Abstract
Purpose of this study
To elucidate the origin of cell populations that contribute to rotator cuff healing, we developed a mouse surgical model where a full-thickness, central detachment is created in the supraspinatus.
Materials and Methods
Three different inducible Cre transgenic mice with Ai9-tdTomato reporter expression (PRG4-9, αSMA-9, and AGC-9) were used to label different cell populations in the shoulder. The defect was created surgically in the supraspinatus. The mice were injected with tamoxifen at surgery to label the cells and sacrificed at 1, 2, and 5 weeks post-operatively. Frozen sections were fluorescently imaged then stained with Toluidine Blue and re-imaged.
Results
Three notable changes were apparent post-operatively. 1) A long thin layer of tissue formed on the bursal side overlying the supraspinatus tendon. 2) The tendon proximal to the defect initially became hypercellular and disorganized. 3) The distal stump at the insertion underwent minimal remodeling. In the uninjured shoulder, tdTomato expression was seen in the tendon midsubstance and paratenon cell on the bursal side in PRG4-9, in paratenon, blood vessels, and periosteum of acromion in SMA-9, and in articular cartilage, unmineralized fibrocartilage of supraspinatus enthesis, and acromioclavicular joint in AGC-9 mice. In the injured PRG4-9 and SMA-9 mice, the healing tissues contained an abundant number of tdTomato+ cells, while minimal contribution of tdTomato+ cells was seen in AGC-9 mice.
Conclusions
The study supports the importance of the bursal side of the tendon to rotator cuff healing and PRG4 and αSMA may be markers for these progenitor cells.
Keywords: tendon healing, supraspinatus tendon, rotator cuff, lineage tracing, alpha smooth muscle actin, Prg4, lubricin
INTRODUCTION
Rotator cuff pathology is a common source of shoulder pain, estimated to account for over 4.5 million clinician visits and 250,000 surgeries annually in the United States [1]. The burden will likely continue to increase with the aging population, as the incidence and prevalence of rotator cuff ailments increases with age [2].
Despite advances in imaging technology, operative technique, and surgical hardware, rotator cuff injuries are still challenging to treat. Partial-thickness tears are difficult to manage, often do not heal [3], and can progress to full-thickness tears. In addition, the appropriate timing and indications for surgical intervention are under debate [4]. Many patients with full-thickness tears that fail conservative management undergo surgical repair, but retears are common, especially with large-sized tears [5,6].
To address these limitations, tissue engineering and biologic augmentations to improve rotator cuff healing have gained much interest [1,7,8]. It is generally thought the key to success of these tissue engineering strategies lies in utilizing and/or stimulating the appropriate target population of cells. In a successful biological augmentation, the tissue engineered construct should support and stimulate the surrounding resident cells to heal the rotator cuff tendons. However, surprisingly little is known about the types and sources of cells that have the potential to directly contribute to rotator cuff healing.
There is still debate regarding whether tendon healing is an intrinsic or extrinsic process [9,10]. The supraspinatus tendon has a number of surrounding tissues that could harbor cells that contribute to the healing response. These tissues include the joint capsule and synovial fluid on the articular side of the tendon and the overlying paratenon, sub-acromial bursa, deltoid muscle, and acromion on the bursal side of the tendon. The paratenon has been shown to be a source of cells that contributes to healing [11–13] as alpha smooth muscle actin (αSMA-CreERT2 x Rosa26-tdTomato) positive paratenon cells turn on Scleraxis (ScxGFP+) during patellar tendon healing [11]. However, the contribution of SMA+ cells to supraspinatus healing has not been demonstrated in this mouse model. The Proteoglycan 4 (Prg4) gene, which encodes for lubricin among other proteins, may have a role in fascicle gliding [14] in tendon, but is also strongly expressed in the synovium of the joint capsule and in articular cartilage [15]. While lubricin expression has been shown during healing of the supraspinatus tendon [16], the contribution of Prg4-expressing cells to the healing response, particularly from the tendon fascicle or underlying synovium, remains largely unknown. In addition, rotator cuff tendon injuries typically occur near the fibrocartilage of the enthesis and the ability of these cells to contribute to the injury response has not been fully studied. Therefore, this study will utilize the inducible Cre mouse driven by the gene aggrecan [17] to label fibrocartilage cells within the enthesis. Thus, while the stages of tendon healing (inflammation, repair, remodeling) have been characterized in numerous pre-clinical studies [18–21], the true origin of cells that contribute to the tendon healing response and the markers that define these cells are lacking because of limited fate-mapping models.
The purpose of the current study was to address this fundamental gap in knowledge by using lineage tracing models to identify the origin of cells that contribute to rotator cuff healing. We developed a mouse surgical model where a central detachment is created in the supraspinatus, and the response of the different cell populations were studied after the surgery. To label distinct cell populations in the shoulder and trace their contribution to healing over time, we used three different inducible Cre transgenic mouse models that controlled CreERT2 expression from the gene regulatory elements Prg4, αSMA, and Aggrecan (Acan).
