Abstract
In immunocompromised patients, infection with Kaposi's sarcoma-associated herpesvirus (KSHV) can give rise to Kaposi's sarcoma and several lymphoproliferative disorders. In these tumors, KSHV establishes a latent infection in many of the rapidly proliferating and morphologically abnormal cells. Only a few viral gene products are expressed by the latent virus, and one of the best characterized is the latency-associated nuclear antigen (LANA), a nuclear protein required for the maintenance of viral episomal DNA in the dividing host cell. LANA can also activate or repress an assortment of cellular and viral promoters and may contribute to pathogenesis by allowing the proliferation and survival of host cells. Here we show that activation of the human E2F1 and cyclin-dependent kinase-2 (CDK2) promoters requires elements from both the N- and C-terminal regions of LANA. Deletion of the first 22 amino acids, which are necessary for episome tethering, does not affect nuclear localization but significantly reduces transactivation. Within the deleted peptide, we have identified a short sequence, termed the chromatin-binding motif (CBM), that binds tightly to interphase and mitotic chromatin. A second chromatin-binding activity resides in the C terminus but is not sufficient for optimal transactivation. Alanine substitutions within the CBM reveal a close correlation between the transactivation and chromatin binding activities, implying a mechanistic link. In contrast to promoter activation, we find that the 223 amino acids of the LANA C terminus are sufficient to inhibit p53-mediated activation of the human BAX promoter, indicating that the CBM is not required for all transcription-related functions.
Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL) are life-threatening proliferative diseases that result from the unchecked growth of endothelial- and lymphoid-derived cells, respectively (12). The common denominator between these diseases is the presence of latent Kaposi's sarcoma-associated herpesvirus (KSHV, also known as human herpesvirus 8) in the majority of abnormal cells. Variants of multicentric Castleman's disease, a rare angioproliferative disorder, are also associated with KSHV infection but differ from KS and PEL in the extent of active viral replication (4, 53). KSHV, with a ∼140-kb double-stranded DNA genome, is a member of the γ2-herpesviruses and, as with all other herpesviruses, exploits two distinct modes of replication, referred to as lytic (productive) and latent (nonproductive). KSHV latency involves expression of only a few of the more than 85 viral genes (51, 65). The majority of cells forming KS or PEL lesions harbor latent KSHV, leading to the hypothesis that latency-associated viral gene products drive the proliferation and survival of these cells. In this respect, KSHV follows a paradigm set by other tumor viruses that establish persistent infections, such as Epstein-Barr virus and the papillomaviruses. That said, there is compelling evidence that lytic products expressed by a much smaller fraction of the infected cells or through abortive entry into the lytic replication play a critical role in the disease process (21, 22).
The most prominent latency product is the latency-associated nuclear antigen (LANA, LANA-1, LNA-1), encoded by open reading frame 73 and transcribed as part of a multicistronic mRNA. LANA is localized to the cell nucleus, where it is distributed throughout the nucleoplasm and also accumulates in speckles referred to as LANA bodies (28, 29, 49, 58). Based on the primary amino acid sequence, LANA can be divided into three discrete regions, a proline and basic residue-rich N terminus, a central region composed of a highly variable number of acidic repeats, and a C-terminal region that shares significant homology to proteins encoded by other γ2-herpesviruses (54). The C terminus acts as a multimerization domain, enabling LANA to form stable oligomers, most likely dimers, independent of other viral gene products or DNA (54). The N- and C-terminal regions each contain a putative nuclear localization sequence (NLS) and independently localize to the nucleus (46, 54, 56). LANA body formation requires the LANA C terminus (46, 54).
To establish and maintain latency, KSHV must (i) ensure propagation of the viral genome, (ii) suppress the lytic program, (iii) stimulate host cell proliferation, (iv) interfere with cellular tumor suppressor functions, and (v) block proapoptotic pathways. LANA has been implicated in each of these tasks, and its important role in promoting proliferation is underscored by the finding that cultures of human primary endothelial cells expressing the LANA protein double at a faster rate and live much longer than control cells (15, 62). Several studies have shown that LANA regulates the expression of a number of viral and cellular genes (18, 33, 50, 62). Autonomous transcriptional repression domains have been identified in the N- and C-terminal regions, and there is evidence that LANA can repress promoter activity by using a variety of mechanisms (14, 17, 32-34, 36, 54). Presumably these repression functions help negate the cellular antiviral response, overcome cell cycle checkpoints, and possibly suppress transcription of viral lytic genes.
The mechanism and specificity of gene activation by LANA is less clearly understood. Microarray studies have identified a number of cellular genes that are stimulated by persistent expression of LANA and include genes involved in cell proliferation and modulation of the antiviral response (50, 62). Transactivation is also observed in transient assays that use a variety of natural and synthetic promoters (1, 31, 48, 50). Knowing how individual genes, especially those involved in growth control and signaling, are regulated by LANA is important for a full understanding of KSHV pathogenesis. There is no evidence for a discrete LANA-responsive sequence or element in promoters that respond to LANA, suggesting that activation does not involve direct binding of LANA to promoter DNA. Rather, LANA appears to act in a more promiscuous manner by modulating the activity of a variety of cellular activator proteins, including AP1 and Sp1 (1, 31). LANA contributes to activation of Tef/Lef-responsive genes by preventing glycogen synthase kinase-3β from degrading the coactivator β-catenin (15).
Another major role for LANA is the maintenance of the nonintegrated viral episome. In PEL-derived cell lines, some 30 to 100 copies of the episome are maintained in the nucleus of each infected cell as circular minichromosomes. The LANA C terminus binds to two closely spaced DNA sequences within each terminal repeat, and this is sufficient to promote DNA replication during S phase (2, 8, 17). Longer-term maintenance requires the LANA N terminus, specifically the first 22 residues, which are capable of tethering LANA, and therefore the viral DNA, to the mitotic host chromosomes (3, 56). Presumably, tethering ensures that newly replicated KSHV genomes are passed to the daughter cells in roughly equal numbers in the dividing host. Direct association with the host chromosomes may also contribute to the efficiency of DNA replication through recruitment of cellular DNA replication proteins.
The relationship between chromosome tethering and other known LANA functions, such as regulation of gene expression, has not been clearly elaborated. Here we show that chromosome tethering is also important for transcriptional activation of cellular promoters. Transactivation requires sequences in the N and C termini of LANA, and deletion of the first 22 residues, which are necessary for association with mitotic chromosomes, substantially reduces promoter activation. Higher-resolution analysis of the N terminus identified a block of seven amino acids, termed the chromatin-binding motif (CBM), sufficient for association with both mitotic and interphase chromatin. Mutations of individual residues within the CBM revealed a striking parallel between the ability to activate transcription and associate with interphase chromatin. Lastly, we also show that both the N terminus and central repetitive region of LANA are completely dispensable for inhibition of p53-mediated transactivation, suggesting that not all transcriptional functions require the CBM.
MATERIALS AND METHODS
Plasmid constructs.
