Abstract
Viroids are highly structured plant pathogenic RNAs that do not code for any protein, and thus, their long-distance movement within the plant must be mediated by direct interaction with cellular factors, the nature of which is presently unknown. In addition to this type of RNAs, recent evidence indicates that endogenous RNAs move through the phloem acting as macromolecular signals involved in plant defense and development. The form in which these RNA molecules are transported to distal parts of the plant is unclear. Viroids can be a good model system to try to identify translocatable proteins that could assist the vascular movement of RNA molecules. Here, we demonstrate by use of immunoprecipitation experiments, that the phloem protein 2 from cucumber (CsPP2) is able to interact in vivo with a viroid RNA. Intergeneric graft assays revealed that both the CsPP2 and the Hop stunt viroid RNA were translocated to the scion. The translocated viroid is symptomatic in the nonhost scion, indicating that the translocated RNA is functional. The CsPP2 gene was cloned and sequenced. The analysis of its primary structure revealed the existence of a potential double-spaced-RNA-binding motif, previously identified in a set of proteins that bind to highly structured RNAs, which could explain its RNA-binding properties. The possible involvement of this phloem protein in assisting the long-distance movement of the viroid RNA within the plant is discussed.
It is presently accepted that in all host-pathogen interactions the pathogens (generally with reduced genetic information) are intimately dependent on host factors (mainly proteins) that assist the normal development of their biological processes such as replication and movement (reviewed in references 25 and 26). This host factor dependence is extreme in viroids because of their particular properties. Viroids are single-stranded, covalently closed, circular pathogenic RNAs with genomes ranging in size from 246 to 401 nucleotides that can infect plants (reviewed in references 9 and 15). Since viroids have no apparent coding capacity, they must exclusively use host factors (probably proteins) to assist most biological functions like replication, processing, or transport. The search for cellular factors that could mediate the regulation of biological functions of viroid diseases has been mainly addressed by studying the RNA-binding properties of host proteins (7, 20, 29, 38, 44). At least in two cases, their involvement with the replication process has been suggested and/or demonstrated (7, 29). On the other hand, the putative host factors involved in the long-distance movement of viroid RNA have been studied comparatively less. Very recently it has been demonstrated that specific nucleotides in a viroid RNA have a profound effect in altering distinct cellular responses, which then lead to well-defined alterations in plant growth and developmental patterns (35). In addition, Vir P1, a protein from tomato, has been suggested to be involved in the systemic spread of viroids (27).
Several lines of evidence suggest that different RNA molecules travel through the phloem: cellular mRNAs (23, 37, 45), small RNAs related to the gene silencing phenomenon (33), and viral RNAs (21). The delivery of RNA to distant tissues may reflect a mechanism used by plants to regulate developmental and defense processes (11, 22, 26). An important question that still remains unanswered is how these RNA molecules are translocated (43). It has recently been suggested that translocatable phloem proteins acting as chaperone-like proteins could be involved in this process (31, 42, 43). In addition, most of the progress made regarding the transport of RNA molecules has come from the study of the translocation of plant viruses. These studies have revealed that some plant viruses are translocated as ribonucleoprotein (RNP) complexes, and recently, it has been proposed that this function could be assisted by existing transport routes and also commander host proteins that have the ability to move through plasmodesmata (8, 26).
Long-distance movement of viroids occurs, as for most plant viruses, by translocation via the phloem (34, 46). Consequently, it is reasonable to assume that phloem protein(s) could interact with viroid RNA to assist its translocation to distal regions of the plant. Consistent with this idea, it has recently been demonstrated that the phloem protein 2 of Cucumis sativus (CsPP2) forms an RNP complex with Hop stunt viroid (HSVd) RNA in vitro (19, 32). However, it remains unsolved whether or not this function operates also in vivo due, in part, to the unavailability of an appropriate antiserum.
The PP2 is a dimeric protein of approximately 48 kDa that behaves upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) predominantly as a 24- to 26-kDa monomer (36). Interestingly, the PP2 of the Cucurbitaceae has the capacity to increase the size exclusion limits of the plasmodesmata (2) and move from the companion cells, where it is synthesized, into sieve tubes (17). In addition, PP2 of Cucurbita maxima has been shown to translocate in intergeneric grafts and suggested to move within the assimilated stream toward sink tissues (13, 17, 18). The functions of this protein are not yet clear (5, 17, 18). Recent analyses have led to the identification of PP2-like genes in species from 17 angiosperm and gymnosperm genera, indicating that these proteins are ancient and common in vascular plants (10).
Here, by using a combined approach of immunoprecipitation assays and grafting experiments, we present direct evidence that the cucumber phloem protein 2 (CsPP2) forms an RNP complex in vivo with HSVd RNA in the phloem exudates of infected cucumber plants. Also, we demonstrate that the CsPP2 and HSVd are simultaneously translocated through intergeneric grafts. The CsPP2 was cloned, sequenced, and partially characterized, which allowed the identification of a potential double-stranded-RNA (dsRNA)-binding motif (dsRBM). The participation of translocatable CsPP2 as a component of the plant information superhighway (22) probably involved in the long-distance movement of HSVd is discussed.