MATERIALS AND METHODS
Transgenic mice
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. An animal care protocol was approved by the Institutional Animal Care and Use Committee of the University of Connecticut Health Center. Cre recombinase fused to a modified estrogen receptor ligand binding domain (ERT2) allowed for temporal activation of the enzyme by tamoxifen injection [22]. The three CreERT2 transgenic mouse models used in this study used DNA transcriptional regulatory elements derived from the following genes: 1) PRG4-CreERT2 (detailed description of this mouse is detailed in another manuscript currently under review), 2) αSMA-CreERT2 [11,23–25], and 3) Aggrecan (AGC)-CreERT2 [17]. The three Cre driver mice were crossed with Ai9 Rosa26-tdTomato reporter mice [26] to generate dual transgenic offspring (PRG4-9, SMA-9, and AGC-9, respectively) that would allow the activation of Cre reporter expression upon Tamoxifen injection to label cells tdTomato+ (tdTom+) at the time of surgery and then trace their contribution to tendon healing over time.
Experimental Design
The limbs from the Cre-Ai9 mice were harvested at 1, 2, and 5 weeks post-surgery similar to our previous studies carried out in the patellar tendon [11,18,27]. The limbs were assigned to groups based on transgenic strain, time point post-surgery, and surgical treatment and sectioned in either the coronal or sagittal plane (n=2–4 per group).
Supraspinatus central detachment
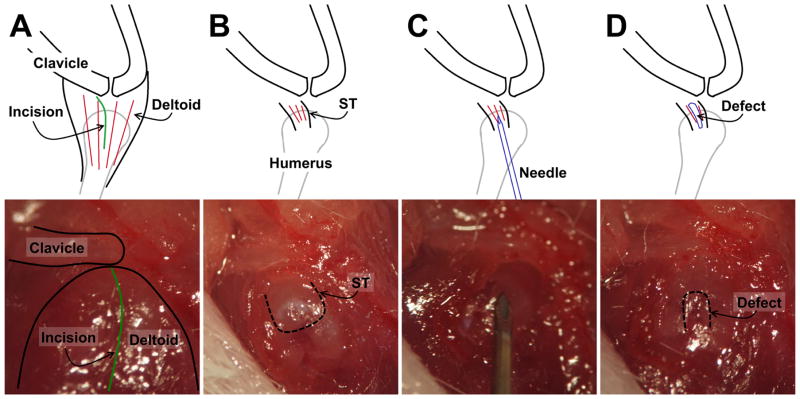
The Cre-Ai9 mice underwent surgery at 15.2±5.1 (mean±SD) weeks of age. The mice were anesthetized with isoflurane gas and received a single intraperitoneal injection of tamoxifen in corn oil (80 μg/g). The shoulder was shaved with clippers and prepped with alcohol and betadine. An anterolateral incision was made over the left shoulder followed by a small longitudinal incision in the lateral deltoid to expose the supraspinatus. A 29G needle was slid along the center of the supraspinatus insertion from lateral to medial to detach the central portion of the tendon (Fig. 1). The articular surface of the humeral head was visualized to confirm that a full-thickness detachment was created. The deltoid and the skin were closed with a 6–0 nylon suture. Half of the mice received a sham procedure on the contralateral shoulder where the supraspinatus was exposed through the deltoid but not injured while the remaining mice had intact contralateral shoulders.
Figure 1. Full-thickness, central width supraspinatus tendon detachment procedure.
A) The lateral deltoid was identified after opening the skin. B) The deltoid was split (green line in A) to expose the supraspinatus tendon (dotted line indicates tendon border). C) A 29-gauge needle was inserted in a lateral-to-medial motion to create the full-thickness, central-width detachment (D, dotted line indicates defect border).
Histology
Mice were euthanized at 1, 2, or 5 weeks after surgery. The upper extremities were harvested and fixed in 10% formalin for 2 days at 4ºC and then placed in PBS with 30% sucrose overnight. The specimens were embedded in Cryomatrix (Thermo Scientific) and 7–8 μm tissue sections were created using a cryofilm technique [28–31]. Tissue sections were rehydrated with PBS and imaged on the Axio Scan.Z1 microscope (Zeiss) for tdTomato expression and tissue architecture via darkfield imaging. Following fluorescent imaging, cover slips were removed and tissue sections were washed in dH2O and stained with 0.025% toluidine blue O (Sigma Aldrich) in ddH2O for one minute. Then the same tissue sections were imaged a second time under brightfield optics using a Axio Scan.Z1 microscope.
Cell quantification
Overlay images of the two rounds of scans were created using Photoshop CS2 (Adobe). The areas with healing tissue were determined using toluidine blue images. The areas were measured and the number of total cells in each area was counted from the DAPI image using the particle analyzer in Fiji image analysis software [32]. The percentage of tdTomato+ positive nuclei was determined based on equivalent minimum thresholds. The percentage of tdTom+ cells in the healing tissue was compared between the PRG4-9, SMA-9, and AGC-9 mice at 2 weeks post-surgery.