Amplification of LANA coding sequences has been described previously (54). To add the FLAG epitope tag, the complete open reading frame or fragments corresponding to the nonrepetitive N- and C-terminal domains were shuffled into the cytomegalovirus-driven expression vector pCGFLAG by using the unique XbaI and BamHI sites (38). To generate green fluorescent protein (GFP)-LANAN Δ22, the N terminus of LANA was amplified by high-fidelity PCR (Roche Applied Science) using the following primers: 5′-GTCTAGATGGTAGGAAAGGAAACAGGTC-3′ and 5′-CGGATCCTAAAGCTTATTGTCATTGTCATCCTTGTC-3′ (XbaI and BamHI sites are underlined). GFP-LANAFL Δ22 was constructed by subcloning a BamHI fragment spanning the central repeat region and C terminus into GFP-LANAN Δ22. Wild-type human p53, pRb, and human papilloma virus (HPV) E6 were expressed with the plasmids pXp53 (a gift of Michele Pagano, New York University), pCMV-pRb (a gift of Brian Dynlacht), and pcDNA3 HPV16 E6 (a gift of Karl Münger, Harvard Medical School), respectively.
Site-directed mutagenesis was performed using the Quikchange Mutagenesis protocol (Stratagene) and custom-synthesized oligonucleotides. Sequences of all truncation and substitution mutants were confirmed by DNA sequencing. Luciferase reporter constructs used were as follows: pBAX-luc (19) (gift of Jennifer Nyborg, Colorado State University), pE2F1-luc, and pCDK2-luc (59) (gift of Ravi Tikoo and Moses Chao, New York University).
Cell culture, transfections, and luciferase assays.
HeLa and SAOS-2 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, antibiotics, and glutamine. HeLa cells were transfected with Polyfect (QIAGEN) or by electroporation (Bio-Rad Genepulser) and were assayed for protein expression or luciferase activity as described previously (54). SAOS-2 cells (a gift of James Borowiec, New York University) were seeded at a density of 105 cells per well in 6-cm-diameter dishes and were transfected using Effectene (QIAGEN).
Immunoblotting.
Immunoblotting was performed with wet transfer and was detected by enhanced chemiluminescence (SuperSignal; Pierce). Incubations with primary antibodies against the T7 epitope (αT7, diluted 1:10,000; Novagen), FLAG epitope (M2, diluted 1:10,000; Sigma), GFP (diluted 1:1,000; Molecular Probes), Rho-GDI (polyclonal antibody A-20, diluted 1:2,000; Santa Cruz), HCF-1 (αH12, diluted 1:1,000), and p53 (polyclonal antibody 6243, diluted 1:2,000; Santa Cruz) were conducted for 60 min at room temperature or overnight at 4°C, followed by incubation with appropriate horseradish peroxide-conjugated secondary antibodies (Amersham).
Small-scale isolation of chromatin.
Biochemical fractionation of transiently transfected HeLa cells was performed as described previously (40, 64). Briefly, 24 h after transfection 2 × 107 cells were collected, washed with phosphate-buffered saline (PBS), and resuspended at 4 × 107 cells/ml in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Triton X-100 was added (0.1% final concentration), the cells were incubated on ice for 8 min, and nuclei (fraction P1) were collected by centrifugation (5 min, 1,300 × g, 4°C). The supernatant (fraction S1) was clarified by high-speed centrifugation (5 min, 20,000 × g, 4°C), and the supernatant (fraction S2) was collected. The P1 nuclei were washed once in buffer A and were lysed for 30 min in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, and PMSF), and insoluble chromatin (fraction P3) and soluble (fraction S3) fractions were separated by centrifugation (5 min, 1,700 × g, 4°C). The P3 fraction was washed once with buffer B and resuspended in sodium dodecyl sulfate (SDS)-Laemmli buffer and was boiled for 10 min. Immunoblots were probed with antisera against human Rho-GDI (polyclonal antibody A-20; Santa Cruz) and the C terminus of human HCF-1 (polyclonal antibody α-H12) (63). For extraction of nuclear proteins under different salt concentrations, P1 nuclei were prepared as described above and then were resuspended in buffer A supplemented with various concentrations of NaCl in place of KCl. After incubation for 30 min on ice, nuclei were separated from solubilized proteins by centrifugation.
Immunofluorescence and green fluorescence microscopy.
HeLa cells were seeded onto sterile coverslips in a 24-well plate and were transfected with 100 ng of expression plasmids encoding T7-LANANC and T7-LANANC Δ22. After 24 h the cells were fixed and probed as described previously (38). Samples were incubated with αT7 antibody (diluted 1:600) followed by fluorescein isothiocyanate-conjugated α-mouse immunoglobulin G secondary antibody (diluted 1:600; Vector Laboratories) and visualized by laser scanning confocal microscopy using a Zeiss LM 510 Meta microscope. Images were captured using Zeiss software and were exported into Adobe Photoshop 5.0 for further processing. For analysis of mitotic cells, transfected cultures were harvested by trypsinization after 24 h, washed with PBS, fixed with 3.7% formaldehyde for 15 min at 25°C, washed again with PBS, and stained with Hoechst 33342 (0.1 μg/ml) for 5 min. Unincorporated stain was removed with water, and the fixed cells were dropped onto slides in 30-μl aliquots. After drying, coverslips were applied to the slides using Dako Fluorescent Mounting Media (DAKO Corporation). Fluorescence was recorded using a Zeiss Axioplan epifluorescence microscope equipped with a digital camera.
RESULTS
LANA functions as a promiscuous activator of transcription.
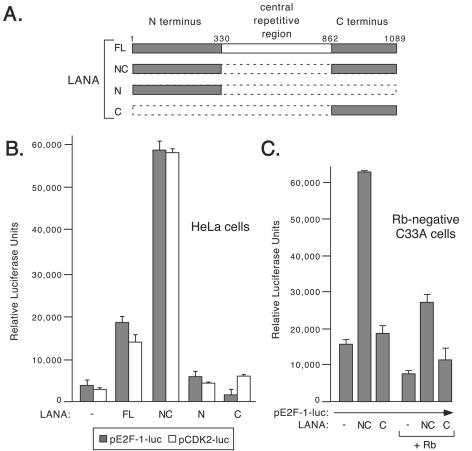
Introduction of LANA into primary endothelial cells leads to a significant increase in the number of cells entering S phase and extends the survival of these cells (62). These changes involve the coordinated expression of many cell-cycle-regulated cellular genes, possibly as a direct consequence of transactivation by LANA. To understand how LANA activates cellular promoters, we investigated the regions of LANA required to transactivate the cell-cycle-regulated human E2F1 and cyclin-dependent kinase-2 (CDK2) promoters (24, 44, 55). In addition to full-length LANA (LANAFL, shown schematically in Fig. 1A) we tested a derivative lacking the central repetitive region (LANANC) as well as variants that contain each of the terminal regions separately (LANAN and LANAC).
FIG. 1.
Both the N- and C-terminal regions of LANA are required for transactivation of the human E2F1 and CDK2 promoters. (A) Schematic showing the general structure of LANA and engineered fragments used in this study. Note that the amino acid numbering corresponds to a previously sequenced KSHV strain (GenBank accession no. AAB626557). (B) E2F1-luc or pCDK2-luc reporters (0.1 μg) were introduced into HeLa cells by electroporation together with an empty expression plasmid (1.0 μg) or one encoding T7-epitope-tagged LANAFL, LANANC, LANAN, or LANAC. Extracts were prepared after 24 h and were assayed for luciferase activity. Each transfection was performed in triplicate, and the mean value and standard deviation are shown. (C) C33A cells (5 × 105) were transiently transfected using Lipofectamine 2000 (Invitrogen). The E2F1-luc reporter plasmid (0.025 μg) was transfected with 0.2 μg of an expression vector encoding LacZ, LANANC, or LANAC with or without 0.05 μg of pCMV-pRb expression plasmid. Extracts were prepared after 24 h and were assayed for luciferase activity.
