Abstract
Proteins belonging to the highly conserved RecQ helicase family are essential for the maintenance of genomic stability. Here, we describe the biochemical properties of the human RECQ5β protein. Like BLM and WRN, RECQ5β is an ATP-dependent 3′–5′ DNA helicase that can promote migration of Holliday junctions. However, RECQ5β required the single-stranded DNA-binding protein RPA in order to mediate the efficient unwinding of oligonucleotide-based substrates. Surprisingly, we found that RECQ5β possesses an intrinsic DNA strand-annealing activity that is inhibited by RPA. Analysis of deletion variants of RECQ5β revealed that the DNA helicase activity resides in the conserved N-terminal portion of the protein, whereas strand annealing is mediated by the unique C-terminal domain. Moreover, the strand-annealing activity of RECQ5β was strongly inhibited by ATPγS, a poorly hydrolyzable analog of ATP. This effect was alleviated by mutations in the ATP-binding motif of RECQ5β, indicating that the ATP-bound form of the protein cannot promote strand annealing. This is the first demonstration of a DNA helicase with an intrinsic DNA strand-annealing function residing in a separate domain.
Keywords: DNA helicase, genomic instability, Holliday junctions, RecQ, single-strand annealing
Introduction
Helicases are ubiquitous enzymes that use the free energy of nucleotide triphosphate hydrolysis to separate nucleic acid duplexes into the constituent single strands (reviewed in Lohman and Bjornson, 1996; Hall and Matson, 1999; von Hippel and Delagoutte, 2001). They play essential roles in nearly all DNA metabolic processes, including DNA replication, recombination and repair. Helicases often contain additional functional domains or interact with other proteins to mediate complex DNA transactions.
DNA helicases of the RecQ family, named after the 3′–5′ DNA helicase RecQ of Escherichia coli, unwind a wide variety of potentially recombinogenic DNA structures, including four-way junctions, D-loops and G-quadruplex DNA. RecQ helicases are highly conserved from bacteria to humans, but while prokaryotes and unicellular eukaryotes such as Saccharomyces cerevisiae or Schizosaccharomyces pombe possess only a single RecQ homolog, multicellular organisms have several (for recent reviews, see Bachrati and Hickson, 2003; Hickson, 2003; Khakhar et al, 2003). In humans, five RecQ homologs have been identified to date: RECQ1, BLM/RECQ2, WRN/RECQ3, RECQ4 and RECQ5. In all organisms, defective RecQ helicase function is associated with genomic instability, which is generally manifested as an increase in the frequency of inappropriate recombination events. Mutations in genes encoding the human RecQ helicases BLM, WRN and RECQ4 give rise to the hereditary disorders Bloom's syndrome, Werner's syndrome and Rothmund–Thomson syndrome, respectively (Bachrati and Hickson, 2003). These diseases are associated with cancer predisposition and variable aspects of premature aging. Although the precise DNA transactions mediated by RecQ helicases remain elusive, the enzymes have been implicated in the processing of aberrant DNA structures arising during DNA replication and repair (Bachrati and Hickson, 2003).
RecQ helicases possess the so-called DExH helicase and RecQ-Ct (RecQ C-terminal) regions, which form the catalytic core of the enzyme (Bachrati and Hickson, 2003; Bernstein and Keck, 2003). The former domain is homologous to the superfamily 2 helicases, while the latter is unique to the RecQ family. Recent structural studies on the E. coli RecQ helicase have shown that the RecQ-Ct region forms two subdomains (Bernstein et al, 2003). The proximal part of this region folds into a platform of four helices containing a Zn2+-binding site and the distal part forms a specialized helix–turn–helix motif, called the winged-helix (WH) domain, which serves as a double-stranded DNA (dsDNA)-binding motif in several proteins including the catabolite gene activator protein (Bernstein et al, 2003). Some RecQ helicases also contain the so-called HRDC (Helicase and RNAseD C-terminal) region, which may serve as an auxiliary DNA-binding domain (Liu et al, 1999; Bernstein and Keck, 2003). The eukaryotic members of the RecQ family usually have additional N- and C-terminal domains flanking the RecQ core, which are involved in protein–protein interactions (Bachrati and Hickson, 2003).
Although defects in WRN, BLM and RECQ4 are associated with heritable human disease, such an association has not been demonstrated for RECQ5. Targeted disruption of RECQ5 in chicken DT40 cells, which have proven to be a valuable model system to study the cellular functions of BLM and WRN, does not result in genomic instability (Imamura et al, 2002; Wang et al, 2003). However, a RECQ5−/− BLM−/− DT40 double mutant exhibits much higher levels of sister chromatid exchanges (SCEs) than a BLM−/− mutant, suggesting that RECQ5 may serve as backup for BLM (Wang et al, 2003). RECQ5 deficiency in Caenorhabditis elegans reduces lifespan and increases cellular sensitivity to ionizing radiation (Jeong et al, 2003). Together, these findings support the notion that the product of the human RECQ5 gene may be important for the maintenance of genomic stability.
The human RECQ5 protein exists in at least three different isoforms, which result from alternative splicing of the RECQ5 transcript (Shimamoto et al, 2000). In addition to the conserved helicase and RecQ-Ct regions, the largest isoform, RECQ5β, contains a long C-terminal region that displays no homology to the other family members (Figure 7A). Also, in contrast to the other RecQ helicases, RECQ5β lacks the WH domain. The other two isoforms, RECQ5α and RECQ5γ, are almost identical. RECQ5α terminates inside the RecQ-Ct region, just upstream of the putative Zn2+-binding site. RECQ5γ contains 25 additional amino acids at the C-terminus that are not present in RECQ5β. The RECQ5β isoform localizes to the nucleus, whereas the two smaller isoforms are cytoplasmic (Shimamoto et al, 2000).
Figure 7.
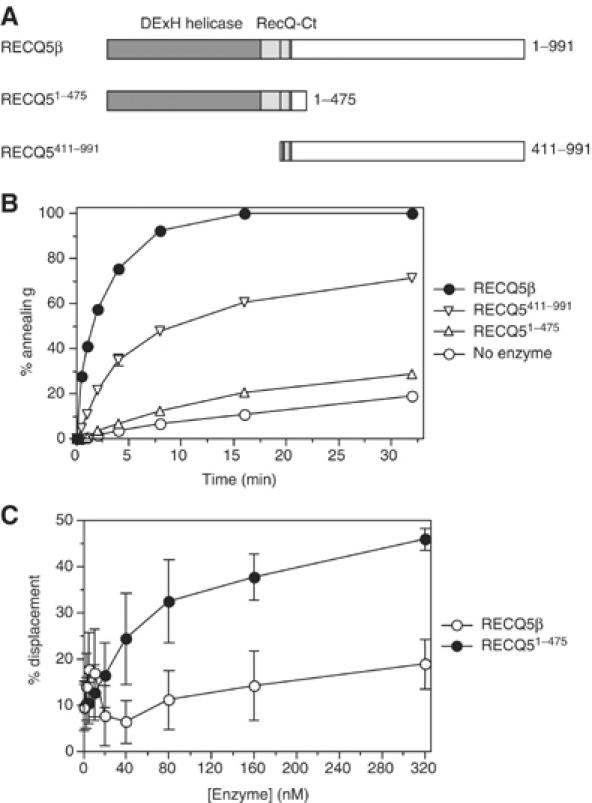
The DNA strand-annealing activity resides in the unique C-terminal portion of the RECQ5β polypeptide. (A) Schematic diagrams indicating RECQ5β and the variant proteins used in this study. The DExH helicase and RecQ-Ct domains conserved among the members of the RecQ family are shown as dark- and light-gray boxes, respectively. The location of the putative Zn2+-binding site within the RecQ-Ct region is indicated as a checkered box. The remaining portion represents the region that is not conserved in the RecQ family. The numbers on the right refer to the primary amino-acid sequence of RECQ5β. (B) Kinetics of annealing of 1 nM 50-mer complementary oligonucleotides A20 and B20 (A20 was radioactively labeled at its 5′-end) in the presence of 20 nM RECQ5β, 20 nM RECQ51–475, 20 nM RECQ5411–991 or without protein. Reactions were carried at 37°C. The relative concentration of the strand-annealing product was determined as described in Materials and methods. (C) Unwinding of a 30-bp forked duplex by RECQ5β and RECQ51–475 as a function of enzyme concentration. Reactions were carried out at 37°C for 20 min and contained 1 nM DNA, 2 mM ATP and varying amounts of enzyme. The percentage of unwound DNA was determined as described in Materials and methods.