Statistical analysis
Image quantification results were evaluated via one way ANOVA with mouse strain as the fixed factor. Tukey’s HSD post-hoc comparisons were made between groups. The significance level was set to p < 0.05.
RESULTS
Healing response to full-thickness, central-width supraspinatus detachment
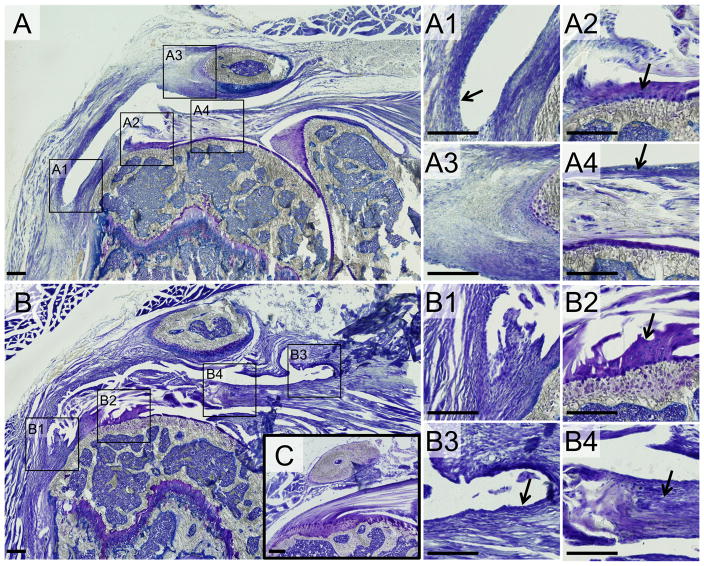
Three notable changes were consistently observed in the mouse shoulder post-operatively. First, a long thin layer of tissue formed on the bursal side overlying the supraspinatus tendon (Fig. 2A–B). This continuous structure was adhered to the surface of the humerus distal to the enthesis (Fig. 2A1), to the surface of the acromion (Fig. 2A3), and to the proximal surface of the tendon and/or epimysium of the supraspinatus (arrow in Fig. 2B3). The tissue had a joint capsule-like appearance as well. Second, the tendinous region proximal to the detachment underwent structural changes, becoming disrupted and even hypocellular at 1 week post-surgery (Fig. 2A4) then transitioning to highly disorganized and cellular at 2 weeks (arrow in Fig. 2B4). These structural changes were often associated with a thickened paratenon (arrows in Fig. 2A4 and 3B). Finally, the remaining tendon stump at the supraspinatus insertion site was mostly unchanged during the healing process with little remodeling of the fibrocartilage tissue (arrows in Fig. 2A2,B2).
Figure 2. The injured shoulder displayed three notable changes.
(A) 1 week post-surgery and (B) 2 weeks post-surgery compared to uninjured control (C). 1) A thin layer of connective tissue formed between the bursal surface of the tendon and the inferior surface of the deltoid and acromion (A1, B1, B3). 2) The region of the supraspinatus tendon proximal to the detachment became disorganized and hyper-cellular (arrow in B4) and the paratenon on the bursal surface of the tendon was thickened (arrow in A4). 4) The remaining tendon tissue near the enthesis was largely unchanged with minimal remodeling (arrows in A2, B2). All images are from coronal sections in the central region of the tendon where the tendon was detached. Scale bars = 200μm.
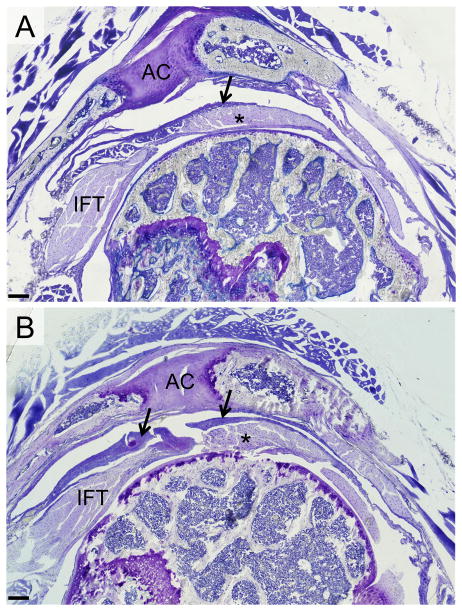
Figure 3. The healing tissue extends beyond the surface of the supraspinatus tendon.
The shoulders were also sectioned in the sagittal plane to assess whether the healing tissue extended over to adjacent rotator cuff tendons. Indeed, the healing tissue on the bursal side of the tendon extended over to the surface of the infraspinatus tendon (IFT) (arrows in B). Panel A is the normal shoulder displaying the IFT, supraspinatus tendon (*), and AC joint (AC). Panel B is the injured shoulder at 2 weeks post-surgery. Scale bars = 200μm.