Human HeLa cells were transiently cotransfected with a luciferase reporter gene fused to either the E2F1 promoter (pE2F1-luc) or CDK2 promoter (pCDK2-luc) together with expression plasmids encoding full-length LANA or various truncations (Fig. 1B). Reporter activity was measured after 24 h and was compared to the activity of the reporter cotransfected with an empty expression vector. Full-length LANA (LANAFL) activated the E2F1 and CDK2 reporters five- to sixfold above the level of the reporter alone. Deletion of the central repetitive region (LANANC) resulted in a greater level of activation (17-fold for E2F1 and 24-fold for CDK2); however, immunoblotting of extracts from transfected cells revealed that the LANANC protein is expressed at higher levels than LANAFL and most likely accounts for the greater activity of LANANC (data not shown; see also Fig. 2C). Expression of the N terminus alone (LANAN) gave only a modest increase in reporter activity, whereas expression of the C terminus (LANAC) resulted in a small but reproducible decrease for the E2F1 reporter and a small increase for the CDK2 reporter. Taken together, these results show that sequences in the N- and C-terminal regions of LANA are required for transactivation of cellular promoters and that neither region is sufficient on its own.
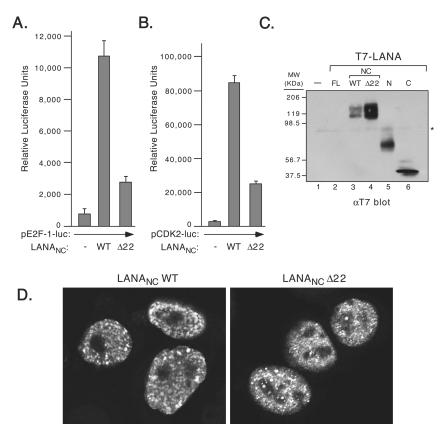
FIG. 2.
Deletion of the first 22 residues of LANA reduces transactivation. (A) The E2F1-luc reporter (0.1 μg) was cotransfected with 1.0 μg of expression plasmid encoding T7-LANANC WT or T7-LANANC Δ22. Twenty-four hours posttransfection extracts were prepared and luciferase activity was quantitated. (B) Cotransfection was performed as described for panel A, except pCDK2-luc (0.1 μg) was used as the reporter. (C) T7-epitope-tagged LANANC WT and LANANC Δ22 proteins were detected by immunoblotting. Protein extracts were resolved on a SDS-10% PAGE and were immunoblotted with an αT7 monoclonal antibody (1:10,000 dilution; Novagen Inc.). (D) Proliferating HeLa cells transiently transfected with T7-LANANC WT or T7-LANANC Δ22 were examined by immunofluorescence using αT7 monoclonal antibody (1:600 dilution) with fluorescein isothiocyanate-conjugated anti-mouse secondary antibody (dilution 1:600; Vector Laboratories) and visualized by confocal laser scanning microscopy.
The C terminus of LANA interacts with the retinoblastoma tumor suppressor protein (pRb), and relief of Rb-mediated repression might explain transactivation of these two E2F-regulated promoters (48). To address this we transfected C33A cells, which lack functional pRb protein (52). As shown in Fig. 1C, LANANC was still able to transactivate the E2F1 promoter despite a very high level of baseline activity of the reporter alone. Expression of human wild-type pRb reduced the constitutive promoter activity but did not significantly alter the relative extent of induction by LANANC. As in HeLa cells, expression of LANAC did not activate the promoter in the presence or absence of pRb. Thus, it is unlikely that LANA transactivates the E2F1 and CDK2 promoters by counteracting Rb-mediated repression.
The first 22 residues of LANA are required for transactivation.
Previous studies have shown that the LANA N terminus is capable of associating with mitotic chromosomes and that this function is important for maintenance of the viral episome (46). This property is lost when residues 5 to 24 at the N terminus of LANA, termed the chromosome-binding site, are deleted (46). To determine whether this chromatin association function might also be relevant to transactivation of the E2F1 and CDK2 promoters, we compared the level of transactivation by the LANANC construct containing a wild-type N terminus (LANANC WT) with that of a derivative lacking the first 22 residues (LANANC Δ22). Note that we used the LANANC fusion rather than the full-length protein because this yielded the most robust response from the reporter assay. As shown in Fig. 2A and B, the deletion resulted in a significant decrease in transactivation of both promoters. For pE2F1-luc the 14-fold activation by LANANC WT was decreased to less than 4-fold by LANANC Δ22. For pCDK2-luc, the 30-fold stimulation produced by LANANC WT was reduced to less than 9-fold by LANANC Δ22. Immunoblotting (Fig. 2C) demonstrated that this decrease was not due to differences in protein expression, because LANANC Δ22 was in fact expressed at higher levels than LANANC WT. Thus, a small region at the extreme N terminus of LANA that has been implicated in mitotic chromosome binding is also important for transactivation.
LANA is associated with interphase chromatin.
With a few exceptions, active transcription of eukaryotic genes occurs exclusively during interphase and is rapidly silenced as cells enter mitosis (20). Thus, the requirement for the first 22 residues suggested to us that LANA might associate with interphase as well as mitotic chromatin. As shown in Fig. 2D, microscopic analysis of interphase cells transfected with T7-epitope-tagged proteins revealed no obvious difference between LANANC WT and LANANC Δ22 in terms of their nuclear localization. Both fusion proteins were found exclusively in the nucleoplasm in a strongly speckled pattern and were excluded from the nucleoli. Similar patterns were observed when the same LANA fragments were fused to GFP (data not shown). Thus, deletion of the first 22 residues did not compromise nuclear localization nor speckle formation in interphase cells.
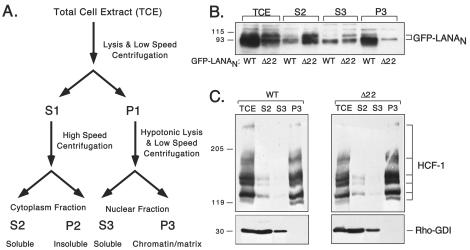
To examine the association with interphase chromatin directly, we used a rapid biochemical fractionation protocol (outlined in Fig. 3A) to prepare protein extracts from transfected HeLa cells (40). For this experiment we used cells transiently expressing the N terminus of LANA fused to GFP. Asynchronously growing cultures expressing GFP-LANAN WT or GFP-LANAN Δ22 were separated into soluble cytoplasmic (S2), soluble nuclear (S3), and chromatin-enriched (P3) fractions. Examination of these proliferating cultures by light microscopy indicated that less than 10% of cells were in metaphase at the time of extract preparation (data not shown). Fractions derived from equivalent numbers of cells were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and were analyzed by immunoblotting using an antibody against GFP (Fig. 3B). The GFP-LANAN WT fusion protein (WT) was found primarily in the chromatin enriched fraction (P3) with a small amount in S3 and S2. In striking contrast, the deletion mutant (Δ22) was preferentially released into the S2 fraction following cell lysis. Thus, important determinants for association with interphase chromatin, but not nuclear localization (Fig. 2D), are contained within the first 22 residues of LANA.
FIG. 3.