MATERIALS AND METHODS
Exudate sampling and PAGE.
Unless otherwise indicated, in all experiments, 10- to 12-day-old plants were cut below the cotyledons, and the exudate was collected and diluted 1/10 in a cooled reduction buffer (RB; 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 10 mM dithiothreitol). The phloem exudate was denatured by heating for 5 min at 95°C and fractionated by SDS-PAGE in 12% gels (24) that were stained with Coomassie blue.
Production of anti-CsPP2 polyclonal antibodies.
The CsPP2 was gel purified after SDS-PAGE separation of phloem exudates (19) and used to raise polyclonal antibodies in rabbits. Before starting the immunization scheme, the rabbit was bled and the serum was tested for nonreactivity to cucumber phloem proteins. Subcutaneous doses (∼5 μg) were injected weekly over a 4-week period, and blood was collected. The protein used in the immunization was free of any other proteins as revealed later by matrix-assisted laser desorption ionization-time of flight analysis (see below).
The phloem exudates from cucumber and five different cultivars of pumpkin (C. maxima) were collected as described above, fractionated on SDS-PAGE 12% gels, and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were treated for 1 h in blocking solution (Tris-buffered saline [TBS; 500 mM NaCl, 20 mM Tris {pH 7.5}], 5% defatted milk, 2% bovine serum albumin, 0.1% Triton X-100) and incubated overnight with the polyclonal antiserum against cucumber PP2 (diluted 1/10,000 in TBS, 2% defatted milk). Membranes were washed (TBS, 0.5% Tween 20), incubated with anti-rabbit immunoglobulin G linked to horseradish peroxidase whole antibody, and revealed by luminescence (ECL+Plus; Amersham-Pharmacia Biotech, Little Chalfont, United Kingdom) according to the manufacturer's instructions.
Immunoprecipitation and reverse transcription (RT)-PCR.
Four weeks after inoculation with HSVd, phloem exudates (250 μl), free of non-phloem RNAs (see Fig. S1 in the supplemental material), were collected from excised petioles of healthy and infected cucumber plants and diluted 1:5 in reduction buffer. The diluted sap was incubated in a gyrator platform with 30 μl of a dilution (1:5) of the CsPP2 antiserum (2 h, 4°C) and then overnight with agarose-conjugated protein A (Roche Diagnostics, Mannheim, Germany). The immunoabsorbed complex was sedimented by centrifugation and washed twice for 30 min at 4°C. Since the in vitro CsPP2-HSVd complex is stable with up to 300 mM NaCl (19), this highly stringent concentration of NaCl was used (instead of the 150 mM suggested by the manufacturer) to eliminate possible nonspecific complexes.
RT-PCR was performed as described previously (1). The oligonucleotides used were the antisense 26-mer VP-19 (5′-dGCCCCGGGGCTCCTTTCTCAGGTAAG-3′, complementary to HSVd residues 60 to 85) and the sense 27-mer VP-20 (5′-dCGCCCGGGGCAACTCTTCTCAGAATCC-3′, residues 78 to 102). Both primers lie in the strictly conserved central region of HSVd. PCR was carried out with one cycle at 95°C for 3 min, followed by 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min. The RT-PCR products were analyzed on nondenaturing 5% polyacrylamide gels.
Intergeneric grafting assays.
Two-week-old pumpkin scions were grafted onto healthy and infected 4-week-old cucumber stocks. Grafted plants were maintained for 2 days at room temperature under controlled humidity conditions and later moved to environmentally controlled growing chambers (34°C, 14 h of light). Approximately 50% of the grafts survived. The phloem exudate (0.5 μl) from cucumber stocks and pumpkin scions, of 6- and 10-day-old grafted plants, was diluted 1:5 in reduction buffer, and proteins were fractionated by SDS-PAGE on 12% gels, blotted, and immunodetected by Western blot assays with CsPP2 antiserum as described above.
Total RNA preparations were obtained as described previously (1) from the excised leaves from scions. Five-microliter aliquots of these preparations were denatured and blotted onto nylon membranes. Hybridization and chemiluminescent detection of HSVd were performed as previously described (3).
Northwestern assays.
The phloem exudate from cucumber (cv. Suyo and cv. Marketer) and pumpkin (cv. Virginia), collected as described before, was denatured by heating for 5 min at 95°C, fractionated by SDS-12% PAGE, and transferred to nitrocellulose membranes (Bio-Rad). Northwestern assays were performed as described previously (28). Membranes were incubated in RN buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 0.05% Triton X-100, 1× Denhardt's reagent) for 2 h at room temperature, followed by a 3-h incubation in RN buffer in the presence of digoxigenin-labeled HSVd-RNA (25 to 50 ng/ml). The detection of hybrid RNA-protein was performed by the colorimetric method as described previously (28). A duplicate SDS-PAGE was Coomassie blue stained and used as a control.
Tissue prints.
Tissue prints were made basically as described previously (30). The upper surfaces of the leaves were printed onto a nylon membrane by using a roller and applying uniform pressure, beginning from the tip of the leaf. Hybridization was performed as described previously (3).