Cell types labeled in the shoulder joint by Prg4, aSMA, and AGC inducible Cre models
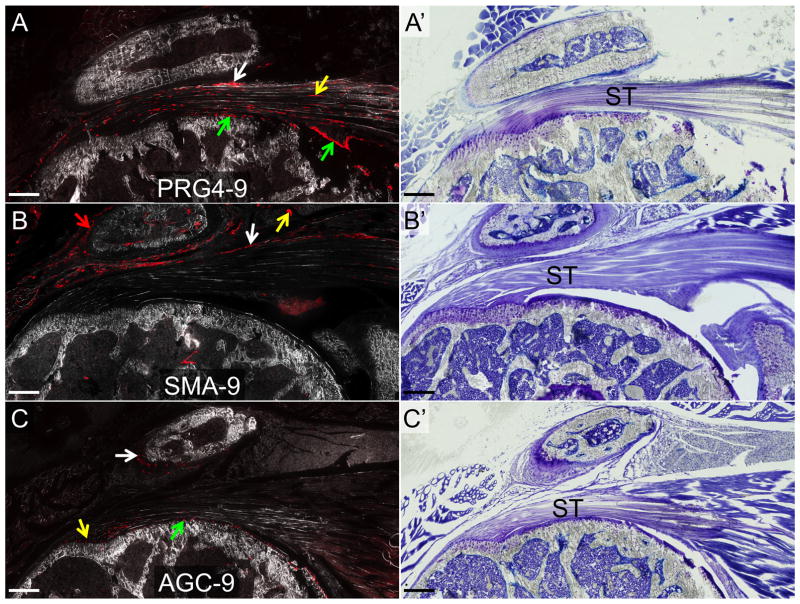
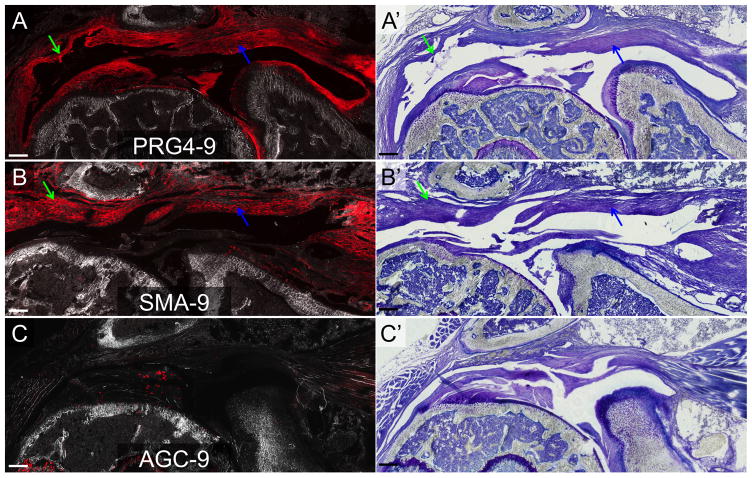
PRG4-9, SMA-9 and AGC-9 models labeled overlapping and distinct cell types in the shoulder joint during the 5-week period. Time post-injection had no effect on the type or number of cells labeled (p > 0.05). PRG4-9 mice showed tdTomato (tdTom) expression in cells in the tendon midsubstance (yellow arrow) and in the paratenon cells (white arrow) on the bursal side of the supraspinatus (Fig. 4A). tdTom+ cells were also evident in the articular cartilage of the humerus and joint capsule (green arrows in Fig. 4A). SMA-9 mice showed tdTom+ cells in the paratenon (white arrow), blood vessels (yellow arrow), and periosteum (red arrow) surrounding the clavicle/acromion (Fig. 4B). In the AGC-9 mice, showed tdTom expression in cells in the articular cartilage of the humerus (green arrow), in unmineralized fibrocartilage at the supraspinatus tendon enthesis (yellow arrow), and in fibrocartilage cells of the acromioclavicular (AC) joint (white arrow). However, no tdTom+ cells were seen elsewhere in the tendon midsubstance, myotendinous junction, or paratenon (Fig. 2C).
Figure 4. tdTom+ cells in the normal shoulder of the PRG4-9, SMA-9, and AGC-9 mice.
A) PRG4-9 mice: tdTom+ cells were evident in the tendon midsubstance, the underlying articular cartilage and synovium as well as the overlying paratenon and deltoid epimysium. B) SMA-9 mice: tdTom+ cells were evident in the overlying paratenon, blood vessels, and deltoid epimysium. C) AGC-9: tdTom+ cells were evident in the supraspinatus tendon (ST) enthesis, acromioclavicular joint, and underlying articular cartilage. Panels A–C are composite images of tdTom fluorescence and darkfield. Panels A′, B′, and C′ are toluidine blue images from the same section as panels A–C. Scale bars = 200μm.
