Summary
The cytokine IL17, and signaling via its heterodimeric IL-17RA/IL-17RC receptor, is critical for host defense against extracellular bacterial and fungal pathogens. Polarized lung epithelial cells express IL-17RA and IL-17RC basolaterally. However, their contribution to IL-17-dependent pulmonary defenses in vivo remains to be determined. To address this, we generated mice with conditional deletion of Il17ra or Il17rc in Scgb1a1-expressing club cells, a major component of the murine bronchiolar epithelium. These mice displayed an impaired ability to recruit neutrophils into the airway lumen in response to IL17, a defect in bacterial clearance upon mucosal challenge with the pulmonary pathogen Klebsiella pneumoniae, and substantially reduced epithelial expression of the chemokine Cxcl5. Neutrophil recruitment and bacterial clearance were restored by intranasal administration of recombinant CXCL5. Our data show that IL-17R signaling in the lung epithelium plays a critical role in establishing chemokine gradients that are essential for mucosal immunity against pulmonary bacterial pathogens.
Graphical abstract
Introduction
The IL-17 family of cytokines includes 6 members, IL-17 (IL-17A), IL-17B, IL-17C, IL-17D, IL-17E, and IL-17F, which are produced by multiple cell types (Pappu et al., 2011). Signaling by IL-17 family cytokines is mediated by members of the IL-17 receptor family, IL-17RA, IL-17RB, IL-17RC, IL-17RD, and IL-17RE (Gaffen, 2009). While IL-17RA is shared among many IL-17 family members, IL-17RC is the unique receptor for IL-17 and IL-17F. These two members share the highest degree of homology and show similar functions. Numerous studies have demonstrated that IL-17 and IL-17F are critical molecules in mucosal host defense and inflammatory diseases (Chen and Kolls, 2013; Kumar and Subramaniyam, 2015; Way et al., 2013). Infections and inflammatory diseases increase the accumulation of IL-17-producing cells in lung tissue. Polarized airway epithelial cells respond to IL-17 via the basolateral expression of IL-17RA (McAllister et al., 2005) and IL-17RC (Kuestner et al., 2007), and these cells can produce antimicrobial proteins and several neutrophil chemoattractants including several ELR+ C-X-C chemokines. These pathways are thought to be critical for the function of IL-17 in mucosal immunity. Consistent with this, germ line IL-17RA KO show reduced CXCL2 and G-CSF expression in a model of intrapulmonary K. pneumoniae infection (Ye et al., 2001). Using bone marrow chimeric mice, several groups have shown that radioresistant tissue resident cells rather than radiosensitive cells of hematopoietic origin are essential for IL-17R signaling in various tissues (Ge et al., 2014; Linden et al., 2005; Meng et al., 2012; Oh et al., 2011). However, this bone marrow chimeric approach does not address what specific cell type is responsible for IL-17 mediated responses. Furthermore, a subset of alveolar macrophages has also been shown to be radioresistant (Guilliams et al., 2013), which complicates the interpretation of data generated from bone marrow chimeras. To study the function of epithelial IL-17RA signaling, we generated lung epithelial specific conditional IL-17RA KO mice using Scgb1a1-cre mice which deletes in club cells, a major component of the bronchiolar epithelium in mice (Simon et al., 2006). Deletion of Il17ra or Il17rc in lung epithelium resulted in compromised clearance of K. pneumoniae in the lung, including the drug resistant isolate expressing New Delhi Metallo-beta-lactamase-1 (2010). This impairment was associated with reduced neutrophil recruitment and a substantial reduction in epithelial expression of Cxcl5, a gene encoding the neutrophil chemoattractant CXCL5 (LIX). The mucosal immune defect in conditional Il17ra mice could be restored with mucosal administration of CXCL5. Our data underscore the essential role of epithelial IL-17R expression in mucosal immunity against K. pneumoniae by controlling mucosal chemokine gradients that are essential for optimal neutrophil recruitment.
