Abstract
The kinetochore, which consists of DNA sequence elements and structural proteins, is essential for high-fidelity chromosome transmission during cell division. In budding yeast, Sgt1, together with Skp1, is required for assembly of the core kinetochore complex (CBF3) via Ctf13 activation. Formation of the active Ctf13-Skp1 complex also requires Hsp90, a molecular chaperone. We have found that Sgt1 interacts with Hsp90 in yeast. We also have determined that Skp1 and Hsc82 (a yeast Hsp90 protein) bind to the N-terminal region of Sgt1 that contains tetratricopeptide repeat motifs. Results of sequence and phenotypic analyses of sgt1 mutants strongly suggest that the N-terminal region containing the Hsc82-binding and Skp1-binding domains of Sgt1 is important for the kinetochore function of Sgt1. We found that Hsp90's binding to Sgt1 stimulates the binding of Sgt1 to Skp1 and that Sgt1 and Hsp90 stimulate the binding of Skp1 to Ctf13, the F-box core kinetochore protein. Our results strongly suggest that Sgt1 and Hsp90 function in assembling CBF3 by activating Skp1 and Ctf13.
The centromere is essential for chromosome inheritance and requires DNA sequence elements and structural and regulatory proteins (the structural protein complex is called the kinetochore) for its activity and coordination within the cell cycle. In budding yeast (Saccharomyces cerevisiae), the kinetochore consists of more than 30 different proteins, and several of these proteins are conserved in mammals (21). The budding yeast centromere DNA is a 125-bp region that contains three conserved regions, CDEI, CDEII, and CDEIII (8, 13). CDEIII (25 bp) is essential for centromere function (12) and was the site shown to be bound by CBF3, a protein complex that contains the major proteins p110 (encoded by NDC10/CTF14/CBF2), p64 (encoded by CEP3/CBF3b), p58 (encoded by CTF13) (4, 5, 11, 17, 23, 24, 37, 39), and Skp1 (4, 37). All four are essential for viability, and mutations in any one abolish the CDEIII-binding activity of CBF3 (18, 36). No known kinetochore protein, except for CDEI-binding Cbf1, can localize to kinetochores when the CBF3 complex is disrupted. Therefore, the CBF3 complex is the fundamental structure of the kinetochore, and the mechanism of CBF3 assembly is of great interest.
SGT1 was isolated as a dosage suppressor of skp1-4, a kinetochore-defective mutant (22). By activating Ctf13, Sgt1 and Skp1 are required for assembly of the CBF3 complex (22). However, the mechanism of activation is not well characterized. Stemmann et al. showed that formation of the active Ctf13-Skp1 complex requires Hsp90, a molecular chaperone (38). Sgt1 has highly conserved tetratricopeptide repeat (TPR) (22) and CS (CHORD protein and Sgt1-specific) motifs—in other proteins these motifs are required for interaction with Hsp90 (2, 6). Here we report that molecular chaperones from the Hsp90 and Hsp70 families form a complex with Sgt1 in yeast. Our results strongly suggest that Hsp90's interaction with Sgt1 is required for kinetochore assembly in yeast. On the basis of our findings, we propose a model in which Sgt1 and its interaction with Hsp90 are required for assembly of the core kinetochore complex, an initial step in kinetochore activation.
MATERIALS AND METHODS
Yeast strains and medium.
Table 1 lists the genotypes of the yeast strains used in this study. The medium used for yeast growth and sporulation was described previously (33). Yeast transformation was done as described by Ito et al. (16). Strains that expressed Sgt1-20myc or Sgt1-13myc were generated in accordance with the procedure of Longtine et al. (26). Sgt1-13myc contains 13 myc tags; Sgt1-20myc contains 20. The numbers of tags were confirmed by direct sequencing. Regions encoding the myc tags were inserted at the 3′ end of the endogenous SGT1 locus.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| YPH499 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | 34a |
| YKK54 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgtI-3::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | 22 |
| YKK45 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-5::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | 22 |
| Y37R | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-1::LEU2 | 22 |
| Y40R | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-2::LEU2 | 22 |
| Y36R | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-4::LEU2 | 22 |
| Y310 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 SGT1-20Myc::H1S3-MX6 | This study |
| Y1019 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 SGT1-13Myc::HIS3-MX6 | This study |
| Y1015 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-6::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1016 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-7::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1134 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-8::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1135 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-9::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1136 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-10::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1137 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-11::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| Y1018 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-12::LEU2 CFIII(CEN3.L.YPH983) TRP1 SUP11 | This study |
| YPH972R | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ctf13-30 | 5 |
| PY694 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 met30-6::KAN | 17a |
| K699 | mataade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+psi+ | 29b |
| MTY871 | mataura3 trp1-1 his3-11 15 can1-100 leu2-3 112 GAL psi+cdc53-1 | 41 |
| US104 | mataura3 trp1-1 his3-11 leu2-3 112 cdc4-1 | 11a |
| US504 | mataa ura3 trp1-1 his3-11 leu2-3 112 cdc34-1 | 11a |
| US581 | mataura3 trp1-1 his3-11 leu2-3 sic1Δ::LEU2 | 11a |
| US893 | mataura3 trp1-1 his3-11 leu2-3 112 cdc4Δ::URA3 pGAL-CDC4/TRP1/CEN | 11a |
| US1198 | mataura3 trp1-1 his3-11 leu2-3 112 cdc4Δ::URA3 pcdc4-12/TRP1/CEN | 11a |
| YPH1161 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 skp1-4::LEU2 CFIII(CEN3.L.YPH983) HIS3 SUP11 | 4 |
| YPH1172 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 skp1-3::LEU2 CFIII(CEN3.L:YPH983) HIS3 SUP11 | 4 |
| ΔPLCDa(T1011) | mataade2-1 can1-100 his3-12,16 leu2-3,112 trp1-1 ura3 hsp82::LEU2 hsc82::LEU2 pRS314-hsp82(T1011) | 29c |
| Y1124 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 pYEp24-HSC82.2μm/URA | This study |
| Y1125 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsp82Δ::TRP1 pYEp24-HSC82 2μm/URA | This study |
| Y1126 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsc82Δ::KAN pYEp24-HSC82 2μm/URA | This study |
| Y1127 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-3::LEU2 pYEp24-HSC82 2μm/URA | This study |
| Y1128 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 sgt1-5::LEU2 pYEp24-HSC82 2μm/URA | This study |
| Y1129 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsc82Δ::KAN hsp82Δ::TRP1 pYEp24-HSC82 2μm/URA | This study |
| Y1130 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsp82Δ::TRP1 sgt1-3::LEU2 pYEp24-HSC82 2μm/URA | This study |
| Y1131 | matα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsp82Δ::TRP1 sgt1-5::LEU2 pYEp24-HSC82 2μm/URA | This study |
| Y1132 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsc82Δ::KAN sgt1-3::LEU2 pYEp24-HSC82 2μm/URA | This study |
| Y1133 | mataura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 hsc82Δ::KAN sgt1-5::LEU2 pYEp24-HSC82 2μm/URA | This study |
Plasmid construction and primers.
Table 2 lists the plasmids used in this study. Details of their construction (19) and primer sequences are available upon request.
TABLE 2.
Plasmids used in this study
Generation and integration of temperature-sensitive alleles.