The biochemical functions of these isoforms are unknown. To gain an insight into the potential role of RECQ5 in the maintenance of genomic stability, we have carried out an extensive biochemical analysis of the RECQ5β protein. We found that RECQ5β is monomeric and contains two separate functional domains: the N-terminal half of the protein comprising the conserved DExH and Zn2+-binding domains functions as a DNA-dependent ATPase and an ATP-dependent 3′–5′ DNA helicase. The unique C-terminal portion possesses an efficient DNA strand-annealing activity. To our knowledge, this is the first demonstration of a DNA helicase with an intrinsic DNA strand-annealing function residing in a separate domain of the same polypeptide.
Results
DNA-dependent ATPase and 3′–5′ DNA helicase activities of RECQ5β
RECQ5β was overproduced in E. coli as a fusion protein with a self-cleaving affinity tag composed of an intein fragment and a chitin-binding domain (CBD), and purified to more than 95% homogeneity (Figure 1A). To determine the quaternary structure of the RECQ5β protein in solution, analytical ultracentrifugation experiments were conducted with 0.5 μM protein. The sedimentation velocity profile of RECQ5β preparation revealed the presence of a single species with an S20,W value of 5.6. Sedimentation equilibrium measurements yielded an apparent molecular mass of 134 kDa indicating that RECQ5β exists as a monomer in solution (the predicted value for the monomer is 108.9 kDa). Size-exclusion chromatography on a Superdex 200 column revealed no change in the quaternary structure of the protein upon addition of a 30-mer oligonucleotide and/or ATPγS-Mg2+ (data not shown).
Figure 1.
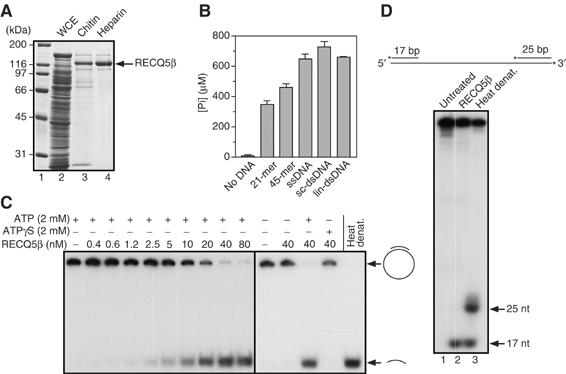
ATPase and DNA helicase activities of RECQ5β. (A) A 10% SDS–polyacrylamide gel showing individual RECQ5β purification steps. RECQ5β was overproduced in E. coli as a fusion with a self-cleaving affinity tag. Lane 1, molecular size marker; lane 2, soluble fraction of whole-cell extract (WCE); lane 3, pooled fractions from the chitin column; lane 4, pooled fractions from the heparin column. (B) ATPase activity of RECQ5β in the presence of the indicated DNA effectors. Reactions were carried out at 37°C for 30 min and contained 20 nM RECQ5β, 2 mM ATP and 25 μg/ml DNA effector. The amount of inorganic phosphate (Pi) released by ATP hydrolysis was determined as described in Materials and methods. sc-dsDNA, supercoiled dsDNA; lin-dsDNA, linear dsDNA. (C) Conventional DNA helicase assay using a 44 bp M13mp18-based duplex radiolabeled at the 3′-end. Reactions were carried out at 37°C for 30 min and contained 0.5 nM DNA, varying concentrations of RECQ5β and 2 mM ATP (or ATPγS) as indicated. The reaction products were analyzed by 10% nondenaturing PAGE. Radiolabeled species were visualized by autoradiography. The last lane contains heat-denatured substrate. (D) Helicase polarity assay using a linearized M13mp18-based DNA substrate with partial-duplex termini. Radiolabeled 3′-ends in this DNA substrate are indicated by asterisks (top panel). Reactions were carried out at 37°C for 30 min and contained 0.5 nM DNA substrate, 40 nM RECQ5β and 2 mM ATP. The products were analyzed as in (C). Lane 1, substrate incubated without enzyme; lane 2, substrate plus enzyme; lane 3, heat-denatured substrate.
The ATPase activity of RECQ5β was analyzed in the presence of various DNA molecules, including short oligonucleotides, circular single-stranded DNA (ssDNA), linear dsDNA or supercoiled plasmid DNA. RECQ5β was found to exhibit robust ATPase activity with both ssDNA and dsDNA, whereas little ATPase activity was observed in the absence of DNA (Figure 1B). In addition to ATP, RECQ5β could hydrolyze dATP with a specific activity similar to that observed with ATP (data not shown). We also found that GTP and dGTP could be hydrolyzed in the presence of ssDNA and dsDNA, but the specific activities were approximately 10-fold lower than that observed with ATP (data not shown). No significant hydrolytic activity was observed with other nucleotides.
To determine whether RECQ5β possesses a DNA helicase activity, we first tested its ability to disrupt an M13mp18-based partial duplex of 44 bp. Using 0.5 nM DNA substrate and increasing amounts of RECQ5β in the presence of ATP, we observed that RECQ5β displaced the annealed oligonucleotide in a concentration-dependent manner (Figure 1C). At a protein concentration of approximately 15–20 nM, 50% strand displacement occurred. DNA helicase activity was dependent on ATP hydrolysis, as strand displacement was not observed when ATP was replaced with poorly hydrolyzable ATP analog, ATPγS (Figure 1C). We found that dATP could substitute for ATP, but failed to observe DNA helicase activity when other nucleotides were used.
To determine the polarity of the RECQ5β helicase, we employed a linear DNA substrate consisting of M13mp18 ssDNA with short 32P-labeled (17 and 25 bp) duplex regions at the 5′- and 3′-ends, respectively (Figure 1D, top panel). We found that RECQ5β could only displace the 17-mer, indicating 3′–5′ polarity, which is a general characteristic of the RecQ helicase family (Figure 1D, lower panel).
RECQ5β requires RPA to unwind oligonucleotide-based partial duplexes
Further analysis of the DNA helicase activity of RECQ5β revealed that the unwinding of oligonucleotide-based substrates was poor compared with the M13-based partial duplexes. Using a 30-bp forked duplex with single-stranded splayed arms, we observed that the formation of unwound products increased proportionally with protein concentration, peaking at 10 nM with about 20% of the substrate dissociated (Figure 2A). Interestingly, at higher protein concentrations (10–40 nM), the strand displacement activity dropped below the level of spontaneous dissociation of the substrate. At even greater protein concentrations (40–320 nM), the activity again increased gradually with protein concentration (Figure 2A). The inability of RECQ5β to promote efficient unwinding of an oligonucleotide-based fork structure was surprising since other RecQ helicases such as BLM or WRN exhibit a preference for this substrate (Mohaghegh et al, 2001).