Results
Phenotypic characterization of IL-17Rfl/fl mice
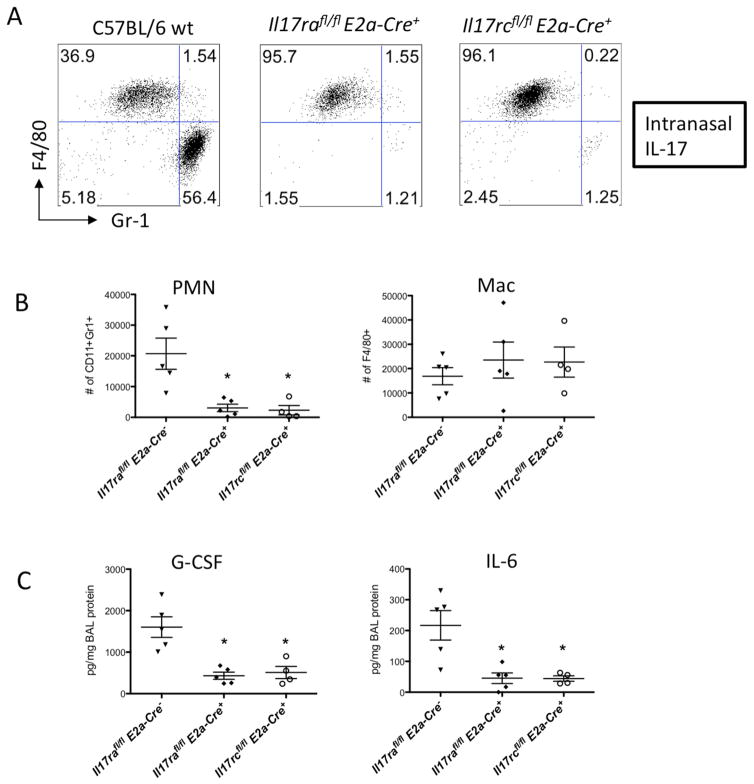
Targeting of Il17ra was achieved by introducing LoxP sites flanking exon 3 of the Il17ra gene, and targeting of Il17rc was accomplished by adding LoxP sites flanking exons 2 and 3 of the Il17rc gene (Kumar et al., 2016). To demonstrate that this would generate a functional null allele, skin fibroblasts from homozygous Il17rafl/fl mice were transduced with adenoviruses encoding the Cre recombinase (AdCRE). As shown in (Supplementary Fig. 1A), fibroblasts treated with AdCRE showed substantially reduced expression of IL-17 induced genes such as Csf3 and Il6, which was also associated with diminished Il17ra expression. Importantly, the expression of Il17rc was not affected. Cre-mediated recombination was also observed in vivo by standard PCR analysis in the spleen of Il17rafl/fl mice receiving intravenous AdCRE (Supplementary Fig. 1B). To further validate the functional deletion of the Il17ra gene in vivo, we crossed the Il17rafl/fl mice to E2a-Cre mice (Lakso et al., 1996). This line carries a Cre transgene under the control of the adenovirus E2a promoter that targets expression of Cre to the early mouse embryo, resulting in Cre-mediated recombination in a wide range of tissues. Flow cytometry analysis on peripheral blood confirmed the absence of surface IL-17RA expression in the Il17rafl/flE2a-cre+ mice (Supplementary Fig. 1C) while the deletion of Il17rc in the Il17rcfl/flE2a-cre+ mice did not seem to affect IL-17RA expression on these cells (Supplementary Fig. 2A). However, the percentage of Gr-1+ neutrophils in the peripheral blood was reduced in the Il17rafl/flE2a-cre+ mice (Supplementary Fig. 2B). This is consistent with the phenotype observed in the global Il17ra−/− mice (Kelly et al., 2005; Smith et al., 2008). To study the functional loss of Il17ra in the lungs of these mice, we used an intranasal delivery model (Herjan et al., 2013) where application of IL-17 in wild type mice induces a massive neutrophil influx into the airways (Fig. 1A&B). Global deletion of Il17ra abolished IL-17-induced pulmonary neutrophilia in the Il17rafl/flE2a-cre+ compared to wild type mice (Fig. 1A&B). Nearly identical defects in IL-17-induced neutrophil influx to the airways were observed in Il17rcfl/flE2a-cre+ mice (Fig. 1A&B). This defect was also accompanied by a significant reduction in IL-17-dependent cytokines in the BAL such as G-CSF and IL-6 (Fig. 1C). The Il17rcfl/flE2a-cre+ mice were still able to respond to intranasal CXCL1, an IL-17 regulated neutrophil chemoattractant, suggesting deletion of IL-17RC did not affect CXCR2 ligand mediated recruitment (Supplementary Fig. 2C). Taken together, these data demonstrate that functional deletion of the Il17ra and Il17rc alleles is successfully achieved in vivo, and these floxed mice can be used in the generation of tissue specific conditional KO mice.
Fig. 1. Characterization of the IL17Rfl/fl mice following IL-17 intranasal administration.
(A) C57BL/6 mice with indicated genotypes were challenged with recombinant IL-17 intranasally (300ng/mouse) for 24h and BAL cells were harvested. Representative FACS plots of neutrophil (PMN) and Macrophage (Mac) determined by Gr-1 and F4/80 staining were shown. (B) Il17rafl/fl and Il17rcfl/fl mice with indicated genotypes were challenged with IL-17 and total numbers of BAL neutrophils and macrophages were enumerated. G-CSF and IL-6 in the BAL fluid were measured by Luminex (C). Data are represented as mean +/− SEM. See also supplementary Fig. 1–2 for further characterization of these mice.
Defective PMN responses and bacterial clearance in the IL17RA lung conditional KO mice
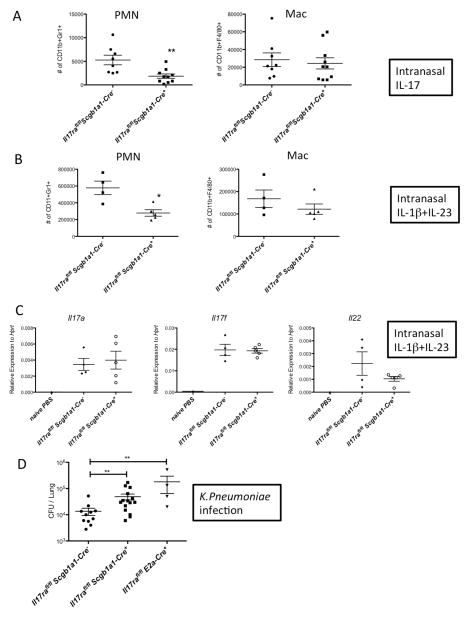
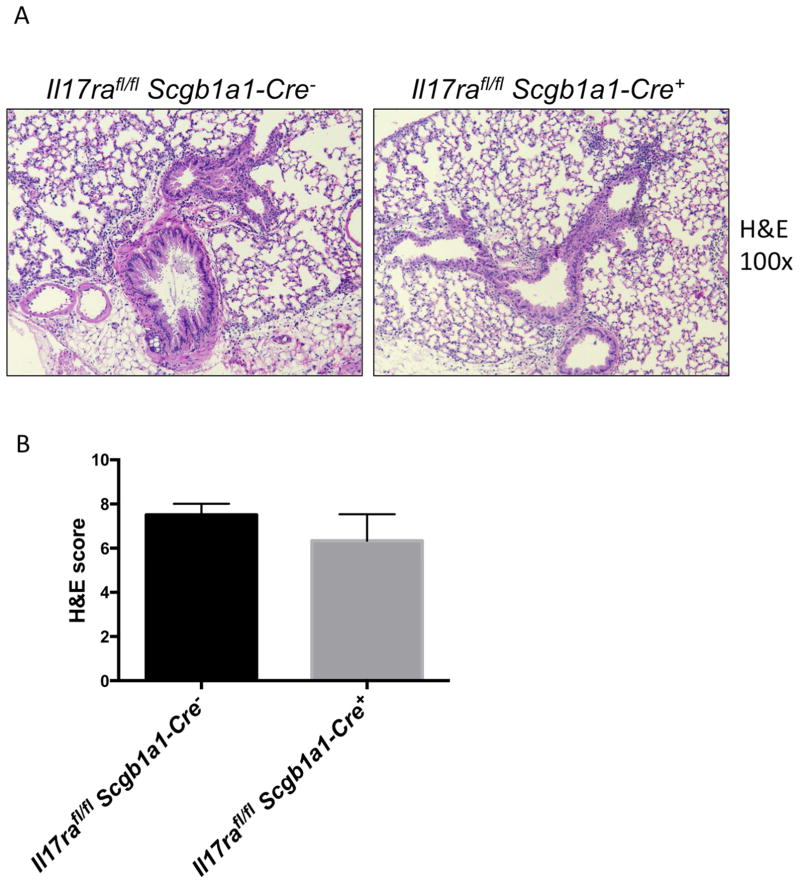