Temperature-sensitive mutant forms of sgt1 were generated by a PCR-based method as described previously (22). New temperature-sensitive alleles (sgt1-6, sgt1-7, sgt1-8, sgt1-9, sgt1-10, sgt1-11, and sgt1-12) were isolated, and the recovered mutated plasmids were retested to determine their effect on cell viability at 37°C. All alleles were integrated into the SGT1 locus in the yeast genome (Table 1). All nucleotides of the mutant alleles in plasmids were sequenced. All portions of the mutant alleles that were integrated into the yeast genome were amplified by PCR, and the PCR fragments were sequenced. We did not find any additional mutations after integration (Table 3).
TABLE 3.
sgt1 mutations
| Allele | No. of mutations | Codon | DNA position | Sequence change |
|---|---|---|---|---|
| sgt1-1 | 2 | 119 | 356 | Y (TAC) to C (TGC) |
| 378 | 1132 | S (TCT) to P (CCT) | ||
| sgt1-2 | 2 | 243 | 728 | L (CTT) to H (CAT) |
| 378 | 1133 | S (TCT) to F (TTT) | ||
| sgt1-3 | 3 | 31 | 92 | L (CTC) to P (CCC) |
| 99 | 295 | F (TTC) to L (CTC) | ||
| 213 | 638 | N (AAT) to I (ATT) | ||
| sgt1-4 | 2 | 360 | 1078 | K (AAA) to E (GAA) |
| 385 | 1153 | S (TCA) to T (ACA) | ||
| sgt1-5 | 2 | 220 | 659 | D (GAT) to V (GTT) |
| 364 | 1090 | E (GAA) to K (AAA) | ||
| sgt1-6 | 2 | 26 | 77 | L (CTT) to P (CCT) |
| 99 | 295 | F (TTC) to L (CTC) | ||
| sgt1-7 | 2 | 117 | 350 | L (CTT) to P (CCT) |
| 191 | 571 | Q (CAA) to K (AAA) | ||
| sgt1-8 | 3 | 360 | 1078 | K (AAA) to E (GAA) |
| 135 | 403 | L (TTG) to L (CTG) | ||
| 288 | 864 | S (TCA) to S (TCG) | ||
| sgt1-9 | 1 | 360 | 1078 | K (AAA) to E (GAA) |
| sgt1-10 | 1 | 378 | 1132 | S (TCT) to P (CCT) |
| sgt1-11 | 3 | 378 | 1132 | S (TCT) to P (CCT) |
| 390 | 1168 | M (ATG) to V (GTG) | ||
| 196 | 588 | V (GTC) to V (GTT) | ||
| sgt1-12 | 2 | 74 | 220 | A (GCT) to T (ACT) |
| 151 | 451 | I (ATT) to V (GTT) |
Synthetic lethality.
Strain Y1129 (matα hsc82Δ::KAN hsp82Δ::TRP1 pYEp24-HSC82 2μm/URA) and strain YKK54 (mata sgt1-3::leu2) or YKK45 (mata sgt1-5::leu2) were mated. After diploid selection and sporulation, Ura+ spores were isolated. From these selected spores, those representing all possible single or double combinations of G418+, Trp+, or Leu+ were selected. To test synthetic lethality, these haploid strains harboring the pYEp24-HSC82 2μm/URA plasmid were restreaked on synthetic medium that contained 5-fluoroorotic acid (Sc+5-FOA) and on Sc that lacked uracil (Sc−Ura). The plates were incubated at 25°C for 2 days.
Mass spectrometry.
Mass spectrometry (under the direction of Clive Slaughter) was performed by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital. Immunopurified proteins (see description of immunoprecipitation below) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins in excised gel pieces were reduced, alkylated with iodoacetamide, and digested with trypsin. For Hsc82 identification, the unfractionated digest was subjected to mass spectrometry, and the mass values of the peptides were matched with the expected values obtained by analysis of the protein sequence. Mass spectrometry of the digested protein in a crystalline matrix of α-cyano-4-hydroxycinnamic acid was performed by matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry in a model 4700 Proteomics analyzer from Applied Biosystems (Foster City, Calif.). The MS-Fit program of Protein Prospector was used for the database search. The program and the database that were accessed are located at http://hc-appc1.stjude.org/index-pp.html. NCBInr. was used for protein identification.
For Ssa1 and Ssb1 identification, tryptic peptides were extracted and subjected to combined capillary liquid chromatography-tandem mass spectrometry by a ThermoQuest LCQ-DECA ion trap mass spectrometer with a nanoelectrospray ion source. Fragment ion (MS2) spectra were subjected to a search by the SEQUEST program of Eng and Yates (ThermoQuest). The National Center for Biotechnology Information nonredundant protein database was used.
Antibodies.
The anti-Hsc82 antibody used was a gift from Suzan Lindquist (Whitehead Institute for Biomedical Research). The anti-Skp1 and anti-Sgt1 antibodies used were previously described (20, 22). The anti-hemagglutinin (anti-HA; Roche), anti-myc (Roche), and anti-His6 (QIAGEN) antibodies used were purchased.
Protein expression and immunoprecipitation.
Immunoprecipitation with yeast lysates was performed as described previously, except that antibodies were cross-linked to Sepharose by using the Seize Primary Immunoprecipitation kit (Pierce) (20, 22).
For in vitro expression of proteins (Sgt1 deletion proteins [Sgt1-Ds] and HA-Hsp82), PCR products that encoded polypeptides of interest were used as templates in the TnT T7 Quick for PCR DNA in vitro transcription-translation system with rabbit reticulocyte lysates (Promega), and Sgt1 deletion proteins were labeled with [35S]methionine. HA-tagged Skp1 was expressed in insect cells as previously described (20, 22). For Skp1- and Hsc82-binding domain analyses, approximately equal amounts of deletion proteins were used for immunoprecipitation with an anti-HA antibody. Immune complexes were washed five or seven times with 1.5 ml of buffer A (50 mM Tris-HCl [pH 7.5], 0.5% NP-40, 150 mM NaCl, 10 mM β-glycerophosphate, protease inhibitor cocktail [Roche]) before they underwent SDS-PAGE and immunoblotting. Total reticulocyte lysates (T) were represented by material in a volume that was equivalent to 2 to 6% of that of the starting material.
For protein expression in bacteria, BL21(DE3) cells were transformed with pGEX (a glutathione S-transferase [GST] gene fusion vector; Amersham Biosciences) or with pGEX-Skp1; expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (1 mM) and incubation for 3 h at 37°C. The GST-tagged proteins were affinity purified in accordance with the manufacturer's instruction and then subjected to immunoprecipitation studies.
Expression and purification of the His6-Sgt1, His6-Sgt1(59-395), and HA-His6-Ctf13 proteins were done in accordance with the manufacturer's (QIAGEN) instructions. Human Hsp90 protein purified from HeLa cells was purchased from Stressgen. Approximately 200 ng of the purified proteins was mixed with 500 μl of buffer B (50 mM Tris-HCl [pH 7.5], 0.2% Triton X-100, 50 mM NaCl, protease inhibitor cocktail [Roche]) and incubated at 30°C for 1 h. The proteins were then mixed with an anti-His6 or anti-HA antibody and Sepharose A and incubated at 4°C for 2 h. Immune complexes were washed five or seven times with 1.5 ml of lysis buffer before they underwent SDS-PAGE and immunoblotting. Total protein mixtures (T) were represented by material in a volume that was equivalent to 5% of that of the starting material.