Figure 2.
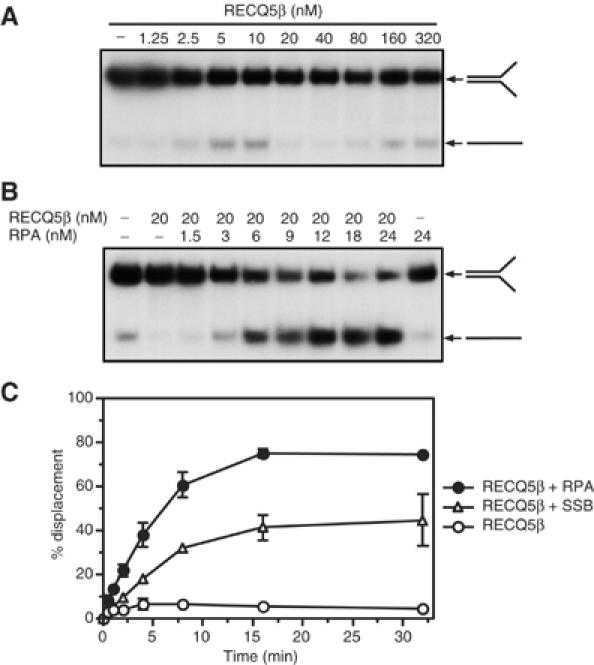
Stimulation of RECQ5β helicase activity on oligonucleotide-based partial duplexes by ssDNA-binding proteins. (A) Unwinding of 1 nM 30-bp forked duplex, 32P-end labeled in the 5′-ssDNA arm, by RECQ5β at concentrations ranging from 0 to 320 nM. Reactions were incubated at 37°C for 20 min, and the products were analyzed by 10% nondenaturing PAGE. Radiolabeled species were visualized by autoradiography. (B) Unwinding of 1 nM 30-bp forked duplex by 20 nM RECQ5β in the presence of the indicated concentrations of RPA. Reactions were carried out and analyzed as in (A). (C) Kinetics of unwinding of 1 nM 30-bp forked duplex by 20 nM RECQ5β in the presence of 24 nM RPA, 120 nM E. coli SSB or without any ssDNA-binding protein. The percentage strand displacement was estimated as described in Materials and methods.
To explore the possibility that RECQ5β requires additional factors to mediate unwinding of oligonucleotide-based substrates, we first examined the effect of the human replication protein A (RPA), an ssDNA-binding protein, on this reaction. RPA has been shown to enhance specifically the unwinding of long DNA duplexes by WRN and BLM (Brosh et al, 1999; 2000). We observed that addition of RPA greatly stimulated RECQ5β-mediated unwinding of the splayed-arm structure in a concentration-dependent manner (Figure 2B). Time-course studies, carried out at 1 nM DNA and 20 nM RECQ5β, showed that in the presence of 24 nM RPA (trimer) the unwinding reaction was completed in <16 min (Figure 2C). The E. coli ssDNA-binding (SSB) protein (120 nM) could partially substitute for RPA, but the efficiency of unwinding was reduced (Figure 2C). These results indicate that the stimulatory effect of RPA may involve specific interactions between RPA and RECQ5β, in addition to its effect in binding the ssDNA products.
RECQ5β promotes Holliday junction branch migration
The BLM and WRN proteins interact with Holliday junctions (HJs) and promote their migration (Constantinou et al, 2000; Karow et al, 2000). To determine whether RECQ5β could catalyze similar reactions, we analyzed its activity with the synthetic HJ substrate X26 that contains a 26-bp core of homologous sequences flanked by heterologous arm sequences. In the presence of ATP, we observed concentration-dependent branch migration that generated splayed-arm products (Figure 3A, lanes 2–6). Branch migration was stimulated by the presence of RPA (Figure 3A, lanes 8–11, and Figure 3B).
Figure 3.
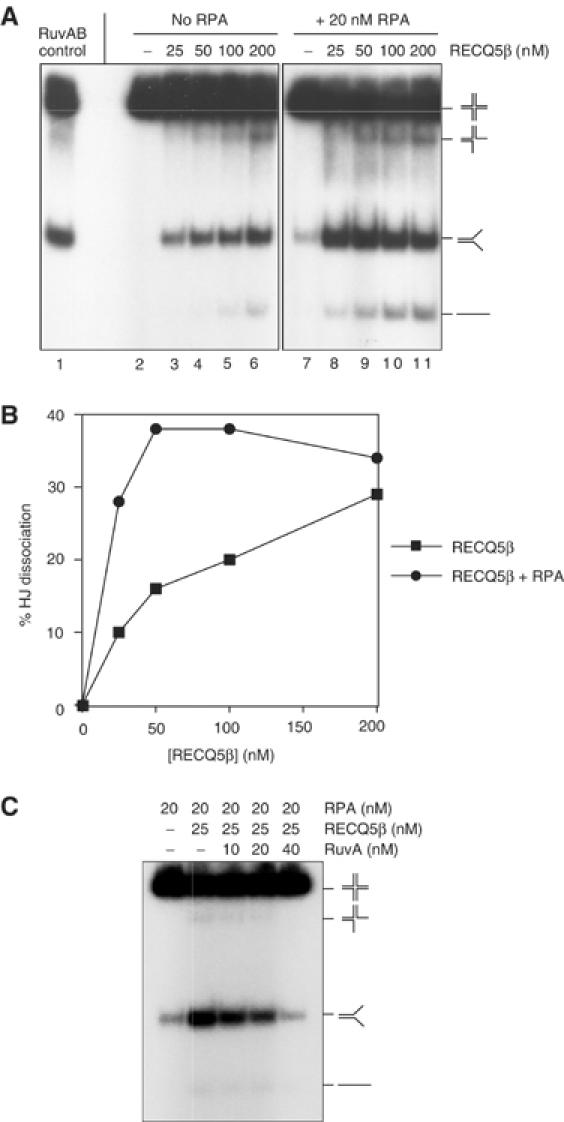
Branch migration of HJs by RECQ5β. (A) RECQ5β-mediated branch migration in the presence or absence of RPA. Reactions contained 0.5 nM synthetic junction X26, and the indicated amounts of RECQ5β protein in the presence or absence of 20 nM RPA. Reactions were carried out at 37°C for 15 min, and the DNA products were analyzed by neutral PAGE. Lane 1, control reaction with E. coli RuvA (25 nM) and RuvB (50 nM). (B) Quantification of the data shown in (A). The relative concentration of the dissociated products is expressed as percentage of total DNA. The dissociated products include three-way junctions, splayed arms (the products of branch migration) and ssDNA. Background levels of dissociated species have been subtracted. (C) Inhibition of RECQ5β branch migration activity by the E. coli HJ-binding protein RuvA. RuvA was preincubated with the junction in reaction buffer for 1 min prior to the addition of the RECQ5β protein. Incubation was then continued, and the products were assayed as described in (A).
In these reactions, comparatively few three-strand junctions or single-stranded products were observed, indicating that the primary RECQ5β-mediated reaction involved recognition of the HJ followed by its movement and dissociation into splayed-arm products. This proposal was supported by the analysis of RECQ5β-mediated branch migration reactions in which the DNA substrate was preincubated with E. coli RuvA, a protein known to bind HJs with high specificity. We found that the presence of 40 nM RuvA imposed a complete block to branch migration mediated by 25 nM RECQ5β (Figure 3C).
