Abstract
Twinfilin and capping protein (CP) are highly conserved actin-binding proteins that regulate cytoskeletal dynamics in organisms from yeast to mammals. Twinfilin binds actin monomer, while CP binds the barbed end of the actin filament. Remarkably, twinfilin and CP also bind directly to each other, but the mechanism and role of this interaction in actin dynamics are not defined. Here, we found that the binding of twinfilin to CP does not affect the binding of either protein to actin. Furthermore, site-directed mutagenesis studies revealed that the CP-binding site resides in the conserved C-terminal tail region of twinfilin. The solution structure of the twinfilin–CP complex supports these conclusions. In vivo, twinfilin's binding to both CP and actin monomer was found to be necessary for twinfilin's role in actin assembly dynamics, based on genetic studies with mutants that have defined biochemical functions. Our results support a novel model for how sequential interactions between actin monomers, twinfilin, CP, and actin filaments promote cytoskeletal dynamics.
Keywords: actin, capping protein, twinfilin, yeast
Introduction
The actin cytoskeleton plays a central role in many cellular processes such as morphogenesis, cell migration, cytokinesis, and endocytosis. The structure and dynamics of the actin cytoskeleton are regulated by a large number of proteins that interact with actin filaments and/or monomeric actin (Pollard et al, 2000). Six classes of actin monomer-binding proteins are conserved in evolution from yeast to mammals. WASP/WAVE, verprolin/WIP, and Srv2/CAP are large, multifunctional proteins that link signaling pathways to the actin cytoskeleton (reviewed in Carlier et al, 1999; Higgs and Pollard, 2001; Takenawa and Miki, 2001; Hubberstey and Mottillo, 2002). The three other ubiquitous proteins, ADF/cofilin, profilin, and twinfilin, are smaller (<40 kDa) and they directly regulate the size, localization, and dynamics of the cellular actin monomer pool. ADF/cofilins contribute to actin dynamics by severing actin filaments and enhancing depolymerization from the pointed end, and thus provide new monomers to the cytoplasmic pool (reviewed in Bamburg et al, 1999; Carlier et al, 1999). Profilins catalyze the nucleotide exchange on actin monomers, and promote assembly at the barbed end of the filaments (Pantaloni and Carlier, 1993; Wolven et al, 2000; Lu and Pollard, 2001).
Twinfilin is a small actin monomer-binding protein originally identified from budding yeast (Goode et al, 1998). In cells, twinfilin shows diffuse cytoplasmic localization, but is also concentrated to cortical actin filament structures. Mutations of twinfilin in budding yeast and Drosophila result in an enlargement of cortical actin patches and defects in actin-dependent developmental processes, respectively (Goode et al, 1998; Vartiainen et al, 2000; Wahlström et al, 2001). Furthermore, deletion of twinfilin in budding yeast is synthetically lethal with certain cofilin and profilin mutations, suggesting that it is intimately involved in the regulation of actin dynamics in vivo (Goode et al, 1998; Wolven et al, 2000).
Twinfilin is composed of two ADF/cofilin-like domains (ADF-H domain), which are separated by a short (∼30-residue) linker region and followed by an ∼35-residue C-terminal tail region. Despite the structural homology to ADF/cofilins, twinfilins do not bind or depolymerize actin filaments, but are instead strictly actin monomer-binding proteins (reviewed in Palmgren et al, 2002). Twinfilin forms a stable, high-affinity complex with ADP-actin monomers, and inhibits their nucleotide exchange and filament assembly (Goode et al, 1998; Palmgren et al, 2001; Ojala et al, 2002). Twinfilin also interacts with phosphatidylinositol-diphosphate (PI(4,5)P2) in vitro, but the possible physiological role(s) of this interaction is not known (Palmgren et al, 2001).
In addition to actin monomers and phospholipids, twinfilin also interacts with capping protein (CP) (Palmgren et al, 2001; Vartiainen et al, 2003). CP is a conserved heterodimeric protein that regulates actin dynamics in a wide variety of organisms (Amatruda et al, 1990; Hug et al, 1995; Hopmann et al, 1996; Hart and Cooper, 1999; Yamashita et al, 2003). It binds to filament barbed ends with high affinity (Kd∼0.1–5 nM) and prevents loss and addition of actin subunits to the filament (Caldwell et al, 1989; Schafer et al, 1996; Kim et al, 2004). CP thus blocks a large fraction of actin filaments and directs actin polymerization to certain regions of the cell (DiNubile et al, 1995; Loisel et al, 1999). Although the interaction between twinfilin and CP is conserved in evolution from yeast to mammals, the structural mechanism and biological role(s) of this interaction are not known.
Here, we examined the mechanism and biological function of twinfilin–CP interaction by a combination of biochemical, genetic, and structural methods. We show that the CP-binding site resides in the C-terminal tail region of twinfilin. Although the interaction between twinfilin and CP does not affect either protein's interaction with actin, it is essential for twinfilin's correct localization and role in actin dynamics in yeast cells.
Results
Interaction with twinfilin does not affect the activity of CP
To investigate whether twinfilin influences the activity of CP, we carried out barbed-end seeded actin assembly and steady-state critical concentration assays, comparing the ability of CP to cap filament barbed ends in the presence and absence of twinfilin. As wild-type twinfilin prevents actin filament assembly through its actin monomer-binding activity, we first carried out this assay by using a specific yeast twinfilin mutant, Twf1–3p, that does not interact with actin monomers but binds CP with similar affinity to wild-type twinfilin (Palmgren et al, 2001). Yeast CP (Cap1/2p) efficiently inhibited barbed-end polymerization at low nM concentrations, and addition of a high concentration (11.6 μM) of Twf1–3p had no significant effect on CP's activity (Figure 1A and B). The equilibrium dissociation constants, obtained by kinetic modeling of the data (Wear et al, 2003), for CP binding to barbed ends (Kcap) in the absence and presence of Twf1–3p were 3.7±1.1 and 3.3±1.4 nM (mean±s.e., n=3), respectively. The addition of high concentrations of Twf1–3p (7.8 μM) also had no effect on CP's activity in steady-state assays (Figure 1C). Kcap values measured in the absence or presence of Twf1–3p were 4.3±1.1 nM (mean±s.e., n=4) and 5.2±2.4 nM (mean±s.e., n=3), respectively.
Figure 1.