For CsPP2 detection, the same leaves used for RNA detection were printed as described above onto PVDF membranes. The membranes were blocked for 3 h and then incubated overnight with the polyclonal antiserum against cucumber PP2 as described above. Membranes were washed, incubated, and revealed by luminescence as described previously (3).
Protein sequencing.
The phloem exudate was collected from 10- to 12-day-old cucumber plants and diluted 1:10 in reduction buffer. The phloem exudate was denatured for 5 min at 95°C, fractionated by SDS-PAGE on 15% gels, and stained with Coomassie blue. The CsPP2 band (of approximately 26 kDa) was excised from the gel and washed several times in acetic acid and methanol. Protein analysis by tandem mass spectrometry was performed by The UVic Proteomic Service (http://www.proteincentre.com; University of Victoria, Victoria, British Columbia, Canada). Characterization by mass spectrometry was performed on an Applied Biosystems/MDS Sciex Qstar hybrid LC/MS/MS quadropole time of flight system and an Applied Biosystems Voyager DE-STR matrix-assisted laser desorption ionization-time of flight system.
CsPP2 gene cloning and sequencing.
Poly(A+) RNA preparations were obtained from hypocotyls of 12-day-old cucumber plants. The first-strand cDNA was synthesized by RT (cDNA synthesis kit; Stratagene, San Diego, Calif.). This cDNA was used as a template for PCR, and degenerated primers were designed based on the sequences of the peptides obtained by microsequencing. The cDNA was cloned and sequenced. The partial CsPP2 sequence obtained was used to design primers to amplify the 3′ [using an oligo(dT)] and 5′ ends (5′ rapid amplification of cDNA end system; GIBCO). The DNA fragments corresponding to the 3′ and 5′ ends were cloned and sequenced, and then two additional primers were designed to obtain the full cDNA sequence. The 675-bp cDNA fragment amplified by PCR was cloned and sequenced again.
RESULTS
Specificity of the CsPP2 antiserum.
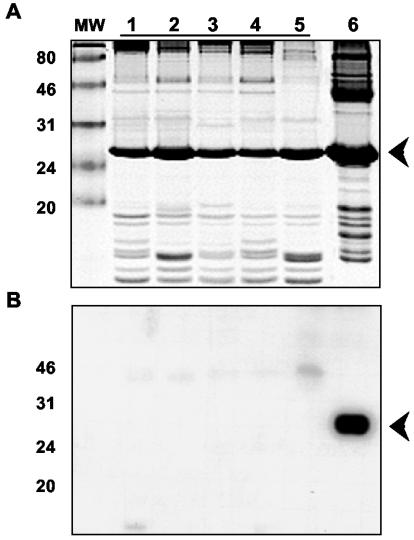
Western blot analysis clearly demonstrated the capacity of the polyclonal antiserum obtained for detecting the phloem protein 2 from cucumber plants (CsPP2) in the phloem exudate (Fig. 1). Interestingly, this antiserum did not recognize any PP2 of the five different pumpkin cultivars analyzed (Fig. 1). This high specificity opened the possibility of studying the translocation of the CsPP2 in intergeneric grafts (see below).
FIG. 1.
Specificity of the CsPP2 antiserum. (A) SDS-PAGE and Coomassie blue staining of phloem exudate from different pumpkin cultivars (lanes 1 to 5: cv. Romani, cv. Tanda chiaro, cv. Virginia, cv. Black beauty, and cv. Argelia temprano, respectively) and cucumber (lane 6). The arrowhead indicates the position at which the different PP2s migrate. (B) The proteins were transferred to a PVDF membrane and incubated with the polyclonal CsPP2 antiserum. CsPP2 was detected, and cross-reaction was not observed with the pumpkin PP2s. Standard marker proteins (in kilodaltons) are indicated (lane MW).
CsPP2 is the only cucumber phloem protein able to bind HSVd.
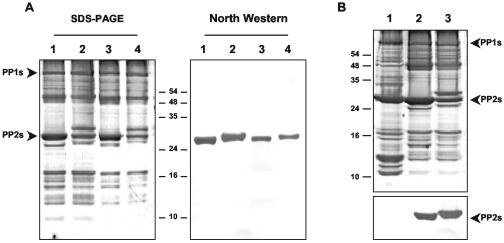
It was previously reported that CsPP2 and HSVd form an RNP complex in vitro (19, 32). However, it could not be excluded that other phloem proteins could also interact directly with HSVd RNA. Northwestern assays were used to determine whether, in addition to CsPP2, there exist other phloem proteins able to bind HSVd-RNA. The results obtained in this experiment demonstrated that, under these conditions, CsPP2 is the only phloem protein that binds the HSVd RNA (Fig. 2A). The two cucumber cultivars analyzed showed similar RNA-binding activities for their corresponding PP2s (Fig. 2A, compare lanes 1 and 2). In addition, this analysis also revealed that the phloem protein 1 (PP1), an abundant phloem protein that cross-links with PP2, does not interact with viroid RNA. Interestingly, when this type of analysis was extended to pumpkin plants, it was observed that its corresponding PP2 (CmPP2) lacks RNA-binding capability (Fig. 2B, lane 1).