The ability of RPA to stimulate branch migration could be a consequence of its ability to bind the single-stranded arms of reaction products thereby (i) stimulating the forward reaction or (ii) blocking a reverse reaction in which the splayed-arm structures reannealed to reform the HJ structure. To distinguish between these possibilities, time-course studies were carried out in the presence and absence of RPA. Without RPA, formation of the splayed-arm branch migration products was greater after 5 min than at later time points (Figure 4A, lanes 3–6, and Figure 4B). This surprising result contrasts with data obtained with reactions carried out in the presence of RPA, in which splayed-arm products accumulated in a time-dependent manner (Figure 4A, lanes 7–11, and Figure 4B). As observed in the helicase assays described earlier, SSB could only partially substitute for RPA (Figure 4C). Taken together, these results indicate that RPA plays a specific role in the branch migration reaction, by specifically inhibiting the reannealing of the splayed-arm products. RPA stimulation therefore involves inhibition of annealing, rather than stimulation of branch migration.
Figure 4.
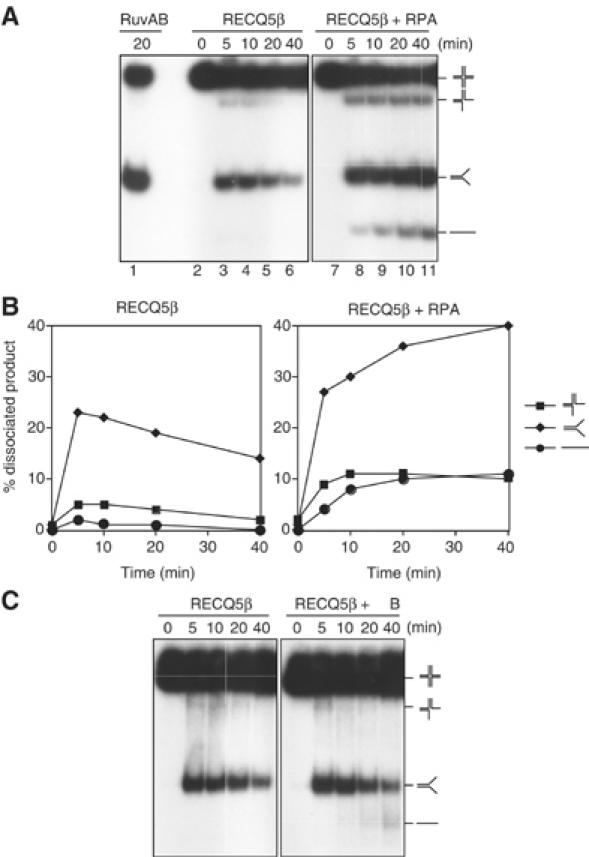
RPA stimulates RECQ5β-mediated branch migration by inhibiting DNA reannealing. (A) Time course of branch migration of 0.5 nM X26 mediated by 50 nM RECQ5β either in the presence or absence of 20 nM RPA. Reactions were carried out at 37°C, and the DNA products were analyzed by neutral PAGE. (B) Quantification of products (three-way junction, splayed-arm branch migration products and single strands) formed during the reactions shown in (A). The relative concentration of the products is expressed as percentage of total dissociated products. (C) Time course of branch migration of 0.5 nM X26 mediated by 50 nM RECQ5β in the presence or absence of excess E. coli SSB protein (1 μM). Reactions were carried out and analyzed as in (A).
RECQ5β possesses DNA strand-annealing activity
The finding that the RECQ5β-mediated dissociation of four-way junctions to splayed-arm products was followed by a slow reannealing reaction (Figure 4A) encouraged us to test whether RECQ5β itself possesses the ability to promote the annealing of complementary single strands. We therefore incubated two partially complementary oligonucleotides (those used to make the splayed-arm helicase substrate) with increasing amounts of RECQ5β in the absence of ATP, and subsequently analyzed the reaction products by 10% nondenaturing polyacrylamide gel electrophoresis (PAGE). We found that RECQ5β promoted an efficient annealing reaction (Figure 5). Using 1 nM ssDNA, we found that annealing was dependent on protein concentration, with the reaction being most efficient at around 40 nM RECQ5β (Figure 5A). A large excess of RECQ5β over DNA slightly inhibited single-strand annealing, possibly as a consequence of protein aggregation (Figure 5A). Time-course experiments conducted at the optimal concentration of RECQ5β (40 nM) revealed that the RECQ5β-promoted annealing reaction was rapid, with 50% of the single-stranded substrate being converted into the double-stranded product within 2 min (Figure 5B). In contrast, spontaneous DNA annealing was slow, with only 15% of the substrate being converted to dsDNA after 32 min (Figure 5B). These findings provide an explanation for the apparent poor helicase activity displayed by RECQ5β on oligonucleotide-based substrates.
Figure 5.
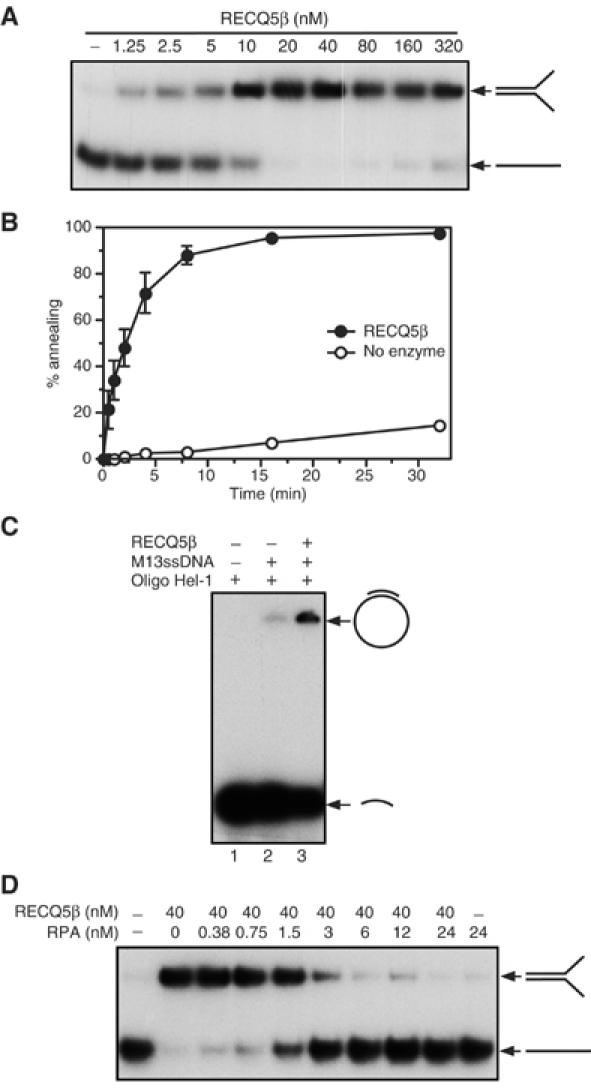
RECQ5β promotes DNA strand annealing. (A) Formation of 30-bp forked duplex in the presence of varying concentrations of RECQ5β. The A20 and B19 complementary oligonucleotides (1 nM), of which A20 was radiolabeled at its 5′-end, were incubated with the indicated concentrations of RECQ5β for 20 min at 37°C. The reaction products were separated by 10% nondenaturing PAGE and visualized by autoradiography. (B) Kinetics of RECQ5β-mediated and spontaneous formation of 30-bp forked duplex. Reactions were carried out at 37°C. The component strands A20 and B19 were present at a concentration of 1 nM and RECQ5β was at a concentration of 40 nM. Reactions were initiated by the addition of unlabeled B19 oligonucleotide. The relative concentration of the strand-annealing product was determined as described in Materials and methods. (C) Annealing of 1 nM 32P-end-labeled Hel-1 oligonucleotide (43-mer) to 2 nM M13mp18 ssDNA in the absence or presence of 100 nM RECQ5β. Reactions were carried out at 37°C for 20 min and analyzed as in (A). RECQ5β was preincubated with Hel-1 for 2 min at room temperature prior to the addition of M13mp18 ssDNA. Lane 1, Hel-1 alone; lane 2, Hel-1 and M13mp18 ssDNA; lane 3, Hel-1, M13mp18 ssDNA and RECQ5β. (D) Effect of RPA on RECQ5β-mediated DNA strand annealing. The complementary oligonucleotides A20 and B19 (1 nM), of which A20 was radiolabeled, were incubated with or without RPA for 5 min at room temperature. RECQ5β was added to a final concentration of 40 nM and incubation was continued at 37°C for 20 min. The reaction products were analyzed as in (A).