LANA associates with interphase chromatin. (A) Flowchart showing small-scale fractionation protocol for isolation of interphase chromatin from transiently transfected cells. (B) HeLa cells expressing GFP-LANAN WT or GFP-LANAN Δ22 were harvested 24 h posttransfection and were fractionated according to the scheme shown in panel A. The resulting extracts were resolved by SDS-12% PAGE and were immunoblotted using antisera to GFP. (C) The fractions shown in panel B were probed for chromatin-associated HCF-1 and cytoplasmic Rho-GDI.
As controls for the fractionation procedure, parallel samples were probed with an antibody against HCF-1, a known chromatin-associated nuclear factor (64), and with an antibody against Rho-GDI, a known cytoplasmic protein (41). As expected, the majority of HCF-1 was found in the P3 chromatin fraction, with a small amount in the S2 fraction. Note that HCF-1 is composed of multiple subunits generated through proteolytic processing and migrates as a series of immunoreactive bands (63). The cytoplasmic marker Rho-GDI was found predominantly in the S2 or cytoplasmic fraction, with a smaller amount in S3. These profiles confirm that we achieved significant enrichment for chromatin-associated proteins in the P3 fraction and, more importantly, that the fractionation was similar between the two populations of transfected cells.
LANA C terminus facilitates the association with interphase chromatin.
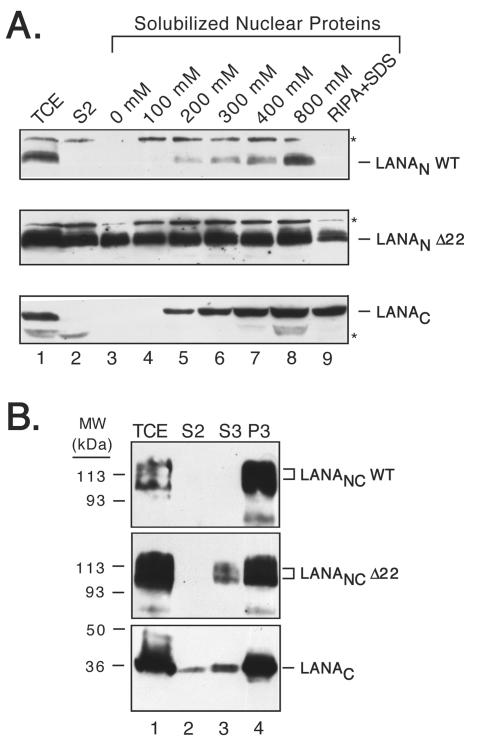
To further compare the chromatin association properties of the wild-type N terminus with those of the 22-residue deletion, P1 nuclei were isolated from transfected cells and separate aliquots were incubated for 30 min in extraction buffers of different ionic strength. Solubilized proteins were detected by immunoblotting (Fig. 4A). Note that in this experiment we used LANA fragments carrying an N-terminal FLAG epitope tag rather than the larger GFP moiety used for Fig. 3B and were thus detected using an α-FLAG antibody. Consistent with the previous fractionation protocol, LANAN WT was relatively resistant to salt extraction, with the bulk of the protein being extracted only in buffer supplemented with 800 mM NaCl. In contrast, LANAN Δ22 could be more readily solubilized, even in the absence of added salt (lane 3), implying a very weak association.
FIG. 4.
Independent association of LANA C terminus with interphase chromatin. (A) Intact nuclei were prepared from HeLa cells expressing FLAG-tagged LANAN WT, LANAN Δ22, and LANAC, incubated in buffer A supplemented with different salt concentrations (lanes 3 to 8, 0 to 800 mM NaCl). Solubilized proteins were analyzed by immunoblotting with αFLAG M2 monoclonal antibody. In addition, nuclei were extracted with RIPA buffer containing 0.1% SDS (lane 9). Asterisks indicate a nonspecific cross-reacting band. Equivalent volumes of total cell (TCE) and cytoplasmic (S2) extracts are also shown. (B) Small-scale chromatin extraction of HeLa cells expressing FLAG-LANANC WT (upper panel), FLAG-LANANC Δ22 (middle panel), and FLAG-LANAC (lower panel). TCE and the S2, S3, and P3 fractions were resolved by SDS-12% PAGE (15% for LANAC) and immunoblotted as described for panel A.
As a control, we included the isolated C terminus (LANAC), which has previously been shown to also localize to the nucleus, where it accumulates in ill-defined speckles reminiscent of the LANA bodies formed by the full-length LANA protein (54). This fragment showed an extraction profile similar to that of LANAN, with little or no extraction below 200 mM NaCl (lane 5) and maximal extraction at 800 mM NaCl (lane 8). There was, however, a profound difference in their sensitivity to extraction by detergent (Fig. 4A, lane 9). When the nuclei were incubated in radioimmunoprecipitation assay (RIPA) buffer supplemented with 0.1% SDS, only LANAC was efficiently released from the P1 nuclei. This suggests that the N and C termini can independently associate with cellular chromatin.
To determine whether the C terminus contributes to chromatin association in the context of the NC fusion, we repeated the small-scale chromatin isolation assay depicted in Fig. 3A, using cells expressing FLAG-tagged LANANC WT and LANANC Δ22 (Fig. 4B). In this context, both proteins were found predominantly in the P3 fraction (lane 4), indicating that addition of the C terminus was able to compensate for deletion of the first 22 residues from the N terminus. Only a trace amount of LANANC Δ22 can be detected in the S3 fraction on this relatively long exposure of the immunoblot. The role of the C terminus in compensating for the Δ22 deletion is further substantiated by the fractionation properties of the isolated C terminus (LANAC). As with the preceding proteins, LANAC was found predominantly in the P3 fraction (lane 4) with only small amounts in the S2 and S3 fractions (lanes 2 and 3, respectively). Thus, two independent domains mediate association with interphase chromatin, one located at the extreme N terminus and the other within the C terminus.
Identification of specific residues required for mitotic chromosome association.
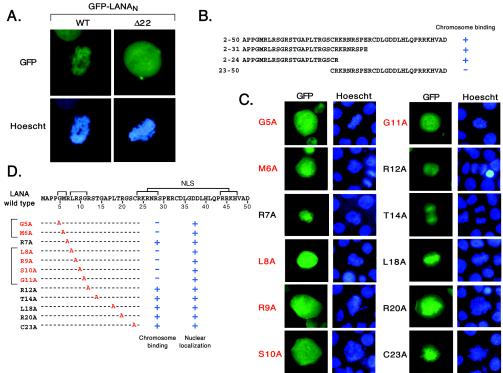
Thus far, the exact relationship between binding to interphase and mitotic chromatin has not been addressed. To identify specific residues required for association with mitotic chromosomes we established a transient assay using GFP-LANAN WT and GFP-LANAN Δ22 expressed in proliferating HeLa cells. After fixation, fields of cells were searched for GFP-expressing cells that were undergoing mitosis at the time of fixation as judged by Hoechst 33342 DNA staining and thereby avoiding the use of spindle inhibitors, such as Colcemid, to artificially arrest cells in metaphase (Fig. 5A). Consistent with previous reports, both fusions were localized to the nucleus. GFP-LANAN WT was closely associated with the condensed chromosomes with a uniform distribution that mirrored that of the Hoechst DNA stain. In contrast, GFP-LANAN Δ22 was spread throughout the remnants of the nuclear compartment without any obvious concentration in the proximity of the mitotic chromosomes. Thus, in line with previous studies, the association of the LANA N terminus with condensed mitotic chromatin is dependent on residues located within the first 22 amino acids (3, 46).