FIG. 2.
Specificity of binding of CsPP2 protein with HSVd RNA. (A) Determination of cucumber phloem proteins with RNA-binding activity. Phloem exudate proteins from two different C. sativus cultivars (cv. Suyo, lanes 1 and 3, and cv. Marketer, lanes 2 and 4) were subjected to electrophoresis in SDS-12% PAGE and silver stained (left panel). A duplicate gel was transferred to a nitrocellulose membrane, renaturalized, and hybridized with a digoxigenin-labeled HSVd-RNA probe (right panel). Only one phloem protein, CsPP2, was able to bind HSVd-RNA. The samples in lanes 3 and 4 are 1:3 dilutions of phloem sap from lanes 1 and 2. (B) The pumpkin phloem protein 2 (CmPP2) is unable to bind HSVd-RNA. Phloem proteins from C. maxima cv. Virginia (lane 1), and two different C. sativus cultivars (cv. Suyo, lane 2, and cv. Marketer, lane 3) were subjected to electrophoresis and silver stained (upper panel). A duplicate gel was transferred and incubated as described for panel A (lower panel). Only the PP2 from both cucumber cultivars was able to bind HSVd-RNA, demonstrating that the CmPP2 is incapable of binding HSVd-RNA. The positions of PP1 and PP2 are indicated.
Detection of an in vivo RNP complex in cucumber phloem.
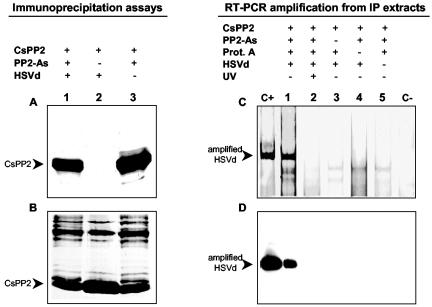
To demonstrate the existence in vivo of the RNP complex (CsPP2-HSVd), immunoprecipitation assays were carried out with the phloem exudate of HSVd-infected cucumber plants. The use of the immunoprecipitation technique to demonstrate in vivo RNA-protein interactions was previously reported (16, 39). The CsPP2 was the only protein recovered from the immunoprecipitate (Fig. 3A), whereas the rest of the proteins remained in the corresponding supernatants (Fig. 3B). Note that the CsPP2 antiserum was able to immunoprecipitate approximately half of the preexisting CsPP2 (Fig. 3B, compare lanes 1 and 3 versus lane 2).
FIG. 3.
Detection in vivo of an RNP complex between the CsPP2 and HSVd RNA by immunoprecipitation assays. (A) Phloem exudates (200 μl) were collected from HSVd-infected (lanes 1 and 2) or noninfected (lane 3) cucumber plants and immunoprecipitated with CsPP2 antiserum (lanes 1 and 3). (B) Immunoprecipitates and supernatants were analyzed by SDS-PAGE and stained with Coomassie blue. Note that approximately 50% of the CsPP2 was immunoprecipitated (compare lanes 1 and 3 with lane 2 in panel B). (C) Nucleic acids were recovered from the immunoprecipitate, subjected to RT-PCR with two HSVd-specific primers, and analyzed by PAGE on 5% gels. Samples were blotted and probed with an HSVd-specific riboprobe (D). Lanes C+ and C−, positive (HSVd-RNA) and negative (water) RT-PCR controls. Total nucleic acids were extracted from the immunoprecipitate (IP) obtained in the presence (+) (lane 1) or absence (−) of CsPP2 As (lane 3), protein A (lane 4), or HSVd-RNA (lane 5, uninfected plant). Lane 2 is equivalent to lane 1, except that the phloem exudate was previously UV cross-linked.
The presence of HSVd RNA in the immunoprecipitate was established after resuspending it and removing the proteins by phenol extraction. When the resulting nucleic acid preparation was subjected to RT-PCR, a band of the expected size was detected only when the immunoprecipitate originated from HSVd-infected exudate (Fig. 3C, lane 1). Interestingly, no amplification was obtained when RT-PCR was carried out with the immunoprecipitate from UV cross-linked sap exudate from infected plants (Fig. 3C, lane 2). This result can be explained by considering that the HSVd RNA covalently bound to CsPP2 is not an appropriate target for RT-PCR, as previously shown with some viral systems (41). The nature of the amplified products was confirmed by Southern blot analysis (Fig. 3D).
CsPP2 and HSVd are translocated between intergeneric grafts.
It is well known that cucumber plants are an appropriate host for HSVd infections, whereas pumpkin plants are not susceptible to HSVd infection (40). We have also determined, by injection of infectious transcripts in stem tissue and by agroinoculation in cotyledons, that the HSVd-RNA is, under these conditions, unable to move within the phloem of pumpkin plants (see Fig. S2 in the supplemental material). Additionally, Northwestern assays clearly demonstrated that CmPP2 is unable to bind HSVd-RNA (Fig. 2B). These observations, in addition to the lack of serological cross-reaction between CsPP2 and CmPP2, determined that pumpkin was selected to be the scion in intergeneric graft assays.