RECQ5β was also tested for its ability to promote the annealing of a short oligonucleotide to a target sequence in M13mp18 ssDNA, which contains a large excess of heterologous sequences. In this experiment, a 32P-end-labeled 43-mer oligonucleotide (also used in the helicase assays shown in Figure 1C) was preincubated with RECQ5β followed by the addition of M13mp18 ssDNA. RECQ5β promoted the annealing of the oligonucleotide to the M13mp18 ssDNA (Figure 5C). However, the yield of the annealed product in this reaction was significantly lower than that of the RECQ5β-promoted annealing of complementary oligonucleotides (compare Figure 5A and C). This negative effect of heterologous DNA sequences on RECQ5β-mediated strand annealing correlates with our earlier studies in which it was shown that RECQ5β efficiently unwinds M13mp18-based partial duplexes (Figure 1C).
To gain further insight into the mechanism of RECQ5β-mediated DNA strand annealing, we investigated whether coating of the oligonucleotides with RPA would affect the annealing reaction. Assuming that the size of the DNA-binding site for RPA is about 30 nucleotides (nt) (Kim et al, 1992), we anticipated that the two partially complementary oligonucleotides (with lengths of 50 and 49 nt) would each be occupied by 1–2 RPA heterotrimers. The oligonucleotides (1 nM) were first preincubated with RPA at a concentration ranging from 0 to 24 nM for 2 min. After this DNA-binding reaction, RECQ5β was added to a final concentration of 40 nM and incubation was continued for 20 min. This protein titration experiment revealed that the presence of RPA at concentrations above 3 nM completely inhibited the DNA strand-annealing activity of RECQ5β (Figure 5D). This result correlates with the observation that RPA enhances the DNA helicase activity of RECQ5β on the oligonucleotide-based substrates (Figure 2C) and also blocks the reassociation of splayed-arm products resulting from RECQ5β-promoted HJ branch migration on the X26 structure (Figure 4A and B).
ATP binding to the helicase domain of RECQ5β suppresses strand annealing
We next sought to examine the effect of ATP on the strand-annealing activity of RECQ5β. Since ATP can be hydrolyzed by RECQ5β under the conditions of the strand-annealing assay, we also employed the poorly hydrolyzable analog of ATP, ATPγS, in order to trap RECQ5β in its ATP-bound form. Using the two partially complementary oligonucleotides, we found that the presence of ATP or ADP had little or no effect on the RECQ5β-mediated strand-annealing reaction (Figure 6A). In contrast, ATPγS dramatically inhibited the efficient strand-annealing activity of RECQ5β (Figure 6A). These data imply that the observed inhibitory effect is the consequence of ATP binding rather than hydrolysis.
Figure 6.
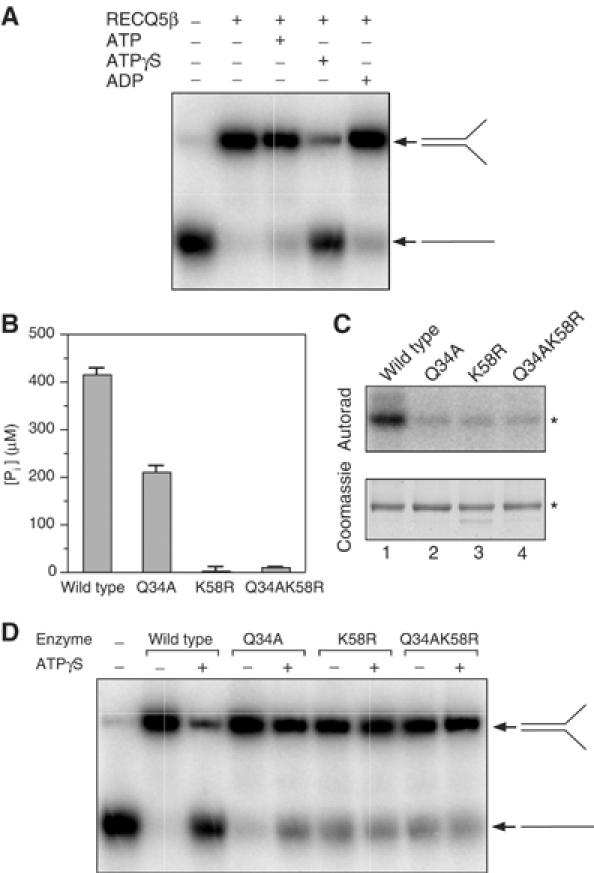
The ATP-bound form of RECQ5β does not promote DNA strand annealing. (A) Effect of ATP, ATPγS and ADP on RECQ5β-promoted annealing of the A20 and B19 oligonucleotides. Reactions were carried out at 37°C for 20 min and contained 40 nM RECQ5β, 1 nM DNAs and 2 mM nucleotides as indicated. The reaction products were analyzed by 10% nondenaturing PAGE. Radiolabeled species were visualized by autoradiography. (B) Comparative ATPase assay for wild-type, Q34A, K58R and Q34AK58R RECQ5β. ATPase reactions were carried out essentially as described in Figure 1B using A20 oligonucleotide as a DNA effector. (C) Comparative ATP-binding assay for wild-type, Q34A, K58R and Q34AK58R RECQ5β. ATP binding was measured using the ATP photo-crosslinking assay described in Materials and methods. The material was resolved on a 10% SDS–polyacrylamide gel, stained with Coomassie brilliant blue (bottom panel) and subjected to autoradiography (top panel). The asterisks indicate the position of the RECQ5β protein. (D) Effect of ATPγS on DNA strand-annealing activities of the wild-type, Q34A, K58R and Q34AK58R RECQ5β. Reaction conditions and analyses were as described in (A).
We next used a mutational approach to prove that the inhibitory effect of ATPγS on RECQ5β-mediated strand annealing was due to its binding to the ATP-binding site of the RECQ5β helicase domain. In the crystal structure of the complex of the E. coli RecQ catalytic core with ATPγS and Mn2+, the adenine moiety forms hydrogen bonds with Gln-30 of motif 0 while the triphosphate is bound to Lys-53 of motif I (Bernstein et al, 2003). We therefore generated mutants in the corresponding residues of RECQ5β, Gln-34 and Lys-58, by replacing them with alanine and arginine, respectively. Both single and double mutants were generated, purified essentially as the wild-type enzyme and tested for their ability to bind and hydrolyze ATP. We found that the Q34A mutant retained partial ATPase activity (about 50% of wild type), whereas the K58A mutant and the double mutant failed to show any significant ATPase activity (Figure 6B). Using a UV crosslinking assay with [γ-32P]ATP, we found that all three mutants exhibited reduced ATP binding relative to the wild-type enzyme (Figure 6C).