Contribution of different Cre-Ai9+ populations to the healing tissues
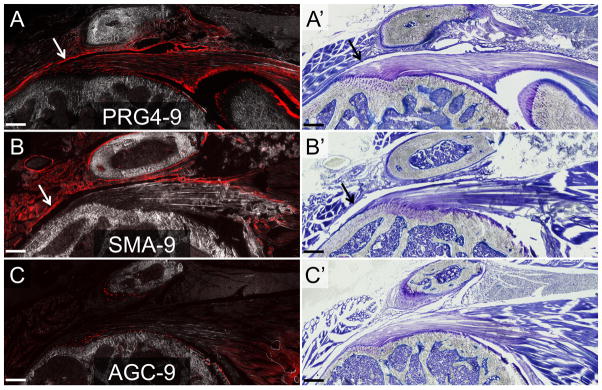
In the PRG4-9 and SMA-9 mice, the overlying layer of capsule-like tissue that forms in response to the tendon injury contained an abundant number of tdTom+ cells (green arrows in Fig. 5A–B), indicating that progeny of these cell populations contributed to this tissue. The density of tdTom+ cells in this tissue in the PRG4-9 and SMA-9 mice was significantly higher than in the AGC mice (p < 0.05), while the total cell density was not different between the groups (p > 0.05). The percentage of tdTom+ cells in this capsule-like tissue was also significantly higher in the PRG4-9 and SMA-9 mice compared to the AGC-9 mice (55.7±14.0% (mean±SD), 43.5±10.9%, and 1.0±0.4% of total healing cells, respectively; p < 0.05). Time post-surgery did not affect the percentage of tdTom+ cells in this tissue (p > 0.05).
Figure 5. PRG4-9+ and SMA-9+ cells were significant contributors to the healing tissue following supraspinatus detachment.
The healing tissue, which was predominately located on the bursal side of the tendon, was created largely by progeny of PRG4-9+ and SMA-9+ cells (A–B). In contrast, the AGC-9+ cells showed minimal expansion following injury (C). All images are from two weeks post-surgery. Panels A–C are composite images of tdTom fluorescence and darkfield. Panels A′, B′, and C′ are toluidine blue images from the same section as panels A–C. Scale bars = 200μm.
In addition, the number of tdTom+ cells in the supraspinatus tendon proximal to the detachment site increased after surgery in both the PRG4-9 (84.5±19.2% vs 35.8±23.0%) and SMA-9 (41.0±22.0% vs 3.7±3.9%) mice (blue arrows in Fig. 5A–B) compared to normal controls. In contrast, the tdTom+ cell population in AGC-9 mice showed no expansion after the surgery and minimal contribution to the proximal tendon (0.6±1.0% vs 0.03±0.13%). Finally, the percentage of tdTom+ cells in the detached tendon was significantly higher in the PRG4-9 than SMA-9 mice (p < 0.05). However, there were significantly more tdTom+ cells in the normal PRG4-9 tendons compared to the SMA-9 tendons (p < 0.05), likely contributing to the increase seen in the injured tendon.
Sham procedure elicited a modest response in the deltoid muscle
Because an injury was created to the deltoid muscle to expose the supraspinatus tendon prior to creating the detachment, we also conducted a sham procedure to determine the healing response of the deltoid injury alone. There was a modest reaction in the shoulder to the sham procedure. The inferior epimysium of the deltoid was thickened in the sham limbs (arrows in Fig. 6). The cells in this tissue were tdTom+ in both the PRG4-9 and SMA-9 mice (Fig. 6A–B) but not in the AGC-9 mice (Fig. 6C). tdTom+ muscle fibers were also seen in the SMA-9 mice because this model is known to label a satellite cell population [25]. While the epimysium thickened in response to the sham procedure, there was no evidence of the capsule-like healing tissue in the subacromial space as seen in the limbs that underwent supraspinatus detachment (Fig. 2).
Figure 6. PRG4-9+ and SMA-9+ cells contributed to a thickened deltoid epimysium following the sham procedure.
The epimysium lining over the undersurface of the deltoid muscle became thickened in response to the sham procedure. The thickened epimysium was predominantly composed of PRG4-9+ (A) and SMA-9+ (B) cells (see arrows) but had minimal AGC-9+ (C) cells. The images are from two weeks post-surgery. Panels A–C are composite images of tdTom fluorescence and darkfield. Panels A′, B′, and C′ are toluidine blue images from the same section as panels A–C. Scale bars = 200μm.
