Abstract
Increasing ammonia emissions could exacerbate air pollution caused by fine particulate matter (PM2.5). Therefore, it is of great importance to investigate ammonia oxidation in PM2.5. This study investigated the diversity, abundance and activity of ammonia oxidizing archaea (AOA), ammonia oxidizing bacteria (AOB) and complete ammonia oxidizers (Comammox) in PM2.5 collected in Beijing-Tianjin-Hebei megalopolis, China. Nitrosopumilus subcluster 5.2 was the most dominant AOA. Nitrosospira multiformis and Nitrosomonas aestuarii were the most dominant AOB. Comammox were present in the atmosphere, as revealed by the occurrence of Candidatus Nitrospira inopinata in PM2.5. The average cell numbers of AOA, AOB and Ca. N. inopinata were 2.82 × 104, 4.65 × 103 and 1.15 × 103 cell m−3 air, respectively. The average maximum nitrification rate of PM2.5 was 0.14 μg (NH4+-N) [m3 air·h]−1. AOA might account for most of the ammonia oxidation, followed by Comammox, while AOB were responsible for a small part of ammonia oxidation. Statistical analyses showed that Nitrososphaera subcluster 4.1 was positively correlated with organic carbon concentration, and Nitrosomonas eutropha showed positive correlation with ammonia concentration. Overall, this study expanded our knowledge concerning AOA, AOB and Comammox in PM2.5 and pointed towards an important role of AOA and Comammox in ammonia oxidation in PM2.5.
Ammonia (NH3), the primary alkaline gas in atmosphere, contributes to the formation of fine particulate matter (PM2.5, aerodynamic diameter less than or equal to 2.5 μm). PM2.5, the main air pollutant in urban cities, can decrease visibility, modify radiation balance of the Earth, reduce lung function and increase pulmonary disease1. PM2.5 is a complex mixture of different inorganic and organic substances. Especially, a large part (20–80%) of the total PM2.5 mass is secondary inorganic aerosol (SIA)2, which is responsible for regional-scale pollution3. SIA is predominantly in the form of ammonium sulfate ((NH4)2SO4), ammonium bisulfate (NH4HSO4), and ammonium nitrate (NH4NO3)4. The formation of these aerosols is mainly due to neutralization of ammonia by sulphuric acid and nitric acid4. Therefore, controlling ammonia emissions will effectively reduce PM2.5 pollution.
Ammonia is also the most abundant reduced form of reactive nitrogen (Nr) in the atmosphere. Nr is highly mobile and widely distributed, the deposition of Nr can reduce the biodiversity and disturb the global biogeochemical cycling of nitrogen5. The air pollution caused by PM2.5 and Nr is in part closely linked to each other. The inorganic species of Nr are important precursors of the secondary inorganic ions of PM2.55. Thus, ammonia plays an important role in air quality, human health, ecosystem health and global biogeochemical cycling of nitrogen. Rebalancing the global nitrogen cycle is the key to controlling Nr and PM2.5 pollutions.
Nitrification, microbial oxidation of ammonia to nitrate, is a central step of the global nitrogen cycle, which links the gain and loss of bioavailable nitrogen6. This process was thought to involve two separate steps: ammonia oxidation (NH3-NO2−) and nitrite oxidation (NO2−-NO3−). Ammonia oxidation is the rate-limiting step of nitrification. Before 2015, this process is considered to be primarily performed by two main distinct ammonia-oxidizing microorganisms (AOMs): ammonia-oxidizing bacteria (AOB) and ammonia oxidizing archaea (AOA). The phylogenetic distribution of AOB is limited to Betaproteobacteria (Beta-AOB) and Gammaproteobacteria (Gamma-AOB), and AOA fall within Thaumarchaeota. AOB fall into three major clusters: Nitrosococcus, Nitrosomonas and Nitrosospira7. AOA are classified into five major clusters: Nitrososphaera, Nitrosopumilus, Nitrosotalea, Nitrosocaldus and Nitrososphaera sister cluster8. Numerous surveys about the diversity, abundance and distribution of AOA and AOB have been undertaken in a variety of environments, such as sediments9, soils10, estuaries11 and wastewater treatment plants (WWTPs)12. Recently, the discovery of complete ammonia oxidizers (Comammox), i.e., bacteria within the genus Nitrospira that completely oxidize ammonia to nitrate, has significantly expanded our understanding of nitrification13,14. Until now, there are three Comammox enrichment cultures: Candidatus Nitrospira inopinata, Candidatus Nitrospira nitrosa and Candidatus Nitrospira nitrificans13,14. Comammox amoA gene sequences fall into two clades (clade A and clade B), and Ca. N. inopinata amoA gene is classified into Comammox amoA clade A13. The studies of van Kessel et al. and Daims et al. suggest that Comammox are widely distributed, including agricultural soils, freshwater environments, WWTPs and drinking water systems13,14. Overall, AOA, AOB and Comammox are widely distributed in nature and engineered environments. Moreover, AOA may easily outcompete AOB under oligotrophic conditions based on the physiological properties of the available AOA cultures15,16. Comammox may also have an advantage over AOA and AOB under substrate-limiting conditions, such as biofilms and microbial aggregates where the ammonia concentration is low6,14. Therefore, atmosphere may be another habitat for AOA and Comammox due to limited nutrient availability. However, very little is known about AOA, AOB and Comammox in the atmosphere, especially in PM2.5.
The occurrence and diversity of AOA in the coarse particulate matter (>3 μm) samples collected in Mainz, Germany are reported in a previous study17. Moreover, in the supplementary material of the study of Cao et al.18, AOA and AOB are found to be included in the total airborne archaea and bacteria identified at the genus level in PM2.5 and PM10 in Beijing during a severe smog event using metagenomic methods18. In the other two studies, AOB are discovered in the total bacterial communities in the total suspended particulate matter (TSP) in an urban area of Northern Italy and in the coarse particular (>3 μm) in a metropolitan subway system based on high throughput sequencing targeting 16S rRNA genes19,20. These three studies suggest the occurrence of AOA and AOB in the total archaea and bacteria in atmospheric particulates with different aerodynamic diameters using high - throughput sequencing. Based on the above arguments, ammonia compounds, AOA and AOB are present in PM2.5, while, the presence of Comammox in PM2.5 remains unknown.
The present study aimed to investigate the diversity, abundance and activity of AOMs in PM2.5. The detailed objectives were 1) to investigate the diversity and abundance of AOA and AOB in PM2.5; 2) to detect the occurrence of Comammox and to determine their abundance; 3) to estimate AOMs’ activity and contributions to ammonia oxidation in PM2.5; and 4) to investigate the relationships between environmental factors and AOMs. Until now, only the primer set for amplification of Ca. N. inopinata amoA gene is reported13, therefore, in this study, the Comammox were limited to Ca. N. inopinata. In this study, PM2.5 samples were collected in Beijing-Tianjin-Hebei (BTH) megalopolis. Diversity, abundance and activity of AOMs associated with PM2.5 were investigated using cloning, quantitative polymerase chain reaction (qPCR) and nitrification potential test, respectively. Multivariate statistical analyses were carried out to assess the potential impact of environmental factors on AOMs in PM2.5.