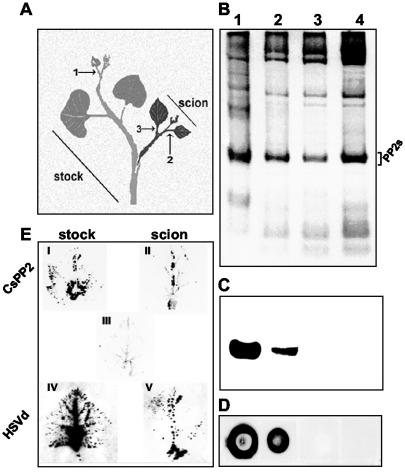
To determine whether or not CsPP2 and HSVd-RNA are translocated between intergeneric grafts, pumpkin plants were grafted onto HSVd-infected cucumber plants (Fig. 4A). One week after grafting, the leaves of pumpkin scions showed discoloration of the veins, epinasty, and necrosis on their edges. In most cases, the grafts become stunted, petioles necrosed, and scions finally died at 4 weeks postgrafting. None of these symptoms were observed on scions grafted onto noninfected cucumber plants. Phloem exudates from C. sativus (stock) and C. maxima (scion) were then analyzed. Endogenous CsPP2 was detected on the cucumber stock and in the pumpkin scion 10 days after grafting (DAG) (Fig. 4B and C, lanes 1 and 2) but not 6 DAG or in nongrafted pumpkin plants (Fig. 4B and C, lanes 3 and 4, respectively), demonstrating the capability of CsPP2 to move through intergeneric grafts as previously described for other phloem proteins (17, 18, 45). Dot blot hybridization showed that the HSVd RNA was only detected on the infected cucumber stock or in the pumpkin scions at 10 DAG (Fig. 4D, dots 1 and 2, respectively) but not at 6 DAG or in nongrafted pumpkin plants (Fig. 4D, dots 3 and 4, respectively). Thus, the translocation time pattern of HSVd parallels that of the CsPP2. Unfortunately, due to the syndrome developed in the scions grafted onto HSVd-infected cucumber plants, insufficient amounts of phloem exudate were recovered from scions and no immunoprecipitation experiments could be carried out to demonstrate the existence of the CsPP2-HSVd complex in the scion. The presence of CsPP2 and HSVd in the scion was confirmed by tissue printing analysis (Fig. 4E). These experiments revealed that CsPP2 and HSVd that moved from the cucumber rootstock into a pumpkin scion are largely confined within the vascular tissue in the scion (Fig. 4E, II and V) strongly suggesting that additional host factors are required for HSVd to efficiently exit the phloem and that the presence of HSVd in the vascular tissue is sufficient to cause symptoms in the scion.
FIG. 4.
The CsPP2 and HSVd-RNA are translocated to intergeneric grafts. (A) Nonhost pumpkin plants (scions) were grafted onto HSVd-infected cucumber plants (stocks). Numbers indicate the sections where the phloem exudate was collected. (B) Total phloem proteins (0.5 μl of exudate) were analyzed by SDS-12% PAGE and Coomassie blue stained. PP2s are indicated. (C) Immunoblot of the gel shown in panel B with an antiserum raised against CsPP2. (B and C) Lane 1, HSVd-infected cucumber stock; lanes 2 and 3, pumpkin scions analyzed at 10 and 6 DAG, respectively; lane 4, ungrafted pumpkin control. (D) Total nucleic acids from the corresponding leaves were analyzed by dot blot hybridization with an HSVd-specific riboprobe. (E) Leaves from cucumber stocks and pumpkin scions were printed onto PVDF and nylon membranes to confirm the translocation of CsPP2 and HSVd-RNA, respectively. I and IV, infected stock; III, ungrafted pumpkin analyzed with CsPP2 antiserum (negative control); II and V, grafted pumpkin scion.
Characterization of CsPP2.
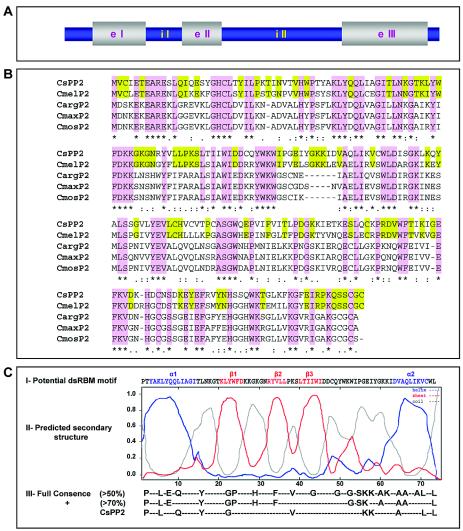
Gel-purified CsPP2 was subjected to characterization by mass spectrometry. Using this technique, 18 peptides were obtained, 3 of which were homologous to others from previously characterized PP2s (see Fig. S3 in the supplemental material). The remaining 15 peptides were later identified as specific for CsPP2. Degenerated primers designed from the sequences of the homologous peptides were used to amplify CsPP2 mRNA by RT-PCR, and the resulting CsPP2 cDNA (675 bp) was cloned and sequenced. By PCR amplification of genomic DNA, we identified two introns within the CsPP2 gene (Fig. 5A).
