Abstract
The human granulocyte-macrophage colony-stimulating factor (GM-CSF) gene is activated by an NFAT-dependent enhancer forming an inducible DNase I hypersensitive (DH) site. The enhancer core comprising the DH site contains the GM330 and GM420 elements that bind NFAT and AP-1 cooperatively. Here we demonstrate that both elements are essential for enhancer activity and that Sp1 and AML1 sites in the enhancer become occupied in vivo only after activation. Chromatin structure analysis revealed that the GM-CSF enhancer core elements are divided between two adjacent nucleosomes that become destabilized and highly accessible after activation. Inducible chromatin reorganization was not restricted to the enhancer core but extended across a 3-kb domain of mobilized nucleosomes, within which the nucleosome repeat length was compressed from approximately 185 to 150 bp. The GM420 element is a high-affinity site that binds NFAT independently of AP-1 but depends on the linked AP-1 site for enhancer function. Nevertheless, just the NFAT motif from the GM420 element was sufficient to form a DH site within chromatin even in the absence of the AP-1 site. Hence, NFAT has the potential to cooperate with other transcription factors by promoting chromatin remodelling and increasing accessibility at inducible regulatory elements.
Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a cytokine that regulates the proliferation and differentiation of the myeloid compartment during hematopoiesis and that mediates the activation and survival of monocytes/macrophages and granulocytic cells at sites of inflammation (23, 44). Because GM-CSF functions as a powerful proinflammatory cytokine, its expression is by necessity very tightly regulated and highly inducible. GM-CSF is produced by activated T cells in response to activation of the T-cell receptor (TCR), and its expression is inhibited by the immunosuppressive drug cyclosporine A (CsA) which specifically blocks the Ca2+/calcineurin signaling pathway (15, 40). CsA suppresses expression of GM-CSF and many other cytokines primarily by inhibiting the Ca2+/calcineurin-dependent induction of the NFAT family of transcription factors (12, 13, 15-18, 25, 29, 35, 40, 41, 45-50). NFAT typically acts synergistically with other families of transcription factors, such as AP-1, Oct, Egr, Maf, and GATA to regulate inducible tissue-specific gene expression (4, 17, 29). NFAT is found as a preexisting protein in the cytoplasm of inactive cells and rapidly translocates to the nucleus upon activation of calcineurin by Ca2+ flux (17, 29, 45, 46). The predominant species of NFAT present in T cells are NFATc1 (also known as NFATc or NFAT2) and NFATc2 (also known as NFATp or NFAT1) (17, 29, 45, 46).
Studies of transgenic mice and cell lines showed that efficient activation of the human GM-CSF gene is dependent upon a highly inducible enhancer located 3 kb upstream that contains an array of NFAT sites (12, 13, 15). Enhancer activity is induced in NFAT-expressing cell types, such as T cells, myeloid cells, and endothelial cells by Ca2+ and kinase signaling pathways which in T cells are coupled to the TCR (13). The GM-CSF enhancer was initially identified as an inducible CsA-sensitive DNase I hypersensitive (DH) site spanning at least 250 bp of DNA (12, 13, 15). The DH site appears in every cell type in which the enhancer is known to function, and to a large degree, this correlates with NFAT expression (4, 13). Many other cytokine genes similarly contain NFAT-dependent promoters and enhancers that exist as inducible CsA-sensitive DH sites (4, 18, 25, 35, 48, 49, 50). Hence, NFAT is likely to be required for both the chromatin remodelling and function of promoters and enhancers in cytokine genes, such as the GM-CSF gene.