Results
Diversity of AOA and AOB in PM2.5
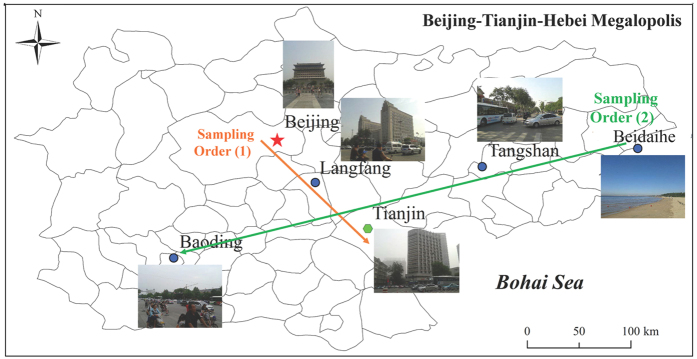
In this study, PM2.5 sampling was carried out from 21 May 2014 to 1 June 2014 at six cities in BTH, including Beijing (BJ), Langfang (LF), Tianjin (TJ), Beidaihe (BDH), Tangshan (TS) and Baoding (BD) (Fig. 1). Four types of cities were selected: two megacity sites (BJ and TJ), two industrial urban sites (TS and BD), a suburban site (LF) and a coastal site (BDH). The surroundings of sampling sites are Residents-Commercial-Transportation Mixed Districts, except BDH, which is on the coast of Bohai Sea (Fig. 1 and Table 1). Longer sampling time (24 h for each site) was used to minimize the impact of unreplicated sampling design and to account for atmospheric movements.
Figure 1. Map showing the sampling orders and sampling sites in Beijing-Tianjin-Hebei (BTH) megalopolis, China.
(Figure created by the authors using MapInfo Pro v15.2 free trial, http://www.pitneybowes.com/us/miprov15-2.html and Microsoft Office 2013, https://products.office.com/zh-cn/buy/office. The photos in the figure were taken by the authors).
Table 1. Description of geography information of sampling sites, samples collected and corresponding meteorological conditions in BTH.
| Cities | City type | Longitude and latitude of locations | Sampling date* |
PM2.5 (μg m−3) | NH3 (μg m−3) | T (°C) | RH(%) | AP (Hpa) | AQI | |
|---|---|---|---|---|---|---|---|---|---|---|
| Start time | End time | |||||||||
| Beijing | Megacity | 39°53′28″N | 5/21 8:00 | 64.58 | 0.903 ± 0.140 | 24 | 52 | 1005.25 | 183 | |
| (BJ) | 116°23′29″E | 5/22 11:30 | 27 | 48 | 1006.11 | 188 | ||||
| Langfang | Semi-urban area | 39°31′16″N | 5/23 8:00 | 83.93 | 0.740 ± 0.068 | 31 | 34 | 1009.64 | 138 | |
| (LF) | 116°43′15″E | 5/24 11:30 | 23 | 75 | 1007.89 | 104 | ||||
| Tianjin | Megacity | 39°7′14″N | 5/25 8:00 | 170.14 | 0.947 ± 0.718 | 24 | 54 | 1006.31 | 134 | |
| (TJ) | 117°10′40″E | 5/26 11:30 | 26 | 20 | 1000.35 | 82 | ||||
| Beidaihe | Coastal city | 39°48′58″N | 5/27 8:00 | 35.42 | 0.169 ± 0.240 | 17 | 62 | 1008.00 | 79 | |
| (BDH) | 119°31′12″E | 5/28 11:30 | 19 | 60 | 1007.00 | 54 | ||||
| Tangshan | Industrial city | 39°39′29″N | 5/29 8:00 | 194.44 | 0.363 ± 0.412 | 23 | 51 | 1004.00 | 192 | |
| (TS) | 118°10′13″E | 5/30 11:30 | 32 | 54 | 1004.00 | 201 | ||||
| Baoding | Industrial city | 38°51′58″N | 5/31 8:00 | 138.19 | 0.000 | 30 | 56 | 1004.00 | 158 | |
| (BD) | 115°29′32″E | 6/1 11:30 | 23 | 57 | 1005.00 | 73 | ||||
Abbreviations: T: Temperature; RH: Relative Humidity; AP: Atmospheric Pressure.
*PM2.5 collection was carried out at a flow rate of 100 l min−1 for 24 h (from 08:00 a.m. to 11:30 a.m. the next day with procedure of 3 h collection and 0.5 h interval to prevent the overheating of the pump).
To investigate the presence of AOA and AOB in PM2.5, the specific primer sets targeting AOA amoA gene and AOB 16S rRNA gene were applied for PCR amplification. For all the six samples, desired single bands of PCR products were observed, which were subsequently purified and cloned. Sequencing results further confirmed that the genes were AOA amoA gene and AOB 16S rRNA gene, suggesting the presence of AOA and AOB in PM2.5 in BTH.
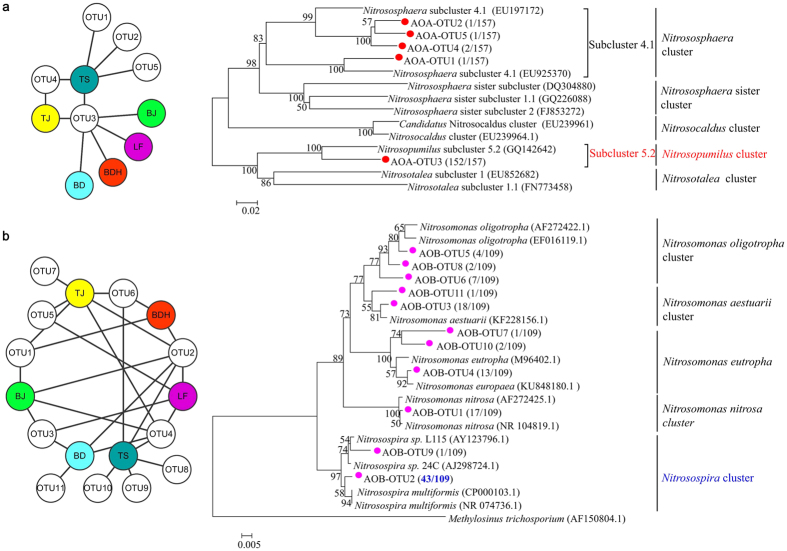
A total of 157 AOA amoA gene sequences were retrieved, and five unique operational taxonomic units (OTUs) were observed at 97% sequence similarity. As shown in Fig. 2a, within each individual clone library, 1–5 OTUs occurred. OTU3 was omnipresent, occurring at all the six PM2.5 samples (Fig. 2a). The diversity of AOA in PM2.5 was low based on the diversity indexes (see Supplementary Table S1). Phylogenetic analyses showed that the five OTUs fell into Nitrosopumilus cluster and Nitrososphaera cluster (Fig. 2a). Specifically, the dominant and omnipresent OTU3 (152 of 157 sequences, 96.82%) was affiliated to Nitrosopumilus subcluster 5.2, indicating its widely distribution in PM2.5 samples. The other four OTUs (5 of 157 sequences, 3.18%) fell into Nitrososphaera subcluster 4.1. For sample BJ, LF, BDH and BD, Nitrosopumilus subcluster 5.2 was the only phylogenetic AOA group. Nitrosopumilus subcluster 5.2 and Nitrososphaera subcluster 4.1 coexisted in sample TS and TJ.
Figure 2.
Distributions of OTUs and NJ phylogenetic tree: (a) AOA amoA gene; (b) AOB 16S rRNA gene. Sequences retrieved in this study are shown in red and purple with “OTU” in the names. The ratios of amounts of sequences within each OTU to the total AOA or AOB sequences are displayed by the number in parentheses.
For six PM2.5 samples, a total of 109 AOB 16S rRNA gene sequences were analyzed. 11 OTUs were recovered at 97% sequence similarity, which were higher than those of AOA amoA gene. The diversity indexes further suggest higher diversity of AOB than AOA (see Supplementary Table S1). As depicted in Fig. 2b, there were 2–6 OTUs in each sample. OTU2 was shared by all the samples. OTU4 occurred at five cities and the other OTUs only appeared in one to three samples. Phylogenetic analysis revealed the co-occurrence of Nitrsomonas and Nitrosospira in PM2.5. Nine OTUs (65 sequences, 59.63%) were affiliated to Nitrsomonas cluster, including Nitrosomonas aestuarii (17.43%), Nitrosomonas nitrosa (15.60%), Nitrosomonas eutropha (14.68%) and Nitrosomonas oligotropha (11.93%). Two OTUs (44 sequences, 40.37%) fell into Nitrosospira cluster. Especially, the predominant OTU2 was affiliated to Nitrosospira multiformis, accounting for 39.45% of total AOB 16S rRNA sequences. These results suggest that N. multiformis and N. aestuarii were the most dominant AOB in PM2.5 in BTH.