FIG. 5.
Properties of the CsPP2 gene and its predicted protein product. (A) Schematic representation of CsPP2 gene (GenBank accession number AF527536) with its two introns. e, exon; i, intron. (B) Alignment of CsPP2 with other PP2s: C. melo (Cmel; GenBank accession number AAM74921), C. maxima (Cmax; GenBank accession number AAA92465), C. moschata (Cmos; GenBank accession number AAF74345), and C. argirosperma (Carg; GenBank accession number AAB33116). The homologous regions between the different PP2s are shaded in purple. Green shading indicates the specific homologous blocks between PP2s from the genus Cucumis. The potential dsRBM motif within the NH2-terminal region from CsPP2 is shown in panel C. (C) For the potential dsRBM sequence (I), the α1 and α2 helices are shown in blue; the β1, β2, and β3 sheets are represented in red; and coils are shown in grey. (II) Stability levels of the predicted secondary structure were determined by multivariate linear regression combination (SOPMA-GOR-SIMPA), with the high degree of stability (greater than 0.8) of the predicted motif being evident. (III) Residues conserved in RNA-binding motifs. Full consensus is defined as residues that are >50% conserved among all dsRBM sequences, and positive (+) consensus is defined as the residues that are >70% conserved among those structures that strongly bind dsRNA (45). The specific residues conserved in the CsPP2 potential dsRBM are shown.
Protein sequence comparison with other recently characterized PP2s revealed a high level of diversity. The predicted CsPP2 sequence exhibited identities of 67% with Cucumis melo PP2 and 46 to 47% with PP2s from diverse Cucurbita sp. (Cucurbita argirosperma, Cucurbita moschata, and C. maxima) (Fig. 5B). It is worthy noting that, whereas the PP2s of the genus Cucurbita exhibited a very high identity among them (∼96%) (4), those of the genus Cucumis presented a significantly lower identity among them.
Interestingly, analysis of the predicted secondary structure of CsPP2, obtained by multivariate linear regression combination (6), showed the presence of a potential dsRBM (12) in its NH2-terminal region (Fig. 5C). The dsRBMs (∼70 amino acids) are characterized by a fold comprising (from the NH2 terminus) α-helix 1, β-strand 1, β-strand 2, β-strand 3, and α-helix 2 and are present in a high number of proteins that interact with dsRNAs or partially duplexed (highly structured) RNAs, generally without obvious RNA sequence specificity (12). CsPP2 has not only the same secondary structure organization but also most of the amino acids described as necessary for RNA-binding activity (Fig. 5C).
DISCUSSION
Viroids are small pathogenic RNAs of higher plants that do not code for any protein (reviewed in references 9 and 15). Thus, all of the critical steps of their biological cycle must be assisted by host factors.
In general, the viroid infection process can be divided into two major steps: replication in an individual plant cell (14) and movement throughout the plant (46). The existence of host proteins able to interact in vivo with viroid RNAs that could participate in the viroid replication process has been previously described (7, 29). However, little is known about the host factors that assist the movement of the viroid RNA through the plant.
The viroid long-distance movement was previously demonstrated to occur, as for most plant viruses, by translocation via the phloem (34, 46). Consequently, it is reasonable to assume that this process could be mediated by interactions with preexisting host proteins, probably involved in the systemic transport of endogenous RNAs. The long-distance transport of endogenous RNAs through the phloem has been proposed to constitute a mechanism likely used by higher plants to integrate developmental and physiological processes on a whole-plant basis (reviewed in references 11, 22, and 43). The mechanism by which these RNAs are translocated through the plant remain unclear, but several previous reports are coincident in suggesting that translocatable phloem proteins forming RNP complexes, resembling the mechanisms that plant viruses use to move systemically, could be involved in this process (26, 42). Indeed, a plant RNA-binding protein, CmPP16, identified in pumpkin sap has been proposed to function as a paralog of viral movement proteins, assisting RNA translocation between companion cells and sieve elements of the phloem (45).
We (19) and others (32) previously reported that a cucumber phloem protein (CsPP2) forms an RNP complex in vitro with HSVd RNA. However, the existence of a similar complex in the phloem sap of infected plants could not be demonstrated. In this work, a CsPP2 polyclonal antiserum was obtained that allowed immunoprecipitation of the RNP complex CsPP2-HSVd RNA from phloem exudate, demonstrating for the first time the existence in vivo of a viroid-host protein RNP complex in the vascular tissue of infected plants. The identification of a potential dsRBM in the predicted structure of the CsPP2, previously reported to be present in dsRNA-binding proteins which bind to highly structured RNA without apparent sequence specificity (12), would explain the RNA-binding properties of the CsPP2.
Grafting experiments revealed that the CsPP2 is a translocatable phloem protein able to move through intergeneric grafts, consistent with previous results obtained with other phloem proteins (13, 17, 18). In addition, the same graft assays showed that the HSVd is also translocated through intergeneric grafts and that the translocated HSVd RNA is able to develop symptoms in pumpkin scion.