DISCUSSION
There was a consistent healing response in these transgenic mice following full-thickness, partial-width supraspinatus detachment injury (Figs. 2–3). A long, thin layer of capsule-like tissue formed on the bursal side of the supraspinatus tendon. This tissue formed adhesions to the proximal humerus just distal to the supraspinatus tendon insertion, to the proximal supraspinatus tendon and epimysium, and to the undersurfaces of the acromion and deltoid muscle. A gap was usually seen between this layer of tissue and the torn supraspinatus tendon. This capsule-like structure likely formed because the glenohumeral joint space was compromised via the supraspinatus detachment. Therefore, synovial fluid likely filled the subacromial space and the cells responded by creating a new joint capsule in this region. Secondly, the midsubstance of the supraspinatus tendon proximal to the detachment became hypercellular and disorganized. These changes may have reflected the altered loading from the injury as well as partial retraction of the injured tendon. Thirdly, the tendon stump leftover at the enthesis distal to the detachment was largely unchanged during the healing period with minimal remodeling of the fibrocartilage at the enthesis.
Most rotator cuff tears occur as chronic degenerative tears, which may or may not be symptomatic. Our animal model was dissimilar to the typical degenerative rotator cuff in that it was a surgically induced acute injury, which may not model the biology and pathogenesis of rotator cuff tendinosis. However, the model had several advantages. First, the detachment was a consistent size (29G needle) through the full thickness of the tendon. The typical width of the supraspinatus tendon of our mice was between 0.7 and 0.8mm, so the detachment was slightly less than 50% of the tendon width, which corresponds to the size of a moderate full-thickness tear seen clinically. Second, limiting the detachment to the central portion of the tendon prevented the supraspinatus from completely retracting. Similar to humans, a large rotator cuff tear in mice is known to lead to retraction and muscle atrophy [33]. While such a model may be useful for studying chronic changes after massive rotator cuff tear, the cuff tear is unlikely to heal. With our model, the detachment did not propagate leading to full retraction of the tendon. Third, care was taken during the procedure to avoid compromising the cortex of the humerus during surgery. In clinical practice, the cortex is typically compromised during surgical repair when inserting suture anchors or creating bone tunnels. Many surgeons actually debride the bone to create a bleeding surface to improve tendon-to-bone healing. However, the purpose of this model was to understand the autologous healing response to rotator cuff tears, not surgically-repaired rotator cuff. Therefore, the cortex was not compromised in this study.
To delineate the origin of cells, we used three different inducible Cre transgenic mice that labeled three different, yet overlapping, populations. To our knowledge, this is the first study to trace the contributions of cells from both the bursal and articular surfaces to healing of the supraspinatus tendon. Both SMA-9 and PRG4-9 mice, which labeled tdTom+ cells on the bursal side of the supraspinatus underneath the acromion in the normal shoulder, showed robust expansion of the tdTom+ population following injury (Fig. 5A–B). In contrast, AGC-9 mice, which labeled tdTom+ cells in the enthesis and articular cartilage in the normal shoulder, showed essentially no expansion of tdTom+ cells, suggesting minimal healing contribution from the enthesis and adjacent articular cartilage (Fig. 5C).
A limitation of our study was that the Cre-Ai9 mice used in the study were not cell-type-specific. Because each transgenic mouse labeled tdTom+ cells in several different cell populations, we could not isolate a single cell population responsible for the healing response. For example, the PRG4-9 mice labeled cells within the joint capsule, tendon midsubstance, and even a small proportion of the paratenon in the normal shoulder. Ideally, there would be a transgenic mouse and an injection protocol that would allow us, for example, to label only the cells in the subacromial bursa to determine the contribution from that specific population. Therefore, we can only conclude that the healing cells are derived from the bursal side of the rotator cuff tendon, likely from the paratenon, the bursa, and the overlying muscle. To further delineate the origin of healing cells, a more specific labeling system will be necessary.
Another potential limitation was that a single dose of tamoxifen was given at the time of surgery via IP injection in corn oil, which may lead to labeling of cells that turn on these genes in response to the injury. Previous work demonstrated that plasma levels of 4-hydroxytamoxifen were down to baseline levels by 72 hours following a single IP injection of tamoxifen in corn oil (0.8 mg in 8–15 week old mice) [34] and activation of CreERT2-responsive cells implanted one week following a 3 mg IP dose of tamoxifen was minimal [22]. Therefore, we injected a subset of PRG4-9 and SMA-9 mice (n=2 per mouse model) with tamoxifen (80 μg/g) one week prior to injury. The contribution of tdTom+ cells in these mice were comparable to mice injected at the time of surgery with similar contribution from both PRG4-9+ and SMA-9+ cells (Fig. S1).
Nevertheless, the study supports the importance of the bursal side of the tendon to rotator cuff healing, which is supported by previous work [13,35]. While these cells were unable to fully heal this moderately sized full-thickness tear, there was significant expansion of this cell population. There have also been several studies that highlight the importance of subacromial bursa for robust scar tissue formation [36,37]. In fact, many surgeons perform a wide bursectomy and debridement when repairing the rotator cuff to obtain good visualization of the rotator cuff tear, but also to potentially reduce pain. However, this debridement may remove a cell population that could improve rotator cuff repairs. Further study is needed to identify analogous αSMA and Prg4 populations in patients and determine whether these cell populations can be stimulated to improve healing of these injuries.