Abundance of AOA and AOB in PM2.5
The qPCR results of AOA and AOB in PM2.5 are shown in Fig. 3a. The abundance of AOA ranged from 1.89 × 103 ± 3.10 × 102 (BD) to 1.14 × 105 ± 5.31 × 103 cell m−3 air (LF), averaged 2.82 × 104 ± 1.94 × 103 cell m−3 air. The abundance of AOA was higher in BJ and LF then in the other four cities. The abundance of AOB of PM2.5 in BTH was in the same order of magnitude, and the average abundance of them was 4.65 × 103 ± 4.20 × 102 cell m−3 air. The abundance of AOA was higher than that of AOB in BJ, LF and TS with the ratio of AOA to AOB ranging from 1.88 to 22.22 (Fig. 3b), while the abundance of AOA was lower than AOB in the other three cities with the ratio ranging from 0.40 to 0.68.
Figure 3.
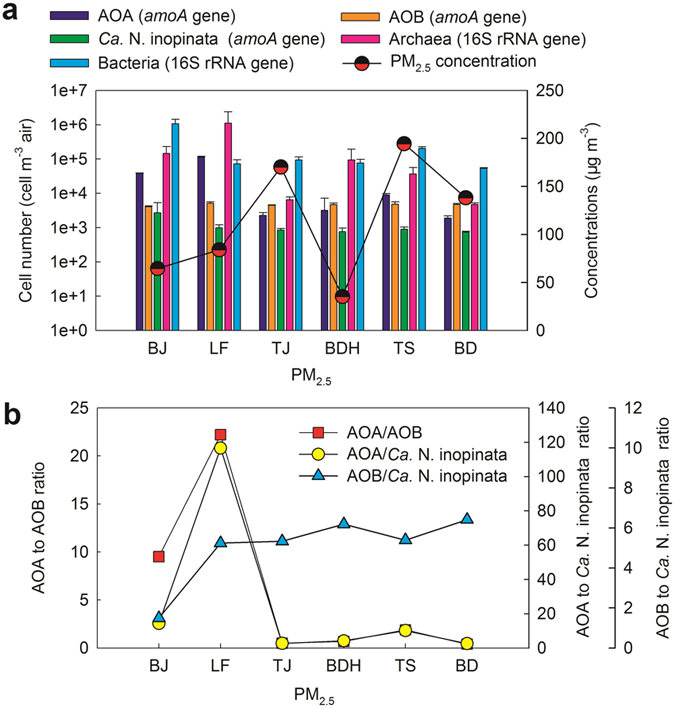
Concentrations of PM2.5 and quantitative analysis of AOA, AOB and Ca. N. inopinata in PM2.5: (a) concentrations of PM2.5, and abundance of AOA amoA gene, AOB 16S rRNA gene and Ca. N. inopinata amoA gene in PM2.5; (b) ratios of AOA to AOB, AOA to Ca. N. inopinata and AOB to Ca. N. inopinata. Ca. N. inopinata is a Comammox enrichment culture.
In order to investigate the proportions of AOA and AOB to total archaea and bacteria in PM2.5, abundances of archaea and bacteria were further investigated (Fig. 3a). Assuming that there are 3.6 copies of 16S rRNA gene per average prokaryotic cell21, the abundance of archaea and bacteria was in the range of 4.72 × 103 ± 5.13 × 102 (BD) to 1.10 × 106 ± 1.28 × 106 (LF) cell m−3 air and 5.36 × 104 ± 1.75 × 103 (BD) to 1.06 × 106 ± 3.85 × 105 (BJ) cell m−3 air, respectively. The proportions of AOA to archaea ranged from 3.38% (BDH) to 40.12% (BJ) with the average of 23.34%. The proportions of AOB to bacteria were in the range of 0.38% (BJ) to 8.79% (BD).
Moreover, as shown in Fig. 3a, concentrations of PM2.5 varied greatly (35.42 μg m−3 in BDH to 194.44 μg m−3 in TS) at six cities in BTH, which were higher in TJ, TS and BD than BJ, LF and BDH. However, the abundances of AOA, AOB, archaea and bacteria were relatively higher in the latter three cities, suggesting that higher concentrations of PM2.5 did not mean higher abundance of microorganisms. Cities TS and BD are the industrial urban sites, the emissions of toxic substances into atmosphere by industrial processes might be much higher than those in BJ, LF and BDH, which might be harmful for the survival of microorganisms, resulting in the relative low abundances.
Presence and abundance of Ca. N. inopinata in PM2.5
The specific primer set targeting Ca. N. inopinata amoA gene was applied to investigate its presence and abundance in PM2.5. The expected single bands of PCR products were observed for each PM2.5 sample. The PCR products were purified, cloned and sequenced. In total, 63 sequences were obtained, which were further aligned with MEGA 5.0 software, and compared with those in the database of National Center for the Biotechnology Information. The results suggest that all the sequences showed 100% similarity with Ca. N. inopinata amoA gene, a Comammox enrichment culture, suggesting the presence of Comammox in PM2.5.
Figure 3a depicts the qPCR results of Ca. N. inopinata amoA gene in PM2.5. The abundance of Ca. N. inopinata amoA gene showed slight variations among six samples in BTH. The highest abundance of Ca. N. inopinata was observed in BJ (2.71 × 103 ± 2.62 × 103 cell m−3 air), and lowest abundance occurred in BD (7.35 × 102 ± 4.53 × 101 cell m−3 air). Compared with AOA and AOB, the abundance of Ca. N. inopinata was low. The ratios of AOA and AOB to Ca. N. inopinata ranged from 2.58 to 116.57 and 1.41 to 6.46, respectively (Fig. 3b).