35S-labeled proteins were detected by autoradiography and quantified with a Molecular Dynamics Phospho-Imager with Image Quant software. Nonradiolabeled proteins were detected by immunoblotting with ECL reagents (Amersham) and quantified with a Chemidoc EQ system (Bio-Rad) with Quantity One 1-D Analysis Software (Bio-Rad). All signals were measured within the linear ranges of detection.
RESULTS
Association of Sgt1 with Hsp90 in budding yeast.
Sgt1, together with Skp1, is required for formation of CBF3 (the core kinetochore complex) in budding yeast (22). To further investigate the function of Sgt1, we performed immunoprecipitation-mass spectrometry analysis of strains expressing myc-tagged Sgt1. Sgt1 is essential for viability, and strains expressing the myc-tagged proteins showed no growth defect or chromosome missegregation (data not shown); therefore, we concluded that myc-tagged Sgt1 was functional. Hsc82 (a yeast Hsp90 protein) and other cochaperones were found specifically in precipitates of myc-tagged Sgt1 (unpublished data). The specificity of the immunoprecipitation was confirmed by Western blot analysis with anti-Hsc82 antibody (Fig. 1A). Skp1 was also present in the myc-tagged Sgt1 immunoprecipitates (unpublished data), a result consistent with our previous finding (22). These results indicate that Sgt1 forms a complex with Hsp90 in vivo.
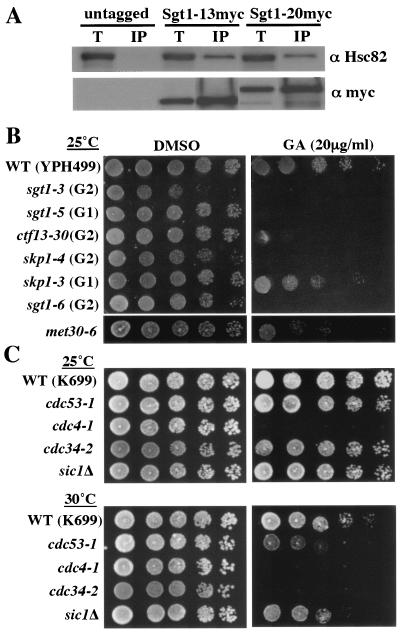
FIG. 1.
Sgt1 associates with Hsp90 in yeast. (A) Proteins from strains expressing untagged Sgt1 or myc-tagged Sgt1 (Sgt1-13myc and Sgt1-20myc) were immunoprecipitated with anti-myc antibody and then immunoblotted with anti-Hsc82 antibody. T, total lysate; IP, immunoprecipitate. (B) The sgt1 mutants and SCF mutants are sensitive to geldanamycin. The indicated strains were grown on yeast extract-peptone-dextrose plates containing 20 μg of geldanamycin per ml (GA) or dimethyl sulfoxide (DMSO) only. The numbers of cells that were spotted onto each plate (left to right) were approximately 5 × 104, 1 × 104, 2 × 103, 4 × 102, and 8 × 101. The plates were incubated at the indicated permissive temperatures for 3 days. (B) Strains in the YPH499 background. (C) Strains in the K699 background. (D) Strains in the YM4575 or US893 background. WT, wild type.
Sensitivity of sgt1 mutants to a specific inhibitor of Hsp90.
Five sgt1 temperature-sensitive mutants were isolated from the initial screen as described previously (22). One mutant, sgt1-3, was arrested in the G2 phase at the nonpermissive temperature (therefore, sgt1-3 is called a G2 allele), and this mutant showed substantial chromosome missegregation (22). The other mutants, sgt1-1, sgt1-2, sgt1-4, and sgt1-5, underwent arrest in the G1 phase at the nonpermissive temperature (therefore, sgt1-1, sgt1-2, sgt1-4, and sgt1-5 are called G1 alleles) (22). Genetic and biochemical experiments indicated that sgt1-3 is defective in kinetochore (CBF3) assembly and that sgt1-5 is deficient in SCF ubiquitination and adenylyl cyclase activity (6, 22). To further characterize the role of Sgt1 in kinetochore function, we obtained seven new temperature-sensitive mutants: sgt1-6, sgt1-7, sgt1-8, sgt1-9, sgt1-10, sgt1-11, and sgt1-12.
To examine whether the function of Sgt1 involves Hsp90, we tested whether sgt1 temperature-sensitive mutants are sensitive to geldanamycin, an Hsp90 inhibitor. All 12 sgt1 temperature-sensitive mutants showed substantial sensitivity to geldanamycin (Fig. 1B and unpublished data). These results are consistent with the previous finding that skp1 temperature-sensitive mutants and ctf13-30 have marked sensitivity to geldanamycin (38). Together with the interaction between Sgt1 and Hsp90, these results suggest that Hsp90 is required for the function of Sgt1.
Because Sgt1 is involved in the function of the SCF ubiquitin ligase complex that targets cell cycle regulators, i.e., Sic1 (a yeast CDK inhibitor) and Clns (yeast G1 cyclins), we tested the sensitivity of other SCF mutants to geldanamycin. Our observation that skp1-3 cells (deficient in SCF function) are moderately sensitive to geldanamycin (Fig. 1B) is consistent with the findings of Stemmann et al. (38). However, in contrast to their finding that cdc34-1, cdc4-1, and cdc4-2 did not show any growth defects on geldanamycin-containing medium (38), the SCF mutants, especially the F-box protein mutants cdc4-1, cdc4-12, met30-6, and grr1Δ, showed substantial sensitivity to geldanamycin at the permissive temperatures (Fig. 1C and D). Our results indicate that Hsp90 is required for cell viability when the SCF activity is compromised.
Genetic interaction between SGT1 and HSP90.
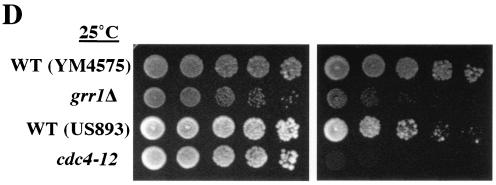
Because inhibition of yeast Hsp90 by geldanamycin resulted in a lethal phenotype in sgt1 mutants, we examined whether SGT1 and HSP90 show genetic interaction. The two budding yeast Hsp90 proteins, Hsc82 and Hsp82, are functionally equivalent (3), and Hsc82 is expressed 10 times as highly as Hsp82 at 25°C (3). The sgt1-3 hsp82Δ, sgt1-3 hsc82Δ, sgt1-5 hsp82Δ, and sgt1-5 hsc82Δ double mutants and the single mutants harbored YEP-HSC82 (URA HSC82). All of the mutants were streaked onto 5-FOA plates to select cells that could lose the HSC82 plasmid. When the cells were incubated at 25°C, sgt1-3 hsc82 and sgt1-5 hsc82 did not grow, although all of the other types of cells grew (Fig. 2A). These results indicate that sgt1 mutations exert synthetic lethality with hsc82Δ.
FIG. 2.
(A) The sgt1 and hsc82 mutations exert synthetic lethality. The indicated single and double mutants carrying YEP24-HSC82 were streaked onto plates containing Sc+5-FOA and Sc−Ura, respectively. The plates were incubated at 25°C for 2 days. (B) Hsp82 overexpression suppressed the temperature sensitivity of sgt1-3. The sgt1-3 cells containing p413-GPD-Hsp82 (GPD promoter/CEN/HIS3) and the vector alone were streaked onto Sc-His plates and incubated at the indicated temperatures for 3 days. (C) Hsp82 overexpression suppressed the chromosome missegregation phenotype of sgt1-3 cells. Shown are the results of a colony color-sectoring assay to analyze sgt1-3 cells that contained the vector alone and that contained p413-GPD-Hsp82. WT, wild type.