Supplementary Material
Acknowledgments
FUNDING
This study was supported by NIH K99-AR067283, R01-AR063702, and R21-AR064941.
Footnotes
DECLARATION OF INTEREST
Augustus D. Mazzocca has received research support from Arthrex, Inc, Naples, Florida.
References
- 1.Amini MH, Ricchetti E, Iannotti J, Derwin K. Rotator cuff repair: challenges and solutions. Orthop Res Rev. 2015;7:57–69. [Google Scholar]
- 2.Sher J, Uribe J, Posada A, Murphy B, Zlatkin M. Abnormal findings on magnetic resonance images of asymptomatic shoulders. J Bone Joint Surg. 1995;77(1):10–15. doi: 10.2106/00004623-199501000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Ellman H. Diagnosis and treatment of incomplete rotator cuff tears. Clin Orthop. 1990;5(254):64–74. [PubMed] [Google Scholar]
- 4.Fukuda H. The management of partial-thickness tears of the rotator cuff. J Bone Joint Surg Br. 2003;85(1):3–11. doi: 10.1302/0301-620x.85b1.13846. [DOI] [PubMed] [Google Scholar]
- 5.Le BTN, Wu XL, Lam PH, Murrell GAC. Factors predicting rotator cuff retears: an analysis of 1000 consecutive rotator cuff repairs. Am J Sports Med. 2014;42(5):1134–1142. doi: 10.1177/0363546514525336. [DOI] [PubMed] [Google Scholar]
- 6.Duquin TR, Buyea C, Bisson LJ. Which method of rotator cuff repair leads to the highest rate of structural healing? A systematic review. Am J Sports Med. 2010;38(4):835–841. doi: 10.1177/0363546509359679. [DOI] [PubMed] [Google Scholar]
- 7.Ratcliffe A, Butler DL, Dyment NA, Cagle PJ, Proctor CS, Ratcliffe SS, Flatow EL. Scaffolds for Tendon and Ligament Repair and Regeneration. Ann Biomed Eng. 2015;43(3):819–831. doi: 10.1007/s10439-015-1263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derwin K, Baker A, Iannotti J, McCarron J. Preclinical models for translating regenerative medicine therapies for rotator cuff repair. Tissue Eng. 2010;16(1):21–30. doi: 10.1089/ten.teb.2009.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nourissat G, Berenbaum F, Duprez D. Tendon injury: From biology to tendon repair. Nat Rev Rheumatol. 2015;11(4):223–233. doi: 10.1038/nrrheum.2015.26. [DOI] [PubMed] [Google Scholar]
- 10.Dyment NA, Galloway JL. Regenerative Biology of Tendon: Mechanisms for Renewal and Repair. Curr Mol Biol Rep. 2015;1(3):124–131. doi: 10.1007/s40610-015-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyment NA, Hagiwara Y, Matthews BG, Li Y, Kalajzic I, Rowe DW. Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS ONE. 2014;9(4):e96113. doi: 10.1371/journal.pone.0096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchis-Alfonso Healing of the patellar tendon donor defect created after central-third patellar tendon autograft harvest. Knee Surg Sports Traumatol Arthrosc. 1999;7(6):340–348. doi: 10.1007/s001670050177. [DOI] [PubMed] [Google Scholar]
- 13.Hirose K, Kondo S, Choi HR, Mishima S, Iwata H, Ishiguro N. Spontaneous healing process of a supraspinatus tendon tear in rabbits. Arch Orthop Trauma Surg. 2004;124(6):374–377. doi: 10.1007/s00402-004-0663-8. [DOI] [PubMed] [Google Scholar]
- 14.Kohrs RT, Zhao C, Sun YL, Jay GD, Zhang L, Warman ML, et al. Tendon fascicle gliding in wild type, heterozygous, and lubricin knockout mice. J Orthop Res. 2011;29(3):384–389. doi: 10.1002/jor.21247. [DOI] [PubMed] [Google Scholar]
- 15.Jay GD, Tantravahi U, Britt DE, Barrach HJ, Cha CJ. Homology of lubricin and superficial zone protein (SZP): products of megakaryocyte stimulating factor (MSF) gene expression by human synovial fibroblasts and articular chondrocytes localized to chromosome 1q25. J Orthop Res. 2001;19(4):677–687. doi: 10.1016/S0736-0266(00)00040-1. [DOI] [PubMed] [Google Scholar]
- 16.Funakoshi T, Martin SD, Schmid TM, Spector M. Distribution of lubricin in the ruptured human rotator cuff and biceps tendon: a pilot study. Clin Orthop Relat Res. 2010;468(6):1588–1599. doi: 10.1007/s11999-009-1108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry SP, Jang C, Deng JM, Zhang Z, Behringer RR, De Crombrugghe B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47(12):805–814. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyment NA, Kazemi N, Aschbacher-Smith LE, Barthelery NJ, Kenter K, Gooch C, et al. The relationships among spatiotemporal collagen gene expression, histology, and biomechanics following full-length injury in the murine patellar tendon. J Orthop Res. 2012;30(1):28–36. doi: 10.1002/jor.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma P, Maffulli N. Basic biology of tendon injury and healing. Surgeon. 2005;3(5):309–316. doi: 10.1016/s1479-666x(05)80109-x. [DOI] [PubMed] [Google Scholar]