Nitrification potential of PM2.5
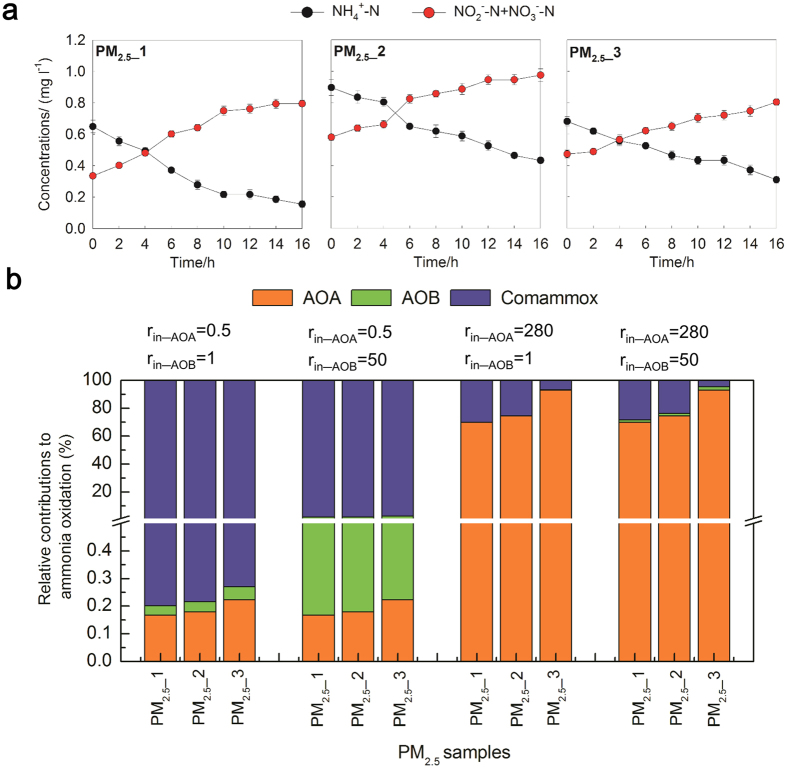
The nitrification potential of PM2.5 was evaluated by incubating PM2.5 suspensions with inorganic medium for 16 h. Changes in net ammonia and nitrite plus nitrate concentrations during the incubations for PM2.5_1, PM2.5_2 and PM2.5_3 are depicted in Fig. 4a. The average net concentrations of NH4+-N were low throughout the incubation period, decreasing from an initial value of 0.65 mg l−1 to 0.15 mg l−1, 0.90 mg l−1 to 0.43 mg l−1 and 0.68 mg l−1 to 0.31 mg l−1 for PM2.5_1, PM2.5_2 and PM2.5_3, respectively. Despite the dissimilar net concentrations of NH4+-N, the maximum nitrification rates were close, which were 0.16 μg (NH4+-N) [m3 air·h]−1 (PM2.5_1), 0.15 μg (NH4+-N) [m3 air·h]−1 (PM2.5_2) and 0.12 μg (NH4+-N) [m3 air·h]−1 (PM2.5_3), respectively, averaged with 0.14 μg (NH4+-N) [m3 air·h]−1. Moreover, the decrease in ammonia concentrations was largely consistent with increase in nitrite plus nitrate concentrations, suggesting ammonia and nitrite plus nitrate balanced.
Figure 4.
Nitrification potential test of PM2.5 and estimation of AOA, AOB and Comammox contributions to ammonia oxidation: (a) changes in net ammonia (NH4+-N) and nitrite (NO2−-N) plus nitrate (NO3−-N) during 16 h incubation of the nitrification potential test; (b) relative contributions of AOA, AOB and Comammox to ammonia oxidation estimated based on the in situ cell-specific ammonia oxidation activity (rin) for AOA and AOB are set at 0.5 or 280 fmol cell−1 h−1 and 1 or 50 fmol cell−1 h−1.
Correlations between environmental factors and community and abundance of AOMs
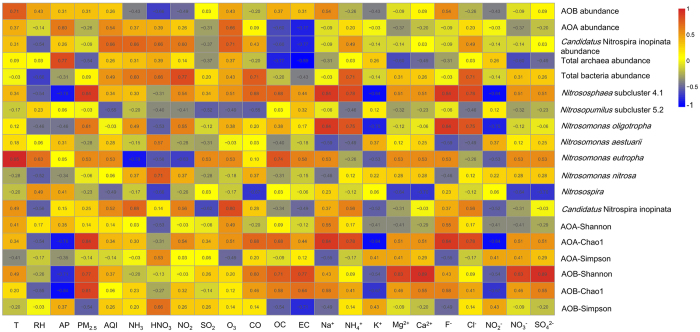
Spearmans’ rank correlation coefficients (SRCCs) were calculated to investigate the relationships between environmental factors (Table 1 and Supplementary Table S2) and diversity and abundance of AOMs. The results are shown in Fig. 5. Only Nitrososphaera subcluster 4.1 was positively correlated with PM2.5, Na+ and F− (SRCC = 0.845, p = 0.034 for the three factors), and negatively correlated with K+ and NO2− (SRCC = −0.845, p = 0.034 for the two factors). However, no significant correlations were observed between environmental factors and Nitrosopumilus subcluster 5.2. N. eutropha showed positive correlation with T (SRCC = 0.949, p = 0.014). N. oligotropha was positively correlated with Na+ and F− (SRCC = 0.841, p = 0.036 for the two factors). Chao 1 of AOA was positively correlated with PM2.5, Na+ and F− (SRCC = 0.845, p = 0.034 for the three factors), and negatively correlated with K+ and NO2− (SRCC = −0.845, p = 0.034 for the two factors). Shannon index of Beta-AOB was positively correlated with Mg2+ (SRCC = 0.829, p = 0.042), NO3− (SRCC = 0.829, p = 0.042), Ca2+ (SRCC = 0.886, p = 0.019) and SO42− (SRCC = 0.886, p = 0.019). Chao 1 of Beta-AOB was negatively correlated with AP (SRCC = −0.878, p = 0.021) and positively correlated with PM2.5 (SRCC = 0.812, p = 0.05). However, no significant correlations were observed between environmental factors and the abundance of AOMs.
Figure 5. Heatmap analysis of the relationships between 23 environmental factors and diversity and abundance of AOA, AOB and Ca. N. inopinata based on SRCCs.
SRCCs between −1 and 1 are shown in the rectangle.
The results of principal components analysis (PCA) and redundancy analysis (RDA) for AOA and Beta-AOB are shown in Fig. 6. As shown in Fig. 6a, the principal component 1 (PC1) and PC2 explained 81.09% and 18.91% of the variance in overall community structure, respectively. Cities BDH, LF and TJ were located close to each other, indicating the similar occurrence of AOA species. However, AOA genera in BJ, BD and TS might be different from others. RDA analysis was carried out to further explore relationships between four factors selected by Monte Carlo permutation tests and the dominant AOA genera (Fig. 6a). The results showed that Nitrososphaera subcluster 4.1 had positive correlations with Na+, PM2.5, organic carbon (OC) and element carbon (EC). However, Nitrosopumilus subcluster 5.2 was negatively related with these environmental factors. OC showed significant positive correlation with Nitrososphaera subcluster 4.1.
Figure 6.
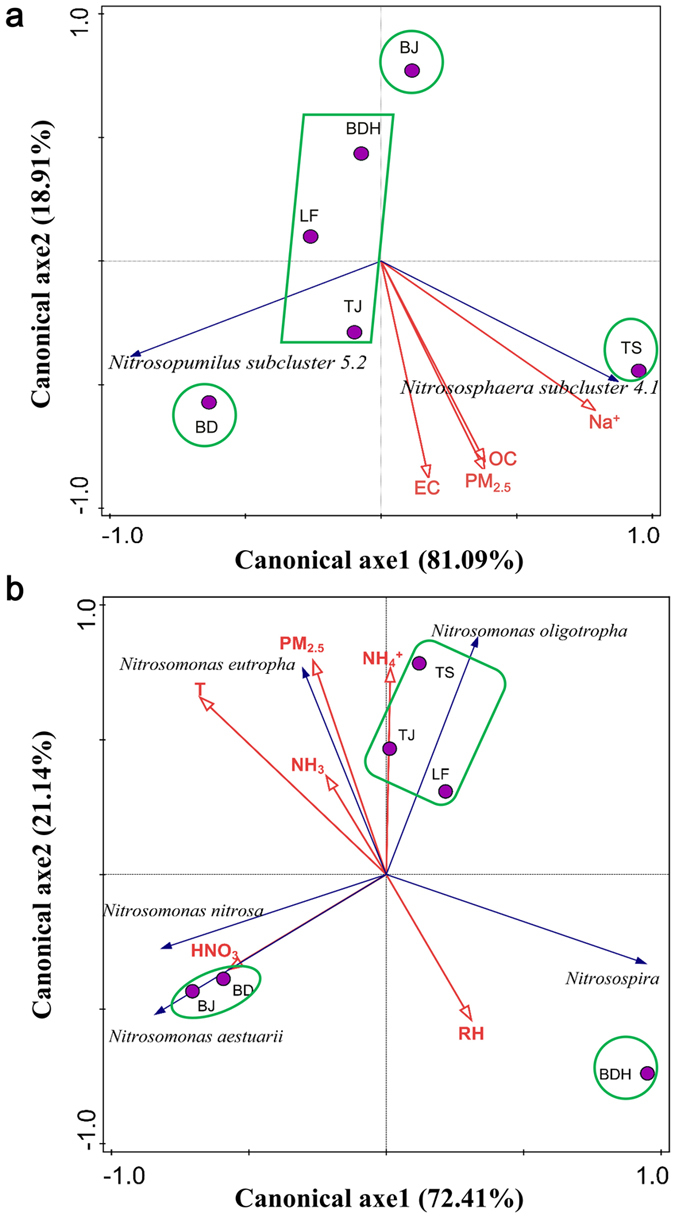
PCA and RDA ordination plots for the first two principal dimensions of the relationship between AOA and AOB community compositions and environmental factors: (a) AOA; (b) AOB.