When we investigated the reciprocal phenotypes, we found that overexpression of Hsp82 can suppress the temperature sensitivity of sgt1-3 (a G2 allele, G2/M arrest at the nonpermissive temperature, kinetochore deficient) but not that of sgt1-5 (a G1 allele, G1 arrest at the nonpermissive temperature) (Fig. 2B and results not shown). Note that overexpression of yeast Hsp90 does not protect cells from heat stress (15). Importantly, overexpression of Hsp82 can suppress the chromosome missegregation phenotype of sgt1-3 (Fig. 2C). These results strongly suggest that Hsp90 activity is relevant to the functions of Sgt1, including its kinetochore function.
Binding of Skp1 and Hsc82 to the N-terminal domain of Sgt1.
Previous analyses of amino acid sequences revealed that Sgt1 has three conserved domains (Fig. 3C and 4A). The first is located at the N-terminal end (amino acids 13 to 141) and contains TPR motifs that are homologous (27% similarity) to those of HOP (Sti1 in budding yeast), an Hsp90-Hsp70-organizing protein (22). The second domain is the CS motif (amino acids 184 to 279), which is homologous (18% similarity) to a domain in the cochaperone p23 (6, 9). The TPR domain of HOP and the CS motif of p23 are required for each protein's interaction with Hsp90. Therefore, we hypothesized that one or both of these domains in Sgt1 are responsible for the association between Hsp90 and Sgt1.
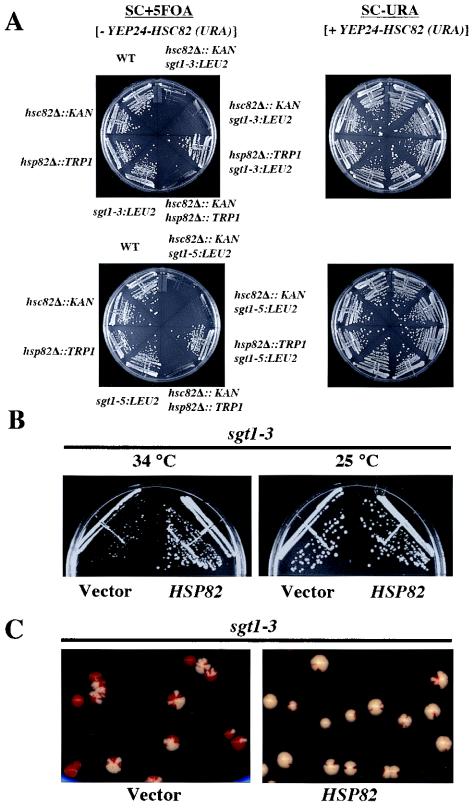
FIG. 3.
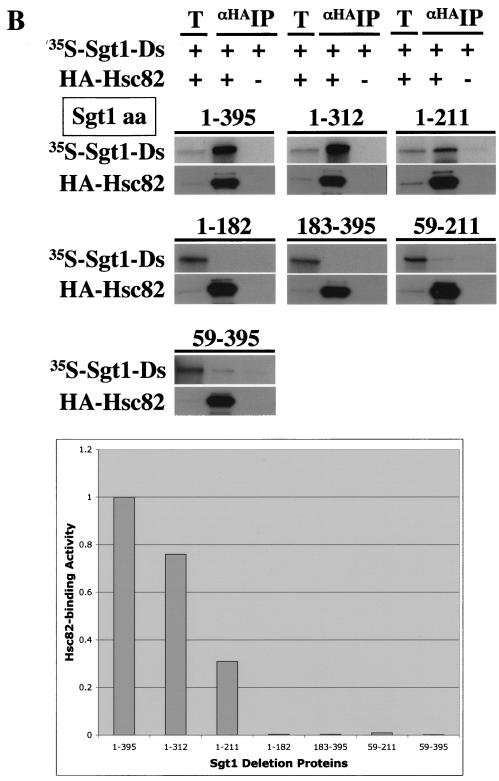
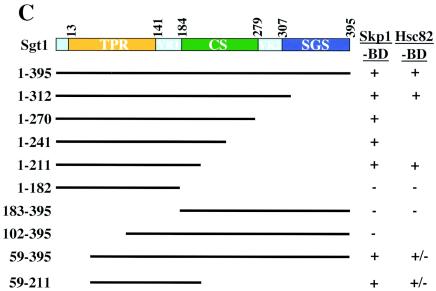
Skp1-binding and Hsc82-binding domains of Sgt1. (A, top) The indicated proteins were expressed and labeled with [35S]methionine in the in vitro translation system with rabbit reticulocyte lysates, and the radiolabeled proteins were mixed and incubated for 1 h at 30°C with an extract containing HA-tagged Skp1 that was not radiolabeled (20, 22). HA-tagged Skp1 was immunoprecipitated by using anti-HA-Sepharose, the immunoprecipitates were eluted and subjected to SDS-PAGE, and the radioactive bands were identified by autoradiography. HA-tagged Skp1 (HA-Skp1) was detected by immunoblotting with anti-HA antibody. T, total lysates (6% of the starting material); IP, immunoprecipitate; Ds, deletion proteins. The numbers above the lanes indicate the positions of the N- and C-terminal amino acids (aa) in each Sgt1 deletion protein. (A, bottom) Relative Skp1-binding activities of Sgt1 mutant proteins. Binding activity was defined as the ratio of the amount of coprecipitated protein to the amount of input protein. The binding activity of Sgt1 (wild type) was defined as 1, and the binding activities of the mutants were normalized to that value. (B, top) Essentially the same experiments as in panel A but with HA-tagged Hsc82 in place of HA-tagged Skp1. HA-tagged Hsc82 was expressed in the in vitro translation system but not radiolabeled. HA-tagged Hsc82 was detected by immunoblotting with anti-HA antibody. The numbers in parentheses indicate the positions of the N- and C-terminal amino acids in each Sgt1 deletion protein. (B, bottom) Relative Hsc82-binding activities of Sgt1 mutant proteins. Binding activity was calculated as described for panel A. (C) Illustration of the Sgt1 deletion proteins used in the Skp1- and Hsc82-binding assays and the results of these assays. The results of the Skp1-binding domain (Skp1-BD) and the Hsc82-binding domain (Hsc82-BD) analyses are summarized. Plus signs indicate significant binding activity. Minus signs indicate no significant binding activity. Plus-minus signs indicate weak binding activity.
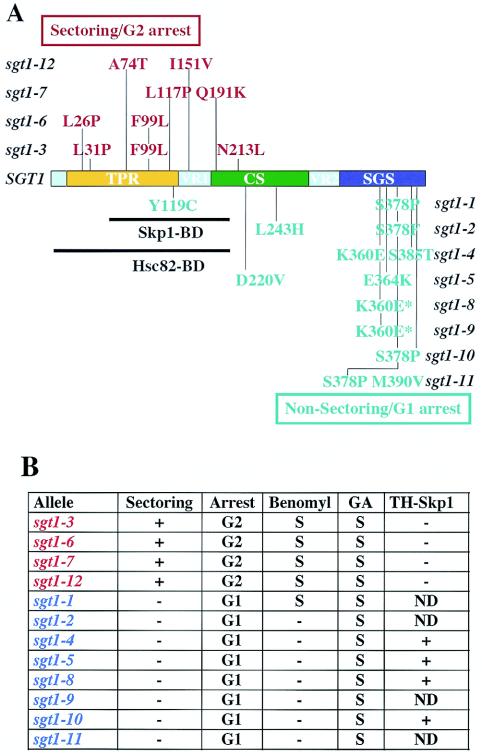
FIG. 4.