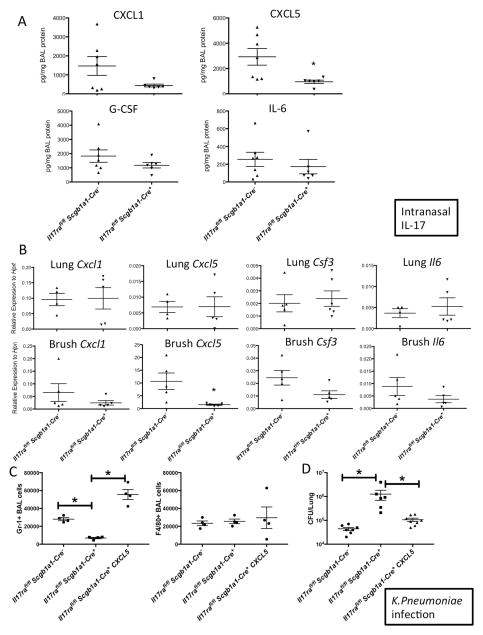
We have previously reported that polarized lung epithelium only respond to IL-17 applied basolaterally due to the site-specific expression of IL-17RA and IL-17RC (Kuestner et al., 2007; McAllister et al., 2005). To determine if the lung epithelium is required for IL-17-mediated responses in vivo, we crossed the Il17rafl/fl and Il17rcfl/fl mice to Scgb1a1-cre transgenic mice with the transgene expression specifically targeting the airway epithelium (Simon et al., 2006). Other groups have also reported that Scgb1a1-Cre is expressed in non-ciliated epithelial cells (Bertin et al., 2005; Rawlins et al., 2009). Scgb1a1-Cre mediated recombination in the lungs of the mice was confirmed by conventional PCR detecting the recombined allele (Supplementary Fig. 3A). In addition, there was significantly reduced expression of Il17ra transcript from bronchial brush samples of these mice (Supplementary Fig. 3B). The Il17rafl/flScgb1a1-cre+ mice displayed significantly reduced neutrophil numbers in the BALs after administration of recombinant IL-17 intranasally compared to littermate Il17rafl/flScgb1a1-cre− controls (Fig. 2A). To assess the neutrophil recruitment in response to endogenous IL-17, we administered intranasal IL-1β+IL-23, a cytokine combination that triggers potent IL-17 release from lung γδ T-cells (Dubin et al., 2012; Price et al., 2012). The Il17rafl/flScgb1a1-cre+ mice also displayed significantly reduced neutrophil numbers in the BAL fluid (Fig. 2B) after intranasal IL-1β+IL-23 treatment. The defects in neutrophil recruitment in the Cre+ mice were not due to a lack of IL-17 production, since type 17 cytokines including IL-17A, IL-17F and IL-22 were equally induced in the Cre+ and Cre− mice (Fig. 2C). These data suggested that epithelial IL-17R is critical for IL-17 mediated neutrophil influx in the lungs. Furthermore, the Il17rafl/flScgb1a1-cre+ mice were unable to control bacterial infection in the lungs following mucosal challenge with K. pneumoniae (Fig. 2D), demonstrating that epithelial IL-17RA signaling is required for bacterial clearance in in this model. We also performed hematoxylin and eosin (H&E) staining of the lungs to assess histologically the impact of defective IL-17 signaling in the Scgb1a1 expressing cells but found similar degrees of tissue inflammation in both the Cre+ and Cre− mice (Fig. 3A & B), suggesting IL-17R signaling in the epithelium is more important in recruiting bacterial killing neutrophils than in promoting epithelial integrity.
Fig. 2. Defective PMN responses and bacterial clearance in the IL-17RA epithelial conditional KO mice.
Il17rafl/fl Scgb1a1-Cre− and Il17rafl/fl Scgb1a1-Cre+ mice were challenged with recombinant IL-17 intranasally (300ng/mouse) for 24h and BAL neutrophils and macrophage numbers were determined by FACS (A). Scgb1a1-Cre mediated recombination was shown in supplementary Fig. 3. Il17rafl/fl Scgb1a1-Cre− and Il17rafl/fl Scgb1a1-Cre+ mice were challenged with recombinant IL-1β (10ng/mouse) plus IL-23 (500ng/mouse) intranasally for 24h and BAL neutrophils and macrophage numbers were determined by FACS (B). Gene expression of Th17 canonical cytokines from the lungs of IL-1β+IL-23 treated mice were determined by real-time RT-PCR (C). Il17rafl/fl Scgb1a1-Cre−, Il17rafl/fl Scgb1a1-Cre+, and Il17rafl/fl E2a-Cre+ mice were infected with 104 KP-43816 intranasally and sacrificed 24h post infection. Bacterial burden in the lungs were determined by standard CFU assay (D). Data are represented as mean +/− SEM.
Fig. 3. Histopathology of the IL-17RA epithelial conditional KO mice after K. pneumoniae infection.
Il17rafl/fl Scgb1a1-Cre− and Il17rafl/fl Scgb1a1-Cre mice were infected with 104 KP-43816 intranasally and sacrificed 24h post infection. Representative H&E stainings were shown (A) and pathology scores were graphed (B). Data are represented as mean +/− SEM.
