnology Information with accession Nos. SRP062035 and SRP063916.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Ruskin B, Krainer AR, Maniatis T, Green MR (1984) Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38: 317–331. [DOI] [PubMed] [Google Scholar]
- 2.Nam K, Lee G, Trambley J, Devine SE, Boeke JD (1997) Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol 17: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karst SM, Rutz ML, Menees TM (2000) The yeast retrotransposons Ty1 and Ty3 require the RNA Lariat debranching enzyme, Dbr1p, for efficient accumulation of reverse transcripts. Biochem Biophys Res Commun 268: 112–117. 10.1006/bbrc.1999.2048 [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Hill K, Perry SE (2004) An Arabidopsis RNA lariat debranching enzyme is essential for embryogenesis. J Biol Chem 279: 1468–1473. 10.1074/jbc.M309106200 [DOI] [PubMed] [Google Scholar]
- 5.Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, et al. (2015) Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell 161: 762–773. 10.1016/j.cell.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooi SL, Dann C 3rd, Nam K, Leahy DJ, Damha MJ, et al. (2001) RNA lariat debranching enzyme. Methods Enzymol 342: 233–248. [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, Kim HC, Kim GM, Yang JM, Boeke JD, et al. (2000) Human RNA lariat debranching enzyme cDNA complements the phenotypes of Saccharomyces cerevisiae dbr1 and Schizosaccharomyces pombe dbr1 mutants. Nucleic Acids Res 28: 3666–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han MH, Goud S, Song L, Fedoroff N (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci U S A 101: 1093–1098. 10.1073/pnas.0307969100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823. 10.1016/j.cub.2007.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y (2007) Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol 48: 1243–1253. 10.1093/pcp/pcm099 [DOI] [PubMed] [Google Scholar]
- 12.Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212. 10.1261/rna.2146906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Han MH, Lesicka J, Fedoroff N (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci U S A 104: 5437–5442. 10.1073/pnas.0701061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100. 10.1016/j.cell.2007.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86. 10.1038/nature05983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng Y, Shao C (2012) Large-scale identification of mirtrons in Arabidopsis and rice. PLoS One 7: e31163 10.1371/journal.pone.0031163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petfalski E, Dandekar T, Henry Y, Tollervey D (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol 18: 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ooi SL, Samarsky DA, Fournier MJ, Boeke JD (1998) Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4: 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, et al. (1997) Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J 16: 5742–5751. 10.1093/emboj/16.18.5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, et al. (1999) Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant Mol Biol 39: 1091–1100. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, et al. (2013) Circular intronic long noncoding RNAs. Mol Cell 51: 792–806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 22.Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, et al. (2012) Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet 44: 1302–1309. 10.1038/ng.2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng B, Chen X, McCormick S (2011) The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of Cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046. 10.1105/tpc.111.083980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, et al. (2014) Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1. Nucleic Acids Res 42: 10845–10855. 10.1093/nar/gku725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351. 10.1016/j.cub.2004.01.035 [DOI] [PubMed] [Google Scholar]
- 26.Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WR, et al. (2009) Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res 37: 3083–3093. 10.1093/nar/gkp189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bielewicz D, Kalak M, Kalyna M, Windels D, Barta A, et al. (2013) Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep 14: 622–628. 10.1038/embor.2013.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhir A, Proudfoot NJ (2013) Feed backwards model for microRNA processing and splicing in plants. EMBO Rep 14: 581–582. 10.1038/embor.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Liu Z, Lu F, Dong A, Huang H (2006) SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850. 10.1111/j.1365-313X.2006.02835.x [DOI] [PubMed] [Google Scholar]