Mutations in temperature-sensitive sgt1 mutants. (A) The mutations in sgt1 G2 alleles are in red; the mutations in sgt1 G1 alleles are in blue. “Arrest” indicates the cell cycle stage at which the mutant cells accumulate when incubated at the nonpermissive temperature (unpublished data). Skp1-BD, Skp1-binding domain of Sgt1; Hsc82-BD, Hsc82-binding domain of Sgt1. The mutants indicated by the asterisk in panel A differ: sgt1-8 has two additional silent mutations at nucleotide positions 403 and 864. (Table 3 contains details of the sgt1 mutations.) (B) Summary of the phenotypes of sgt1 mutants. Plus signs in the sectoring column indicate that the mutant cell showed a chromosome missegregation defect that was shown by a colony color-sectoring assay. Minus signs in the sectoring column indicate that the cells did not show substantial chromosome missegregation. The letter S indicates sensitivity to benomyl or geldanamycin (GA); a minus sign indicates resistance. Because the sgt1-10 mutant, which has only the S378P mutation, is not sensitive to benomyl, we believe that the Y119C mutation is probably responsible for the benomyl sensitivity of the sgt1-1 mutant, which has two mutations (Y119C and S378P). Also shown are results of the two-hybrid assay (TH-Skp1) to assess the ability of sgt1 mutant proteins to bind to Skp1 (unpublished data). A plus sign in the TH-Skp1 column indicates that the mutant protein bound to Skp1 in the two-hybrid system. A minus sign indicates that the mutant protein did not do so. ND, not done.
We performed immunoprecipitation experiments with truncated Sgt1 proteins to identify the Skp1-binding and Hsc82-binding domains. Various deletion proteins (Fig. 3A and B) were expressed and labeled with [35S]methionine by the in vitro translation system, and the reticulocyte lysates containing the deletion proteins were mixed with a protein extract containing HA-tagged Skp1 that was expressed separately. HA-tagged Skp1 was immunoprecipitated by using Sepharose to which anti-HA antibody was conjugated (Fig. 3A). Analysis of the binding activity of the deletion proteins revealed that there is an inhibitory region between amino acids 270 and 312 (Fig. 3A). The Skp1-binding activity of the deletion protein (amino acids 1 to 270) is approximately twice that of the shorter deletion proteins (amino acids 1 to 241 or amino acids 1 to 211), a result that suggests that part of the CS domain contributes to the binding activity (Fig. 3A). Because the absence of the first 58 amino acids did not affect the binding activity (Fig. 3A and B), we conclude that the Skp1-binding domain of Sgt1 is located between amino acids 59 and 211 (Fig. 3C). The Skp1-binding domain contains two of the three repeats of the putative TPR domain and a short part (amino acids 184 to 211) of the CS domain (Fig. 3C).
By using the same immunoprecipitation method and HA-tagged Hsc82 instead of HA-tagged Skp1, we found that the N-terminal region (amino acids 1 to 211) of Sgt1 is able to bind to Hsc82 (Fig. 3B). However, the binding activity of the deletion protein (amino acids 1 to 211) is less than that of the longer deletion protein (amino acids 1 to 312) (Fig. 3B), a result that suggests that the CS domain contributes to the binding activity. Furthermore, a deletion protein lacking amino acids 1 through 58 but containing the Skp1-binding domain (amino acids 59 to 211) or the rest of the protein (amino acids 59 to 395) bound weakly to Hsc82 (Fig. 3B). Thus, we conclude that the full-length TPR domain and a short part (amino acids 184 to 211) of the CS domain are required for Sgt1's binding to Hsc82 (Fig. 3C). Together, these results indicate that the Hsc82-binding domain and the Skp1-binding domain are located within the N-terminal region of Sgt1.
Requirement of the N-terminal domain of Sgt1 for kinetochore function.
To assess the biological significance of the domains of Sgt1, we analyzed the phenotypes and sequences of sgt1 temperature-sensitive mutants. We previously showed that sgt1-3 cells, which undergo arrest in the G2/M phases at the nonpermissive temperature, lack the ability to assemble CBF3 and thus exhibit a chromosome-missegregation phenotype; in addition, sgt1-5 cells, which undergo arrest in the G1 phase at the nonpermissive temperature, are deficient in the function of the SCF complex and in activation of adenylyl cyclase (6, 22). Therefore, we categorized all 12 mutants on the basis of their chromosome stability.
The colony color-sectoring assay, which measures the stability of a marker chromosome fragment, revealed that only sgt1-3 (22), sgt1-6, sgt1-7, and sgt1-12 showed increased chromosome missegregation (Fig. 4A and B; unpublished data). These mutants consistently exhibited sensitivity to benomyl, a microtubule-destabilizing agent; this phenotype is typical of a kinetochore mutant. Except for sgt1-1, the other temperature-sensitive mutants were not sensitive to benomyl and underwent cell cycle arrest in the G1 phase at a nonpermissive temperature (Fig. 4A and B; unpublished data).
Analysis of the sgt1 sequences in the temperature-sensitive mutants revealed that mutations in the strains with the colony color-sectoring and G2 arrest phenotypes (G2 alleles) are in the N-terminal region of Sgt1, which contains the Skp1-binding and Hsc82-binding domains (Fig. 4A and B). In contrast, mutations in the strains with the nonsectoring and G1 arrest phenotypes (G1 alleles) are in the C-terminal region of Sgt1, which contains the SGS motif (2) that is highly conserved in Sgt1 homologs of eukaryotes (Fig. 4A and B). These results are consistent with our previous observation that Skp1 binds to sgt1-5 (G1 allele) protein but not to sgt1-3 (G2 allele) protein in vitro (22) and with our two-hybrid finding that Skp1 binds to G1 allele proteins but not to G2 allele proteins (Fig. 4B and unpublished data).
These results strongly suggest that the N-terminal region of Sgt1 containing the Skp1- and Hsc82-binding domains is important for kinetochore function and that the C-terminal region containing the SGS domain appears to be important for the G1 function of Sgt1.
Hsp90 stimulates Sgt1's binding to Skp1.
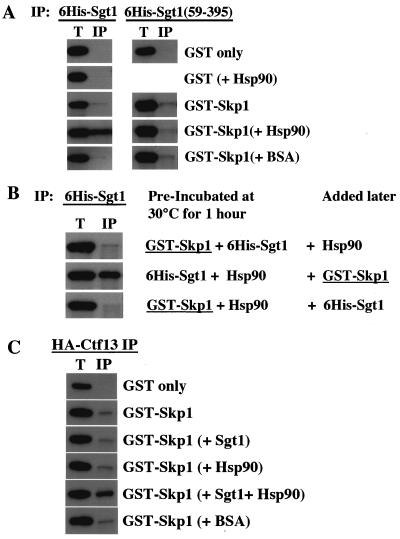
The analyses of the Skp1- and Hsc82-binding domains of Sgt1 raised the possibility that the chaperone protein Hsp90 may stimulate Sgt1's binding to Skp1. To test this hypothesis, we determined whether the addition of purified human Hsp90 protein stimulates Sgt1's binding to Skp1. GST-Skp1 and His6-Sgt1 were expressed in Escherichia coli and partially purified. These proteins were mixed together and incubated at 30°C for 1 h, and the interaction was tested by immunoprecipitation. We observed a weak but specific interaction between GST-Skp1 and His6-Sgt1 (Fig. 5A, left side; unpublished data). The addition of human Hsp90 stimulated Sgt1's binding to Skp1 (Fig. 5A, left side). The stimulation was approximately 4.8-fold (unpublished data). To confirm the specificity of this stimulation, we used a deletion protein that lacks amino acids 1 through 58 and has lost the ability to bind to Hsp90 substantially; the deletion protein was expressed in E. coli and partially purified. As expected on the basis of results of the binding domain analyses (Fig. 3A), we observed a weak but specific interaction between GST-Skp1 and His6-Sgt1(59-395). However, addition of human Hsp90 did not stimulate the binding of Sgt1(59-395) to Skp1 (Fig. 5A, right side, and unpublished data).