Loss of Cxcl5 expression in the IL17RA lung conditional KO mice
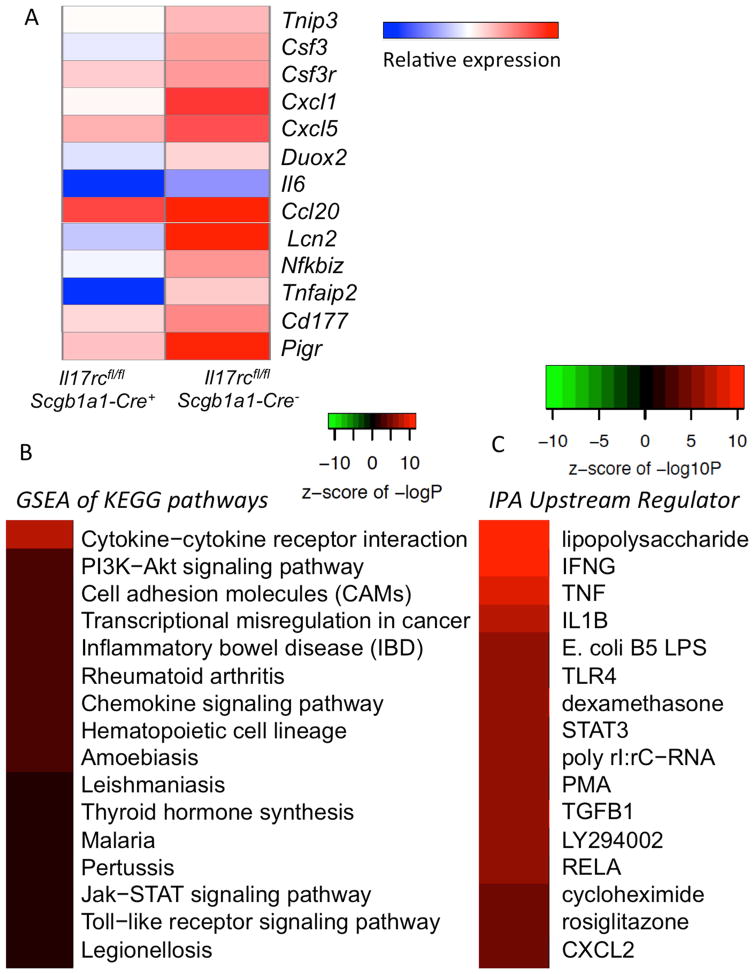
To explore the mechanism that leads to the impaired neutrophil recruitment and consequent mucosal bacterial clearance defect in the IL-17R epithelial conditional KO mice, we first performed RNA-seq analysis on the bronchial epithelium of the Il17rcfl/flScgb1a1-cre− and the Il17rcfl/flScgb1a1-cre+ mice after intranasal IL-17 challenge. Our analysis identified 694 genes that were differentially expressed in the Il17rcfl/flScgb1a1-cre− mice when compared to the Il17rcfl/flScgb1a1-cre+ mice, defined as those with a minimum of a 1.2-fold change in expression (p-value <0.05). Genes that have been reported to be regulated by IL-17 in vitro were highly up-regulated in vivo including chemokines (Ccl20, Cxcl1, Cxcl5), cytokines (Csf3, Il6), and other genes controlled by IL-17 (Lcn2, Nfkbiz, Pigr) (Fig. 4A). Gene set enrichment analysis (GSEA) of the data showed enrichment of the Cytokine-cytokine receptor interaction and Chemokine signaling KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (Fig. 4B). Using the same dataset and the Upstream Regulator Analytic from the Ingenuity Pathway Analysis software, we identified a numbers of signaling pathways related to innate immunity (Lipopolysaccharide, TLR4, RELA) and inflammation (IFNG, TNF, IL1B, TGFB1) (Fig. 4C). Real-time RT-PCR analysis on Cxcl1, Cxcl5, Csf3 and Il6 transcription in mice received the same treatment validated the RNA-seq results (Supplementary Fig. 4). To confirm the findings on chemokines and cytokines related to neutrophil recruitment, we assayed cytokines and chemokines related to neutrophil recruitment in the BAL fluid 24 hours post IL-17 challenge. The levels of G-CSF and IL-6 production in the BAL were reduced in the conditional knockout mice (Fig. 5A). Additionally, ligands for CXCR2, an essential receptor for neutrophil recruitment, CXCL1 (KC) and CXCL5 (LIX) were also analyzed. CXCL1, encoded by Cxcl1, was reduced while CXCL5, encoded by Cxcl5, was substantially reduced in the Il17rafl/flScgb1a1-cre+ mice. To determine if these genes are transcriptionally regulated as suggested by our RNA-seq analysis, we performed real-time RT-PCR but found all were equally expressed in RNA samples obtained from whole lung homogenate (Fig. 5B). However, when gene expression was specifically assayed in the lung epithelium using an optimized mouse bronchial brushing technique, Cxcl5 expression was found to be significantly reduced in Il17rafl/flScgb1a1-cre+ mice compared to littermate Il17rafl/flScgb1a1-cre− controls (Fig. 5B). These data suggest that epithelial Cxcl5 expression is critically regulated by epithelial Il17ra expression (Fig. 5B). To investigate the functional relevance of CXCL5 we designed a CXCL5 rescue experiment in the conditional knockout mice. To assess early neutrophil recruitment in K. pneumoniae infection, we chose an earlier time point, 4h after infection, as the bacterial burden in the conditional KO mice were substantially higher 24h after infection, which could confound interpretations. At 4h post infection, the Il17rafl/flScgb1a1-cre+ had significantly less neutrophils in the BAL as compared to the Il17rafl/flScgb1a1-cre− mice while administration of rCXCL5 intranasally 2h after infection substantially increased the neutrophil counts in the BAL (Fig. 5C). Additionally, at 24h after infection, administration of rCXCL5 at two hours post-infection substantially improved the bacterial clearance of K. pneumoniae in the Il17rafl/flScgb1a1-cre+ mice (Fig. 5D and Supplementary Fig. 5).
Fig. 4. RNA-seq analysis on mouse bronchial brushes from the IL-17RC epithelial conditional KO mice.
Il17rcfl/fl Scgb1a1-Cre− and Il17rcfl/fl Scgb1a1-Cre+ mice were challenged with recombinant IL-17 intranasally (300ng/mouse) for 6h and bronchial brushings were harvested for RNA extraction and RNA-seq analysis. (A). Heat map of selected known IL-17R-dependent genes. (B). Enriched KEGG pathways from the gene set enrichment analysis (GSEA). (C). Enriched pathways using the Upstream Regulator Analytic from the Ingenuity Pathway Analysis software. Real-time RT-PCR validation of these RNA-seq findings was shown in supplementary Fig. 4.
Fig. 5. Loss of Cxcl5 expression in the IL-17RA epithelial conditional KO mice.
Il17rafl/fl Scgb1a1-Cre− and Il17rafl/fl Scgb1a1-Cre+ mice were challenged with recombinant IL-17 intranasally (300ng/mouse) for 24h and IL-17 downstream chemokines and cytokines in the BAL fluids were measured by Luminex (A). Gene expression in whole lung homogenates and bronchial brushings were determined by real-time RT-PCR (B). Il17rafl/fl Scgb1a1-Cre− and Il17rafl/fl Scgb1a1-Cre+ mice were infected with 104 KP43816 intranasally. 2h post infection, a subgroup of Il17rafl/fl Scgb1a1-Cre+ the mice received 1ug recombinant CXCL5 (LIX). Mice were sacrificed at 4h after infection for BAL cell enumeration by FACS (C). A separate cohort of mice were sacrificed at 24h after infection and bacterial burden in the lungs were determined by CFU (D). See also the pathology of these mice in supplementary Fig. 5. Data are represented as mean +/− SEM.