For Beta-AOB species, PC1and PC2 explained 72.41% and 21.14% of the variance in overall community structure, respectively (Fig. 6b). Three groups of six PM2.5 samples could be plotted off: the first group contained LF, TJ and TS, the second group contained BJ and BD, and there was only one sample (BDH) in the third group. The selected environmental factors were further used for RDA analysis (Fig. 6b). The results suggest that PM2.5, NH3, T and NH4+ showed positive correlations with N. eutropha. NH4+ was also positively correlated with N. oligotropha. HNO3 showed significant positive correlation with N. aestuarii and N. nitrosa. RH was positively correlated with Nitrosospria.
Discussion
AOA or AOB are present in particulate matter with different aerodynamic diameters, e.g., PM2.5 and PM10 in Beijing18, coarse particulate (>3 μm) in Germany17, TSP in an urban area of Northern Italy19, and coarse particular (>3 μm) in a metropolitan subway system of New York20. This study provided fundamental information regarding AOA, AOB and Comammox in PM2.5 in BTH. The newly discovered Comammox are widely distributed in a variety of environments, including natural and man-made ecosystems6,13,14,22. In this study, the occurrence of Ca. N. inopinata, a Comammox enrichment culture, in PM2.5 expanded our knowledge of nitrification. The diversity of AOA and AOB was scarce in PM2.5, which might be due to the extremely low level of nutrient in the atmosphere23.
In China, the total ammonia emission was 16.55 Tg for 2005 and keeps an increase trend24. Ammonia plays a significant role in the neutralization of acid species to form SIA and PM2.5 pollution4. Ammonia is also the energy source for AOA and AOB, and plays an important role in the niche separations of different species of them16. In this study, Nitrosopumilus subcluster 5.2, was found to be the dominant AOA species (96.82%) in PM2.5, and Nitrososphaera cluster only accounted for a small percentage (3.18%). Nitrosopumilus maritimus, the cultivated representative of Nitrosopumilus, possesses high affinity to ammonia with low half-saturation constant (Km = 0.133 μmol l−1)16. In this study, the low ammonia concentrations in the atmosphere (0.363–0.947 mg m−3) might be a reason for the dominance of Nitrosopumilus. Also, Nitrosopumilus is a marine AOA clade16, indicating that AOA observed in PM2.5 might be similar to those in marine. Furthermore, the air movement might make Nitrosopumilus cluster prevail in a broader area, not only in BTH.
The previous studies suggest that Nitrosomonas and Nitrosospira were the main AOB in particulate matters18,19,20, which was in agreement with our study. In this study, 59.63% of AOB in PM2.5 fell into Nitrosomonas cluster and 40.37% of them fell into Nitrosospira cluster. N. multiformis and N. aestuarii were the most dominant AOB in PM2.5 in BTH. N. multiformis is a commonly used model organism for soil AOB, as it is a representative of Nitrosospira cluster 3, which is widespread in agricultural soils25,26. N. aestuarii, one of the marine AOB species, is retrieved from marine environments27. These results suggest that AOB observed in PM2.5 might be similar to those in agricultural soils and marine. Moreover, N. nitrosa (15.60%), N. eutropha (14.68%) and N. oligotropha (11.93%) were also the main AOB in PM2.5. Members of N. nitrosa and N. oligotropha exhibit relatively low Km values28. Therefore, the low ammonia concentration in the atmosphere is in favor of their survival. N. nitrosa and N. eutropha are also the main AOB in eutrophic freshwater. Members of N. eutropha are common in eutrophic freshwater habitats27.
Occurrence of Ca. N. inopinata in PM2.5 in BTH, suggesting that atmosphere (a substrate-limiting environment) maybe a common habitat for Comammox. Previous studies suggest that Comammox may have an advantage over AOA and AOB under substrate-limiting environments6,14. Low substrate is in favor of the enrichment of Comammox. van Kessel et al. enrich the biofilm samples collected from a recirculation aquaculture system in the presence of low ammonia concentration for one year, after which the cultures are capable of complete nitrification and are mostly composed of microorganisms belonging to Comammox Nitrospira bacteria14. Daims et al. enrich the biofilm samples collected from a pipe in a deep oil exploration well under low ammonia concentration for four years, after which most of the microorganisms within the culture are Comammox Nitrospira bacteria13. Overall, the low ammonia concentration in the atmosphere might be the main reason for occurrence and ecological niche distribution of AOMs in PM2.5.
Relationships between environmental factors and the communities of AOA and AOB were further investigated by SRCC, PCA and RDA. Some interesting relationships were observed. OC showed significant positive correlation with Nitrososphaera subcluster 4.1, suggesting that some compounds of OC could stimulate their growth. Kim et al.29 has confirmed the activity of AOA strain DDS1 isolated from seawater can be enhanced by adding α-keto acids (e.g., pyruvate, oxaloacetate). These organic carbon substrates are not assimilated as a carbon source but act as chemical scavengers, suggesting that AOA broadly feature strict autotrophic nutrition29. RDA results also indicate that NH3 showed positive correlation with N. eutropha cluster. Previous studies suggest that high ammonia concentration is in favor of N. eutropha cluster16,30, which was in accordance with our study.
The average abundance of AOA and AOB was 2.82 × 104 and 4.65 × 103 cell m−3 air, respectively. Comparisons of AOA and AOB cell numbers of PM2.5 with different types of samples (soils, compost samples, activated sludge samples and sediment samples) from previous studies are summarized in Supplementary Table S3. AOA abundance in PM2.5 was close to the quantities of AOA reported for soils collected from a large geographical scale across North to South China with different pH values31, activated sludge treating domestic wastewater32 and sediments in the hyporheic zone of a eutrophic river in North China33. The abundance of AOB in PM2.5 was comparable with the compost samples collected from the suburb of Changsha, China34 and the activated sludge samples treating municipal wastewater35. The average abundance of AOA was one order of magnitude higher than that of AOB, which was consistent with most of the studies mentioned in Supplementary Table S3. The average abundance of Ca. N. inopinata was 1.15 × 103 cell m−3 air, which was in the same order of magnitude with AOB, but one to two orders of magnitude lower than AOA. However, the abundance of total Comammox in PM2.5 is still unknown. Further investigations are needed to investigate the abundance of Comammox in PM2.5 with suitable primer set.
Thus far, the actual contributions of AOA, AOB and Comammox to ammonia oxidation in PM2.5 remain unknown. The maximum nitrification rate (NNR_max), in situ cell-specific ammonia oxidation activity (rin, fmol cell−1 h−1) for AOA and AOB, and the abundances of AOA and AOB were used to estimate their relative contributions to ammonia oxidation with formulas (3), (4) and (5) mentioned in methods. Ammonia assimilation of heterotrophic bacteria could remove 10–30% of the ammonia36,37,38. However, in this study, the ammonia assimilation of heterotrophic bacteria was not considered in the calculation of relative contributions of AOMs to nitrification because the ammonia assimilation was insignificant in low ammonia concentration environment39. On the one hand, in direct nutrient-limited competition, the ammonium turnover per unit biomass of Nitrosopumilus-like AOA would be at least 5 times higher than of oligotrophic heterotrophs16. On the other hand, the half-saturation constant (Km) of Nitrosopumilus cluster for ammonia is much lower than the lowest Km of ammonia assimilation of heterotrophic bacteria16.