FIG. 5.
The first 24 residues of LANA are sufficient for association of the N terminus with mitotic chromosomes. (A) Deletion of residues 2 to 22 prevents chromosome association of the LANA N terminus with mitotic chromosomes. HeLa cells were electroporated with plasmids encoding GFP-LANAN WT or GFP-LANAN Δ22 and after 24 h were fixed, dropped onto glass slides, and examined by epifluorescence microscopy. GFP-positive cells (green) undergoing mitosis at the time of fixation were identified by Hoechst 33342 staining (blue) to detect DNA in the condensed chromosomes. (B) Determinants for nuclear localization and chromatin binding lie within the first 24 amino acids. Peptides corresponding to LANA residues 2 to 50, 2 to 31, 2 to 24, and 23 to 50 were fused to GFP and assayed for mitotic chromosome association in HeLa cells. (C) Representative mitotic cells expressing 1 of 12 alanine substitution mutants at selected residues within the first 24 residues of LANA (assayed in the context of GFP-LANAN2-50). The names of mutants showing no colocalization with the condensed chromosomes are labeled in red. (D) Summary of the alanine substitution mutant data based on analysis of 10 or more cells per mutant. Nuclear localization was scored in interphase cells. The putative bipartite NLS corresponds to two clusters of basic residues (Arg-24 to Arg-28 and Arg-44 to His-47).
To further localize the sequences required for mitotic chromosome association, we tested shorter peptides corresponding to residues 2 to 50, 2 to 31, 2 to 24, and 23 to 50. The peptides were expressed as GFP fusions and were tested for mitotic chromosome association as well as nuclear localization in transiently transfected HeLa cells. The results of this analysis are summarized in Fig. 5B. All four fusions were localized to the nucleus, contrasting with the broader distribution of GFP alone throughout the cell (data not shown). Although the putative NLS (residues 24 to 47) is disrupted, these fusions are small enough to enter the nucleus by diffusion rather than active import and accumulate through association with nuclear components. In transfected cells undergoing mitosis at the time of fixation, residues 2 to 31 (GFP-LANAN2-31) and 2 to 24 (GFP-LANAN2-24) were sufficient to tether GFP to the condensed cellular chromosomes. Deletion of these residues (GFP-LANAN23-50) abolished chromosome binding but not nuclear accumulation. These results indicate that mitotic chromosome-binding function lies between residues 2 and 24.
We next used alanine-scanning mutagenesis to identify individual residues critical for mitotic chromosome binding (Fig. 5C and D). Mutations were generated in the context of GFP-LANAN2-50, because this fusion showed the most robust association and included a putative bipartite NLS comprising two clusters of basic residues separated by 15 residues. In interphase cells, all 12 mutants were shown by immunoblotting to be expressed at similar levels and localized to the nucleoplasm in a pattern indistinguishable from that of the wild type (data not shown). Examination of mitotic cells revealed that six mutants (G5A, M6A, L8A, R9A, S10A, and G11A) were not obviously associated with the mitotic chromosomes in any of the transfected cells examined. The sensitive residues comprise two clusters separated by a single residue (Arg-7), which could be replaced by alanine without preventing mitotic chromosome binding. Thus, the N-terminal function required for tethering the LANA N terminus to mitotic chromosomes maps to a heptameric sequence between residues Gly-5 and Gly-11.
The CBM mediates association with interphase chromatin.
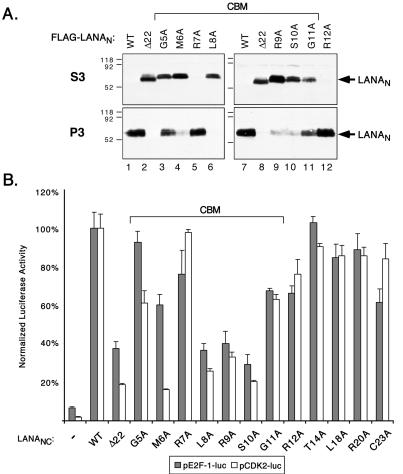
Having identified specific residues required for association with mitotic chromatin, we asked whether these same residues were required for binding to interphase chromatin. The alanine substitution mutants were introduced into the intact N terminus (residues 2 to 330) carrying a FLAG epitope tag and were tested for chromatin association by using the biochemical fractionation scheme described above. Immunoblots showing only the more informative S3 and P3 fractions are presented in Fig. 6A. As shown in Fig. 3, the wild-type N terminus (FLAG-LANAN WT), which is shown in duplicate, was almost exclusively found in the P3, or chromatin-enriched, fraction (lanes 1 and 7), whereas the equivalent fragment lacking the first 22 residues (FLAG-LANAN Δ22) is found in the S3 fraction (lanes 2 and 8). For reasons that are unclear, GFP-LANAN Δ22 (Fig. 3B) accumulated in the S2 rather than the S3 fraction. Mutations of Met-6 (lane 4), Leu-8 (lane 6), Arg-9 (lane 9), and Ser-10 (lane 10) have a significant effect on chromatin association, such that the majority of the protein is found in S3 rather than P3. Conspicuously, substitutions of the two glycine residues (Gly-5 and Gly-11, lanes 3 and 11, respectively) had a more modest effect, such that roughly equal fractions of the tagged protein remained in the P3 fraction. In this experiment, very little transfected protein was detected in S2. Consistent with the analysis in mitotic cells, substitution of Arg-7 (lane 5) and Arg-12 (lane 12) had no effect on chromatin binding. Thus, we observed a perfect overlap between the residues required for association with interphase chromatin (defined by biochemical fractionation) and mitotic chromosome association (defined by microscopy). It is reasonable to suppose that the limited sensitivity of the mitotic assay obscured the intermediate phenotypes of Gly-5 and Gly-11. Based on the close parallels between interphase and mitotic chromatin association, we can conclude that the LANA N terminus contains a single functional element, which we rename the chromatin-binding motif (CBM) to distinguish it from the less precisely mapped but probably equivalent chromosome-binding site (46). Importantly, these results show that the CBM mediates chromatin association throughout the cell cycle and is not specific for mitotic chromatin.
FIG. 6.
The CBM is important for association with interphase chromatin and transactivation. (A) Association with interphase chromatin by individual alanine substitution mutants. HeLa cells expressing wild-type or mutant versions of T7-LANAN were fractionated according to the scheme shown in Fig. 3A, resolved by SDS-PAGE, and immunoblotted with αT7 monoclonal antibody. Only the S3 and P3 samples are shown. (B) Activation of the E2F1 (filled bars) and CDK2 (open bars) promoters by wild-type or mutant versions of T7-LANANC. To simplify the comparisons, values and standard deviations were normalized to the mean of activity of the reporter cotransfected with wild-type LANANC (expressed as 100%).
The CBM is important for transactivation.