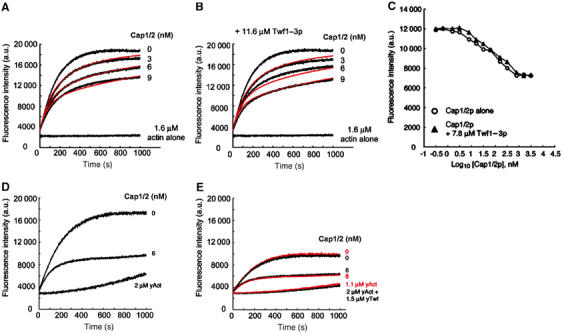
Twinfilin does not affect the filament barbed-end capping activity of capping protein. The effect of Cap1/2p on actin polymerization (1.6 μM actin, 5.2% pyrene labeled) seeded by the addition of spectrin-F-actin seeds was monitored by the increase in pyrene fluorescence, with [Cap1/2p] as indicated. The experiment was carried out in the absence (A) and presence (B) of 11.6 μM Twf1–3p. The barbed-end capping activity of Cap1/2p is not affected by the presence of Twf1–3p. Cap1/2p and Twf1–3p were incubated together for 45 min at 25°C prior to addition to the actin mixture. Red lines are the best fits of the data to a capping model, and yield a Kcap (equilibrium dissociation constant for capping the barbed end) of 3.7±1.06 nM in the absence and 3.3±1.4 nM in the presence of Twf1–3p. (C) Effect of Cap1/2p on the steady-state actin filament concentration (2 μM actin, 5% pyrene labelled), in the absence (open circles) or presence of 7.8 μM Twf1–3p (closed triangles). (D) Inhibition of 2 μM yeast actin polymerization from the barbed end (nucleated by addition of spectrin-actin seeds) by 6 nM Cap1/2p. The spontaneous nucleation and polymerization of the same concentration of yeast actin alone are shown for comparison. (E) The effect of 1.5 μM wild-type yeast twinfilin (in the presence of 0 or 6 nM Cap1/2p) on barbed-end polymerization (black time courses), and a similar set of assays where the amount of ATP-G-actin sequestered by wild-type twinfilin (with a Kd of 0.6 μM) is corrected for and performed with 1.1 μM actin alone (red time courses). The spontaneous nucleation and polymerization of 2 μM actin+1.5 μM twinfilin and that of 1.1 μM actin are also shown. The corrected time courses lie essentially on top of those performed in the presence of 1.5 μM twinfilin, indicating that wild-type twinfilin does not affect Cap1/2p's activity.
Similar results were obtained when the kinetic assembly assay was performed with wild-type yeast twinfilin and CP (Cap1/2p), and the data were corrected for the amount of actin monomers sequestered by twinfilin (Figure 1D and E). The amount of actin monomers sequestered by twinfilin in this assay was calculated from the equilibrium dissociation constant determined for the twinfilin–ATP-G-actin complex by a pyrene-actin fluorescence titration assay (Kd=0.6 μM; Supplementary Figure 1). The corrected time courses of actin polymerization lie essentially on top of those performed in the presence of 1.5 μM twinfilin, indicating that wild-type yeast twinfilin does not have detectable effect on Cap1/2p's activity (Figure 1E). Very similar results were also obtained when barbed-end assembly assays were carried out with ADP-actin or ATP-actin in the presence of mouse CP and mouse twinfilin-1 (Supplementary Figures 2 and 3). Together, these results demonstrate that twinfilin (either alone or in complex with ADP- or ATP-G-actin) does not modulate the activity of CP.
CP-binding site is located at the conserved C-terminal tail region of twinfilin
To map the CP-binding site on twinfilin, we mutated those evolutionarily conserved residues that, based on previous mutagenesis studies, are not involved in interactions with actin monomers (Palmgren et al, 2001; Paavilainen et al, 2002). Seven mutants were alanine substitutions of conserved residues (Twf1–4p (K68A, E70A); Twf1–5p (P139A, T141A); Twf1–6p (D143A, E144A, E145A); Twf1–7p (K152A, Q153A, Q154A); Twf1–8p (D203A, N206A, E207A); Twf1–9p (D221A, E222A); Twf1–11p (R328A, K329A, R330A, R331A)) and one mutant was a deletion of twinfilin's 10 C-terminal residues (Twf1–10p (ΔP323–T332)). Five of the mutant proteins (Twf1–5p, Twf1–6p, Twf1–7p, Twf1–10p, Twf1–11p) were soluble and showed similar stability to wild-type twinfilin. Native page electrophoresis assays demonstrated that Twf1–5p, Twf1–6p, and Twf1-7p interact normally with purified CP. It is important to note that, although Twf1–6p alone migrates as a smear on native gel, it shifts the mobility of CP when these proteins are mixed with each other before loading on the gel. In contrast, Twf1–10p and Twf1–11p do not display detectable binding to purified yeast CP (Figure 2A). This binding site is also conserved in mammalian proteins, because a deletion at the C-terminal region of mouse twinfilin-1 (ΔK332-D350) disrupts CP binding (Figure 2B). Interestingly, the CP-binding site appears to be entirely located at the C-terminal half of the twinfilin molecule. Twinfilin's isolated N-terminal domain (residues 1–142) does not interact with CP on a native gel-electrophoresis assay, whereas a construct that contains twinfilin's C-terminal ADF-H domain and tail region (residues 169–350) binds CP similarly to full-length twinfilin (Figure 2B).
Figure 2.
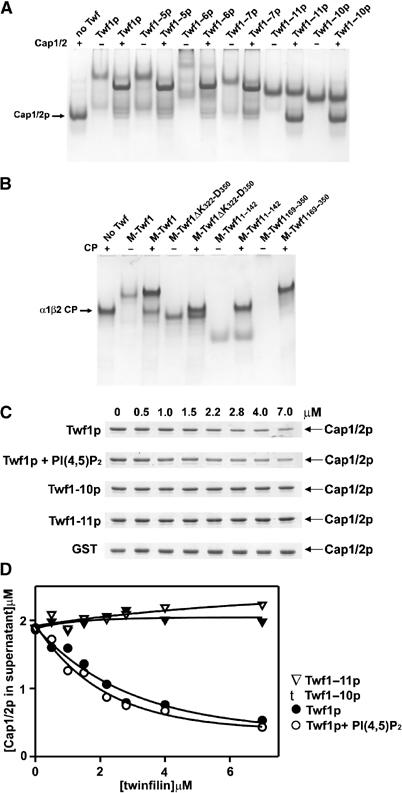
Identification of the capping protein-binding site on twinfilin. (A) Native gel-electrophoresis assay for determining the interaction between yeast twinfilin mutants and capping protein. Twf1–10p and Twf1–11p do not interact with Cap1/2p but instead migrate as separate bands on the gel, while wild-type twinfilin (Twf1p) and the remaining mutants (Twf1–5p, Twf1–6p, and Twf1–7p) form complexes with Cap1/2p and migrate at different mobilities than either protein alone. The concentrations of Cap1/2p and twinfilin were 5 and 4 μM, respectively. (B) The C-terminal region is essential for capping protein binding also in mouse twinfilin-1. Wild-type twinfilin-1 (Twf1) and the construct containing twinfilin's C-terminal half (M-Twf1169–350) interact with α1β2 CP, whereas twinfilin's N-terminal ADF-H domain (M-Twf11–142) and a protein carrying mutations at its C-terminal tail region (M-Twf1ΔK322-D350) do not bind CP in vitro. Note that, due to its high isoelectric point, the isolated M-Twf1169–350 alone does not enter the gel. The final concentrations of both mouse twinfilin and α1β2 CP were 4 μM. (C) The ability of wild-type yeast twinfilin, Twf1–10p, and Twf1–11p mutant proteins to deplete yeast CP from the supernatant in a pull-down assay under physiological ionic conditions. The CP concentration in this assay was 2 μM and the concentration of twinfilin was varied from 0 to 7 μM. Wild-type GST-twinfilin efficiently decreases the amount of capping protein from the supernatant, and this activity is not significantly inhibited by 25 μM PI(4,5)P2. In contrast, GST-Twf1–10p and GST-Twf1–11p mutant proteins or GST alone are not able to deplete CP from the supernatant. Note that the molecular weights of yeast Cap1p (α-subunit) and Cap2p (β-subunit) are 32.1 and 33.6 kDa, respectively, and thus the subunits are not resolved in the gel. (D) Graphical representation of CP binding activities of the wild-type twinfilin and twinfilin mutants as quantified from the intensities of the Coomassie-stained SDS gel bands by TINA software.