Defective PMN responses and bacterial clearance in IL17RC lung conditional KO mice
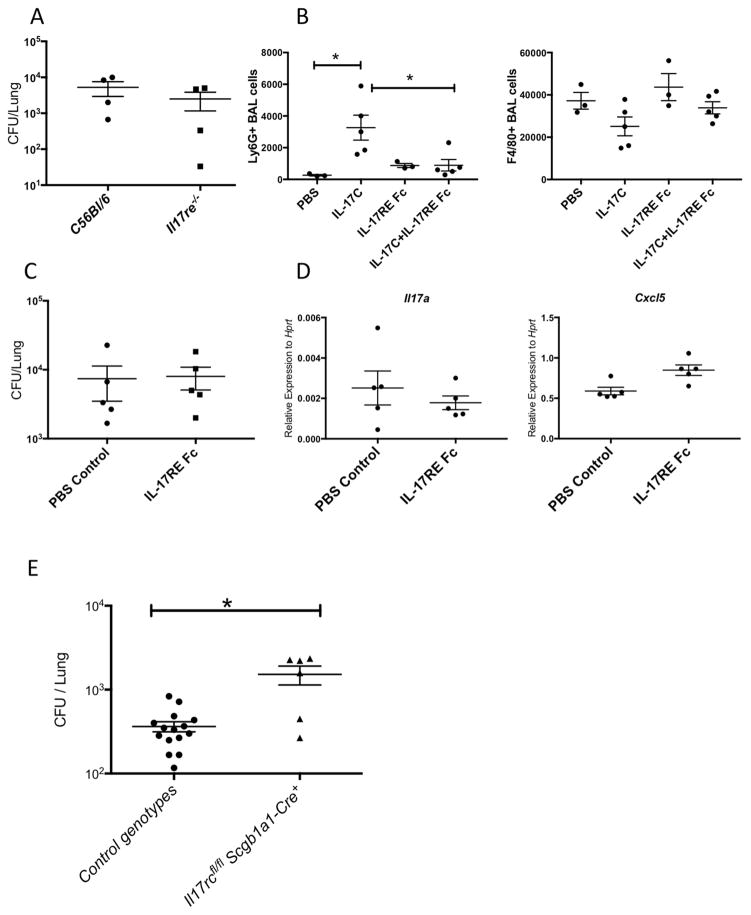
IL-17RA is utilized by several IL-17 family members, thus the defects observed in the Il17rafl/flScgb1a1-cre+ mice could be due defective signaling of several IL-17 family members other than IL-17 or IL-17F. IL-17C, which signals through IL-17RA and IL-17RE, was indeed induced in normal human bronchial epithelial (NHBE) cells in response to IL-17 stimulation (Ray et al., 2015) (Supplementary Fig. 6A). Transcription of Il17c was also induced in vivo in both the intranasal IL-17 challenge as well as K. pneumoniae infection (Supplementary Fig. 6B). However, IL-17C/IL-17RE signaling did not seem to contribute to the induction of chemokine responses and cytokines in response to intranasal IL-17, since neutrophil recruitment and the expression of Cxcl1, Cxcl2, Cxcl5 and Il6 did not change in Il17re−/− mice (Supplementary Fig. 6C&D). IL-17RE is the unique receptor subunit for IL-17C signaling (Swamy and Hayday, 2011). When infected with live K. pneumoniae, the Il17re−/− mice had a similar bacterial burden in the lungs as the wild type controls (Fig. 6). To assess the IL-17C/IL-17RE signaling in the lungs specifically, we sought to block IL-17C in vivo with recombinant IL-17RE Fc protein. To assess the specificity of IL-17RE Fc, mice were treated with IL-17RE Fc in addition to IL-17C. IL-17C was sufficient to induce neutrophil recruitment into the BAL fluid and this was reduced in mice administered IL-17RE Fc, validating that IL-17RE Fc effectively blocks the IL-17C signaling (Fig 6B). However, blocking IL-17C with IL-17RE Fc showed no significant impact on bacteria burden or IL-17 related gene expression in the lungs after K. Pneumoniae infection (Fig. 6C&D). These data suggest that IL-17C signaling is not required for the clearance of K. pneumoniae, emphasizing the important role of IL-17A, the other ligand for IL-17RA, in host defense against K. pneumonia.
Fig. 6. Defective PMN responses and bacterial clearance in the IL-17RC epithelial conditional KO mice.
WT or Il17re+/− as well as littermates Il17re−/− mice were infected with 104 KP-43816 intranasally and sacrificed 24h post infection. Bacterial burden in the lungs were determined by standard CFU assay (A). See also supplementary Fig. 6. C57BL/6 mice were challenged with recombinant IL-17C (500ng/mouse) with or without IL-17RE Fc (500ng/mouse) intranasally and sacrificed at 24h. BAL neutrophils and macrophage numbers were determined by FACS (B). C57BL/6 mice were infected with 104 KP43816 intranasally with or without IL-17RE Fc (500ng/mouse) intranasally and sacrificed at 24h. Bacterial burden in the lungs were determined by CFU (C) and gene expression in the lungs were analyzed by real-time RT-PCR (D). Littermate controls as well as Il17rcfl/fl and Il17rcfl/fl Scgb1a1-Cre+ mice were infected with 105 KP-2146 intranasally and sacrificed 24h post infection. Bacterial burden in the lungs were determined by CFU (E). Data are represented as mean +/− SEM.
In contrast to IL-17RA, IL-17RC is used only by IL-17A and IL-17F (Toy et al., 2006). To confirm the requirement of epithelial IL-17RC in host defense against bacterial infection, we challenged the Il17rcfl/flScgb1a1-cre+ mice with a drug resistant K. pneumoniae isolate expressing New Delhi Metallo-beta-lactamase-1 intranasally. The Il17rcfl/flScgb1a1-cre+ mice indeed demonstrated a defect in controlling of the bacterial burden in the lungs (Fig. 6E).
Discussion
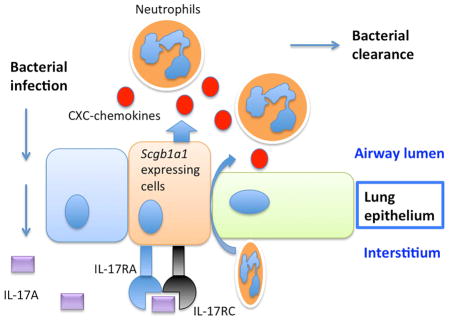
The potential role of lung epithelium in orchestrating chemokine gradients and subsequent recruitment of neutrophils to the airways has been hypothesized for several years (Dubin and Kolls, 2008). This hypothesis is supported by in vitro data showing that the lung epithelial cells are capable of producing various chemokines including IL-8 and CXCL5 upon IL-17 stimulation (Manni et al., 2014). In situ hybridization staining on the lungs from K. pneumoniae infected mice also demonstrated that the mRNA expression of Cxcl1 and Cxcl2 are co-localized within the lung epithelium and administration of IL-17 augmented hybridization signals (Aujla et al., 2008). Moreover, recent data using bone marrow chimeras in Cxcl5−/− mice also demonstrated that non-myeloid cells are the cellular source of CXCL5 (Balamayooran et al., 2012; Liu et al., 2011). These two studies using two different models, smoke induced lung inflammation and E. coli infection respectively, demonstrated that radioresident cells but not hematopoietic cell-driven CXCL5 is important for mediating lung inflammation. Experiments with bone marrow chimeric mice in an M. tuberculosis model also demonstrated that the radioresistant lung-resident cells but not the hematopoietic lineage are responsible for CXCL5-dependent PMN recruitment (Nouailles et al., 2014).