The same 12 mutants were tested in the context of LANANC for the ability to transactivate the E2F1 and CDK2 promoters (Fig. 6B). Substitutions at Leu-8, Arg-9, and Ser-10 had the most dramatic effect, reducing transactivation to a level comparable to that of the Δ22 deletion. Substitutions at Gly-5, Met-6, Gly-11, and Arg-12 had measurable but slightly smaller effects. In general, the behaviors of the two promoters to each of the mutations were similar; however, we found CDK2 to be significantly more sensitive to the substitutions at Gly-5 and Met-6 than E2F1. The intermediate transactivation phenotypes of Gly-5 and Gly-11 correlate well with the partial association of these mutants with interphase chromatin (Fig. 6A, lanes 3 and 11). Oddly, substitution of Cys-23, which lies outside of the CBM and consequently was not tested for interphase binding, gave a moderate reduction in E2F1 activation but had no effect on CDK2. Overall, these results show that there is a close correlation between chromatin association properties of the N terminus and the ability of the larger LANANC fusion to activate transcription.
The N terminus is dispensable for inhibition of p53-mediated transactivation.
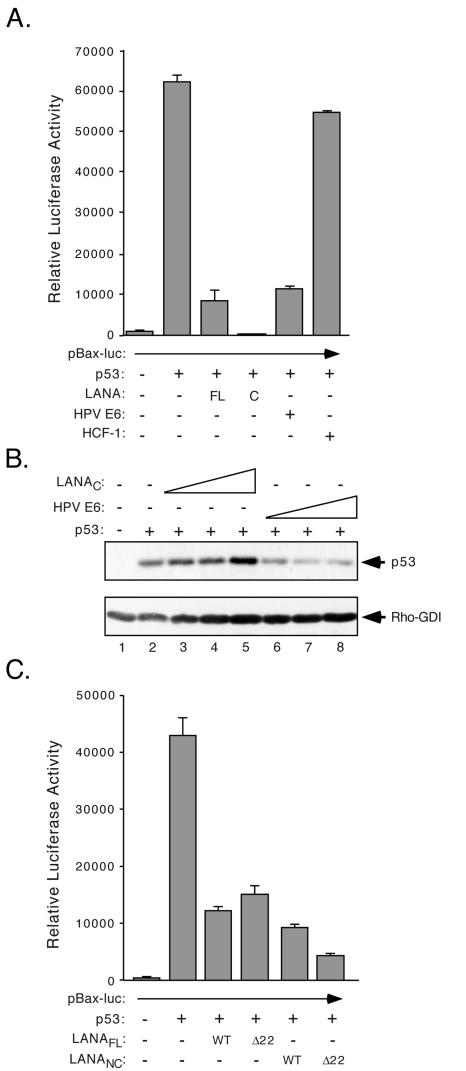
Several studies have shown that LANA is able to repress transcription (14, 18, 33, 36, 50, 54, 62). One potentially important example is its ability to suppress transcriptional activation by the cellular p53 protein and thereby contribute to the blockade of host cell death by apoptosis (14). This raises the question of whether chromatin binding is important for inhibition of p53 as well as transactivation. To address this, we assayed p53-mediated transactivation of BAX, a well-characterized p53 target (19, 43). The human BAX promoter (−340 to +31) fused to luciferase was transiently cotransfected in the presence or absence of LANA fragments into p53-null SAOS-2 osteosarcoma cells (5). The results of this experiment are shown in Fig. 7A. Transfection of wild-type p53 expression plasmid resulted in a ∼75-fold activation of the pBAX-luc reporter. This increase was greatly reduced by cotransfection of 0.275 μg of expression plasmid encoding full-length LANA (LANAFL) and expression of an equivalent amount of the C terminus (LANAC) reduced activation by p53 to an even greater extent. As a positive control we tested the E6 protein encoded by HPV-16, which promotes the ubiquitin-mediated degradation of p53. As expected, this reduced pBAX-luc activation. As a negative control, we expressed human HCF-1, a large nuclear protein that is not known to regulate p53, and this did not significantly affect pBAX-luc activation.
FIG. 7.
CBM is dispensable for inhibition of p53-mediated activation. (A) SAOS-2 cells (105) (p53 null) were transiently transfected using Effectene (QIAGEN). The pBAX-luc reporter plasmid (0.25 μg) was transfected alone or with 0.075 μg of an expression vector encoding wild-type p53 with or without plasmids (0.275 μg) encoding LANAFL, LANAC, HPV E6, and HCF-1. Luciferase activity was measured after 24 h. (B) Expression of LANAC does not alter the steady-state levels of p53 protein. SAOS-2 cells (4 × 105) were transfected with 1.5 μg of LacZ expression vector (lane 1) or 0.5 μg of p53 expression plasmid (lanes 2 to 8) plus 0.25 (lane 3), 0.5 (lane 4), and 1.0 μg (lane 5) of LANAC plasmid or 0.25 (lane 6), 0.5 (lane 7), and 1.0 μg (lane 8) of HPV E6 plasmid. After 24 h, cells were lysed by boiling in SDS sample buffer and proteins were resolved by SDS-10% PAGE and probed for p53 or by SDS-15% PAGE and probed for Rho-GDI. (C) The Δ22 deletion does not prevent LANA from inhibiting p53. SAOS-2 cells were transfected as described for panel A using the wild-type and Δ22 versions of LANAFL and LANANC.
Immunoblotting of protein extracts from the transiently transfected SAOS-2 cells (Fig. 7B) using a α-p53 polyclonal antibody showed that the suppression of p53-mediated transactivation by LANAC was not mediated by changes in the steady-state levels of p53 protein (compare lane 2 with lanes 3 to 5). In contrast, expression of HPV E6 reduces p53 levels, presumably by promoting increased turnover (lanes 6 to 8). These results indicate that the entire N terminus and central repetitive region of LANA are dispensable for inhibition of p53-mediated transactivation and that LANA does this without altering the levels of p53 protein.
To ask specifically whether the CBM contributes to p53 inhibition in the context of the intact protein, we compared LANAFL WT with LANAFL Δ22 (Fig. 7C). As before, the reporter was induced strongly (93-fold over reporter alone) by expression of p53, and this activation was blocked to similar extents by the two versions of LANA. Comparable results were obtained with LANANC WT and LANANC Δ22. Taken together these results clearly demonstrate that a functional element(s) located within the first 22 residues of LANA is not required for p53 inhibition. Thus, the CBM is not required for all transcription-related functions of LANA.
DISCUSSION
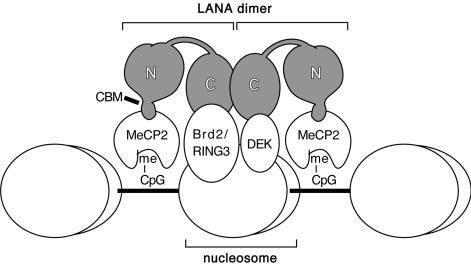
This study reveals a new role for the N-terminal chromatin-binding function of LANA, namely in facilitating transcriptional activation. Optimal transactivation of the cellular E2F1 and CDK2 promoters by LANA requires both the N-terminal CBM and the C terminus. We find that the central repetitive region is dispensable for both transactivation and chromatin association. In fact, deletion of this segment produces a more potent activator than full-length LANA. Although this may be due to the improved expression of the smaller protein, we cannot yet exclude the possibility that the deletion removes inhibitory sequences. Biochemical fractionation showed that the N- and C-terminal regions are independently capable of forming a tight association with interphase chromatin that is resistant to extraction in buffer of medium ionic strength. In the case of the N terminus, this interaction is mediated exclusively by the CBM, while the C-terminal activity has not been pinpointed with any precision. In contrast to chromatin binding, transactivation requires both the CBM and the C terminus, arguing that chromatin tethering is not sufficient per se and that transactivation requires specific molecular interactions mediated by sequences from both ends of the protein. Figure 8 presents a model conceptualizing the multivalent interactions of a single LANA dimer with components of cellular chromatin.