The native gel-electrophoresis assay was carried out at pH 8.5 under low ionic strength. Therefore, we next examined the twinfilin–CP interaction under physiological ionic conditions by a pull-down assay. Wild-type yeast glutathione-S-transferase (GST)-twinfilin efficiently decreased the concentration of yeast CP in the supernatant, and this activity was not significantly affected by 25 μM PI(4,5)P2 (Figure 2C and D). However, Twf1–10p and Twf1–11p mutant twinfilins did not display detectable CP binding in the pull-down assay (Figure 2C and D).
The two yeast twinfilin mutants Twf1–10p and Twf1–11p, unable to bind CP, were next analyzed for twinfilin's other known activities: actin monomer and PI(4,5)P2 binding. Fluorometric 7-chloro-4-nitrobenz-2-oxa-1,3-diazole (NBD)-actin assay showed that Twf1–10p and Twf1–11p bind ADP-actin monomers with an affinity similar to wild-type twinfilin (Figure 3A). The Kd values obtained for wild-type twinfilin (Kd≈0.04 μM), Twf1–10p (Kd≈0.06 μM), and Twf1–11p (Kd≈0.05 μM) are within experimental error. We also examined whether binding to CP influences twinfilin's affinity for actin monomers. In the presence of 3 μM yeast CP, twinfilin bound ADP-G-actin with an affinity similar (Kd≈0.04 μM) to that in the absence of CP, suggesting that at least in vitro CP does not affect twinfilin's actin-binding activity (Figure 3A).
Figure 3.
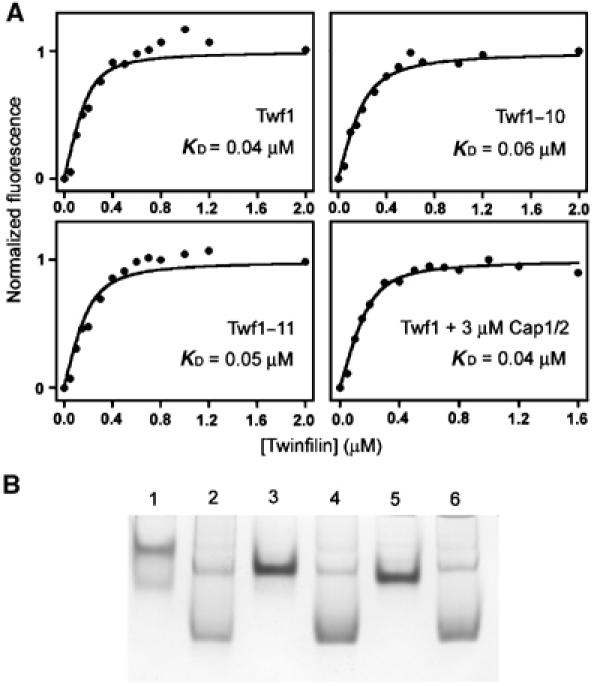
Twf1–10p and Twf1–11p bind actin and PI(4,5)P2 similarly as wild-type twinfilin. (A) The change in fluorescence of 0.2 μM NBD-labelled MgADP-actin was measured at different twinfilin concentrations under physiological conditions. Symbols are data and the lines are calculated binding curves. Twf1–10p and Twf1–11p bind ADP-G-actin with affinities similar to wild-type twinfilin. Cap1/2p does not have significant effect on twinfilin's actin-binding ability in solution. (B) Twf1–10p and Twf1–11p interact with PI(4,5)P2 in a native gel electrophoresis assay. Lane 1, 4 μM wild-type yeast twinfilin; lane 2, 4 μM wild-type twinfilin+24 μM PI(4,5)P2; lane 3, 4 μM Twf1–11p; lane 4, 4 μM Twf1–11p+24 μM PI(4,5)P2; lane 5, 4 μM Twf1–10p; lane 6, 4 μM Twf1–10p+24 μM PI(4,5)P2. In the absence of phospholipid, these proteins migrate as sharp bands on the gel, but when incubated with PI(4,5)P2 before loading on a gel these proteins showed smear-like migration patterns.
The PI(4,5)P2 binding of wild-type twinfilin, Twf1–10p, and Twf1–11p was measured by a native gel-electrophoresis assay. In the absence of phospholipids, wild-type twinfilin, Twf1–10p, and Twf1–11p migrate as sharp bands on native gels. In the presence of PI(4,5)P2, there is a clear shift in their mobilities, and instead of sharp bands these proteins show a smear-like migration pattern, demonstrating that they interact with PI(4,5)P2 in vitro (Figure 3B). Taken together, these biochemical data show that Twf1–10p and Twf1–11p mutants have specific defects in CP binding, but interact with PI(4,5)P2 and actin monomers similarly to wild-type twinfilin.
Interaction with CP is essential for twinfilin's correct localization in cells
To examine the in vivo role of twinfilin–CP interaction, we expressed wild-type twinfilin, Twf1–10p, and Twf1–11p in Δtwf1 yeast cells under their own promoters from CEN plasmids. A Western blot assay demonstrated that the cellular levels of both mutant proteins were similar to those of the wild-type protein (Figure 4A). Interaction of wild-type and mutant twinfilins with CP in vivo was next examined by a co-immunoprecipitation assay. Wild-type twinfilin efficiently co-immunoprecipitated with an anti-Cap2 antibody, whereas no detectable amounts of Twf1–10p or Twf1–11p co-immunoprecipitated with CP (Figure 4B). Similarly, CP co-immunoprecipitated with anti-Twf1p antibody from wild-type but not from the mutant yeast extracts (Figure 4C). These data show that Twf1–10p and Twf1–11p do not interact with CP in vivo. Furthermore, immunofluorescence microscopy showed that in wild-type yeast cells twinfilin localizes to cortical actin patches, whereas twf1–10 and twf1–11 cells show a diffuse cytoplasmic localization of twinfilin (Figure 4D). Direct interaction with CP is thus essential for correct localization of twinfilin in yeast cells. However, twinfilin is not required for the localization of CP, because Cap1/2p shows normal cortical actin patch localization in Δtwf1 yeast cells (Figure 4E).
Figure 4.