The partially complementary oligonucleotides were then used to assess the effect of ATPγS on the strand-annealing activity of the mutant RECQ5β proteins. We found that, in the absence of ATPγS, all mutant proteins promoted DNA annealing, although the activities of the K58R mutant and the double mutant were slightly reduced compared to that of the wild-type enzyme (Figure 6D). However, in contrast to the dramatic inhibitory effect seen with the wild-type enzyme, ATPγS only partially inhibited the strand-annealing activity of the Q34A mutant and had no effect on the strand-annealing activities of the K58R and Q34AK58R mutants (Figure 6D). These results correlate nicely with the observed ATPase activities of these proteins (Figure 6B), and show that the ATP-bound form of RECQ5β does not possess the ability to promote the reannealing of two complementary DNA strands.
DNA strand-annealing activity resides in the C-terminal region of RECQ5β
To define the region of RECQ5β responsible for the observed strand-annealing activity, the RECQ51–475 and RECQ5411–991 deletion variants (Figure 7A) were tested for their ability to promote this reaction. The former mutant consists of the conserved portion of RECQ5β including the DExH and the Zn2+-binding domains while the latter includes the unique C-terminal half of RECQ5β. In these reactions, we used two fully complementary 50-mer oligonucleotides. We found that the annealing activity promoted by RECQ51–475 was only slightly greater than that observed in the absence of added protein and 30-fold lower than that of the wild-type enzyme (Figure 7B). In contrast, RECQ5411–991 displayed a significant strand-annealing activity although the rate of this reaction was reduced (four-fold) with respect to the wild-type enzyme (Figure 7B). From these data, we suggest that the observed DNA strand-annealing activity resides in the unique C-terminal part of the RECQ5β polypeptide, although it is evident that the helicase domain increases this activity possibly by enhancing the affinity of the enzyme for ssDNA.
Because RECQ51–475 does not display a significant DNA strand-annealing activity and contains the entire helicase catalytic core, we anticipated that it would unwind oligonucleotide substrates more efficiently than the full-size protein. To address this question, we compared the helicase activities of RECQ5β and RECQ51–475 on the 30-bp splayed-arm substrate at a wide range of protein concentrations (0–320 nM). We found that RECQ51–475 exhibited a significantly greater DNA helicase activity than the wild-type enzyme at protein concentrations >20 nM, at which RECQ5β displayed efficient strand-annealing activity (compare Figures 5A and 7C). As mentioned above, the helicase activity of RECQ5β displayed a biphasic dependence on protein concentration. In contrast, the helicase activity of RECQ51–475 gradually increased within the protein concentration range used (Figure 7C). The increase in helicase activity of RECQ5β at enzyme concentrations above 40 nM presumably reflects an inhibitory effect of ATP on the RECQ5β-mediated reannealing reaction, which we have observed to become more pronounced at elevated protein concentrations (data not shown).
We conclude that RECQ5β contains two separate functional domains: a DNA helicase domain and a DNA strand-annealing domain.
Discussion
In this work, we have defined the biochemical characteristics of the human RECQ5β protein, a member of the RecQ family of DNA helicases. Our work revealed that RECQ5β functions as an ssDNA/dsDNA-dependent ATPase and an ATP-dependent 3′–5′ DNA helicase with the ability to promote branch migration of HJs. These characteristics are shared by other members of the RecQ helicase family (Bachrati and Hickson, 2003). However, in contrast to BLM and WRN that form oligomeric structures (Karow et al, 1999; Xue et al, 2002), the RECQ5β helicase exists as a monomer both in free and DNA/ATP-bound forms. Most importantly, our analyses revealed that RECQ5β exhibits an efficient DNA strand-annealing activity, residing in the unique C-terminal half of the protein. Such a feature has not been seen for any DNA helicase characterized so far.
The observed inhibition of RECQ5β-mediated DNA strand annealing by RPA indicates that the annealing mechanism is likely to be distinct from the mode of action of classical single-strand-annealing proteins such as RAD52, which involves a cooperative interaction with the complex of the cognate ssDNA-binding protein and ssDNA (Sugiyama et al, 1998). Moreover, classical strand-annealing proteins are known to form oligomeric ring structures (Van Dyck et al, 1998; Passy et al, 1999; Stasiak et al, 2000), whereas RECQ5β protein is monomeric. Structural analyses of the mechanism of RAD52-mediated strand annealing revealed that multiple RAD52 rings appear to be the active catalytic species in the reaction, with ssDNA bound around each ring with the bases exposed on the surface of the protein (Van Dyck et al, 2001; Singleton et al, 2002).
The strand-annealing mechanism exhibited by RECQ5β may be more closely related to that of MRE11, which is also strongly inhibited by RPA (de Jager et al, 2001). MRE11 is thought to promote DNA annealing by bridging two complementary DNA segments such that their close proximity results in base pairing and duplex DNA formation. Consistent with this mode of annealing, RECQ5β-mediated annealing of complementary DNA strands was dramatically reduced by the presence of a large excess of heterologous DNA sequence, as found by measuring the annealing of a short oligonucleotide to the complementary region in M13mp18 ssDNA.
We also found that the DNA strand-annealing activity of RECQ5β was inhibited by the binding of ATPγS to the helicase domain of the enzyme. Since helicases are known to translocate along DNA in a reaction driven by ATP binding and hydrolysis, we suggest that ATPγS binding may lead to the formation of an intermediate DNA–protein complex that is incapable of DNA translocation. Alternatively, the binding of ATPγS may induce an allosteric change in RECQ5β leading to inactivation of the putative DNA strand-annealing domain. The fact that ATP did not produce a significant inhibitory effect is presumably due to its hydrolysis by the ssDNA–enzyme complex that drives protein translocation and release from the ssDNA template, allowing the strand-annealing domain to mediate duplex formation. These findings are consistent with a model in which the helicase and the strand-annealing domains of RECQ5β act in a coordinated fashion.
In the case of oligonucleotide-based partial duplexes as well as four-way junctions, the strand-annealing activity of RECQ5β dominated over its helicase activity, unless the annealing reaction was blocked by coating of the displaced DNA strands with RPA. In contrast, RECQ5β displayed an efficient DNA helicase activity with the M13-based partial duplexes. We suggest that the M13-ssDNA substrate may trap the strand-annealing moiety of RECQ5β, thus effectively disabling the annealing activity. Under these conditions, RECQ5β could act as a DNA helicase. With oligonucleotide substrates, the strand-annealing domain is free to mediate DNA reannealing during, or immediately after, the strand displacement reaction.
Previous biochemical analysis of the large RECQ5 isoform of Drosophila melanogaster (DmRECQ5/QE) did not reveal the presence of an intrinsic DNA strand-annealing activity (Kawasaki et al, 2002). However, it should be noted that the helicase activity of DmRECQ5/QE was tested solely on M13-based substrates. In addition, the C-terminal half of human RECQ5β that is responsible for the observed strand-annealing activity shows extensive amino-acid sequence homology to the corresponding part of the DmRECQ5/QE polypeptide (Shimamoto et al, 2000). Therefore, the existence of this activity in the Drosophila protein cannot be excluded.