FIG. 8.
Model of LANA in association with cellular chromatin. In this schematic view, LANA multimers (most likely dimers) establish protein-protein contacts with several different components of host components (see the text for references). The CBM in the LANA N terminus binds to MeCP2, which in turn is bound to methylated CpG dinucleotides in the chromosomal DNA. In the C-terminal domain, LANA complexes with the bromodomain protein Brd2/RING3, which associates with acetylated histone H4 and, to a smaller extent, acetylated histone H2B. The C terminus also associates with DEK, which can bind to DNA that is incorporated into nucleosomes. The LANA C terminus mediates multimerization and, although not shown here, contains a sequence-specific DNA-binding domain that recognizes a GC-rich sequence found twice in each of the KSHV terminal repeats.
To date, searches of the protein sequence databases have not uncovered identical or near-identical matches to the CBM heptamer. This is surprising given the well-established precedent for viral proteins to co-opt cellular motifs for their own use and that the other γ2-herpesviruses express obvious LANA homologues. We hypothesize that despite its small size, there is significant flexibility within the motif in terms of its precise amino acid sequence and/or the spacing of critical residues. Further mutagenesis and mapping of functionally analogous sequences in selected LANA homologues would address this hypothesis.
What is the target for the CBM?
Because of the small size of the CBM and limited requirement for basic residues, it seems unlikely that the CBM functions as a DNA-binding domain contacting the host chromosome directly. Instead we favor the idea that the CBM is the founding member of a new class of protein-protein interaction motif, analogous to LxxLL motifs that mediate interaction between steroid hormone receptors and their cofactors or the HCF-binding motif found in herpes simplex virus transactivator VP16 and a host of cellular transcription factors (13, 23, 37, 60). Again, the discovery of additional examples of the CBM will help to elaborate this idea and offer insight into the sequence requirements.
One likely chromosomal target for the LANA CBM is methyl-CpG-binding protein 2 (MeCP2), a ubiquitous nuclear protein that binds to methylated DNA (Fig. 8). In a recent study, Krithivas and colleagues found that human MeCP2 interacts with LANA in vitro and in vivo and showed that binding is prevented by deletion of the first 15 residues of LANA (32). Whether binding of MeCP2 is sensitive to mutations of individual residues within the CBM remains to be tested. MeCP2 is an excellent candidate for chromosome tethering during mitosis, because (i) it is associated with chromosomes throughout the cell cycle (9) and (ii) tethering of LANA to metaphase chromosomes in murine 3T3 cells is dependent on the expression of recombinant MeCP2 (32). That said, it is harder to reconcile use of MeCP2 with the activation function reported here. Studies with a variety of experimental systems have shown that MeCP2 is preferentially associated with repressed chromatin and functions as a gene-specific transcriptional repressor (39, 57). Various in vitro and in vivo studies have shown that a transcriptional repression domain within MeCP2 recruits corepressors Sin3A and SMRT and that these in turn recruit histone deacetylase, histone methyltransferase, and DNA methyltransferase activities responsible for gene inactivation and assembly of repressed chromatin (16, 30, 57). One possibility is that binding of LANA reprograms the MeCP2/Sin3a complex, converting it from a repressor into an activator. Alternatively, the new complex might sequester these negative regulators so that they cannot repress promoter activity. A derepression mechanism such as this would not require direct association with the responsive promoter and perhaps would account for the promiscuous nature of LANA-mediated transactivation. A third possibility is that the CBM recognizes proteins distinct from MeCP2 that participate in transcriptional activation rather than repression. It is worth mentioning that although LANA has been reported to interact with histone H1, there is no information on the regions of LANA necessary for this association, and, as with MeCP2, it is unclear how histone H1 binding would facilitate gene activation (7, 25).
Sequences required for chromatin association by the C terminus have not been mapped further in this study. The C terminus provides a constitutive dimerization function and contains a sequence-specific DNA-binding domain. Responsive promoters lack obvious homologies to the LANA-binding sites in the terminal repeat elements, and it is unlikely that LANA makes sequence-specific contacts with promoter DNA. Interactions with cellular transcription factors may be more relevant. Known partners include Brd2/RING3 and other members of the BET bromodomain-containing protein family (see Fig. 8) as well as the protooncogene DEK, α and β isoforms of heterochromatin protein 1 (HP1α and HP1β), and pRb protein (32, 35, 47, 61). Both Brd2/RING3 and pRb regulate promoters containing E2F binding sites and would therefore include the E2F1 and CDK2 promoters used in this study, albeit through different mechanisms (11). The twin bromodomains of Brd2/RING3 have a high affinity for acetylated N-terminal tails of histones H2A and H3, and Brd2/RING3 has been identified in macromolecular complexes that contain components of the mammalian Mediator complex as well as E2F proteins (10, 25).
We cannot exclude the possibility that the C terminus also contributes to transactivation by specifying the formation of LANA dimers or other multimers. Truncation analyses within the C terminus have been uninformative in this regard. Deletions that prevent dimer formation simultaneously abolish a variety of functions and reduced protein solubility, consistent with the unfolding of the entire domain. Also, replacement of the C terminus with the DNA-binding and dimerization domain from the yeast Gal4 protein (residues 1 to 94) does not restore transactivation of the E2F1 promoter, supporting the idea that the C terminus participates in promoter activation through specific interactions with cellular transcription factors (L. Y. Wong and A. C. Wilson, unpublished results). With regard to the chromatin-binding function with the C terminus, Brd2/RING3, DEK, and HP1α are known to be bound to chromatin throughout the cell cycle and might therefore also contribute to chromosome tethering (25, 26, 42).
The LANA C terminus is sufficient for inhibition of p53-mediated activation.
Apoptosis and cell cycle arrest triggered by p53 represent important defense mechanisms used by mammalian cells to protect against colonization by persistent viruses. Latently infected PEL lines and advanced cutaneous stages of KS express elevated levels of wild-type p53 protein, suggesting that one or more of the latency products provide an effective block to p53 function (27, 45). Our present data confirms and extends the initial observation by Friborg and colleagues that LANA is sufficient to inhibit p53-mediated transactivation of a reporter composed of 13 reiterated p53 consensus-binding sites (14). Using the promoter from a known p53-responsive gene, we show that LANA is an effective inhibitor and further map the minimal inhibitory domain to the C-terminal 228 residues of LANA. Immunoblotting showed that LANAC does not reduce the levels of p53 protein in the cell and likewise does not alter the distribution of p53 within the nucleus (data not shown). Contrary to the previous report, our own efforts to demonstrate direct association between p53 and LANAC have not been successful, and it remains an open question whether LANA targets p53 itself or cofactors required for transcriptional activation.
Chromatin association and replication of the latent episome.