The human GM-CSF enhancer encompasses four NFAT binding sites termed the GM170, GM330, GM420, and GM550 elements according to their locations within a 717-bp BglII fragment of DNA (GenBank accession no. L07488) that originally defined the enhancer (12, 15) (Fig. 1). Each one of these NFAT sites lies adjacent to an AP-1 binding site (Fig. 1). Three of these NFAT sites exist as true composite elements (GM330, GM420, and GM550) that have the capacity to bind NFAT and AP-1 cooperatively and to function in isolation as enhancer elements (4, 12, 15). These three elements each loosely conform to the consensus sequence GGAAANNNTGAGTCA (Fig. 1) that directs cooperative binding of NFAT and AP-1 (12). These sites, together with the −90, −160, and −280 NFAT/AP-1 sites in the interleukin-2 (IL-2) promoter (12, 31, 41, 47) and a site in the IL-5 promoter (36), define a family of composite elements that bind NFAT and AP-1 cooperatively via protein-protein contacts between the DNA-binding domains (9, 29, 46). The unifying characteristic of this distinctive family of elements is that the spacing between the AP-1 and NFAT elements is perfectly conserved, whereas the individual binding sites are often poorly conserved. Hence, the individual factors typically bind poorly on their own, and in each case, cooperativity increases the binding of both NFAT and AP-1. This family of composite elements does, however, cover the complete spectrum of binding affinities which range from the GM330 element, which is an ideal AP-1 site but cannot bind NFAT alone, to the human IL-2 promoter −280 element which is a moderate-affinity NFAT site but cannot bind AP-1 alone. The GM420 element, however, is a perfect consensus high-affinity NFAT site linked to a moderate-affinity AP-1 site and does not rely on AP-1 for binding. The importance of NFAT/AP-1 cooperativity in gene regulation was revealed by a study that demonstrated that mutations in the AP-1-interacting domain of NFATc2 led to loss of expression of many cytokine genes, including GM-CSF and IL-2 (39).
FIG. 1.
Map of the human GM-CSF enhancer. The locations of regulatory elements and the DH site in the GM-CSF enhancer are shown. Sites are labeled according to their positions in the 717-bp BglII fragment that defines the enhancer. DH sites (vertical arrows) and the probe used to map DH sites (box) are indicated. Composite NFAT/AP-1 sites are depicted below the map. HS, hypersensitive.
The role of the GM-CSF enhancer in GM-CSF gene regulation is now established (13), and the properties of the intact enhancer are well characterized (4, 12, 13, 15). However, the DNA elements required for its activity and the functions contributed by the factors that bind to them have never been determined. Furthermore, the family of composite NFAT/AP-1 elements as a whole has not been sufficiently analyzed to determine the specific contributions of the individual NFAT and AP-1 sites to their function. Although NFAT and AP-1 can each recruit the CBP/p300 family of histone acetyltransferases (2, 21), the frequent conjoining of NFAT and AP-1 elements would suggest that they each also direct some unique function. It remains unknown, however, whether NFAT or AP-1 alone is sufficient to activate any of these composite elements. Only in the case of the IL-2 −280 NFAT/AP-1 element is it established that deletion of the AP-1 site is sufficient to destroy activity (6), but this result could be due to the cooperative nature of NFAT and AP-1 binding to this particular site. Although the specific functions of NFAT are poorly understood, it is clear that composite NFAT/AP-1 elements provide a powerful mechanism for tightly regulating gene expression and serve as a point where the Ca2+ and kinase signaling pathways converge in an interdependent manner in the activation of cytokine genes.
The early events in the process of gene activation are known to be dependent upon chromatin remodelling, but in most cases, the mechanisms underlying the formation of DH sites are not known in any detail. The GM-CSF enhancer DH has been defined as a 250-bp region of DNA, but the underlying chromatin structure has never been investigated, and no in vivo analysis of transcription factor binding to the enhancer has been undertaken. It also remains to be determined which DNA elements are actually required for the formation of the DH site and which factors have the capacity to bind to the enhancer in a chromatin context either before or after induction of the DH site.
In this study, we have addressed some of these issues. Activation of the enhancer was associated with in vivo binding of both inducible and constitutive factors, suggesting that transcription factor recruitment is cooperative. To gain further insights into the molecular basis of inducible enhancer activation, we investigated the functions of individual NFAT elements in the GM-CSF enhancer. We identified a high level of cooperation in the functions of the individual AP-1 and NFAT components of composite elements that was much greater than what might be expected from cooperative binding alone. Enhancer activation appears to rely on an NFAT-dependent chromatin remodelling activity that creates a DH site and thereby promotes binding of other factors required for enhancer function. Surprisingly, we also found that at least 3 kb of the nucleosome array encompassing the enhancer is extensively reorganized upon activation.
MATERIALS AND METHODS
Cell culture.