However, there is a lack of data confirming this idea using targeted genetic approaches that can identify specific cell types responsible for CXCL5 production. In this study, Il17rafl/fl mice were crossed to lung epithelial specific Scgb1a1-cre mice to establish mice with epithelial conditional deletion of Il17ra. These mice displayed reduced neutrophil influx into the airways upon intranasal IL-17 challenge. As a consequence, the lung conditional KO mice were also unable to control mucosal K. pneumoniae infection. Analyses of gene expression in the bronchial epithelium as well as the whole long homogenate revealed that there was a major defect in Cxcl5 expression, which was responsible for the inability of the host to control the bacterial burden. To confirm that this phenotype was IL-17A/IL-17F specific, we crossed the Il17rcfl/fl mice to the Scgb1a1-cre mice and found that these mice phenocopied the Il17rafl/fl mice.
We also found that epithelial IL-17R expression is also required for the controlling the growth of the NDM-1+ strain of K. pneumoniae suggesting that compromised epithelial IL-17R signaling may increase susceptibility to infection caused by extracellular pathogens including the antibiotic resistant strains. Indeed, humans with deficiencies in IL-17RA, IL-17RC and IL-17F have been shown to have increased susceptibility to Candida species and develop Chronic mucocutaneous candidiasis (Ling et al., 2015; Puel et al., 2011). Alternatively perhaps IL-17RA or IL-17RA signaling could be augmented to enhance treatment of multi-drug resistant K. pneumoniae infections. The accompanying paper by Conti et al. also demonstrated a critical role for epithelial IL-17R signaling in oral mucosal immunity against C. albicans. In this study, a new CRE-expressing transgenic mouse line specific for the oral and esophageal epithelium, i.e., the K13CRE line, was crossed to Il17rafl/fl mice to generate mice lacking IL-17RA exclusively in the oral/esophageal epithelium. When challenged with Candida albicans, these conditional IL-17R deficient mice phenocopied global Il17ra−/− mice in many aspects including increased mucosal fungal burden, disease symptoms, and gene expression patterns (Conti, 2016). However, the defective effector molecule in these mice was β-defensin 3, which differs somewhat from CXCL5 observed in the Il17rafl/flScgb1a1-cre+ mice. This may reflect different functions of different types of epithelial cells as K13 expression is restricted to the internal stratified squamous epithelial in the oral cavity (Winter et al., 1990) while Scgb1a1 is mostly expressed in cuboidal, non-ciliated, secretory club cells of the bronchiolar epithelium (Simon et al., 2006).
Several groups have observed that blood neutrophil counts are reduced in IL-17R-deficient mice (Kelly et al., 2005; Smith et al., 2008) and this is consistent with our observations in our Il17rafl/flE2a-cre+ mice. This developmental defect could lead to the reduced neutrophil influx in the lung upon intranasal IL-17 administration. These observations led us to generate the epithelial specific KO mice by crossing the IL-17R flox mice to the Scgb1a1-cre line. Scgb1a1-cre did efficiently mediate recombination of Il17ra in the lungs (Supplementary Fig. 3). However, the lung epithelium is heterogeneous and deletion of IL-17R in only one cell type might be expected to exhibit only a partially impaired response. This may explain why only one principal CXCR2 ligand, Cxcl5, was reduced in the Il17rafl/fl Scgb1a1-cre+ mice following IL-17 challenge (Fig. 5A&B). The gene expression from lung homogenate is comparable between two groups since most cells in the lungs from the epithelial IL-17R KO mice still express the IL-17 receptors. For example airway smooth muscle cells and fibroblasts have been shown to produce chemokines in response to IL-17 (Ivanov and Linden, 2007; Kwofie et al., 2015). This study establishes a critical role of epithelial IL-17RA/RC signaling to achieve the necessary chemokine gradients for lumenal egress of neutrophils in the lung. Further studies crossing the Il17rafl/fl mice with additional epithelial specific CRE lines are needed to determine the exact roles of epithelial IL-17R in pulmonary host defense as well as chronic lung inflammation in other cell lineages. For example, a recently established mouse model incorporated a tamoxifen-inducible Cre recombinase (Cre-ERT2) under the control of the human surfactant protein C (SPC) promoter which deletes in distal lung epithelium (Gui et al., 2012). Crossing this line with the IL-17R flox mice would permit ablation of IL-17R in the distal lung in a temporally controlled manner.
Another study using an E. coli pneumonia model demonstrated that a bacterial clearance defect when ablating Onconstatin-M during pneumonia was also dependent on the epithelial expression of Cxcl5 (Traber et al., 2015). These studies together with the present study emphasize the important role of epithelial derived CXCL5-secreting cells in orchestrating PMN recruitment to bacterial infection in the lung. Indeed, the essential role of CXCL5 in multiple models demands a more comprehensive analysis of this chemokine, which shares receptors with many others such as CXCL1 and CXCL2.
Taken together, our study demonstrated that epithelial IL-17R signaling is essential in regulating chemokine pathways in vivo, particularly Cxcl5 expression and subsequent recruitment of neutrophils for the clearance of bacteria in the lungs. Loss of epithelial IL-17R signaling may compromise host defense against extracellular bacteria and targeting this molecular pathway may provide clinical benefits for patients with pneumonia.
Experimental Procedures
Animal Studies
8–10 week old mice were used for in vivo studies. The Il17rafl/fl and Il17rcfl/fl mice were generated at Ozgene and have been recently described (Kumar et al., 2016). Il17re−/− mice were acquired from Genentech (Conti et al., 2015). Recombinant cytokines or 104 K. pneumoniae were given to isoflurane anesthetized mice in cold sterile PBS (50μL) by intranasal injection. All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Experimental K. pneumoniae Infection
Klebsiella pneumoniae ATCC strain 43816, serotype 2 or BAA-2146 (NDM1+) (ATCC, Rockville, MD) isolates were grown in 100 mL of tryptic soy broth (Difco, Detroit, MI) for 18 h at 37°C. The culture was then diluted at 1:100 and grown for 2 additional hours to reach early logarithmic phase. The concentration of K. pneumoniae was determined by measuring the absorbance at 600 nm. Bacteria were pelleted by centrifugation at 5,000 rpm for 15 min, washed twice in cold PBS, and resuspended at the desired concentration. 24h after infection, mice were sacrificed and left lungs were harvested in TriZol for RNA extraction, right lungs were harvested in sterile PBS for bacterial burden (CFU) and Luminex (Millipore) on the Bioplex reader (Bio-Rad).