At the present time, the biological significance of our findings cannot be readily assessed because of the lack of information on the phenotypic consequences of RECQ5 deficiency in human cells. It is known that targeted inactivation of the chicken RECQ5 homolog results in a dramatic increase in the frequency of SCEs, albeit solely in a BLM−/− background (Wang et al, 2003). BLM and DNA topoisomerase IIIα (TOPOIIIα) have been shown to affect the dissolution of double HJ recombination intermediates using a strand-passage mechanism. This reaction yields only non-crossover products, explaining the observed high levels of SCEs in BLM-deficient cells (Wu and Hickson, 2003). Our observation that the human RECQ5β protein can promote HJ branch migration, an essential step in HJ or replication intermediate processing, in combination with the finding that RECQ5β co-immunoprecipitates with TOPOIIIα and TOPOIIIβ from human cell extracts (Shimamoto et al, 2000) is consistent with a model in which RECQ5β serves as a backup for BLM. However, we show here that RECQ5β has substantially different biochemical properties compared to those of BLM. It is therefore plausible that it has alternative and possibly more specialized cellular roles.
The biochemical properties of RECQ5β led us to suggest that the protein may be involved in a DNA repair pathway that requires the coordinated action of DNA helicase and DNA strand-annealing activities. Depletion of the RECQ5 homolog in C. elegans results in hypersensitivity to γ-radiation (Jeong et al, 2003), indicating that RECQ5β may play a role in the repair of radiation-induced lesions such as DNA double-strand breaks, or may be active at stalled replication forks that arise through radiation damage. One attractive possibility is that the DNA helicase and reannealing activities of RECQ5β are coordinated to mediate fork regression, such that the helicase promotes movement of the branch point while the annealing activity facilitates base pairing of the newly synthesized strands during the regression reaction. In agreement with this hypothesis, the small Drosophila RECQ5 isoform was found to unwind preferentially the ‘lagging-strand arm' in a synthetic DNA molecule resembling a stalled replication fork (Ozsoy et al, 2003). Further analysis of the precise cellular roles of RECQ5β will shed light on the functions of the RecQ helicases in maintenance of genomic stability.
Materials and methods
Plasmid construction
The PP1045 cDNA including the RECQ5β codons 411–991 (GenBank accession no. AF193041, kindly provided by Dr Gu) and the RECQ5γ cDNA (kindly provided by Dr J Sekelsky) were used as PCR templates to reconstruct the full-size RECQ5β coding region. In the first step, the 5′- and 3′-halves of the RECQ5β coding region overlapping in the codons 403–417 were amplified and the resulting DNA fragments were fused in a second round of PCR amplification to yield the entire RECQ5β coding region. The coding region of RECQ5β was then inserted between the NdeI and SapI sites of the plasmid pTXB1 (NEB) to construct a translational fusion between RECQ5β and a self-cleaving affinity tag composed of an Mxe intein fragment and the CBD. The resulting plasmid was named pPG10. An additional methionine codon was placed between RECQ5β and the affinity tag according to the manufacturer's instructions. The plasmids pPG16 and pPG19 encoding the deletion variants of RECQ5β (RECQ5411–991 and RECQ51–475, respectively) were constructed in the same way as pPG10. Site-directed mutagenesis of the ATP-binding motifs of RECQ5β was performed using a QuickChange™ site-directed mutagenesis kit (Stratagene) essentially according to the manufacturer's protocol with pPG10 as template.
Protein purifications and analysis
The RECQ5β protein and its variants were produced as C-terminal fusions with the self-cleaving Mxe-CBD affinity tag in the E. coli BL21-CodonPlus-(DE3)-RIL cells (Stratagene) and purified essentially as previously described for the production and purification of the BLM642–1290 fragment (Janscak et al, 2003). E. coli RuvA and RuvB were purified as described (Eggleston et al, 1997) and E. coli SSB was purchased from USB. Human RPA protein was prepared essentially as described, but the purification procedure also included chromatography on an ssDNA–cellulose column (Henricksen et al, 1994). Concentrations of proteins are expressed in moles of monomer except for RPA, which is expressed in moles of trimeric complex.
Gel filtration chromatography on a Superdex 200 PC3.2/30 column was performed using an ÄKTA system (Amersham Pharmacia Biotech) under the conditions described previously (Janscak et al, 2003). Sedimentation velocity and sedimentation equilibrium runs were carried out in a buffer containing 20 mM Tris–HCl (pH 7.5), 75 mM NaCl, 0.1 mM EDTA and 1 mM DTT as described previously (Janscak et al, 2001).
DNA substrates
All oligonucleotides used in the helicase and strand-annealing assays were purchased from Microsynth (Switzerland) and purified by PAGE. The oligonucleotides A20 (50-mer) and B19 (49-mer) are described elsewhere (Hohl et al, 2003). The 43-mer Hel-1 (5′-CTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCC GGGTACCG-3′) is complementary to the M13mp18 viral strand. The 50-mer B20 (5′-GAGGTCACTCCAGTGAATTCGAGCTCGCAGTGTCT AGGTCGTGACTT TGA-3′) is the complement of A20. The M13mp18-based partial-duplex substrate for conventional helicase assays was prepared by annealing the Hel-1 43-mer to circular M13mp18 ssDNA (NEB) and by extension of this molecule by 1 nt using the Klenow fragment (NEB) and [α-32P]dATP (Amersham Pharmacia Biotech) as described previously (Janscak et al, 2003). The substrate for the helicase polarity assay was prepared by cutting the 43-bp M13mp18/Hel-1 partial duplex with AccI to produce a linear ssDNA molecule with short duplex regions at both ends. All available 3′-ends in this molecule were radioactively labeled using the Klenow fragment, [α-32P]dATP and [α-32P]dCTP. For oligonucleotide-based helicase substrates, one of the oligonucleotides was labeled at the 5′-end using T4 polynucleotide kinase (NEB) and [γ-32P]ATP, and annealed to the appropriate complementary strand as described (Janscak et al, 2003). The synthetic HJ X26, which contains a 26 base pair region of homology flanked by heterologous arms, was made by annealing four oligonucleotides as described (Constantinou et al, 2001).
ATPase assays
ATPase activity was determined by colorimetric estimation of the concentration of inorganic phosphate released by ATP hydrolysis using the malachite green assay in a 96-well microplate setup (Chan et al, 1986; Janscak et al, 1996). Reactions were carried out at 37°C in a buffer containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT and 50 μg/ml BSA. Reaction mixtures (typically 20 μl) contained 20 nM wild-type or mutant RECQ5β in the presence or absence of saturating concentrations of DNA effectors (25 μg/ml). Supercoiled pGEM-13Zf(+) DNA (Stratagene), NdeI-linearized pGEM-13Zf(+) DNA, single-stranded pGEM-13Zf(+) DNA, and 45- and 21-mer oligonucleotides were used as DNA effectors. Reactions were usually initiated by adding ATP to a final concentration of 2 mM and terminated after 30 min by adding one reaction volume of 0.1 M EDTA (pH 8.0).
Helicase assays
Helicase reactions were carried out at 37°C in buffer HA (20 mM Tris-acetate, pH 7.9, 50 mM KOAc, 10 mM Mg(OAc)2, 1 mM DTT, 50 μg/ml BSA). Reaction mixtures (10 μl) contained either 0.5 nM M13mp18-based or 1 nM oligonucleotide-based partial duplexes, 2 mM ATP and indicated concentrations of RECQ5β (or its mutants). Where required, RPA or SSB was added at indicated concentrations. Reactions were started with enzyme and terminated typically after 20 min by adding 0.5 reaction volumes of solution S (150 mM EDTA, 2% (w/v) SDS, 30% (v/v) glycerol, 0.1% (w/v) bromophenol blue). In time-course experiments, the reaction volume was 50 μl, and 5 μl aliquots were removed at 0.5, 1, 2, 4, 8, 16 and 32 min. The reaction mixtures were resolved using a 10% (w/v) polyacrylamide gel (acrylamide to bis-acrylamide ratios 19:1 for oligonucleotide substrates and 37.5:1 for M13mp18-based substrates) run in TBE buffer (90 mM Tris-borate (pH 8.3), 2 mM EDTA) at 100 V and room temperature. Radiolabeled DNA species were visualized by autoradiography and quantified using a Molecular Dynamics Typhoon 9400 scanner with associated IMAGEQUANT software. The relative concentration of displaced products was expressed as percentage of total DNA.