- 30.Yu B, Bi L, Zheng B, Ji L, Chevalier D, et al. (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105: 10073–10078. 10.1073/pnas.0804218105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, et al. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154. 10.1104/pp.105.062943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YJ, Zheng B, Yu Y, Won SY, Mo B, et al. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822. 10.1038/emboj.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquier A, Rosbash M (1986) RNA splicing and intron turnover are greatly diminished by a mutant yeast branch point. Proc Natl Acad Sci U S A 83: 5835–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, et al. (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34: e63 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Yan Q, Liu Y, Hong F, Sun Z, et al. (2013) Complementation of HYPONASTIC LEAVES1 by double-strand RNA-binding domains of DICER-LIKE1 in nuclear dicing bodies. Plant Physiol 163: 108–117. 10.1104/pp.113.219071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kataoka N, Dobashi I, Hagiwara M, Ohno M (2013) hDbr1 is a nucleocytoplasmic shuttling protein with a protein phosphatase-like motif essential for debranching activity. Sci Rep 3: 1090 10.1038/srep01090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haim L, Zipor G, Aronov S, Gerst JE (2007) A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4: 409–412. 10.1038/nmeth1040 [DOI] [PubMed] [Google Scholar]
- 39.Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, et al. (2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870. 10.1016/j.cell.2012.09.039 [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Song X, Gu L, Li X, Cao S, et al. (2013) NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25: 715–727. 10.1105/tpc.112.105882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu X, Shi Y, Li J, Xu L, Fang Y, et al. (2013) A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res 23: 645–657. 10.1038/cr.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiao Y, Shi J, Zhai Y, Hou Y, Ma W (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JW, Marshall DF, Echeverria M (2008) Intronic noncoding RNAs and splicing. Trends Plant Sci 13: 335–342. 10.1016/j.tplants.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 44.Ben Chaabane S, Liu R, Chinnusamy V, Kwon Y, Park JH, et al. (2013) STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res 41: 1984–1997. 10.1093/nar/gks1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szweykowska-Kulinska Z, Jarmolowski A, Vazquez F (2013) The crosstalk between plant microRNA biogenesis factors and the spliceosome. Plant Signal Behav 8: e26955 10.4161/psb.26955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pouch-Pelissier MN, Pelissier T, Elmayan T, Vaucheret H, Boko D, et al. (2008) SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet 4: e1000096 10.1371/journal.pgen.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, et al. (2015) Functional Anatomy of the Human Microprocessor. Cell 161: 1374–1387. 10.1016/j.cell.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 48.Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, et al. (2012) DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol 19: 760–766. 10.1038/nsmb.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, et al. (2014) A variety of dicer substrates in human and C. elegans. Cell 159: 1153–1167. 10.1016/j.cell.2014.10.040 [DOI] [PubMed] [Google Scholar]
- 50.Dumesic PA, Natarajan P, Chen C, Drinnenberg IA, Schiller BJ, et al. (2013) Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell 152: 957–968. 10.1016/j.cell.2013.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jagadeeswaran G, Nimmakayala P, Zheng Y, Gowdu K, Reddy UK, et al. (2012) Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genomics 13: 329 10.1186/1471-2164-13-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- 53.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Partial amino acid sequence alignment of DBR1 from various species. DBR1 from Mouse (Mus musculus, NP_113580.2), Human (Homo sapiens, NP_057300.2), Fish (Danio rerio, NP_955947.1), Plant (Arabidopsis thaliana, NP_567881.1), Fly (Drosophila melanogaster, NP_996022.1), Worm (Caenorhabditis elegans, NP_491868.2), and fission yeast (Schizosaccharomyces pombe, NP_593470.2), were aligned using ClustalW. Identical residues are highlighted in yellow. Asterisk indicates the Gly that was mutated to Arg (G135R) in dbr1-2. (B) The protein structure of DBR1. The closed triangle indicates the T-DNA insertion site in dbr1-1 and dbr1-3. The arrow indicates the location of the mutation of glycine to arginine in dbr1-2. MPE indicates the metallophosphoesterase domain, LRL indicates the lariat recognition loop, and CTD indicates a novel C-terminal domain. (C) Allelic analysis of dbr1-2 and dbr1-3. A small field was shown for 3-week-old growing F1 progeny, and dbr1-2-like plants are highlighted by red circles. (D) Western blot to determine DBR1 levels in wild type (Col-0) and dbr1-2 using anti-DBR1 antibody. # indicates the endogenous DBR1 in Col-0 and dbr1-2, and * indicates DBR1-Flag and DBR1-GFP in 35S::DBR1-Flag or 35S::DBR1-GFP transgenic plants, respectively. Notably, the loading amount in Col-0 and dbr1-2 is half of that of two transgenic plants. Hsc70 served as a loading control.