Luminex
Kits with custom luminex panel including G-CSF, IL-6, CXCL1, CXCL5 and CCL2 were purchased from EMD Millipore Corporation (Billerica, MA) and the assays were performed per manufacturer instructions.
RNA isolation from mouse bronchial brush
The bronchial brushing was performed using a 5–7cm long PE50 tubing (Becton Dickinson), sanding to create roughness (P240 sand paper), and treating with RNaseZap (Ambion). Two sanded tubes were inserted into the right main and left main bronchus separately with gentle brushing (twisting) and immediately placed in the same eppendorff tube containing 1 mL TriZol (life technology). These tubes were vortexed thoroughly before adding chloroform for RNA extraction and RNA carrier Glycoblue (life technology) was added during RNA precipitation. RNA samples were then quantified by spectrophotometer and subjected to cDNA synthesis and gene expression analysis. RNA sequencing data was analyzed using the CLC Genomics Workbench software (QIAGEN) and the raw data have been deposited into the Sequencing Read Archive under SRA accession number SRP087890.
Real Time PCR
RNA was isolated using TriZol reagent (Life Technology) and cDNA was prepared using iScript reverse transcriptase master mix (Bio-Rad, Hercules, CA). Real time PCR was carried out with Bio-rad CFX96 system using TaqMan PCR Master Mix (Life Technologies) and premixed primers/probe sets from Life Technologies.
Flow cytometry
anti-mouse Gr-1, Ly6G, CD11b and F4/80 monoclonal antibodies were purchased from eBioscience. Data were acquired using FACS LSR II (BD Bioscience) and analyzed by FlowJo software (Treestar, CA).
Ear skin fibroblasts isolation and stimulation
Mouse ear skin pieces (app. 3×3mm) were washed with 70% EtOH and transferred into DMEM. Skin pieces were then cut into small pieces using scalpel before incubation for 1hr at 37 °C (water bath) with 0.25% trypsin. Cells were then pelleted and re-suspended in DMEM/10%FCS and fed with fresh medium the next day. IL-17 stimulation was performed once the fibroblasts reached 70% confluence.
Pathology scores
Hematoxylin and eosin–stained slides were coded and scored from 0 (absent) to 4 (severe) for the following parameters: interstitial inflammation, endothelialitis, bronchitis, edema, thrombi, pleuritis and percentage of the lung surface demonstrating confluent (diffuse) inflammatory infiltrate by a pathologist blinded for groups. The total “lung inflammation score” was expressed as the sum of the scores for each parameter.
Statistical Analyses
Unpaired, two-tailed, Student’s t tests, ∝ = 0.05, were used to assess whether the means of two normally distributed groups differed significantly. One-way ANOVA analysis was used to compare multiple means. Significance is indicated as P < 0.05. All error bars represent the SEM.
Supplementary Material
Acknowledgments
We thank Mei Hulver for technical assistance on K. pneumoniae infection in the IL-17RE KO mice. This work was supported by R37-HL079142 (KC and JKK). KC was also supported by the Research Advisory Committee of Children’s Hospital of Pittsburgh of UPMC. SLG was supported by NIH grants AI107825 and DE022550.
Footnotes
Authors Contributions:
KC and JKK designed all the experiments and wrote the manuscript. KC performed all the in vivo experiments with the assistance of GT, CJE, MB and AVG. TE scored the histology. EEW, WE and DMR participated in the characterization of the flox mice and performed in vitro experiments. TW and WC provided assistance on RNA-seq analysis. SLG and JSL provided the reagents for the experiments involving the Il17re−/− mice and helped with manuscript writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase - United States. MMWR Morb Mortal Wkly Rep. 2010;59:750. [PubMed] [Google Scholar]
- 2.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balamayooran G, Batra S, Cai S, Mei J, Worthen GS, Penn AL, Jeyaseelan S. Role of CXCL5 in leukocyte recruitment to the lungs during secondhand smoke exposure. Am J Respir Cell Mol Biol. 2012;47:104–111. doi: 10.1165/rcmb.2011-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertin G, Poujeol C, Rubera I, Poujeol P, Tauc M. In vivo Cre/loxP mediated recombination in mouse Clara cells. Transgenic Res. 2005;14:645–654. doi: 10.1007/s11248-005-7214-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen K, Kolls JK. T cell-mediated host immune defenses in the lung. Annu Rev Immunol. 2013;31:605–633. doi: 10.1146/annurev-immunol-032712-100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conti HR, Bruno VM, Childs EE, Daugherty S, Hunter JP, Mengesha BG, Saevig DL, Hendricks MR, Coleman BM, Brane L, Solis N, Cruz JA, Verma AH, Garg AV, Hise AG, Richardson JP, Naglik JR, Filler SG, Kolls JK, Sinha S, Gaffen SL. IL-17 receptor signaling in oral epithelial cells is necessary and sufficient for protection against oropharyngeal candidiasis. Cell Host & Microbe. 2016 doi: 10.1016/j.chom.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti HR, Whibley N, Coleman BM, Garg AV, Jaycox JR, Gaffen SL. Signaling through IL-17C/IL-17RE is dispensable for immunity to systemic, oral and cutaneous candidiasis. PLoS One. 2015;10:e0122807. doi: 10.1371/journal.pone.0122807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubin PJ, Kolls JK. Th17 cytokines and mucosal immunity. Immunol Rev. 2008;226:160–171. doi: 10.1111/j.1600-065X.2008.00703.x. [DOI] [PubMed] [Google Scholar]
- 9.Dubin PJ, Martz A, Eisenstatt JR, Fox MD, Logar A, Kolls JK. Interleukin-23-mediated inflammation in Pseudomonas aeruginosa pulmonary infection. Infect Immun. 2012;80:398–409. doi: 10.1128/IAI.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge S, Hertel B, Susnik N, Rong S, Dittrich AM, Schmitt R, Haller H, von Vietinghoff S. Interleukin 17 receptor A modulates monocyte subsets and macrophage generation in vivo. PLoS One. 2014;9:e85461. doi: 10.1371/journal.pone.0085461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gui YS, Wang L, Tian X, Feng R, Ma A, Cai B, Zhang H, Xu KF. SPC-Cre-ERT2 transgenic mouse for temporal gene deletion in alveolar epithelial cells. PLoS One. 2012;7:e46076. doi: 10.1371/journal.pone.0046076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, Lambrecht BN. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herjan T, Yao P, Qian W, Li X, Liu C, Bulek K, Sun D, Yang WP, Zhu J, He A, et al. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J Immunol. 2013;191:640–649. doi: 10.4049/jimmunol.1203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov S, Linden A. Th-17 cells in the lungs? Expert Rev Respir Med. 2007;1:279–293. doi: 10.1586/17476348.1.2.279. [DOI] [PubMed] [Google Scholar]