In the present study, the rin for AOA was set at 0.5 or 208 fmol cell−1 h−1 by referring to the following studies on in situ activity of AOA: 0.5 fmol cell−1 h−1 in freshwater sediment40, 0.53 fmol cell−1 h−1 for N. maritimus SCM116, 4.9–80.6 in drinking water treatment processes39, and 0.03–207.97 fmol cell−1 h−1 in WWTPs41. For AOB, the rin was set at 1 or 50 fmol cell−1 h−1 according to the following studies reported the rin values: 4.4–23.0 fmol cell−1 h−1 for AOB isolates42,43, 1.3–8 fmol cell−1 h−1 in freshwater sediment44; and 0–49.6 fmol cell−1 h−1 in WWTPs45.
Figure 4b shows the estimations of relative contributions of AOA, AOB and Comammox to ammonia oxidation based on different rin for AOA and AOB. If rin for AOA and AOB were set as 0.5 and 1 fmol cell−1 h−1 or 0.5 and 50 fmol cell−1 h−1, the relative contributions of AOA and AOB to ammonia oxidation were extremely low, and Comammox accounted for almost 100% of ammonia oxidation for the three PM2.5 samples. Since the ammonia concentrations in the nitrification potential test were low and AOA have lower Km and higher affinity to ammonia16, AOA contribution in the test may be underestimated. If rin values for AOA and AOB were set as 208 and 1 fmol cell−1 h−1, for three PM2.5 samples, AOA, AOB and Comammox were responsible for 69.83–93.10%, 0.04–0.05% and 6.85–30.14% of the ammonia oxidation, respectively. If rin values for AOA and AOB were set as 208 and 50 fmol cell−1 h−1, AOA, AOB and Comammox were responsible for 69.83–93.10%, 1.76–2.35% and 4.55–28.41% of the ammonia oxidation, respectively. These two results suggest that AOA accounted for most of the ammonia oxidation, followed by Comammox, however, the contribution of AOB to ammonia oxidation was low, which might be related to the low ammonia concentration in PM2.5 incubations and the higher abundance of AOA and Comammox in PM2.5. In previous studies, the same calculation methods were used to evaluate the relative contributions of AOA and AOB to the nitrification of activated sludge in full-scale WWTPs41 and granular activated carbon used in a full-scale advanced drinking water treatment plant39. Their results reveal the significant contribution made by AOA to nitrification under low ammonia concentrations, which were consistent with our study. While, their results also suggest AOB play the dominant role of nitrification under higher ammonia concentration conditions41. In fact, a more accurate estimation of their contributions to nitrification should only depend on the active AOMs. RNA-based methods or DNA based stable-isotope probing (DNA-SIP) technique may be more effective and accurate to evaluate the contributions of AOMs to nitrification. Further investigation is still needed to validate the hypothesis that AOA played the predominant role in ammonia oxidation of PM2.5.
In conclusion, Nitrosopumilus subcluster 5.2 was the most dominant AOA. N. multiformis and N. aestuarii were the main AOB. The occurrence of Comammox in PM2.5 was confirmed by the presence of Ca. N. inopinata in PM2.5. The average cell numbers of AOA, AOB and Ca. N. inopinata were 2.82 × 104, 4.65 × 103 and 1.15 × 103 cell m−3 air, respectively. Nitrososphaera subcluster 4.1 was positively correlated with PM2.5 and OC concentrations, and N. eutropha cluster and N. aestuarii cluster showed positive correlations with NH3 and HNO3 concentrations. The average maximum nitrification rate of PM2.5 was 0.14 μg (NH4+-N) [m3 air·h]−1. AOA and Comammox may be the major contributors to ammonia oxidation in PM2.5. However, further investigations regarding Comammox in PM2.5 based on an appropriate primer set are still needed.
Methods
Sample collection, meteorological conditions and chemical analyses
A model KC-6120 comprehensive atmospheric sampler (Laoshan Electronic Instrument Factory, Qingdao, China) was used for the collection of PM2.5, NH3 and HNO3 samples. The glass fiber filters were pre-heated at 450 °C for 4 h to remove organic material and their weight were measured by a microbalance before PM2.5 collection. PM2.5 collection was carried out at a flow rate of 100 l min−1 for 24 h (Table 1). The collections of NH3 and HNO3 were according to the national standard method of the People’s Republic of China (GB/T 18204.25 2000). During summer, the predominant wind direction of BTH is from southeast; therefore, as shown in Fig. 1, the first order of samples collection was from BJ to LF and then to TJ. After sampling from the three cities, samples were collected from the northeast to southwest axis of BTH (namely BDH-TS-BD).
The meteorological data, atmospheric pollutants and air pollution index (AQI) were recorded concurrently with air sampling (Table 1 and Supplementary Table S2). The carbonaceous species (OC and EC) and water-soluble inorganic ions of PM2.5 were analyzed by a thermal/optical carbon aerosol analyzer (DRI Model 2001A, Desert Research Institute, USA) and ion chromatography (ICS-90, Dionex, USA), respectively (see Supplementary Table S2).
DNA extraction, PCR, cloning and sequencing
For PM2.5 samples, 1/4 of the whole glass fiber was cut into pieces using sterilized handling instruments. Genomic DNA was extracted using a Fast-DNA ® SPIN Kit following the manufacturer’s protocol (Qiagen, CA, USA).
Primer sets Arch-amoAF/Arch-amoAR46, CTO189f/CTO645r47, amoA-3F/amoB-4R48 and Nino_amoA_19F/Nino_amoA_252R13 were used to amplify AOA amoA gene, Beta-AOB 16S rRNA gene, Gamma-AOB amoA gene and Ca. N. inopinata amoA gene fragments of PM2.5 samples. The CTO189f was a mixture of CTO189fA/B and CTO189fc at a ratio of 2:1. For AOA amoA gene, the components of PCR mixture and protocols of PCR were followed by the study of Gao et al.35. For Beta-AOB 16S rRNA gene and Ca. N. inopinata amoA gene, the PCR protocols were the same as AOA amoA gene except the annealing temperature (58 °C and 60 °C for Beta- AOB and Ca. N. inopinata, respectively). Gradient PCR was applied to detect Gamma-AOB in PM2.5. However, the amplification of Gamma-AOB amoA gene was failed. PCR products for the other genes were purified, cloned and sequenced. For each sample, 15–30 white colonies for AOA amoA gene, Beta-AOB 16S rRNA gene and Ca. N. inopinata amoA gene were randomly picked for sequencing with ABI 3730 XL capillary sequencers (PE Applied Biosystems, Foster City, USA).
The AOA amoA gene, Beta-AOB 16S rRNA gene and Ca. N. inopinata amoA gene sequences have been deposited in the GenBank library under accession numbers from KM402456 to KM402612, KY008589 to KY008697 and KX273257 to KX273319, respectively.
Phylogenetic analyses
The sequences were grouped into OTUs with a 97% sequence similarity using Mothur 1.28. Cytoscape 2.32 was applied for visualization of the shared OTUs between samples. MEGA 5.0 was used to construct a phylogenetic tree using the neighbor-joining (NJ) method with the Jukes–Cantor correction model. The NJ tree was calculated after bootstrapping with 1000 replicate trees.