The possibility of exploiting existing transport routes and also commander host proteins that have the ability to move through the plasmodesmata was previously suggested as probably related to virus movement (8, 11, 26). Additional evidence, described in this work, for example, that pumpkin PP2 (CmPP2) is unable to bind HSVd and that the HSVd is not translocated as a free molecule through the phloem conduits of pumpkin varieties even after the injection of HSVd into the stem tissue, support this idea (see Fig. S3C and D in the supplemental material).
At least a minimum of three properties must be considered for a chaperone-like host protein to be able to assist the long-distance movement of HSVd through infected plants: (i) RNA-binding activity (to form an RNP complex with viroid RNA), (ii) capability to interact with the plasmodesmata and to increase their size exclusion limit (to permit the exit of the RNP complex from the cell), and (iii) to be a long-distance translocatable molecule (to distribute the viroid RNA in the whole plant). The evidence shown here regarding the in vivo RNA-binding activity of CsPP2 and its capacity to translocate through intergeneric grafts, together with the previously reported capability of increasing the plasmodesmata SEL (2), strongly suggest that CsPP2 posses the minimum properties required to carry out chaperone-like functions. However, the possibility that the binding of CsPP2 to HSVd was an intrinsic host mechanism to sequester a foreign RNA resulting in the HSVd movement with CsPP2 from a fortuitous process cannot be ruled out at this stage. Mutagenesis experiments to inhibit CsPP2 movement or to block interactions between CsPP2 and HSVd are needed to further establish the role of CsPP2 in the viroid long-distance movement.
In summary, in the present work we have demonstrated for the first time that a pathogenic RNA forms an RNP complex in vivo with a translocatable phloem protein from cucumber that, from all the available evidence, can be suggested to assist in the long-distance translocation of HSVd-RNA through the plant.
Supplementary Material
Acknowledgments
We thank R. Flores, A. Granell, and J. A. Daròs for valuable comments and critical reading of the manuscript. We also thank an anonymous referee for valuable suggestions.
This work was supported by grant BIO2002-040099 from the Spanish granting agency DGICYT. G.G. was the recipient of a fellowship from UPV.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amari, K., G. Gómez, A. Myrta, B. Di Terlizzi, and V. Pallás. 2001. The molecular characterization of sixteen new sequence variants of Hop stunt viroid (HSVd) reveals the existence of invariable regions and a conserved hammerhead-like structure on the viroid molecule. J. Gen. Virol. 82:953-962. [DOI] [PubMed] [Google Scholar]
- 2.Balachandran, S., Y. Xiang, C. Schobert, G. Thompson, and W. Lucas. 1997. Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc. Natl. Acad. Sci. USA 94:14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, J. M., P. Más, M. A. Sanchez-Pina, and V. Pallás. 1997. Spatial distribution of acidic chitinases and their messenger RNAs in tobacco plants infected with Cherry leaf roll virus. Mol. Plant-Microbe Interact. 10:784-788. [Google Scholar]
- 4.Bostwick, D. E., M. I. Skaggs, and G. A. Thompson. 1994. Organization and characterization of Cucurbita phloem lectin genes. Plant Mol. Biol. 26:887-897. [DOI] [PubMed] [Google Scholar]
- 5.Bostwick, D. E., J. M. Dannenhoffer, M. I. Skaggs, R. M. Lister, B. A. Larkins, and G. A. Thompson. 1992. Pumpkin phloem lectins genes are specifically expressed in companion cells. Plant Cell 4:1539-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 7.Daros, J. A., and R. Flores. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desvoyes, B., S. Faure-Rabasse, M. Chen, J. Park, and H. Scholthof. 2002. A novel plant homeodomain protein interacts in a functionally relevant manner with a virus movement protein. Plant Physiol. 129:1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener, T. O. 1991. Subviral pathogens of plants: viroids and viroid-like satellite RNAs. FASEB J. 5:2808-2813. [DOI] [PubMed] [Google Scholar]
- 10.Dinant, S., A. Clark, Y. Zhu, F. Vilaine, J. Palauqui, C. Kusiak, and G. A. Thompson. 2003. Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 131:114-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, B., A. Itaya, and Y. Qi. 2003. Symplasmic protein and RNA traffic: regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6:596-602. [DOI] [PubMed] [Google Scholar]
- 12.Fierro-Monti, I., and M. Mathews. 2000. Protein binding to duplexes RNA: one motif, multiple functions. Trends Biochem. Sci. 25:241-246. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, D., Y. Wu, and M. Ku. 1992. Turnover of soluble proteins in the wheat sieve tubes. Plant Physiol. 100:1433-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores, R., F. Di Serio, and C. Hernandez. 1997. Viroids: the non-coding genome. Semin. Virol. 8:65-73. [Google Scholar]
- 15.Flores, R., J. W. Randles, M. Bar-Joseph, and T. O. Diener. 2000. Sub-viral agents: viroids, p. 1009-1024. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the international Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 16.Freire, M. A., and M. Pagés. 1995. Functional characteristics of the maize RNA-binding protein MA16. Plant Mol. Biol. 29:797-807. [DOI] [PubMed] [Google Scholar]