- 16.Kelly MN, Kolls JK, Happel K, Schwartzman JD, Schwarzenberger P, Combe C, Moretto M, Khan IA. Interleukin-17/interleukin-17 receptor-mediated signaling is important for generation of an optimal polymorphonuclear response against Toxoplasma gondii infection. Infect Immun. 2005;73:617–621. doi: 10.1128/IAI.73.1.617-621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, et al. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J Immunol. 2007;179:5462–5473. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity. 2016;44:659–671. doi: 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Subramaniyam G. Molecular underpinnings of Th17 immune-regulation and their implications in autoimmune diabetes. Cytokine. 2015;71:366–376. doi: 10.1016/j.cyto.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Kwofie K, Scott M, Rodrigues R, Guerette J, Radford K, Nair P, Richards CD. Regulation of IL-17A responses in human airway smooth muscle cells by Oncostatin M. Respir Res. 2015;16:14. doi: 10.1186/s12931-014-0164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden A, Laan M, Anderson GP. Neutrophils, interleukin-17A and lung disease. Eur Respir J. 2005;25:159–172. doi: 10.1183/09031936.04.00032904. [DOI] [PubMed] [Google Scholar]
- 23.Ling Y, Cypowyj S, Aytekin C, Galicchio M, Camcioglu Y, Nepesov S, Ikinciogullari A, Dogu F, Belkadi A, Levy R, et al. Inherited IL-17RC deficiency in patients with chronic mucocutaneous candidiasis. J Exp Med. 2015;212:619–631. doi: 10.1084/jem.20141065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Mei J, Gonzales L, Yang G, Dai N, Wang P, Zhang P, Favara M, Malcolm KC, Guttentag S, Worthen GS. IL-17A and TNF-alpha exert synergistic effects on expression of CXCL5 by alveolar type II cells in vivo and in vitro. J Immunol. 2011;186:3197–3205. doi: 10.4049/jimmunol.1002016. [DOI] [PubMed] [Google Scholar]
- 25.Manni ML, Robinson KM, Alcorn JF. A tale of two cytokines: IL-17 and IL-22 in asthma and infection. Expert Rev Respir Med. 2014;8:25–42. doi: 10.1586/17476348.2014.854167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, Finder JD, Pilewski JM, Carreno BM, Goldman SJ, et al. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F, Wang K, Aoyama T, Grivennikov SI, Paik Y, Scholten D, Cong M, Iwaisako K, Liu X, Zhang M, et al. Interleukin-17 signaling in inflammatory, Kupffer cells, and hepatic stellate cells exacerbates liver fibrosis in mice. Gastroenterology. 2012;143:765–776. e761–763. doi: 10.1053/j.gastro.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nouailles G, Dorhoi A, Koch M, Zerrahn J, Weiner J, 3rd, Fae KC, Arrey F, Kuhlmann S, Bandermann S, Loewe D, et al. CXCL5-secreting pulmonary epithelial cells drive destructive neutrophilic inflammation in tuberculosis. J Clin Invest. 2014;124:1268–1282. doi: 10.1172/JCI72030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh K, Park HB, Byoun OJ, Shin DM, Jeong EM, Kim YW, Kim YS, Melino G, Kim IG, Lee DS. Epithelial transglutaminase 2 is needed for T cell interleukin-17 production and subsequent pulmonary inflammation and fibrosis in bleomycin-treated mice. J Exp Med. 2011;208:1707–1719. doi: 10.1084/jem.20101457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pappu R, Ramirez-Carrozzi V, Sambandam A. The interleukin-17 cytokine family: critical players in host defence and inflammatory diseases. Immunology. 2011;134:8–16. doi: 10.1111/j.1365-2567.2011.03465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price AE, Reinhardt RL, Liang HE, Locksley RM. Marking and quantifying IL-17A-producing cells in vivo. PLoS One. 2012;7:e39750. doi: 10.1371/journal.pone.0039750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray M, Horne W, McAleer JP, Ricks DM, Kreindler JL, Fitzsimons MS, Chan PP, Trevejo-Nunez G, Chen K, Fajt M, et al. RNA-seq in Pulmonary Medicine: How Much Is Enough? Am J Respir Crit Care Med. 2015;192:389–391. doi: 10.1164/rccm.201403-0475LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon DM, Arikan MC, Srisuma S, Bhattacharya S, Tsai LW, Ingenito EP, Gonzalez F, Shapiro SD, Mariani TJ. Epithelial cell PPAR[gamma] contributes to normal lung maturation. FASEB J. 2006;20:1507–1509. doi: 10.1096/fj.05-5410fje. [DOI] [PubMed] [Google Scholar]
- 36.Smith E, Stark MA, Zarbock A, Burcin TL, Bruce AC, Vaswani D, Foley P, Ley K. IL-17A inhibits the expansion of IL-17A-producing T cells in mice through “short-loop” inhibition via IL-17 receptor. J Immunol. 2008;181:1357–1364. doi: 10.4049/jimmunol.181.2.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swamy M, Hayday A. Provocative exhibits at the Seventeen Gallery. Nat Immunol. 2011;12:1131–1133. doi: 10.1038/ni.2164. [DOI] [PubMed] [Google Scholar]
- 38.Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, Tocker J, Peschon J. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J Immunol. 2006;177:36–39. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- 39.Traber KE, Hilliard KL, Allen E, Wasserman GA, Yamamoto K, Jones MR, Mizgerd JP, Quinton LJ. Induction of STAT3-Dependent CXCL5 Expression and Neutrophil Recruitment by Oncostatin-M during Pneumonia. Am J Respir Cell Mol Biol. 2015;53:479–488. doi: 10.1165/rcmb.2014-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Way EE, Chen K, Kolls JK. Dysregulation in lung immunity - the protective and pathologic Th17 response in infection. Eur J Immunol. 2013;43:3116–3124. doi: 10.1002/eji.201343713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter H, Rentrop M, Nischt R, Schweizer J. Tissue-specific expression of murine keratin K13 in internal stratified squamous epithelia and its aberrant expression during two-stage mouse skin carcinogenesis is associated with the methylation state of a distinct Cog site in the remote 5′-flanking region of the gene. Differentiation. 1990;43:105–114. doi: 10.1111/j.1432-0436.1990.tb00436.x. [DOI] [PubMed] [Google Scholar]
- 42.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.