The tethering of viral episomes to host chromosomes has emerged as a fundamental property of DNA viruses that establish persistent infections in proliferating cells (6). It is likely that tethering provides a simple mechanism by which these viruses ensure efficient partitioning of newly replicated genomes to each daughter cell at mitosis. Association with host chromatin may also facilitate access to the cellular DNA replication and chromatin assembly machinery. While the manuscript of this study was in preparation, Barbera and colleagues described a similar approach to mapping N-terminal residues involved in mitotic chromosome binding (3). In their study, these authors used clustered rather than single amino acid substitutions; however, our results for chromatin association are in good agreement. Interestingly, transient DNA replication also appeared to be sensitive to mutations that disrupt the CBM, raising the possibility that the transcriptional activation function characterized here may also be required for initiation of DNA synthesis, as has been observed in other DNA replication origin contexts. Curiously, combined mutations of Thr-14 and Gly-15 had a deleterious effect on DNA replication and episome persistence but did not prevent binding to mitotic chromosomes (3). Although we did not test Gly-15, our single alanine substitution at Thr-14 did not affect transactivation or chromatin association, suggesting that the DNA replication function may have additional requirements.
In conclusion, the identification of a relatively small motif that participates in multiple functions, each of which has the potential to be critical for the establishment and maintenance of latency, suggests an opportunity for pharmacological intervention through the design of small molecules that compete with the LANA CBM for binding to cellular targets.
Acknowledgments
We thank Naoko Tanese and Jim Borowiec for discussions and helpful comments on the manuscript. Plasmids, antibodies, and cell lines used in this study were generously provided by Jim Borowiec, Moses Chao, Brian Dynlacht, Michael Garabedian, Stavros Giannakopoulos, Karl Münger, Jennifer Nyborg, Michele Pagano, Laura Su, Ravi Tikoo, and Naoko Tanese. Lastly, we thank Randy Luciano, Karen Allen, and Matt Valento for valuable help.
This study was supported by a Lymphoma Research Foundation Junior Faculty Award (to A.C.W.), funds from the Center for AIDS Research, and by the NIH (GM61139-04 and S10 RR017970-01).
REFERENCES
- 1.An, J., Y. Sun, and M. B. Rettig. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222-228. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbera, A. J., M. E. Ballestas, and K. M. Kaye. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman, E. 2002. The role of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Recent Results Cancer Res. 159:27-37. [DOI] [PubMed] [Google Scholar]
- 5.Chandar, N., B. Billig, J. McMaster, and J. Novak. 1992. Inactivation of p53 gene in human and murine osteosarcoma cells. Br. J. Cancer 65:208-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, C. M., and P. G. Medveczky. 2002. Genetic requirements for the episomal maintenance of oncogenic herpesvirus genomes. Adv. Cancer Res. 84:155-174. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., Jr., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, M. A., Jr., C. Subramanian, and E. S. Robertson. 2001. The Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 9.Craig, J. M., E. Earle, P. Canham, L. H. Wong, M. Anderson, and K. H. Choo. 2003. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 12:3109-3121. [DOI] [PubMed] [Google Scholar]
- 10.Crowley, T. E., E. M. Kaine, M. Yoshida, A. Nandi, and D. J. Wolgemuth. 2002. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 16:1727-1737. [DOI] [PubMed] [Google Scholar]
- 11.Denis, G. V., C. Vaziri, N. Guo, and D. V. Faller. 2000. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 11:417-424. [PMC free article] [PubMed] [Google Scholar]
- 12.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiman, R. N., and W. Herr. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 15.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 16.Fuks, F., P. J. Hurd, D. Wolf, X. Nan, A. P. Bird, and T. Kouzarides. 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278:4035-4040. [DOI] [PubMed] [Google Scholar]
- 17.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 18.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giebler, H. A., I. Lemasson, and J. K. Nyborg. 2000. p53 recruitment of CREB binding protein mediated through phosphorylated CREB: a novel pathway of tumor suppressor regulation. Mol. Cell. Biol. 20:4849-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Grundhoff, A., and D. Ganem. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J. Clin. Investig. 113:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward, G. S. 2003. Initiation of angiogenic Kaposi's sarcoma lesions. Cancer Cell. 3:1-3. [DOI] [PubMed] [Google Scholar]
- 23.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, D. G. 1995. Regulation of E2F-1 gene expression by p130 (Rb2) and D-type cyclin kinase activity. Oncogene 11:1685-1692. [PubMed] [Google Scholar]
- 25.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 13:33-43. [DOI] [PubMed] [Google Scholar]
- 26.Kappes, F., K. Burger, M. Baack, F. O. Fackelmayer, and C. Gruss. 2001. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 276:26317-26323. [DOI] [PubMed] [Google Scholar]
- 27.Katano, H., Y. Sato, and T. Sata. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92:3076-3084. [DOI] [PubMed] [Google Scholar]
- 28.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 30.Kimura, H., and K. Shiota. 2002. Methyl-CpG binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 278:4806-4812. [DOI] [PubMed] [Google Scholar]
- 31.Knight, J. S., M. A. Cotter, Jr., and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 32.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Hum. Herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 35.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 36.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 37.Luciano, R. L., and A. C. Wilson. 2003. HCF-1 Functions as a coactivator for the zinc finger protein Krox20. J. Biol. Chem. 278:51116-51124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahajan, S. S., M. M. Little, R. Vazquez, and A. C. Wilson. 2002. Interaction of HCF-1 with a cellular nuclear export factor. J. Biol. Chem. 277:44292-44299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinowich, K., D. Hattori, H. Wu, S. Fouse, F. He, Y. Hu, G. Fan, and Y. E. Sun. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890-893. [DOI] [PubMed] [Google Scholar]
- 40.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush, and M. R. Philips. 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152:111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 43.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 44.Neuman, E., E. K. Flemington, W. R. Sellers, and W. G. Kaelin, Jr. 1994. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol. Cell. Biol. 14:6607-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noel, J. C., F. De Thier, T. Simonart, J. Andre, P. Hermans, J. P. Van Vooren, and M. Heenen. 1997. p53 protein overexpression is a common but late event in the pathogenesis of iatrogenic and AIDS-related Kaposi's sarcoma. Arch. Dermatol. Res. 289:660-661. [DOI] [PubMed] [Google Scholar]
- 46.Piolot, T., M. Tramier, M. Coppey, J. C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 49.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheffner, M., K. Munger, J. C. Byrne, and P. M. Howley. 1991. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA 88:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz, T. F. 2001. KSHV/HHV8-associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1217-1226. [DOI] [PubMed] [Google Scholar]
- 54.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiffman, D., E. E. Brooks, A. R. Brooks, C. S. Chan, and P. G. Milner. 1996. Characterization of the human cyclin-dependent kinase 2 gene. Promoter analysis and gene structure. J. Biol. Chem. 271:12199-12204. [DOI] [PubMed] [Google Scholar]
- 56.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stancheva, I., A. L. Collins, I. B. Van den Veyver, H. Zoghbi, and R. R. Meehan. 2003. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol. Cell. 12:425-435. [DOI] [PubMed] [Google Scholar]
- 58.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]
- 59.Tikoo, R., G. Zanazzi, D. Shiffman, J. Salzer, and M. V. Chao. 2000. Cell cycle control of Schwann cell proliferation: role of cyclin-dependent kinase-2. J. Neurosci. 20:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 61.Viejo-Borbolla, A., E. Kati, J. A. Sheldon, K. Nathan, K. Mattsson, L. Szekely, and T. F. Schulz. 2003. A domain in the C-terminal region of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus affects transcriptional activation and binding to nuclear heterochromatin. J. Virol. 77:7093-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, T., M. Sugaya, A. M. Atkins, E. A. Aquilino, A. Yang, D. L. Borris, J. Brady, and A. Blauvelt. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J. Virol. 77:6188-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 64.Wysocka, J., P. T. Reilly, and W. Herr. 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]