Quantification of AOA, AOB, Comammox, bacteria and archaea of PM2.5
The abundance of AOA, AOB, Ca. N. inopinata, bacteria and archaea of PM2.5 were quantified by the following primer sets: GenAOAF/GenAOAF49, amoA-1Fmod and GenAOBR49, Nino_amoA_19F/Nino_amoA_252R13, Uni1055F and 1392R50 and 934f/1040r51 on a Stratagene MX3005p thermocycler (Agilent Technologies, USA) in triplicate with a GoTaq® qPCR Master Mix (Promega, USA). The components of qPCR mixture were the same as the previous study35. The qPCR conditions were also followed by this study except different annealing temperature: 56 °C, 58 °C, 60 °C, 53 °C and 59 °C for AOA, AOB, Ca. N. inopinata, bacteria and archaea. The standard curve was generated by using 10-fold serial dilutions of linearized plasmid extracted from the correct insert clones of each target gene. The amplification efficiencies of qPCR assays ranged from 90.6 to 106.0%, and R2 value for each standard curves exceeded 0.996.
Nitrification potential test
In order to investigate the nitrification potential of PM2.5, three PM2.5 samples (PM2.5_1, PM2.5_2 and PM2.5_3) were collected in BJ for 24 h (8 h for each sampling) at a flow rate of 100 l min−1, resulting a total of 48 m3 air collected for each sample. A blank filter (as control) was put at the side of the sampler with each PM2.5 sampling, therefore three blank samples (Blnak_1, Blank_2 and Blank_3) were obtained. After sampling, the PM2.5 and blank filters were immediately wrapped with aluminum foil and taken back to the lab within five minutes.
Technologies for investigation of microorganisms in the atmosphere are still in immature stage of development, and the methods for evaluation of nitrification activity of AOMs in PM2.5 have not been reported. The overall microbial community composition in PM2.5 maybe similar as that in soil because soil is one of the main source for PM2.552. Soil suspension technique is a recommended method for assessing nitrification potential53. In this technique, the substrate and moisture limitations are eliminated, and the changes in AOMs are unlikely to occur after short-term incubation. Therefore, the nitrification rate measured approximates the maximum nitrification rate possible at the specific temperature of the incubation53. Moreover, this technique may be the easiest to interpret and most reproducible for all laboratory nitrification assays53.
In this study, maximum nitrification rate of PM2.5 was investigated according to soil suspension technique with some modification, which may be defined as PM2.5 suspension technique. Briefly, the PM2.5 and blank filters were cut into pieces and put into 50 ml centrifuge tubes filled with sterilized and oxygenated phosphate-buffered saline (PBS) (g l−1: NaCl, 8.0; KCl, 0.2; Na2HPO4, 1.44; KH2PO4, 0.24; pH, 7.4) followed by vortexing for 30 min and sonication for 2 h to generate suspensions of PM2.5 and blank samples. The 50 ml suspensions of PM2.5 and blank samples were incubated individually with 200 ml of inorganic medium in 500 ml Erlenmeyer flasks. The Erlenmeyer flasks were closed with plastic wrap and incubated at 30 °C under agitation at 100 rpm. Compositions of inorganic medium were as follows: 3 ml NH4Cl (10 μg l−1), 3 ml NaHCO3 (20 μg l−1), 0.25 ml trace element and 193.75 ml PBS. The compositions of trace element were according to the previous study54. After 0, 2, 4, 6, 8, 10, 12, 14, 16 h of incubation, supernatant was collected and filtered through 0.2 μm pore size polytetrafluoroethylene membranes. Concentrations of ammonia (NH4+-N), nitrite (NO2−-N) and nitrate (NO3−-N) were analyzed in triplicate in accordance with standard methods55.
The net concentrations of NH4+-N (NH4+-N_net, mg l−1) in PM2.5 sample incubations were calculated using the following formula:
 |
where NH4+-N_PM2.5_measured and NH4+-N_Blank_measured are the measured ammonia concentration in the incubations of PM2.5 and blank samples.
The calculations of net concentrations of NO2−-N and NO3−-N in PM2.5 sample incubations were the same as the NH4+-N. The NNR_max (μg (NH4+-N) [m3 air·h]−1) of PM2.5 samples were calculated by the following formula:
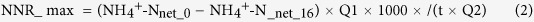 |
where NH4+-N net_0 and NH4+-N net_16 are the net ammonia concentrations in PM2.5 incubations at 0 and 16 h, Q1 is the incubation volume (Q1 = 0.25 l), t is the incubation time (t = 16 h), and Q2 is the volume of sampled air (Q2 = 48 m3).
Estimation of relative contributions of AOA, AOB and Comammox to ammonia oxidation in PM2.5
The estimation of relative contributions of AOA, AOB and Comammox to ammonia oxidation was carried out with some assumptions: (1) only AOA, AOB and Comammox determined were involved in the ammonia oxidation in PM2.5; (2) the ammonia assimilation of heterotrophic bacteria was not considered; (3) there were 1 amoA gene copy per AOA and Comammox, and 2.5 amoA gene copies per AOB13,15,21; (4) all AOA, AOB and Comammox were equally active enough to contribute to ammonia oxidation. Their relative contributions (RC) to ammonia oxidation of PM2.5 were estimated using the following formulas according to previous studies36,39,41:
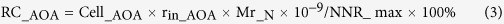 |
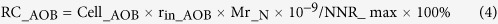 |
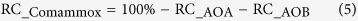 |
where RC_AOA, RC_AOB and RC_Comammox represent the relative contribution of AOA, AOB and Comammox. Cell_AOA and Cell_AOB are the abundance of AOA and AOB (cells m−3 air), and rin_AOA and rin_AOB are the in situ cell-specific ammonia oxidation activity (rin, fmol cell−1 h−1) for AOA and AOB. Mr_N is the relative molecular mass of nitrogen. NNR_max is the maximum nitrification rate (μg (NH4+-N) [m3 air·h]−1).
Statistical analysis
SRCC, PCA and RDA were applied to address the correlations between environmental factors and AOMs (AOA, Beta-AOB and Ca. N. inopinata). The Monte Carlo permutation test (999 replicates) was used to estimate the significance of the correlations. All of the statistical analyses were done using R software version 2.15.
Additional Information
How to cite this article: Gao, J.-F. et al. Diversity, abundance and activity of ammonia-oxidizing microorganisms in fine particulate matter. Sci. Rep. 6, 38785; doi: 10.1038/srep38785 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank the NSFC (51078007, 51378027 and 51578015), BJNSF (8162010) and Beijing Talent Foundation of BJUT (2013-JH-L06) for the financial supports of this study.
Footnotes
Author Contributions J.G. and X.F. conceived the experiments. X.F. and H.L. collected the PM2.5 samples in BTH. X.F. conducted the experiments and analyzed the data. J.G. and X.F. discussed the results and co-wrote the paper. K.P. and L.S. provided the PCR conditions of Comammox amoA gene. All authors read and approved the final manuscript.