- 17.Golecky, B., A. Schulz, and G. A. Thompson. 1999. Translocation of structural P proteins in the phloem. Plant Cell 11:127-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golecky, B., A. Schulz, U. Carstens-Behrens, and R. Kollman. 1998. Evidence for graft transmission of structural phloem proteins or their precursors in heterografts of Cucurbitaceae. Planta 206:630-640. [Google Scholar]
- 19.Gómez, G., and V. Pallás. 2001. Identification of a ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber. Mol. Plant-Microbe Interact. 14:910-913. [DOI] [PubMed] [Google Scholar]
- 20.Hadidi, A. 1988. Synthesis of disease associated protein in viroid infected tomato leaves and binding of viroids to host proteins. Phytopathology 78:575-578. [Google Scholar]
- 21.Heinlein, M. 2002. The spread of Tobacco mosaic virus infection: insights into the cellular mechanism of RNA transport. Cell. Mol. Life Sci. 59:58-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen, R. A., R. G. Atkinson, R. L. Foster, and W. J. Lucas. 1998. An RNA-based information superhighway in plants. Science 279:1486-1487. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M., W. Canio, S. Kessler, and N. Sinha. 2001. Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293:280-289. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Lucas, W. J., B. C. Yoo, and F. Kragler. 2001. RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell. Biol. 2:849-857. [DOI] [PubMed] [Google Scholar]
- 27.Maniataki, E., A. E. Martinez de Alba, R. Sagesser, M. Tabler, and M. Tsagris. 2003. Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9:346-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcos, J. F., M. Vilar, E. Perez-Paya, and V. Pallás. 1999. In vivo detection, RNA-binding properties and characterization of the RNA-binding domain of the p7 putative movement protein from carnation mottle carmovirus (CarMV). Virology 255:354-365. [DOI] [PubMed] [Google Scholar]
- 29.Martinez de Alba, A. E., R. Sagesser, M. Tabler, and M. Tsagris. 2003. A bromodomain-containing protein from tomato specifically binds potato spindle tuber viroid RNA in vitro and in vivo. J. Virol. 77:9685-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Más, P., and V. Pallás. 1995. Non-isotopic tissue-printing hybridization: a new technique to study long-distance plant virus movement. J. Virol. Methods 3:317-326. [DOI] [PubMed] [Google Scholar]
- 31.Mlotshwa, S., O. Voinnet, M. Mette, M. Matzke, H. Vaucheret, S. Ding, G. Pruss, and V. Vance. 2002. RNA silencing and the mobile silencing signal. Plant Cell 14:289-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owens, R. A., M. Blackburn, and B. Ding. 2001. Possible involvement of a phloem lectin in long distance viroid movement. Mol. Plant-Microbe Interact. 14:905-909. [DOI] [PubMed] [Google Scholar]
- 33.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16:4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palukaitis, P. 1987. Potato spindle tuber viroid: investigation of long-distance intra-plant transport route. Virology 158:239-241. [DOI] [PubMed] [Google Scholar]
- 35.Qi, Y., and B. Ding. 2002. Replication of potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Plant Cell 15:1360-1374. [DOI] [PubMed] [Google Scholar]
- 36.Read, S., and D. Northcote. 1983. Chemical and immunological similarities between the phloem proteins of three genera of Cucurbitaceae plants. Planta 158:119-127. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Medrano, R., B. Xoconostle-Cázares, and W. Lucas. 1999. Phloem long distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126:4405-4419. [DOI] [PubMed] [Google Scholar]
- 38.Sagesser, R., E. Martinez, M. Tsagris, and M. Tabler. 1997. Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res. 35:3816-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaoxiong, C., and L. Hyman. 1998. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 26:4965-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shikata, E. 1987. Hop stunt, p. 280-290. In T. O. Diener (ed.), The viroids. Academic Press, San Diego, Calif.
- 41.Simon-Buela, L., and F. Garcia-Arenal. 1999. Virus particles of cucumber green mottle mosaic virus tobamovirus move systematically in the phloem of infected cucumber plants. Mol. Plant-Microbe Interact. 12:112-118. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, G., and A. Schulz. 1999. Macromolecular trafficking in the phloem. Trends Plant Sci. 4:354-360. [DOI] [PubMed] [Google Scholar]
- 43.Ueki, S., and V. Citovsky. 2001. RNA commutes to work: regulation of plant gene expression by systemically transported RNA molecules. Bioessays 23:1087-1090. [DOI] [PubMed] [Google Scholar]
- 44.Wolff, P., R. Gilz, J. Schumacher, and D. Riesner. 1985. Complexes of viroids with histones and other proteins. Nucleic Acids Res. 13:355-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xonocostle-Cazares, B., Y. Xiang, R. Ruiz-Medrano, H. Wang, J. Monzer, B. Yoo, K. McFarland, V. Franceschi, and W. Lucas. 1999. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 238:94-98. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Y., L. Green, Y. Woo, R. Owens, and B. Ding. 2001. Cellular basis of potato spindle tuber viroid systemic movement. Virology 279:69-77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.