FIG. 5.
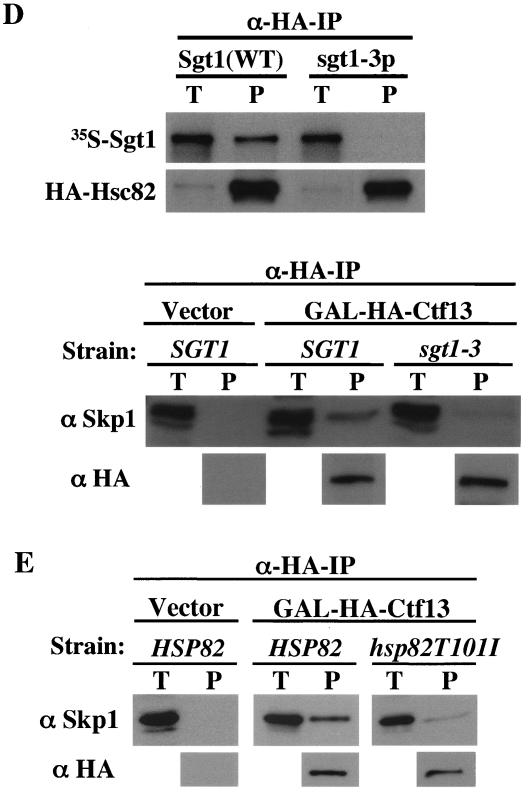
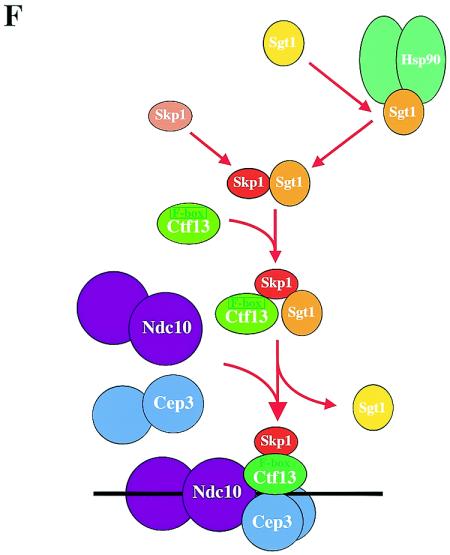
Sgt1, together with Hsp90, stimulates Skp1-Ctf13 binding. (A) Hsp90 requires the N-terminal region of Sgt1 to promote its interaction with Skp1. Approximately 200 ng of each indicated protein was used for immunoprecipitation (IP). After the samples were incubated at 30°C for 1 h, His6-tagged Sgt1 or His6-tagged Sgt1(59-395) was immunoprecipitated with anti-His antibody conjugated to Sepharose. GST-Skp1 and GST proteins were detected by immunoblotting. The bands in the top two panels are GST protein only, and the bands in the bottom three panels are the GST-Skp1 fusion protein. The names of proteins in parentheses indicate that these proteins were present in the reaction mixture but are not shown. BSA, bovine serum albumin. (B) Experiment similar to that described for panel A but with preincubation of two of the three indicated proteins at 30°C for 1 h and addition of the other later (immediately before immunoprecipitation). (C) Sgt1 and Hsp90 stimulate the binding of Ctf13 to Skp1. This experiment was conducted as described for panel A, except that HA-tagged Ctf13 was precipitated with anti-HA antibody conjugated to Sepharose. The bands in the top panel are GST protein only, and the bands in the bottom five panels are the GST-Skp1 protein. (D, top) sgt1-3 protein (sgt1-3p) does not bind to Hsc82 in vitro. Wild-type (WT) Sgt1 protein and sgt1-3 mutant protein were expressed and labeled with [35S]methionine in the in vitro translation system, and the radiolabeled proteins were mixed and incubated for 1 h at 30°C with an extract containing HA-tagged Hsc82 (HA-Hsc82) that was not radiolabeled. HA-Hsc82 was immunoprecipitated by using anti-HA-Sepharose, the immunoprecipitates were eluted and subjected to SDS-PAGE, and the radioactive bands were identified by autoradiography. HA-Hsc82 was detected by immunoblotting with anti-HA antibody. T, total lysates (6% of the starting material); P, immunoprecipitate. (D, bottom) The Skp1-Ctf13 association was reduced in sgt1-3 cells. HA-tagged Ctf13 expression was induced by addition of galactose; 1 h later, the temperature was shifted to 37°C and the cells were incubated for 90 min. HA-tagged Ctf13 was immunoprecipitated by using anti-HA antibody conjugated to Sepharose. Skp1 was detected by Western blot analysis with anti-Skp1 antibody. (E) The Skp1-Ctf13 association was reduced in hsp82T101I cells. HA-tagged Ctf13 expression was induced by addition of galactose; 1 h later, the temperature was shifted to 37°C and the cells were incubated for 90 min. HA-tagged Ctf13 was immunoprecipitated by using anti-HA antibody conjugated to Sepharose. Skp1 was detected by Western blot analysis with anti-Skp1 antibody. (F) Model of CBF3 assembly. Once Sgt1 forms a complex containing Hsp90, Sgt1 is activated to bind to Skp1. By binding to Sgt1, Skp1 is activated to bind to Ctf13 (34).
To confirm the importance of Sgt1-Hsp90 association for Sgt1's binding to Skp1, two of these three proteins were first incubated together and the other was added later. As expected, when His6-Sgt1 and Hsp90 were incubated together before Skp1 was added, the result was stimulated binding of Sgt1 to Skp1 (Fig. 5B). In contrast, no such stimulated binding was observed when either GST-Skp1 and His6-Sgt1 or GST-Skp1 and Hsp90 were incubated together initially. Taken together, these results support the hypothesis that Hsp90's binding to Sgt1 is required for the activation of Sgt1 in binding to Skp1.
Hsp90 and Sgt1 stimulate Skp1's binding to Ctf13.
Next we tested whether this Hsp90-induced activation of Sgt1's binding to Skp1 affects Skp1's binding to Ctf13, which is the F-box protein of the core kinetochore. We observed a weak but specific interaction between GST-Skp1 and His6-Ctf13 (Fig. 5C; unpublished data). The addition of His6-Sgt1 and human Hsp90 stimulated the binding of Skp1 to Ctf13 (Fig. 5C), a finding that strongly suggests that Hsp90 plays a role in promoting the association of the core kinetochore proteins. The stimulation was approximately 3.2-fold (unpublished data). On the basis of these in vitro data, we hypothesized that Hsc82 activates Sgt1 to bind to Skp1, and then Skp1 is activated to bind to Ctf13 (see Discussion; Fig. 5F).