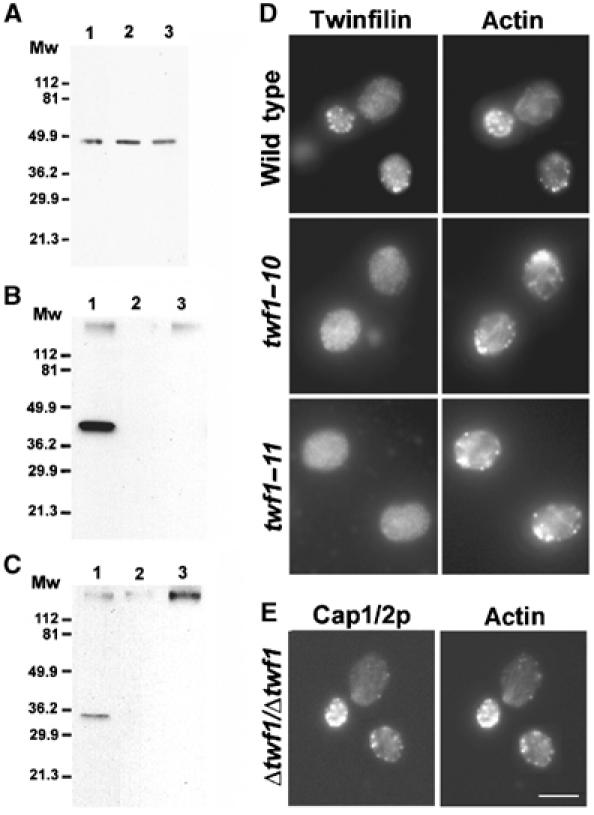
Twinfilin's localization in yeast is dependent on direct interaction with Cap1/2p. (A) Equal amounts of yeast cell extracts from Δtwf (DDY1434) cells expressing wild-type twinfilin (lane 1), Twf1–11p (lane 2), and Twf1–10p (lane 3) were run on a 12% polyacrylamide gel, and twinfilin was subsequently visualized by Western blotting. The mutant proteins, Twf1–10p and Twf1–11p, show normal expression levels when compared to wild-type twinfilin. (B) Immunoprecipitation of Cap1/2p with anti-yeast Cap2p antibody was carried out from Δtwf1/Δtwf1 (DDY1436) cells expressing wild-type twinfilin (lane 1), Twf1–10p (lane 2), or Twf1–11p (lane 3). The blot was detected with anti-Twf1p antibody, and it demonstrates specific co-immunoprecipitation of wild-type twinfilin (lane 1), but no co-immunoprecipitation of Twf1–10p (lane 2) or Twf1–11p (lane 3) with Cap1/2p. (C) Similarly, only wild-type Cap1/2p co-immunoprecipitated with anti-Twf1p antibody. Immunoprecipitation of twinfilin was carried out from DDY1436 strain expressing wild-type twinfilin (lane 1), Twf1–10p (lane 2), and Twf1–11p (lane 3). The blot was detected with an anti-Cap1/2p antibody. (D) Wild-type twinfilin, Twf1–10p, and Twf1–11p were expressed under their own promoters in twfΔ cells (DDY1434), and twinfilin (left) and actin (right) were visualized. Twinfilin localizes to the actin patches in cells expressing wild-type twinfilin, but shows diffuse cytoplasmic staining and does not localize to the actin patches in Twf1–10p- and Twf1–11p-expressing cells. (E) Localization of capping protein in Δtwf1/Δtwf1 yeast strain. Twinfilin is not required for the localization of capping protein (left) to the cortical actin patches (right). Bar, 5 μm.
Interactions with CP and actin monomers are required for twinfilin's role in actin dynamics in vivo
To examine the biological role of twinfilin–CP and twinfilin–actin interactions, we took advantage of the previously identified synthetic lethality between twinfilin deletion and cof1–22, as well as pfy1–4 mutants, in budding yeast (Goode et al, 1998; Wolven et al, 2000). In these experiments, we used two twinfilin mutants: twf1–3 that has a specific defect in actin binding (Palmgren et al, 2001) and twf1–10 that has a specific defect in CP binding (see above). To examine the biological role of twinfilin's CP interaction, haploid Δtwf yeasts carrying either an empty CEN plasmid or one encoding wild-type TWF1 or twf1–10 were crossed with cof1–22 or pfy1–4 haploid yeast strains. Sporulation of the diploid yeast and dissection of ∼20 tetrads per construct revealed that, like Δtwf, twf1–10 also shows synthetic lethality with cof1–22 and pfy1–4 mutants (Figure 5A). Similarly, also twf1–3 that has a specific defect in actin monomer interactions showed synthetic lethality with cof1–22 (Figure 5A). After prolonged (10 days at 24°C) incubation, a few viable cof1–22xtwf1–10, cof1–22xtwf1–3, and cof1–22xtwfΔ cells could be obtained. The phenotypes of these viable cof1–22xtwf1–10, cof1–22xtwf1–3, and cof1–22xtwfΔ double-mutant cells were very similar to each other, with grossly enlarged cells and abnormal accumulation of F-actin. Twf1–10p shows diffuse cytoplasmic localization in these cells, whereas Twf1–3p shows some colocalization with the cortical actin patches (Figure 5B). Taken together, these data demonstrate that twinfilin's abilities to bind both CP and actin monomers are essential for its role in actin dynamics in vivo (Table I).
Figure 5.
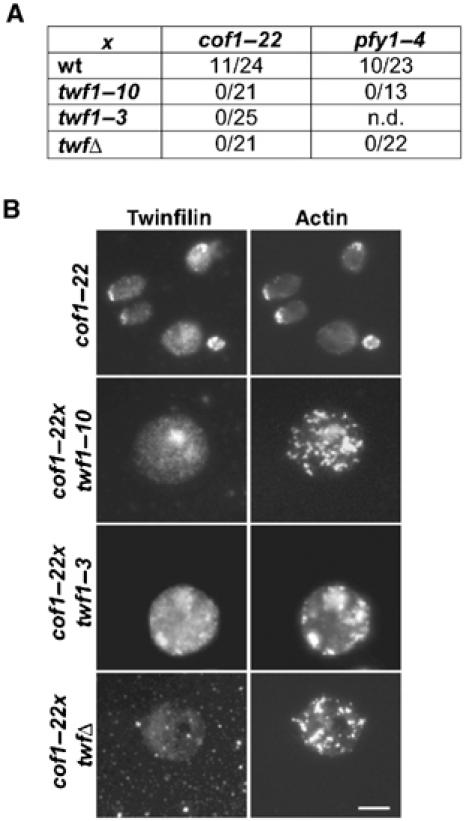
Interactions with Cap1/2p and actin monomers are essential for twinfilin's role in actin dynamics. (A) Genetic interactions between yeast twinfilin, cofilin, and profilin mutants. Double mutants were inferred by marker segregation and colonies were scored 4 days after tetrad dissection at 24°C. The number of double mutants forming colonies over the total number of tetrads is indicated. Similar to Δtwf1, twf1–10 also shows synthetic lethality with cof1–22 and pfy1–4. Furthermore, twf1–3 also shows synthetic lethality with cof1–22, whereas its genetic interactions with pfy1–4 could not be examined due to sporulation problems. (B) After prolonged incubation at +24°C, a few viable cof1–22xtwf1–10, cof1–22xtwf1–3, and cof1–22xtwfΔ cells were obtained and the cells were stained with anti-twinfilin (left) and anti-actin (right) antibodies. cof1–22 cells show relatively normal organization of the actin cytoskeleton, whereas cof1–22xtwf1–10, cof1–22xtwf1–3, and cof1–22xtwfΔ cells are grossly enlarged and show abnormal accumulation of F-actin. Bar, 5 μm.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| DDY1102 | MAT a/MAT α, ade2-1/+, his3Δ200/his3Δ200, leu2–3,112/leu2–3,112, ura3–52/ura3–52, lys2–801/+ |
| DDY1266 | MAT α, ura3–52, his3Δ200, leu2–3,112, lys2–801, cof1–22∷LEU2 |
| DDY1434 | MAT a, ade2-1, his3Δ200, leu2–3,112, ura3–52, Δtwf1∷URA3 |
| DDY1436 | MAT a/MAT α, ade2-1/ade2-1, his3Δ200/his3Δ200, leu2–3,112/leu2–3,112, ura3–52/ura3–52, Δtwf1∷URA3/Δtwf1∷URA3 |
| DDY2009 | MAT α, his3Δ200, leu2–3,112, lys2–801, ura3–52, pfy1–4∷LEU2 |
| YJC0389 | MAT a, rho+, ade2–101, his3–11,15, leu2–3,112, trp1-1, ura3–1, cap2-Δ1∷HIS3 |
| Standard methods were employed for growth, sporulation, and tetrad dissection of yeast (Rose et al, 1989). | |
Structural mechanism of twinfilin–CP interaction
Solution X-ray scattering is a powerful tool in studies of the solution structures of biological macromolecules. Although solution X-ray scattering is not suitable for the analysis of atomic structure, it provides information about gross structural features such as shape, and quartenary and tertiary structures of proteins and protein complexes (Svergun and Koch, 2002). Here we utilized this technique to show the molecular architecture of the complex between twinfilin and CP, as well as the individual protein conformations in solution. Due to better solubility, these assays were carried out with mouse twinfilin-1 and α1β2 CP instead of yeast proteins.