- 20.Thomopoulos S, Parks WC, Rifkin DB, Derwin KA. Mechanisms of tendon injury and repair. J Orthop Res. 2015;33(6):832–839. doi: 10.1002/jor.22806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voleti PB, Buckley MR, Soslowsky LJ. Tendon healing: Repair and regeneration. Annu Rev Biomed Eng. 2012;14:47–71. doi: 10.1146/annurev-bioeng-071811-150122. [DOI] [PubMed] [Google Scholar]
- 22.Reinert RB, Kantz J, Misfeldt AA, Poffenberger G, Gannon M, Brissova M, et al. Tamoxifen-Induced Cre-loxP Recombination Is Prolonged in Pancreatic Islets of Adult Mice. PLoS ONE. 2012;7(3):e33529. doi: 10.1371/journal.pone.0033529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grcevic D, Pejda S, Matthews BG, Repic D, Wang L, Li H, et al. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells. 2012;30(2):187–196. doi: 10.1002/stem.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthews BG, Grcevic D, Wang L, Hagiwara Y, Roguljic H, Joshi P, et al. Analysis of asMA-labeled progenitor cell commitment identifies notch signaling as an important pathway in fracture healing. J Bone Miner Res. 2014;29(5):1283–1294. doi: 10.1002/jbmr.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews BG, Torreggiani E, Roeder E, Matic I, Grcevic D, Kalajzic I. Osteogenic potential of alpha smooth muscle actin expressing muscle resident progenitor cells. Bone. 2016;84:69–77. doi: 10.1016/j.bone.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyment NA, Liu C-, Kazemi N, Aschbacher-Smith LE, Kenter K, Breidenbach AP, et al. The Paratenon Contributes to Scleraxis-Expressing Cells during Patellar Tendon Healing. PLoS ONE. 2013;8(3):e59944. doi: 10.1371/journal.pone.0059944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 2003;66(2):123–143. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- 29.Hagiwara Y, Dyment NA, Jiang X, Jiang Ping H, Ackert-Bicknell C, Adams DJ, et al. Fixation stability dictates the differentiation pathway of periosteal progenitor cells in fracture repair. J Orthop Res. 2015;33(7):948–956. doi: 10.1002/jor.22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyment NA, Breidenbach A, Schwartz A, Russell RP, Aschbacher-Smith L, Liu H, et al. Gdf5 progenitors give rise to fibrocartilage cells that mineralize via hedgehog signaling to form the zonal enthesis. Dev Biol. 2015;405(1):96–107. doi: 10.1016/j.ydbio.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyment NA, Hagiwara Y, Jiang X, Huang J, Adams DJ, Rowe DW. Response of knee fibrocartilage to joint destabilization. Osteoarthr Cartil. 2015;23(6):996–1006. doi: 10.1016/j.joca.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82(7–8):518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Laron D, Natsuhara K, Manzano G, Kim HT, Feeley BT. A mouse model of massive rotator cuff tears. J Bone Joint Surg. 2012;94(7):e41–e41. doi: 10.2106/JBJS.K.00620. [DOI] [PubMed] [Google Scholar]
- 34.Wilson CH, Gamper I, Perfetto A, Auw J, Littlewood TD, Evan GI. The kinetics of ER fusion protein activation in vivo. Oncogene. 2014;33(40):4877–4880. doi: 10.1038/onc.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi K, Hamada K, Gotoh M, Handa A, Yamakawa H, Fukuda H. Healing of full-thickness tears of avian supracoracoid tendons: in situ hybridization of alpha1(I) and alpha1(III) procollagen mRNA. J Orthop Res. 2001;19(5):862–868. doi: 10.1016/S0736-0266(01)00015-8. [DOI] [PubMed] [Google Scholar]
- 36.Chillemi C, Petrozza V, Garro L, Sardella B, Diotallevi R, Ferrara A, et al. Rotator cuff re-tear or non-healing: histopathological aspects and predictive factors. Knee Surg Sports Traumatol Arthrosc. 2011;19(9):1588–1596. doi: 10.1007/s00167-011-1521-1. [DOI] [PubMed] [Google Scholar]
- 37.Uhthoff H, Sarkar K. Surgical repair of rotator cuff ruptures. The importance of the subacromial bursa. J Bone Joint Surg Br. 1991;73(3):399–401. doi: 10.1302/0301-620X.73B3.1670436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.