The CBF3 complex fails to form in sgt1-3 cells, and the sgt1-3 protein cannot bind to Skp1 (22). Therefore, we tested whether the sgt1-3 protein can bind to Hsc82. We performed immunoprecipitation experiments by using Sgt1 (wild-type) and sgt1-3 proteins that were expressed and labeled with [35S]methionine by the in vitro translation system, and we found that the sgt1-3 protein cannot bind to Hsc82 in vitro (Fig. 5D). If our hypothesis (the Ctf13-Skp1 interaction is activated by Sgt1 and Hsp90) is true, the Skp1-Ctf13 interaction should be altered in sgt1-3 cells. In addition, because the CBF3 complex fails to form in hsp82T101I mutant cells (38), the Skp1-Ctf13 interaction should be reduced in hsp82T101I mutant cells. To confirm the biological relevance of our in vitro observations, we tested the Ctf13-Skp1 interaction in sgt1-3 cells and in hsp82T101I cells in vivo. Because the level of endogenous expression of Ctf13 is extremely low in cells, HA-tagged Ctf13 was overexpressed through the use of a galactose-inducible promoter and immunoprecipitated with anti-HA antibody. The amounts of Skp1-Ctf13 complex detected in sgt1-3 and hsp82T101I cells, respectively, were significantly less than that detected in wild-type cells (Fig. 5D, bottom, and E). These results strongly support the biological significance of our in vitro data that indicate the activation of the Ctf13-Skp1 interaction by Sgt1 and Hsp90. Taken together, these results strongly suggest that Sgt1 and its interactor Hsp90 are required for proper interactions among Sgt1, Skp1, and Ctf13; these interactions are the initial steps of assembly of the core kinetochore complex (see Discussion; Fig. 5F).
DISCUSSION
In this study we showed that Sgt1 associates with the molecular chaperone Hsp90 in yeast. Analyses of Sgt1 domains revealed that the N-terminal domain including TRP motifs is able to bind to Skp1 or Hsc82 (a yeast Hsp90 protein), and sequence analysis of sgt1 temperature-sensitive mutants suggested that the N-terminal domain of Sgt1 is important for kinetochore function. On the basis of this work, we have developed a model of CBF assembly mediated by Sgt1 and Hsp90.
Model of CBF3 assembly.
By activating Ctf13, Sgt1, together with Skp1, is required for the formation of the CBF3 complex (22). Hsp90 is also required for activation of the Ctf13-Skp1 complex and thus the formation of the CBF3 complex (38). Here we showed that Sgt1 associates with Hsc82 (a yeast Hsp90 protein) in vivo and that the Hsc82-binding domain of Sgt1 contains its Skp1-binding domain. The addition of Hsp90 stimulated Sgt1-Skp1 binding. Moreover, Sgt1 and Hsp90 stimulated Skp1-Ctf13 binding. Therefore, we propose that in vivo Sgt1 binds to Hsc82 first to form the proper structure for binding to Skp1 and that by binding to Skp1, Sgt1 activates the Skp1-Ctf13 complex for subsequent formation of the CBF3 complex (Fig. 5F) (34). Skp1 can form a complex with Sgt1 and an F-box protein (e.g., Ctf13 is an F-box protein; Fig. 5F, middle) (22). Because Sgt1 is not in the CBF3 complex (22), Sgt1 is expected to dissociate from the complex after assembly (Fig. 5F, bottom).
Possible role of Sgt1 as a cochaperone.
Sgt1 participates in multiple activities: kinetochore complex assembly, SCF activity, adenylyl cyclase activity in yeasts, and R gene-triggered disease resistance in plants (1, 2, 6, 22). Our present results suggest that Hsp90 stimulates the binding of Sgt1 to Skp1 and the binding of Skp1 to Ctf13 via Sgt1, which is essential for the assembly of the CBF3 complex. Sgt1 did not appear to be unstable in geldanamycin-treated cells (results not shown), an observation that suggests that Sgt1 is not solely an Hsp90 substrate (often called a “client” protein) that is stabilized by the chaperone (30). Hsp90 contains three distinct domains: an amino-terminal ATPase domain, a client-binding domain in the middle, and the C-terminal end containing a dimerization domain and an MEEVD motif that binds to TPR domains of many cochaperones (29). The significant homology of Sgt1's TPR domain to that of the Hsp70-Hsp90-organizing protein HOP (Sti1 in yeast) and the stable physical interactions between Sgt1 and Hsp90 in vivo suggest that Sgt1 acts as a cochaperone (22, 38). However, the expression pattern of Sgt1 in yeast under various stress conditions is very different from those of the known cochaperones, Aha1, Hch1, Sti1, and Crp6 (10, 31). Therefore, Sgt1 does not seem to be required for stress responses. We could not detect physical interactions in yeast cells between Sgt1 and the cochaperones Cdc37, Sba1 (p23 in humans), Aha1, and Sti1 (HOP in humans) (data not shown). Cdc37, p23, and HOP inhibit the ATPase activity of Hsp90, but Aha1 stimulates this activity of this chaperone (31, 32). Therefore, it is of interest to examine whether Sgt1 affects the ATPase activity of Hsp90. In addition, we did not observe any difference in the results of our binding assays when ATP was present or absent (data not shown). Lee et al. also reported that human Sgt1's binding to Hsp90 does not depend on ATP (25).
Sensitivity of SCF mutants to geldanamycin.
We found that the N-terminal domain of Sgt1 is required for its interaction with Hsp90 and that the mutations of the sgt1 G2 alleles are located in the region that encodes this domain. These findings imply that only the G2/kineothcore function (but not the G1 function) of Sgt1 is involved in the activity of Hsp90. However, all of the sgt1 G1 alleles are also sensitive to geldanamycin. Because mutations in these strains occurred in the SGS domain, which is located at the C-terminal end, the function of the SGS domain may be related to the function of Hsp90 by an unknown mechanism. Because the sgt1-5 mutant, one of the sgt1 G1 alleles, is deficient in SCF activity (22), we examined whether the SCF mutants are sensitive to geldanamycin. All of the SCF mutants that we tested (skp1-3, cdc53-1, cdc34-2, cdc4-1, cdc4-12, met30-6, and grr1Δ) were sensitive to geldanamycin, and the F-box protein mutants (cdc4-1, cdc4-12, met30-6, and grr1Δ) were especially sensitive to geldanamycin. Skp1 binds to F-box proteins via the F-box motif (7, 35). Because Sgt1 is required for activation of Ctf13, an F-box protein, Sgt1 may also be required for activation or stabilization of other F-box proteins. Recently, Melville et al. reported that Hsp70 and TriC/CCT chaperones are required for assembly of the von Hippel-Lindau tumor suppressor complex, which is structurally analogous to the SCF complex (28). Further investigation is required to address whether the chaperone activity of Hsp90 or another, related, protein is required for the function of the SCF complex.
Role of human SGT1 in kinetochore function.
Human SGT1 (SUGT1) can rescue the sgt1-null mutant (22), a result that suggests that the function of Sgt1 is conserved between humans and yeast. SGT1 associates with Hsp90 in human cells (S. Ohta and K. Kitagawa, unpublished data). Very recently, Lee et al. reported that the CS domain, but not the TPR domain, of human SGT1 binds to Hsp90; this result is not consistent with our observation about yeast Sgt1 (25). We do not know whether this discrepancy reflects differences between the two organisms. Further analysis is required to clarify the discrepancy.