For the X-ray scattering experiments, each protein/protein complex was purified by gel filtration to remove any possible aggregates before the measurements. The increase in the scattering at low angles was monitored at 30 s intervals during the experiment to rule out the possibility of aggregation due to radiation damage. CP and CP–twinfilin complex were highly stable during the data collection, whereas full-length twinfilin showed some aggregation after 30–60 s of exposure. Thus, only the first 30 s of the data were used for the construction of the scattering curves of twinfilin, and this resulted in a small increase in the noise at higher scattering angles. Figure 6 shows the composite experimental scattering profiles along with the experimental distance distribution curves. Radius of gyration and maximum diameter values obtained from these data were 36.6 and 116.0 Å (twinfilin), 30.3 and 95.0 Å (CP), and 45.4 and 150 Å (twinfilin–CP complex). The low-resolution structures of twinfilin, CP, and their complex were then generated as described in Materials and methods. All shape reconstructions resulted in very similar models, representative shapes of which are shown in Figure 7.
Figure 6.
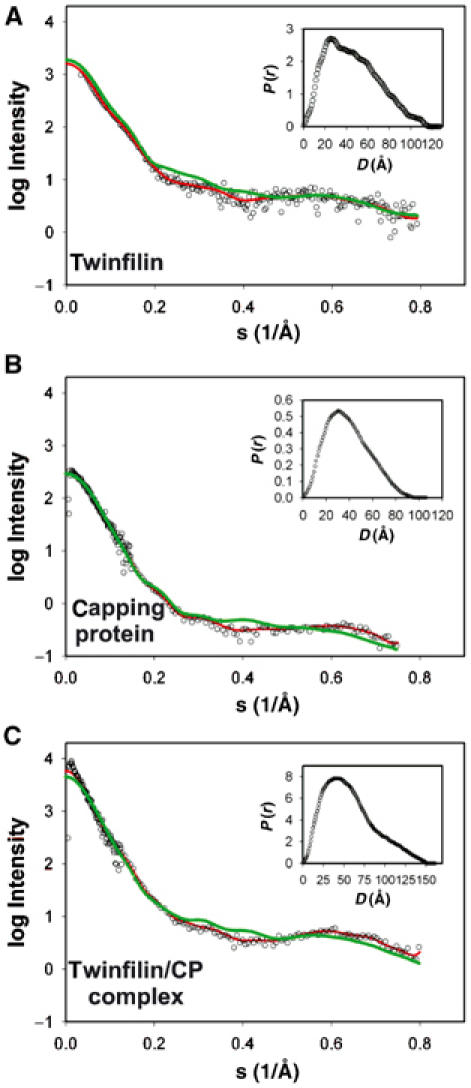
Scattering profiles of full-length twinfilin, capping protein, the twinfilin–capping protein complex, and twinfilin's C-terminal ADF-H domain. Open symbols show the experimental data and the red lines show the fit produced by the restored shape models of full-length twinfilin (A), capping protein (B), and the twinfilin–capping protein complex (C). The theoretical scattering simulations for twinfilin, capping protein, and twinfilin–capping protein complex obtained from the models shown in Figure 7 are indicated with green lines. Inset graphs show the P(r) distance distribution versus D for the best fit to each protein.
Figure 7.
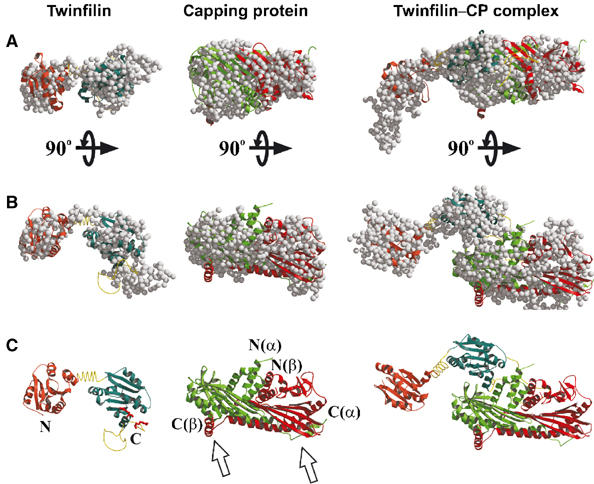
Three-dimensional models for full-length twinfilin, capping protein, and the twinfilin–capping protein complex. (A) Representative dummy residue models for twinfilin (left), capping protein (middle), and twinfilin–capping protein complex (right). (B) Same models rotated 90° relative to their horizontal axes. (C) Structural models for twinfilin (left), capping protein (middle), and twinfilin–capping protein complex (right). These models were obtained by fitting the high-resolution structures of twinfilin's ADF-H domain and capping protein onto the models generated from the scattering data. Red ball-and-sticks indicate the positions of critical capping protein-binding residues (corresponding to Arg328 and Arg 331 of yeast protein) on the twinfilin molecule. The two arrows indicate the positions of the actin-binding tentacles of capping protein. The picture was created with the programs Molscript (Kraulis, 1991) and RASTER3D (Merrit and Murphy, 1994).
These studies revealed that full-length twinfilin is an elongated two-domain structure, with a kinked linker region separating the two structural domains. The dummy residue model of CP displays an asymmetric particle, approximately 9 nm in length. The generated model for the twinfilin–CP complex shows an asymmetric part connected to an elongated two-domain structure. These models were well in accordance with the experimental p(r) curves, which yielded biphasic patterns for full-length twinfilin and CP (as well as for their complex), indicative of elongated structures (Figures 6 and 7).