Jurkat T cells and Ball-1 B cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, penicillin and streptavidin at 500 U/ml each (Gibco), 25 mM HEPES, and 2 mM l-glutamine. Human peripheral blood T cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, 25 mM HEPES, 50 μM β-mercaptoethanol, 10 mM sodium pyruvate (Gibco), penicillin and streptavidin at 500 U/ml each (Gibco), 2 mM l-glutamine, 0.2% glucose, and minimal essential medium (MEM) essential amino acids and nonessential amino acids (Gibco). T cells were prepared from 20- or 50-ml peripheral blood samples obtained from healthy volunteers as follows. Mononuclear cells were purified by gradient centrifugation over Lymphoprep (Gibco) and cultured for 2 to 3 days in the presence of 2 μg of phytohemagglutinin per ml at a density of between 0.5 and 2 × 106 cells/ml. Nonadherent cells were then spun down and cultured for 4 to 7 days in fresh medium in the presence of 10 U of recombinant human IL-2 (Boehringer Mannheim) per ml at a density of 0.5 × 106 to 1.5 × 106/ml. Purity was assessed by staining for CD3. Under these conditions, we typically obtained cultures of approximately 97% CD3-positive T cells. Human GM-CSF transgenic mouse T cells were prepared by a similar procedure, by first culturing splenocytes in the presence of 2 μg of concanavalin A per ml for 2 days and then in the presence of 10 U of recombinant mouse IL-2 (Boehringer Mannheim) per ml at a density of 0.5 × 106 to 1.5 × 106/ml. For this purpose, we bred the previously described transgenic mouse line M268 (13) to homozygosity to create a line carrying approximately 100 copies of the human GM-CSF gene.
mRNA analyses.
GM-CSF mRNA expression relative to glyceraldehyde phosphate dehydrogenase (GAPDH) expression was measured by real-time PCR analysis as follows. To induce GM-CSF expression, cells were stimulated with 20 ng of phorbol 12-myristate 13-acetate (PMA) per ml and 2 μM calcium ionophore A23187. RNA was extracted from cells with TRIzol reagent (Invitrogen) and purified according to the manufacturer's instructions. cDNA was prepared using oligo(dT) and reverse transcriptase. Real-time quantitative PCR was performed using a SYBR green-based approach and detected on an ABI Prism 7700 sequence detection system (Perkin-Elmer) according to the manufacturer's instructions. A preliminary step (heating the mixture to 50°C for 2 min) preceded a 10-min 95°C heating step. Forty PCR cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C, were then performed. Human GAPDH and GM-CSF gene oligonucleotide primers used for PCR were as follows: for GM-CSF, CACTGCTGCTGAGATGAATGAAA and GTCTGTAGGCAGGTCGGCTC; for GAPDH, AACAGCGACACCCACTCCTC and CATACCAGGAAATGAGCTTGACAA.
In vivo footprinting analyses.
The treatment of intact cells with dimethyl sulfate (DMS) and the subsequent ligation-mediated PCR (LM-PCR) were performed by the method of Hershkovitz and Riggs (28) essentially as described by Kontaraki et al. (34). This method can be briefly summarized as follows. Cells were treated with 0.2% DMS in phosphate-buffered saline (PBS) (Invitrogen) at room temperature for 5 min. The reactions were stopped by the addition of 20 volumes of ice-cold PBS, and two subsequent washes with PBS were performed. Cells were placed in lysis buffer overnight at room temperature, and DNA was extracted from the lysed cells. Methylated guanidine residues were cleaved by incubation in 0.1 M piperidine for 10 min at 90°C, and DNA was then butanol extracted and ethanol precipitated. A cleaved methylated guanidine DNA control prepared by in vitro DMS treatment of purified DNA was generated by the method of Maxim and Gilbert (42). DNA fragments were detected by LM-PCR by the method of Hershkovitz and Riggs (28) with the modifications of Kontaraki et al. (34) and using a linker duplex consisting of the 25-base LP25 nucleotide GCGGTGACCCGGGAGATCTGAATTC annealed to the 21-base LP21 nucleotide GAATTCAGATCTCCCGGGTCA. LM-PCR was performed with 1 μg of DNA template by using a biotinylated primer to extend one round of DNA synthesis to the cleavage sites, ligating the LP25-LP21 linker duplex to the extension products, binding the biotinylated products to magnetic beads, performing a nested PCR for 18 to 22 cycles with an enhancer-specific primer and the LP25 primer, performing six cycles of another nested PCR with a 32P-end-labeled primer, and analyzing the products on a 6% polyacrylamide gel under DNA-denaturing conditions with the aid of a phosphorimager. Betaine (1.4 M) and 5% dimethyl sulfoxide were used in the amplifying LM-PCRs in order to increase oligonucleotide primer stringency and specificity.