Branch migration assay
Reactions were carried out at 37°C in a buffer containing 50 mM Tris–HCl (pH 8.0), 2 mM MgCl2, 2 mM ATP, 1 mM DTT and 100 μg/ml BSA. Reaction mixtures (10 μl) contained 0.5 nM 5′-32P-labeled synthetic HJ DNA (X26) and indicated amounts of RECQ5β. RPA (20 nM) or SSB (1 μM) was added as indicated. For the inhibition experiment, RuvA was preincubated with the HJ in reaction buffer for 1 min at room temperature before the addition of RECQ5β protein. The DNA products were deproteinized and electrophoresed through 10% neutral polyacrylamide gels. Control reactions with RuvAB (25 nM RuvA, 50 nM RuvB) were carried out in a buffer containing 20 mM Tris-acetate (pH 7.5), 15 mM Mg(OAc)2, 2 mM ATP, 1 mM DTT and 100 μg/ml BSA.
DNA strand-annealing assays
The DNA strand-annealing activity of RECQ5β was measured using complementary synthetic oligonucleotides (each at a concentration of 1 nM) of which one was labeled at the 5′-end using [γ-32P]ATP and T4 polynucleotide kinase. Annealing of a 32P-labeled oligonucleotide (1 nM) to M13mp18 ssDNA (2 nM) was also measured. Annealing reactions (typically 10 μl) were carried out at 37°C in buffer HA and contained the indicated amounts of RECQ5β (or its mutants). Where required, RPA (0.38–24 nM), ATP (2 mM), ATPγS (2 mM) and ADP (2 mM) were added. Reactions were usually initiated by adding the unlabeled DNA strand and were incubated for 20 min. In time-course experiments, 50 μl reactions were initiated and 5 μl aliquots were removed at 0.5, 1, 2, 4, 8, 16 and 32 min. Reactions were analyzed essentially as the helicase reactions (see above).
ATP photo-crosslinking
Purified protein (1 μg) was incubated in a volume of 10 μl of buffer HA with 25 μM ATP and 2 μCi of [γ-32P]ATP (3000 Ci/mmol; Amersham Pharmacia Biotech) for 5 min at 37°C. Reactions were spotted onto Parafilm, placed on ice and irradiated for 5 min using a UV-Stratalinker 1800 lamp (Stratagene). The covalent protein–ATP complex was separated from free ATP by electrophoresis in a 10% SDS–polyacrylamide gel. Radiolabeled species were visualized by autoradiography. The gel was subsequently stained with Coomassie brilliant blue to determine the position of the protein.
Acknowledgments
We thank Jian-ren Gu for generously providing us with PP1045 cDNA, Jeff Sekelsky for RECQ5γ cDNA, Ariel Lustig for analytical ultracentrifugation measurements, Marcel Hohl for help with preparation of DNA substrates and Christiane Koenig for technical assistance. This work was supported by the Cancer League of Kanton Zürich and Cancer Research UK. YL is a recipient of a postdoctoral fellowship from the American Cancer Society.
References
- Bachrati CZ, Hickson ID (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J 374: 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Keck JL (2003) Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res 31: 2778–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL (2003) High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J 22: 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA (2000) Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem 275: 23500–23508 [DOI] [PubMed] [Google Scholar]
- Brosh RM Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA (1999) Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem 274: 18341–18350 [DOI] [PubMed] [Google Scholar]
- Chan KM, Delfert D, Junger KD (1986) A direct colorimetric assay for Ca2+ -stimulated ATPase activity. Anal Biochem 157: 375–380 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Davies AA, West SC (2001) Branch migration and Holliday junction resolution catalyzed by activities from mammalian cells. Cell 104: 259–268 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC (2000) Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep 1: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, van Gent DC (2001) DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res 29: 1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston AK, Mitchell AH, West SC (1997) In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell 89: 607–617 [DOI] [PubMed] [Google Scholar]
- Hall MC, Matson SW (1999) Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol 34: 867–877 [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS (1994) Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem 269: 11121–11132 [PubMed] [Google Scholar]
- Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3: 169–178 [DOI] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, Scharer OD (2003) Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem 278: 19500–19508 [DOI] [PubMed] [Google Scholar]
- Imamura O, Fujita K, Itoh C, Takeda S, Furuichi Y, Matsumoto T (2002) Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene 21: 954–963 [DOI] [PubMed] [Google Scholar]
- Janscak P, Abadjieva A, Firman K (1996) The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J Mol Biol 257: 977–991 [DOI] [PubMed] [Google Scholar]
- Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA (2003) Characterization and mutational analysis of the RecQ core of the Bloom syndrome protein. J Mol Biol 330: 29–42 [DOI] [PubMed] [Google Scholar]
- Janscak P, Sandmeier U, Szczelkun MD, Bickle TA (2001) Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J Mol Biol 306: 417–431 [DOI] [PubMed] [Google Scholar]
- Jeong YS, Kang Y, Lim KH, Lee MH, Lee J, Koo HS (2003) Deficiency of Caenorhabditis elegans RecQ5 homologue reduces life span and increases sensitivity to ionizing radiation. DNA Repair (Amst) 2: 1309–1319 [DOI] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA 97: 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Newman RH, Freemont PS, Hickson ID (1999) Oligomeric ring structure of the Bloom's syndrome helicase. Curr Biol 9: 597–600 [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Maruyama S, Nakayama M, Matsumoto K, Shibata T (2002) Drosophila melanogaster RECQ5/QE DNA helicase: stimulation by GTP binding. Nucleic Acids Res 30: 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM (2003) RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol 13: 493–501 [DOI] [PubMed] [Google Scholar]
- Kim C, Snyder RO, Wold MS (1992) Binding properties of replication protein A from human and yeast cells. Mol Cell Biol 12: 3050–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M (1999) The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Struct Fold Des 7: 1557–1566 [DOI] [PubMed] [Google Scholar]
- Lohman TM, Bjornson KP (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem 65: 169–214 [DOI] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM Jr, Bohr VA, Hickson ID (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res 29: 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsoy AZ, Ragonese HM, Matson SW (2003) Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res 31: 1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH (1999) Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA 96: 4279–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Nishikawa K, Kitao S, Furuichi Y (2000) Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res 28: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB (2002) Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA 99: 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH (2000) The human Rad52 protein exists as a heptameric ring. Curr Biol 10: 337–340 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA 95: 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E, Hajibagheri NM, Stasiak A, West SC (1998) Visualisation of human rad52 protein and its complexes with hRad51 and DNA. J Mol Biol 284: 1027–1038 [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC (2001) Visualization of recombination intermediates produced by RAD52-mediated single-strand annealing. EMBO Rep 2: 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Delagoutte E (2001) A general model for nucleic acid helicases and their ‘coupling' within macromolecular machines. Cell 104: 177–190 [DOI] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y, Enomoto T (2003) Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol Cell Biol 23: 3527–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Xue Y, Ratcliff GC, Wang H, Davis-Searles PR, Gray MD, Erie DA, Redinbo MR (2002) A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′–5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry 41: 2901–2912 [DOI] [PubMed] [Google Scholar]