The atomic structures of twinfilin's N-terminal ADF-H domain (residues 1–142) and α1β1 CP have been solved by X-ray crystallography (Paavilainen et al, 2002; Yamashita et al, 2003). Based on their sequence homology (∼25% identical at the amino-acid level) and similar actin-binding properties and interfaces (Palmgren et al, 2001; Ojala et al, 2002), we can predict that the two ADF-H domains of twinfilin are structurally very similar to each other. Thus, we were able to fit these atomic models onto the obtained experimental dummy residue models (Figure 7). The missing linker regions of the full-length twinfilin molecule were built either manually or automatically using programs O (Jones et al, 1991) or CHADD (Petoukhov et al, 2002), respectively, and these models were subsequently used for scattering simulations with the program CRYSOL. The simulated scattering profiles were well in accordance with the measured data and yielded χ2 values of 2.4 (CP), 1.7 (twinfilin), and 2.8 (twinfilin–CP complex).
A model based on our scattering data demonstrates that in solution twinfilin is an elongated two-domain protein with a smaller domain composed of the N-terminal ADF-H domain, and a larger domain composed of the C-terminal ADF-H domain and flexible C-terminal tail region. The solution structure of α1β2 CP obtained here by X-ray scattering is very similar to the recently determined crystal structure of α1β1 CP (Yamashita et al, 2003). The model of the complex shows that the N-terminal ADF-H domain of twinfilin does not interact with CP, whereas the lower region of the C-terminal domain (most likely the C-terminal tail region) is in direct contact with CP. This model is also in good agreement with the biochemical binding studies performed with twinfilin deletion constructs and site-directed mutants (see Figure 2). As our biochemical data (see Figure 1) demonstrated that actin filament binding of CP is not affected by twinfilin, CP in this model is oriented such that its α-helical region around the N-termini of the α and β subunits is in contact with twinfilin, and the C-terminal actin-binding sites (Wear et al, 2003) are pointing away from the twinfilin molecule (Figure 7).
Discussion
Here we show that twinfilin is composed of two structurally separable domains, and that its conserved C-terminal tail region is critical for interactions with CP in yeast and mammals. Our biochemical studies demonstrate that twinfilin (either alone or in complex with ADP- or ATP-G-actin) does not regulate the actin filament capping activity of CP. However, genetic studies with mutant yeast twinfilins with specific defects in either CP or actin binding demonstrated that both these activities are essential for twinfilin's role in actin dynamics.
A structural model of the twinfilin–CP complex obtained by solution X-ray scattering studies demonstrates that the CP-binding site is entirely located at only one of the two globular domains and that the other domain of twinfilin does not contribute to this interaction. This model is further refined by our biochemical analysis of mutant twinfilins demonstrating that the CP-binding site is located at the conserved C-terminal tail region of twinfilin and that the C-terminal half of twinfilin alone is sufficient for CP binding. Although our genetic studies showed that interaction with CP is essential for twinfilin's role in actin dynamics in vivo, this interaction does not affect the actin filament barbed-end capping activity of CP. These data suggest that the biological role of twinfilin–CP interaction is to regulate the activity and localization of twinfilin in cells. The observation that twinfilin is not a regulator of CP suggests that the twinfilin-binding site on CP does not overlap with its actin-binding site. Thus, in our model, the C-terminal regions of both the α and β subunits essential for CP's actin interactions (Wear et al, 2003; Kim et al, 2004) are located away from the twinfilin-binding site of the CP molecule.
Our studies also show that in solution CP does not affect the binding of twinfilin to ADP-G-actin. However, it is important to note that for technical reasons this assay was carried out using CP, which is not bound to filament ends. Therefore, it is possible that the filament-bound form of CP may affect the actin binding of twinfilin, for example, by promoting the dissociation of ADP-G-actin from twinfilin. This hypothesis is supported by the fact that twinfilin's high-affinity actin-binding site is located at the C-terminal ADF-H domain (Ojala et al, 2002), which in the solution structure is located close to the CP-binding interface.
Genetic and cell biological studies with specific twinfilin mutants (twf1–10 and twf1–11) demonstrate that interaction with CP is essential for twinfilin's correct localization in cells and for its role in actin dynamics. Similarly, studies using a twinfilin mutant with a specific defect in actin binding (twf1–3; see Palmgren et al, 2001) show that also the ability to interact with actin monomers is essential for the biological role of twinfilin. These data show that in cells twinfilin is not just a simple actin monomer-sequestering protein, but that its ability to localize to actin filament structures through an interaction with CP is central for its biological activity. Furthermore, as described above, twinfilin is also not a regulator of CP, suggesting that it plays a more complex role in actin dynamics.
Studies on budding yeast, Drosophila melanogaster, and mouse demonstrated that twinfilin shows diffuse cytoplasmic localization, but is also concentrated to certain actin filament structures in cells (Palmgren et al, 2001; Wahlström et al, 2001; Vartiainen et al, 2003). It is possible that the biological role of twinfilin is to sequester ADP-actin monomers from the cytoplasm and localize them to relevant regions of cells through direct interaction with CP. After dissociation from the twinfilin–CP complex, the actin monomer would undergo nucleotide exchange (catalyzed, for example, by profilin) and assemble into a nearby uncapped filament barbed end. As twinfilin forms a relatively stable complex with ADP-actin monomers (Ojala et al, 2002) and inhibits their nucleotide exchange (Goode et al, 1998; Vartiainen et al, 2003), it is well suited for preventing actin assembly at undesired cell regions and localizing actin monomers to sites of rapid filament assembly such as cortical actin patches in yeast cells. Both twinfilin and CP are abundant proteins in yeast, found in approximately 1:10 and 1:5 molar ratios, respectively, to actin (Palmgren et al, 2001; Kim et al, 2004). A large fraction of CP in yeast is localized to cortical actin patches and bound to actin (Kim et al, 2004), indicating that yeast actin patches contain a high concentration of barbed ends capped by CP. This suggests that the possible targeting of actin monomers by twinfilin to CP-bound filament ends could have a significant effect on the dynamics and localization of the cytoplasmic actin monomer pool. This hypothesis is also supported by the observation that a strong hypomorphic twinfilin mutation resulted in accumulation of ectopic actin filament structures in Drosophila bristles (Wahlström et al, 2001). However, it is also possible that twinfilin, together with CP, plays a more complex role in regulating actin filament turnover.
Although twinfilin is highly conserved in evolution, it appears to be essential for morphogenesis and viability only under certain specific conditions and cell types. In budding yeast a deletion of twinfilin gene leads to relatively mild defects in actin organization (Goode et al, 1998), and in Drosophila S2 cells the presence of twinfilin is not required for normal lamellipodia formation (Rogers et al, 2003). However, twinfilin is essential for morphogenesis of large and/or elongated cells such as Drosophila ommatidia and bristles (Wahlström et al, 2001). Furthermore, in budding yeast, twinfilin becomes essential when the cytoplasmic actin monomer pool is diminished by a specific mutation in the cofilin gene that decreases filament depolymerization rates (Lappalainen and Drubin, 1997; Goode et al, 1998). Our genetic studies show that twinfilin's ability to localize to cortical actin patches through direct interaction with CP is required for rescuing the synthetic lethality with this cofilin mutation. This suggests that twinfilin's possible role in localizing actin monomers to the sites of actin assembly is specifically required under conditions when the size of the cytoplasmic actin monomer pool is limiting.