Interestingly, Steensgaard et al. found that CENP-I, CENP-F, and Hec1, but not CENP-C, are absent from kinetochores in human Sgt1-depleted cells. This absence suggests that human Sgt1 is a critical factor for assembly of mammalian kinetochores (36a). These data strongly suggest that Sgt1 is functionally conserved between humans and yeast. Additional analysis is needed to investigate the molecular mechanism.
Other functions of the Sgt1-chaperone complex.
In addition to its role in the function of the kinetochore and of the SCF complex, Sgt1 is involved in activation of adenylyl cyclase in budding and fission yeasts and in R gene-triggered resistance in plants (1, 2, 6, 22). Recently, three research groups independently found that Hsp90 is required for R gene-triggered resistance in plants (14, 27, 40). Notably, Sgt1 coimmunoprecipitates with Hsp90 proteins in Arabidopsis and barley (40). Sgt1 is highly conserved in eukaryotes, and the Sgt1-chaperone interaction appears to be conserved in yeast, plants, and humans. In this paper, we provide evidence that suggests an initial activation step in the assembly of the CBF3 complex-Sgt1- and Hsp90-mediated activation of the Skp1-Ctf13 association. Similar molecular mechanisms may be involved in the formation of other complexes in which Sgt1 participates. Moreover, these mechanisms may be conserved in other organisms.
Acknowledgments
We thank V. Measday, A. Musacchio, and K. Shirasu for helpful comments; J. Nitiss and M.-A. Bjornsti for stimulating conversation and advice; A. Mishra, V. Pagala, and C. Slaughter for mass spectrometry; I. Yahara, J. Imai, K. Kohno, S. Lindquist, E. A. Craig, K. B. Kaplan, J. Lechner, K. A. Morano, U. Surana, P. Kaiser, K. Nasmyth, and M. Tyres for generous gifts of reagents; and J. C. Jones for editing the manuscript.
This work was supported by Cancer Center support grant CA21765 from the National Cancer Institute, by NIH grant GM68418, and by the American Lebanese Syrian Association Charities (ALSAC).
REFERENCES
- 1.Austin, M. J., P. Muskett, K. Kahn, B. J. Feys, J. D. Jones, and J. E. Parker. 2002. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295:2077-2080. [DOI] [PubMed] [Google Scholar]
- 2.Azevedo, C., A. Sadanandom, K. Kitagawa, A. Freialdenhoven, K. Shirasu, and P. Schulze-Lefert. 2002. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073-2076. [DOI] [PubMed] [Google Scholar]
- 3.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doheny, K. F., P. K. Sorger, A. A. Hyman, S. Tugendreich, F. Spencer, and P. Hieter. 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell 73:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubacq, C., R. Guerois, R. Courbeyrette, K. Kitagawa, and C. Mann. 2002. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1:568-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29:235-244. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Ranea, J. A., G. Mirey, J. Camonis, and A. Valencia. 2002. p23 and HSP20/α-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529:162-167. [DOI] [PubMed] [Google Scholar]
- 10.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh, P. Y., and J. V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Goh, P. Y., and U. Surana. 1999. Cdc4, a protein required for the onset of S phase, serves an essential function during G2M transition in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5512-5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegemann, J. H., J. H. Shero, G. Cottarel, P. Philippsen, and P. Hieter. 1988. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2523-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hieter, P., D. Pridmore, J. H. Hegemann, M. Thomas, R. W. Davis, and P. Philippsen. 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42:913-921. [DOI] [PubMed] [Google Scholar]
- 14.Hubert, D. A., P. Tornero, Y. Belkhadir, P. Krishna, A. Takahashi, K. Shirasu, and J. L. Dangl. 2003. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imai, J., and I. Yahara. 2000. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell. Biol. 20:9262-9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, W., J. Lechner, and J. Carbon. 1993. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J. Cell Biol. 121:513-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Kaiser, P., R. A. Sia, E. G. Bardes, D. J. Lew, and S. I. Reed. 1998. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swel. Genes Dev. 12:2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa, K., and R. Abdulle. 2002. In vivo site-directed mutagenesis of yeast plasmids using a three-fragment homologous recombination system. BioTechniques 33:288, 290, 292. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa, K., R. Abdulle, P. K. Bansal, G. Cagney, S. Fields, and P. Hieter. 2003. Requirement of Skp1-Bub1 interaction for kinetochore-mediated activation of the spindle checkpoint. Mol. Cell 11:1201-1213. [DOI] [PubMed] [Google Scholar]
- 21.Kitagawa, K., and P. Hieter. 2001. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell. Biol. 2:678-687. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 23.Lechner, J. 1994. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 13:5203-5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64:717-725. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y. T., J. Jacob, W. Michowski, M. Nowotny, J. Kuznicki, and W. J. Chazin. 2004. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 279:16511-16517. [DOI] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Lu, R., I. Malcuit, P. Moffett, M. T. Ruiz, J. Peart, A. J. Wu, J. P. Rathjen, A. Bendahmane, L. Day, and D. C. Baulcombe. 2003. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22:5690-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melville, M. W., A. J. McClellan, A. S. Meyer, A. Darveau, and J. Frydman. 2003. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, P., C. Prodromou, B. Hu, C. Vaughan, S. M. Roe, B. Panaretou, P. W. Piper, and L. H. Pearl. 2003. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11:647-658. [DOI] [PubMed] [Google Scholar]
- 29a.Morano, K. A., N. Santoro, K. A. Koch, and D. J. Thiele. 1999. A transactivation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19:402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29b.Nasmyth, K., G. Adolf, D. Lydall, and A. Seddon. 1990. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell 62:631-647. [DOI] [PubMed] [Google Scholar]
- 29c.Nathan, D. F., and S. Lindquist. 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15:3917-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neckers, L. 2002. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol. Med. 8:S55-S61. [DOI] [PubMed] [Google Scholar]
- 31.Panaretou, B., G. Siligardi, P. Meyer, A. Maloney, J. K. Sullivan, S. Singh, S. H. Millson, P. A. Clarke, S. Naaby-Hansen, R. Stein, R. Cramer, M. Mollapour, P. Workman, P. W. Piper, L. H. Pearl, and C. Prodromou. 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10:1307-1318. [DOI] [PubMed] [Google Scholar]
- 32.Pearl, L. H., and C. Prodromou. 2001. Structure, function, and mechanism of the Hsp90 molecular chaperone. Adv. Protein Chem. 59:157-186. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Russell, I. D., A. S. Grancell, and P. K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 36.Sorger, P. K., K. F. Doheny, P. Hieter, K. M. Kopski, T. C. Huffaker, and A. A. Hyman. 1995. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA 92:12026-12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Steensgaard, P., M. Garre, I. Muradore, P. Transidico, E. A. Nigg, K. Kitagawa, W. C. Earnshaw, M. Farretta, and A. Musacchio. 2004. Human Sgt1 is required for kinetochore assembly. EMBO Rep. 5:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stemmann, O., and J. Lechner. 1996. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 15:3611-3620. [PMC free article] [PubMed] [Google Scholar]
- 38.Stemmann, O., A. Neidig, T. Kocher, M. Wilm, and J. Lechner. 2002. Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. USA 99:8585-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strunnikov, A. V., J. Kingsbury, and D. Koshland. 1995. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128:749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, A., C. Casais, K. Ichimura, and K. Shirasu. 2003. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:11777-11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated GI cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]