LM-PCR analyses were performed with sets of three nested primers specific for the GM-CSF enhancer, plus the linker primer. To cover the entire enhancer, we employed three primer sets which are complementary to the lower strand and amplify in the forward direction (1F, 2F, and 3F) and two primer sets complementary to the upper strand which amplify in the reverse direction (1R and 2R). The three primers in each set are designated B for the biotinylated primer used at the first stage, P for the primer used at the second PCR stage, and L for the primer used at the final labeling stage. The three oligonucleotide primers in each set of primers follow (the sequences of the primers, their coordinates within the enhancer, and the annealing temperatures used for the primers are shown in parentheses): 1R primers, EB1R (ACGGAAGTTATTCAGGAAGAAGAGC, positions 677 to 653, 56°C), EP1R (CTGTGTTTAAGGGCTGGG, positions 637 to 620, 55°C), and EL1R (ATAACAGATGGGCAAGTAACAGGC, positions 617 to 594, 56°C); 2R primers, EB2R (GCCCAAGTCAGCACAAACAGG, positions 507 to 486, 57°C), EP2R (GTCAGCACAAACAGGACAGAAATC, positions 501 to 478, 56°C), and EL2R (GGGTTTGGTGATGAGGTTTGTGGGC, positions 474 to 450, 60°C); 1F primers, EB1F (GTCCCCAGAGATAACAAGAAC, positions 10 to 31, 54°C), EP1F (CAGGTCAAAGAAGGATCAGATCAGC, positions 48 to 71, 57°C), and EL1F (CAAAGCCTGTGCGGGGGTGCC, positions 75 to 86, 62°C); 2F primers, EB2F (TTGTGATAATGTCTCTCGTGATAAGG, positions 134 to 149, 54°C), EP2F (GGGTGGACACGCATAGG, positions 202 to 218, 55°C), EL2F (AAACTCCTTCCAGAGGGTTTTCTCC, positions 219 to 243, 57°C); and 3F primers, EB3F (CCCATCGTGGTCCCCGCCTCTGC, positions 283 to 305, 65°C), EP3F (CCACAGCCCCATCGGAGC, positions 310 to 327, 59°C), and EL3F (GCATGGCTGGCTATCGGTTGACACTG, positions 338 to 363, 62°C).
EMSAs.
Gel electrophoretic mobility shift assays (EMSAs) of nuclear extracts were performed as previously described (4) using the stromelysin gene AP-1 site GCAAGGATGAGTCAAGCTGCGGGTGATCC (4, 15), the GM430 NFAT probe CACACATCTTTCTCATGGAAAGATGA (which represents a truncated form of the GM420 NFAT element lacking the AP-1 binding site) (4, 12), the consensus Sp1 sequence GCTCGCCCCGCCCCGATCGAATG, and the GM-CSF promoter AML1 site CCTCCCTGGCATTTTGTGGTCACCATTAAT (14). (Underlining indicates the locations of binding sites.) EMSAs of recombinant NFAT and AP-1 were performed as previously described using the Rel domain of NFATc2 as a source of NFAT and the DNA-binding and leucine zipper domains of c-Fos and c-Jun as a source of AP-1 (12).
Transient-transfection assays.