Materials and methods
Site-directed mutagenesis and plasmid construction
The site-directed mutations to yeast (pPL61) and mouse (pPL78) twinfilin expression plasmids were introduced by PCR-based overlap extension method (Higuchi et al, 1988). The oligonucleotides used in the amplifications created NcoI and HindIII sites at the 5′ and 3′ ends of the final PCR fragments, respectively. These fragments were digested and ligated into pGAT2 (yeast twinfilin constructs) or pHAT2 (mouse twinfilin constructs) vectors (Peränen et al, 1996). To express wild-type and mutant twinfilins in yeast, a 1.8 kb genomic fragment containing the TWF1 open reading frame and its promoter regions was subcloned into yeast centromere-based LEU plasmid pRS315 to create plasmid pPL87. Site-directed mutations were introduced to this plasmid using a mutagenesis kit (Transformer™, Clontech), generating plasmids pPL178 (twf1–11), pPL179 (twf1–10), and pPL247 (twf1–3). The wild-type TWF1, twf1–10, and twf1–3 genomic fragments were also subcloned to centromere-based HIS vector pRS413 to create plasmids pPL229, pPL230, and pPL248. All constructs were sequenced by the chain-termination method to verify the correct sequences.
Protein expression and purification
Wild-type and mutant yeast twinfilins were expressed and purified as described (Goode et al, 1998). Full-length mouse twinfilin-1 and its individual domains were purified as described (Vartiainen et al, 2003). Mouse α1β2 CP and yeast Cap1/2p were expressed and purified as described (Palmgren et al, 2001; Kim et al, 2004). Rabbit muscle actin was prepared from acetone powder as described previously (Spudich and Watt, 1971). ADP-actin was prepared by incubating NBD-actin with hexokinase-agarose beads (Sigma) and glucose for 3 h at +4°C (Pollard, 1986). Yeast actin was purified as described previously (Goode, 2002).
Actin interaction assays
Twinfilin's influence on the actin filament barbed-end capping ability of Cap1/2p was examined by a seeded actin assembly and steady-state critical concentration assays, and the equilibrium dissociation and capping rate constants were determined as described (Wear et al, 2003). The affinities of wild-type and mutant twinfilins for ADP-actin monomers were determined by measuring the fluorescence of NBD-labeled G-actin as previously described (Ojala et al, 2002).
Native gel electrophoresis assays
Twinfilin–Cap1/2p interaction was studied on 10% native polyacrylamide gels as described (Palmgren et al, 2001). Native polyacrylamide gel electrophoresis for studying twinfilin–PI(4,5)P2 (Sigma) interactions was performed as described (Gungabissoon et al, 1998).
Supernatant depletion pull-down assays
The GST–yeast twinfilin fusion proteins were immobilized on glutathione sepharose 4B beads (Amersham) at a concentration of approximately 5 mg of fusion protein/2 ml prewashed beads. The total amount of protein on beads was quantified from a Coomassie-stained SDS gel of the beads. In all, 250 μl of 2 μM yeast CP (in 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM MgCl2, 0.5 mM DTT) was incubated alone or with increasing concentrations of GST-twinfilin coupled to glutathione sepharose 4B beads for 10 min at 25°C. The samples were centrifuged for 5 min at 13 000 g to pellet the beads and any bound proteins. The amount of CP present in the supernatant was then examined from 12% SDS gels, and quantified by using TINA software.
Immunofluorescence microscopy
Cells were grown in an appropriate medium at 30°C to an optical density of 0.5 at 600 nm and prepared for immunofluorescence as described by Ayscough and Drubin (1998). Twinfilin, actin, and CP were visualized as previously described (Palmgren et al, 2001; Kim et al, 2004). The secondary antibodies used were TRITC anti-rabbit and FITC anti-guinea-pig (Jackson Inc.). Images were acquired through a SenSys (Photometrics Ltd) camera on an AX70 Provis microscope (Olympus).
Western blotting and co-immunoprecipitation
Cells were grown to confluence overnight in YEPD medium (1% (w/v) Bacto-yeast extract, 2% Bacto-peptone, 2% glucose) at 30°C, diluted 1:10, and allowed to grow for ∼3 h. Cells from 3 ml cultures were harvested, resuspended in 100 μl 20 mM Tris–HCl (pH 7.5), 0.6 mM PMSF, and lysed by addition of glass beads in 1:1 and vortexing for 5 min at room temperature. Western blotting was carried out as described previously (Vartiainen et al, 2000) with the following primary antibodies: guinea-pig anti-yeast CP (1:10 000 dilution) or rabbit anti-yeast twinfilin (1:1000 dilution). Horseradish peroxidase-conjugated anti-guinea-pig or anti-rabbit secondary antibodies (Jackson Inc.) were used at 1:10 000 dilutions. Co-immunoprecipitation experiments from 1 × 108 diploid Δtwf1/Δtwf1 cells, transformed with the desired CEN plasmids, were carried out as described (Palmgren et al, 2001).
Solution X-ray scattering experiments
Synchrotron radiation X-ray scattering data were collected with a 2D detector at station 2.1 (Towns-Andrews et al, 1989) of the Synchrotron Radiation Source (SRS), Daresbury Laboratory, UK. The studies were performed in a standard cell with two protein concentrations (1 and 16.5 mg/ml for twinfilin; 2 and 10 mg/ml for CP; 2 and 11.5 mg/ml for twinfilin–CP complex at 1 and 4.25 m detector distances, respectively) according to established procedures (Grossmann et al, 2002; Witty et al, 2002). To verify the sample homogeneity, a gel filtration chromatography (Superdex-200, Amersham) was carried out for each sample before measurements. All proteins eluted as single peaks at the expected elution positions for monomeric proteins or a protein complex (twinfilin–CP complex). All samples were measured at 4°C at a wavelength of 1.54 Å. Full-length twinfilin was highly radiation sensitive and thus only the first 30–60 s were used for averaging. Two sample-to-detector distances (1 and 4.25 m) were used and the corresponding profiles merged so as to cover the momentum transfer interval 0.02 Å−1⩽s⩽0.8 Å−1. The modulus of the momentum transfer is defined as s=4π sin Θ/λ, where 2Θ is the scattering angle and λ is the wavelength used (1.54 Å). The maximum scattering angle corresponds to a nominal Bragg resolution of 8 Å. The radius of gyration, the forward scattering intensity, and the intraparticle distance distribution function p(r) were evaluated with the indirect Fourier transform program GNOM (Semenyuk and Svergun, 1991).