References
- Pope C. R., Ezzati M. & Dockery D. W. Fine-particulate air pollution and life expectancy in the United States. N. Engl. J. Med. 360, 376–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Liu X., Dore A. J. & Li K. Stemming PM2.5 pollution in China: re-evaluating the role of ammonia, aviation and non-exhaust road traffic emissions. Environ. Sci. Technol. 46, 13035–13036 (2012). [DOI] [PubMed] [Google Scholar]
- Guo S., Hu M., Wang Z. B., Slanina J. & Zhao Y. L. Size-resolved aerosol water-soluble ionic compositions in the summer of Beijing: implication of regional secondary formation. Atmos. Chem. Phys. 10, 947–959 (2010). [Google Scholar]
- Baek B. H. & Aneja V. P. Observation based analysis for the determination of equilibrium time constant between ammonia, acid gases, and fine particles. Int. J. Environ. Pollut. 23, 239–247 (2005). [Google Scholar]
- Galloway J. N. et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science. 320, 889–892 (2008). [DOI] [PubMed] [Google Scholar]
- Nunes-Alves C. Microbial ecology: Do it yourself nitrification. Nat. Rev. Microbiol. 14, 61 (2016). [DOI] [PubMed] [Google Scholar]
- Utaker J. B., Bakken L., Jiang Q. Q. & Nes I. F. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA gene sequences. Syst. Appl. Microbiol. 18, 549–559 (1996). [Google Scholar]
- Pester M. et al. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14, 525–539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollmann A., Bullerjahn G. S. & McKay R. M. Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior. Plos One 9, e97068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S. et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442, 806–809 (2006). [DOI] [PubMed] [Google Scholar]
- Caffrey J. M., Bano N., Kalanetra K. & Hollibaugh J. T. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1, 660–662 (2007). [DOI] [PubMed] [Google Scholar]
- Gao J., Luo X., Wu G., Li T. & Peng Y. Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems. Appl. Microbiol. Biotechnol. 98, 3339–3354 (2014). [DOI] [PubMed] [Google Scholar]
- Daims H. et al. Complete nitrification by Nitrospira bacteria. Nature. 528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel M. A. H. J. et al. Complete nitrification by a single microorganism. Nature 528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M. et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546 (2005). [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R. & Stahl D. A. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature 461, 976–979 (2009). [DOI] [PubMed] [Google Scholar]
- Fröhlich-Nowoisky J. et al. Diversity and seasonal dynamics of airborne archaea. Biogeosciences. 11, 6067–6079 (2014). [Google Scholar]
- Cao C. et al. Inhalable microorganisms in Beijing’s PM2.5 and PM10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini V. et al. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl. Microbiol. Biotechnol. 97, 6561–6570 (2013). [DOI] [PubMed] [Google Scholar]
- Robertson C. E. et al. Culture-independent analysis of aerosol microbiology in a metropolitan subway system. Appl. Environ. Microbiol. 79, 3485–3493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klappenbach J. A., Saxman P. R., Cole J. R. & Schmidt T. M. rrndb: the ribosomal RNA operon copy number database. Nucleic. Acids. Res. 29, 181–184 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers M. M. M. A division of labour combined. Nature 528, 487–488 (2015). [DOI] [PubMed] [Google Scholar]
- Brodie E. L. et al. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. 104, 299–304 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Impact assessment of ammonia emissions on inorganic aerosols in east China using response surface modeling technique. Environ. Sci. Technol. 45, 9293–9300 (2011). [DOI] [PubMed] [Google Scholar]
- Mendum T. A. & Hirsch P. R. Changes in the population structure of beta-group autotrophic ammonia oxidising bacteria in arable soils in response to agricultural practice. Soil. Biol. Biochem. 34, 1479–1485 (2002). [Google Scholar]
- Avrahami S. & Bohannan B. J. A. Response of Nitrosospira sp. strain AF-like ammonia oxidizers to changes in temperature, soil moisture content, and fertilizer concentration (vol 73, pg 1166, 2007). Appl. Environ. Microbiol. 73, 3121 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerening-Röser A., Rath G. & Koops H. P. Phylogenetic diversity within the genus Nitrosomonas [J]. Syst. Appl. Microbiol. 19, 344–351 (1996). [Google Scholar]
- Koops H. & Pommerening R. A. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37, 1–9 (2001). [Google Scholar]
- Kim J. G., Park S. J., Damsté J. S., Schouten S. & Rijpstra W. I. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc. Natl. Acad. Sci. 113, 7888–7893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguder T. H., Boon N., Wittebolle L., Marzorati M. & Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol. Rev. 33, 855–869 (2009). [DOI] [PubMed] [Google Scholar]
- Hu H., Zhang L., Dai Y., Di H. & He J. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J. Soil Sediment 13, 1439–1449 (2013). [Google Scholar]
- Bai Y., Sun Q., Wen D. & Tang X. Abundance of ammonia-oxidizing bacteria and archaea in industrial and domestic wastewater treatment systems. FEMS Microbiol. Ecol. 80, 323–330 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang Z., Huang C. & Pei Y. Vertical distribution of ammonia-oxidizing archaea (AOA) in the hyporheic zone of a eutrophic river in North China. World J. Microbiol. Biotechnol. 30, 1335–1346 (2014). [DOI] [PubMed] [Google Scholar]
- Zeng G. et al. Relative contributions of archaea and bacteria to microbial ammonia oxidation differ under different conditions during agricultural waste composting. Bioresource Technol. 102, 9026–9032 (2011). [DOI] [PubMed] [Google Scholar]
- Gao J. F., Luo X., Wu G. X., Li T. & Peng Y. Z. Quantitative analyses of the composition and abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in eight full-scale biological wastewater treatment plants. Bioresource Technol. 138, 285–296 (2013). [DOI] [PubMed] [Google Scholar]
- Daims H., Ramsing N. B., Schleifer K. H. & Wagner M. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67, 5810–5818 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H. et al. Nitrification in sequencing biofilm batch reactors: Lessons from molecular approaches. Water Sci. Technol. 43, 9–18 (2001). [PubMed] [Google Scholar]
- Harms G. et al. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37, 343–351 (2003). [DOI] [PubMed] [Google Scholar]
- Kasuga I., Nakagaki H., Kurisu F. & Furumai H. Predominance of ammonia-oxidizing archaea on granular activated carbon used in a full-scale advanced drinking water treatment plant. Water Res. 44, 5039–5049 (2010). [DOI] [PubMed] [Google Scholar]
- Herrmann M., Saunders A. M. & Schramm A. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74, 3279–3283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayee P., Sonthiphand P., Rongsayamanont C. & Limpiyakorn T. Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microbiol. Ecol. 62, 776–788 (2011). [DOI] [PubMed] [Google Scholar]
- Belser L. & Schmidt E. Growth and oxidation kinetics of three genera of ammonia oxidizing nitrifiers. FEMS Microbiol. Lett. 7, 213–216 (1980). [Google Scholar]
- Laanbroek H. J., Bodelier P. & Gerards S. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch Microbiol. 161, 156–162 (1994). [Google Scholar]
- Altmann D., Stief P., Amann R., De Beer D. & Schramm A. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5, 798–803 (2003). [DOI] [PubMed] [Google Scholar]
- Limpiyakorn T., Shinohara Y., Kurisu F. & Yagi O. Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol. Ecol. 54, 205–217 (2005). [DOI] [PubMed] [Google Scholar]
- Francis C. A., Roberts K. J., Beman J. M., Santoro A. E. & Oakley B. B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. 102, 14683–14688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalchuk G. A. et al. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Microbiol. Biotechnol. 63, 1489–1497 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkhold U. et al. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66, 5368–5382 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt K. A. et al. Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia-oxidizing archaea and bacteria. Environ. Microbiol. Rep. 7, 354–363 (2015). [DOI] [PubMed] [Google Scholar]
- Ferris M. J., Muyzer G. & Ward D. M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Microbiol. Biotechnol. 62, 340–346 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G. et al. Stratification of archaea in the deep sediments of a freshwater meromictic lake: vertical shift from methanogenic to uncultured archaeal lineages. Plos One 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers R. M. et al. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol. 77, 6350–6356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart S. C., Stark J. M., Davidson E. A. & Firestone M. K. Nitrogen Mineralization, Immobilization, and Nitrification. Methods of Soil Analysis: Part 2—Microbiological and Biochemical Properties, (methodsofsoilan2) 985–1018 (1994). [Google Scholar]
- Cua L. S. & Stein L. Y. Effects of nitrite on ammonia-oxidizing activity and gene regulation in three ammonia-oxidizing bacteria. FEMS Microbiol. Lett. 319, 169–175 (2011). [DOI] [PubMed] [Google Scholar]
- APHA. Standard methods for the examination of water and wastewater, 20th ed, (Washington DC, USA 1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.