(TIF)
(A) RT-qPCR analysis of mRNA levels of miRNA biogenesis genes. SE was calculated from three biological replicates. (B) Western blot to determine levels of DCL1, HYL1, SE, AGO1, and NRPB2 in Col-0 and dbr1-2. Hsc70 was the loading control. (C) and (D) ChIP performed with Col-0 (dark gray bars) or dbr1-2 (light gray bars) with anti-NRPB2 antibody. SE was calculated from three biological replicates. (E) Representative GUS staining images of MIR172a::GUS and MIR390b::GUS transgenic plants in Col-0 and dbr1-2 backgrounds. (F) RT-qPCR analysis of the transcript levels of GUS, pri-miR172a, and pri-miR390b in the indicated plants. SE was calculated from three biological replicates.
(TIF)
(A) RIP performed with dbr1-2 inflorescences using anti-DBR1 antibody (light gray bars) or no antibody (dark gray bars) as the negative control. RNA was immunoprecipitated from inflorescences of dbr1-2. The amount of pri-miRNAs was determined by RT-qPCR and normalized to the input. UBQ5 was used as a negative control. Lariat RNAs served as positive controls. Error bars show SE calculated from three biological replicates. (B) Co-immunoprecipitation between DCL1 and DBR1, SE, and HYL1. DCL1-YFP was co-expressed with DBR1-Flag, SE-Flag, or HYL1-Flag in N. benthamiana. HYL1-Flag and SE-Flag were used as positive controls. (C) Schematic flow of RNA-seq analysis. 5 μg total RNA were digested by Dnase I, and then RNA were removed ribosomal RNA using Ribo-Zero kit. The half amount of RNAs were treated by RNase R as R(+) RNA to remove linear RNAs, the other half amount of RNAs were used as R(-) RNA. Those RNA fragments with size larger than 100 nt were used for library construction and sequencing. (D) Chromas figures of partial sequences from lariat7 and lariat24a by Sanger sequencing. PCR products of lariat7 and lariat24a from Fig 3C were sequenced using the primer lariat7R and lariat24aR, respectively. The asterisk represents branch points “A” in the DNA sequence, here it was reversed to “T” after cDNA synthesis. The “GT” downstream of the branch point was indicated the 5’ splice site. The 5’ intron sequence means the partial intron sequences starting with the splice site “GT”. The 3’ intron sequence means the partial intron sequences ending with the branch point.
(TIF)
(A) Recombinant proteins of MBP, MBP-HYL1 (only the N-terminal harboring two dsRNA binding domains, D1D2 was fused to MBP), GST, GST-DBR1. (B) R-EMSA to determine if above recombinant proteins binding Biotin-labeled pri-miR167b. The arrow indicates the HYL1-pri-miR167b complex. (C) R-EMSA to determine if linear RNAs compete for HYL1 binding to pri-miR167b. Cold GAPDH single-stranded linear RNA transcribed by in vitro transcription using the T7 promoter was gradually increased in the reaction, and cold pri-miR167b with corresponding concentrations was used as the positive control. The arrow indicates the HYL1-pri-miR167b complex. (D) R-EMSA to determine if circular RNAs compete for HYL1 binding, using wild type or a dbr1 yeast strain. Experiments were performed as Fig 4C, except total RNAs from WT or dbr1 yeast cells. The arrow indicates the HYL1-pri-miR167b complex. (E) Hybridization intensities were quantified and normalized to the controls (lane 1 in D), and are shown in the line graph. Bars represent the average normalized intensity of three biological replicates.