Three-dimensional modeling and fitting of atomic structures
Low-resolution models of the proteins were generated ab initio from the scattering profiles with the program GASBOR version 18 (Svergun et al, 2001) using the smoothed data set generated by GNOM. A large number (>20) of independent ab initio shape restorations were performed for all the proteins and the reconstructions provided highly consistent shape models. The selected models are representative of the two proteins and their complex. Superimposition of the known atomic structures (twinfilin residues 1–142, CP α1β2) were carried out either manually using the program O (Jones et al, 1991) or automatically using the program SUPCOMB (Kozin and Svergun, 2001). The two twinfilin domains were fitted independently into the low-resolution bead model and the linker regions were then added to the resulting pseudo-atomic model. High-resolution models of twinfilin's N-terminal ADF-H domain (Paavilainen et al, 2002) and α1β1 CP (Yamashita et al, 2003) were used in the theoretical calculation of scattering curves by the program CRYSOL (Svergun et al, 1995). This method takes into account the solvent effect by surrounding the protein with a hydration shell of thickness 3 Å and fitting its excess average scattering density.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Acknowledgments
We aknowledge Michael C Merckel for advice on the X-ray-scattering experiments. This study was supported by grants from Academy of Finland, Sigrid Juselius Foundation, Biocentrum Helsinki, EMBO Young Investigator Program (to PL) and NIH grant GM38542 (to JAC). VOP was supported by a fellowship from the Helsinki Graduate School in Biosciences.
References
- Amatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA (1990) Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature 344: 352–354 [DOI] [PubMed] [Google Scholar]
- Ayscough K, Drubin DG (1998) Immunofluorescence microscopy of yeast cells. In Cell Biology: A Laboratory Handbook, JE Celis (ed), 2nd edn. San Diego, CA: Academic Press [Google Scholar]
- Bamburg JR, McGough A, Ono S (1999) Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol 9: 364–370 [DOI] [PubMed] [Google Scholar]
- Caldwell JE, Heiss SG, Mermall V, Cooper JA (1989) Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry 28: 8506–8514 [DOI] [PubMed] [Google Scholar]
- Carlier MF, Ressad F, Pantaloni D (1999) Control of actin dynamics in cell motility. Role of ADF/cofilin. J Biol Chem 274: 33827–33830 [DOI] [PubMed] [Google Scholar]
- DiNubile MJ, Cassimeris L, Joyce M, Zigmond SH (1995) Actin filament barbed-end capping activity in neutrophil lysates: the role of capping protein-beta 2. Mol Biol Cell 6: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL (2002) Purification of yeast actin and actin-associated proteins. Methods Enzymol 351: 433–441 [DOI] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Lappalainen P (1998) Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J Cell Biol 142: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann JG, Hall JF, Kanbi LD, Hasnain SS (2002) The N-terminal extension of rusticyanin is not responsible for its acid stability. Biochemistry 41: 3613–3619 [DOI] [PubMed] [Google Scholar]
- Gungabissoon RA, Jiang CJ, Drøbak BK, Maciver SK, Hussey P (1998) Interaction of maize-actin-depolymerising factor with actin and phosphoinositides and its inhibition of plant phospholipase C. Plant J 16: 689–696 [Google Scholar]
- Hart MC, Cooper JA (1999) Vertebrate isoforms of actin capping protein beta have distinct functions in vivo. J Cell Biol 147: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD (2001) Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu Rev Biochem 70: 649–676 [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16: 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG (1996) Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J Cell Biol 133: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubberstey AV, Mottillo EP (2002) Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J 16: 487–499 [DOI] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA (1995) Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell 81: 591–600 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kim K, Yamashita A, Wear MA, Maeda Y, Cooper JA (2004) Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J Cell Biol 164: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI (2001) Automated matching of high- and low-resolution structural models. J Appl Cryst 34: 33–41 [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst 24: 946–950 [Google Scholar]
- Lappalainen P, Drubin DG (1997) Cofilin promotes rapid actin filament turnover in vivo. Nature 388: 78–82 [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD (2001) Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell 12: 1161–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrit EA, Murphy MEP (1994) Raster3D Version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr D 50: 873. [DOI] [PubMed] [Google Scholar]
- Ojala PJ, Paavilainen VO, Vartiainen MK, Tuma R, Weeds AG, Lappalainen P (2002) The two ADF-H domains of twinfilin play functionally distinct roles in interactions with actin monomers. Mol Biol Cell 13: 3811–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavilainen VO, Merckel MC, Falck S, Ojala PJ, Pohl E, Wilmanns M, Lappalainen P (2002) Structural conservation between the actin monomer-binding sites of twinfilin and actin-depolymerizing factor (ADF)/cofilin. J Biol Chem 277: 43089–43095 [DOI] [PubMed] [Google Scholar]
- Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P (2001) Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol 155: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren S, Vartiainen M, Lappalainen P (2002) Twinfilin, a molecular mailman for actin monomers. J Cell Sci 115: 881–886 [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF (1993) How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell 75: 1007–1014 [DOI] [PubMed] [Google Scholar]
- Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem 236: 371–373 [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Eady NA, Brown KA, Svergun DI (2002) Addition of missing loops and domains to protein models by X-ray solution scattering. Biophys J 83: 3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 6: 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29: 545–576 [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD (2003) Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol 162: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston FM, Hieter P (1989) Methods in Yeast Genetics: A Laboratory Course Manual, 198pp. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA (1996) Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol 135: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenyuk AV, Svergun DI (1991) GNOM—a program package for small-angle scattering data processing. J Appl Cryst 24: 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S (1971) The regulation of rabbit skeletal muscle contraction. J Biol Chem 246: 4866–4871 [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ (1995) CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst 28: 768–773 [Google Scholar]
- Svergun DI, Koch MH (2002) Advances in structure analysis using small-angle scattering in solution. Curr Opin Struct Biol 12: 654–660 [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Miki H (2001) WASP and WAVE family proteins: key molecules for rapid rearrangement of cortical actin filaments and cell movement. J Cell Sci 114: 1801–1809 [DOI] [PubMed] [Google Scholar]
- Towns-Andrews E, Berry A, Bordas J, Mant PK, Murray K, Roberts K, Sumner I, Worgan JS, Lewis R, Gabriel A (1989) Time-resolved X-ray diffraction station: X-ray optics, detectors, and data acquisition. Rev Sci Instrum 60: 2346–2349 [Google Scholar]
- Vartiainen M, Ojala PJ, Auvinen P, Peränen J, Lappalainen P (2000) Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol Cell Biol 20: 1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Sarkkinen EM, Matilainen T, Salminen M, Lappalainen P (2003) Mammals have two twinfilin isoforms whose subcellular localizations and tissue distributions are differentially regulated. J Biol Chem 278: 34347–34355 [DOI] [PubMed] [Google Scholar]
- Wahlström G, Vartiainen M, Yamamoto L, Mattila PK, Lappalainen P, Heino TI (2001) Twinfilin is required for actin-dependent developmental processes in Drosophila. J Cell Biol 155: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA (2003) How capping protein binds the barbed end of the actin filament. Curr Biol 13: 1531–1537 [DOI] [PubMed] [Google Scholar]
- Witty M, Sanz C, Shah A, Grossmann JG, Mizuguchi K, Perham RN, Luisi B (2002) Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein. EMBO J 21: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven AK, Belmont LD, Mahoney NM, Almo SC, Drubin DG (2000) In vivo importance of actin nucleotide exchange catalyzed by profilin. J Cell Biol 150: 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Maeda K, Maeda Y (2003) Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J 22: 1529–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4