(TIF)
(A) Strategy to over-express lariat42 in tobacco leaves. The intron sequence (indicated as lariat42-OE) was inserted into the split YFP and transiently expressed in tobacco leaves. (B) YFP signals were shown in tobacco leaves infiltrated by the above plasmid. Bright dots indicate YFP signals in nuclei. (C) RT-PCR analysis to detect the expression of lariat42. Total RNA from tobacco leaves infiltrated by the above plasmid or the blank control was used for cDNA synthesis, and lariat42 was amplified to indicate the level of lariat42, Actin7 as the loading control. (D) Northern blot analysis of miR167 in the control and lariat42-OE. U6 was used as a loading control.
(TIF)
(A) Subcellular localization of DBR1 and DBR1-2. 35S::DBR1-YFP and 35S::DBR1-2-YFP were transiently expressed in tobacco leaves and fluorescence signals were observed after 48 hr. (B) Strategy to visualize lariat RNAs in live cells. The MS2 sequence (indicated as stem-loops) was inserted into the lariat41-located intron. A co-expressed GFP-tagged MS2-CP protein was used to visualize lariat41. Grey boxes indicate exons, and lines indicate the intron. (C) HYL1-RFP alone or MS2-CP-GFP and local24-MS2 were transiently expressed in N. benthamiana leaves. Representative images were shown. (D-E) Three additional representative images of lariat24a (D) and two additional representative images of lariat41 (E) were shown. Scale bars = 10 μm.
(TIF)
More than 100 miRNAs with more than 100 RPTM were identified in small RNA sequencing libraries, and are listed with information of their ID, normalized reads (expression, RPTM), and log2 ratio of dbr1-2/Col-0, from two biological replicates. The first sheet lists 57 miRNAs that were significantly reduced in the replicate 1 of dbr1-2, the second sheet lists 56 miRNAs that were significantly reduced in the replicate 2 of dbr1-2, the third sheet lists 54 miRNAs that were commonly reduced in the two replicates of dbr1-2, the fourth sheet lists the information of genome structure of 54 commonly reduced miRNAs in dbr1-2, and the fifth sheet lists miRNAs co-regulated by both DBR1 and HYL1.
(XLSX)
There were 55 and 48 miRNA genes with log2 fold changes of smaller than -1 or larger than 1 identified as deregulated in binding to DCL1 and HYL1, respectively. Most of them, i.e. 47 in the DCL1- and 29 in the HYL1- RIP-seq profiles, had less occupancy of DCL1 and HYL1 in the dbr1-2 mutant. The genomic location, strand information, binding capacity (RPKM), and log2 ratios of dbr1-2/Col-0 from DCL1 and HYL1 RIP-seq were listed in the first and second sheets of the table.
(XLSX)
The genomic location, strand information, intron ID, and normalized abundance (RPKM) in different RNA-seq libraries, log fold change (logFC), log count per million sequencing reads (logCPM), P-value and False Discovery Rate (FDR) are listed. The intron ID is named according to the accession ID of the corresponding gene, followed by "I" and the intron sequential number. The first sheet lists all 1560 intron RNAs that have significantly different expression levels between Col-0 and dbr1-2 in RNase R (-) libraries, the second sheet lists the 1534 intron RNAs whose average abundance in dbr1-2_RNase R (+) libraries are ≥ 5 from 1560 introns, and the third sheet lists the 360 introns whose average abundance values in Col_RNase R (-) libraries are ≥ 5 RPKM from the 1534 introns on the second sheet.
(XLSX)
(DOC)
Data Availability Statement
All RNA sequencing data files are available from the SRA database of National Center for Biotechnology Information with accession Nos. SRP062035 and SRP063916.