Luciferase reporter gene plasmids were transiently transfected into Jurkat T cells approximately 20 h prior to stimulation with 20 ng of PMA per ml and 1 μM calcium ionophore A23187 for 9 h. Transfection assays and luciferase assays were performed as described previously (4, 13, 18), using 5 μg of DNA per transfection. Plasmids were purified via two cycles of CsCl gradient centrifugation, and at least two separate preparations of each plasmid were tested. Each plasmid construct was assayed in several independent series of transfection assays, and in each case, a total of at least 12 individual assays were performed. Plasmids were based on the luciferase reporter gene plasmid pXPG (5), which has a novel high-copy-number origin of replication and the Luc+ gene from pGL3, but it does not suffer from the high background read-through transcription activities associated with pGL3 (5), because it has two upstream simian virus 40 polyadenylation or termination elements. The full-length GM-CSF promoter plasmid pXPG-GM627 (5) contains a segment of the promoter from positions −627 to +28 (this plasmid is referred to as pGM in reference 13), and pXPG-GM627-B717 is the same plasmid with the 717-bp BglII fragment of the GM-CSF enhancer inserted upstream of the promoter (referred to as pGME in reference 13). The minimal GM-CSF promoter plasmid pXPG-GM55 (4, 5) contains a segment of the GM-CSF promoter from positions −55 to +28.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) as outlined in the manufacturer's instructions. Site-directed mutagenesis was performed on a pGEM plasmid containing a 3.7-kb HindIII genomic fragment incorporating the 717-bp BglII GM-CSF enhancer region. Clones of mutated plasmids were sequenced across the enhancer to confirm that the desired mutations were accurately introduced, and the 717-bp BglII fragments of the enhancer were then subcloned upstream of the GM-CSF promoter in the pXPG-GM627 GM-CSF promoter/luciferase reporter gene plasmid to create the pXPG-GM627-B717 family of plasmids. The sites and the sequences of the oligonucleotide primer pairs used for mutagenesis, with the binding sites underlined and the mutations shown in lowercase, are listed below (note that the sequence shown represents just one of the two complementary primer pairs required for the mutagenesis technique): Ets site, GGACTTTGAGGCCCATggCCTTCCCCTTGTGATAATG; GATA sites, CCTTCCCCTTGTctTAATGTCTCTCGTctTAAGGATCCTGGAG; GM170 AP-1 site, GGATCCTGGAGTagCcCAAGCCCCTGTTTCCTAC; GM170 NFAT site, GTGACTCAAGCCCCTGTggCCTACAGAGCCTGTGG;Sp1 site, CCCATCGTGGTCCCCGaaTCTGCCCTGCCACAGC; GM330NFAT site, GCCACAGCCCCATCGGccCCCCTGAGTCAGCATGG; GM330 AP-1 site, CCATCGGAGCCCCTGgGTtAGCATGGCTGGCTATC; GM420 AP-1 site, GGCCTCAGCCCTagTGcgATCTTTCCATGAGAAAG; GM420 NFAT site, GCCCTGATGTCATCTggCCAGAGAAAGATGTGTGGC; GM450 AML1 site, GATGTGTGGCTGCggACAAACCTCATCACC; andGM550 NFAT site, GATTTCTTATTATGACTCTTGCTggCCTCCTggCCTTCCCCTCCTAGGCC. With the exception of the potential Ets site, for which binding data are not available, all of the above sites are known to bind factors, and the mutations are known to disrupt binding.
Stable cell transfections.
Jurkat T cells were cotransfected by electroporation with pHGM0.6 GM-CSF promoter plasmids (15) linearized at a unique XmnI site and the selectable neomycin resistance gene plasmid pSV2Neo (GenBank accession number U02434). Stably transfected cells were selected by culture in the presence of 600 μg of G418 per ml, and single clones of cells were selected after two cycles of cloning by limiting dilutions.
Analysis of nuclease cleavage sites within nuclei.
For all nuclease digestions, nuclei were digested in nucleus digestion buffer, which contains 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, and 10 mM Tris (pH 7.4), and suspended to a final concentration of 0.4 mg of total nucleic acids per ml as described previously (11). DNase I and micrococcal nuclease (MNase) were obtained from Worthington, and nuclei were digested in the presence of 1 mM CaCl2. All imaging and densitometry of hybridization patterns were performed using a Bio-Rad FX phosphorimager and Quantity One software.
DH sites in the GM-CSF locus were mapped as previously described (11, 15). Nuclei were briefly digested with DNase I, purified DNA was digested with the specified restriction enzymes, and DH sites were identified by Southern blot hybridization by indirect labeling of the ends. EcoRI digestion products were assayed using an upstream 1.4-kb BamHI fragment of DNA as a probe (15). BglI digestion products were assayed using either an upstream 0.45-kb BglI-BamHI fragment or a downstream 0.6-kb BglII-BglI fragment of DNA as a probe. DH sites in stably transfected cells were mapped as described above, except that a 0.7-kb SacI-ScaI fragment of pHGM0.6 was used to map sites within ScaI-digested DNA.
MNase digestions were performed essentially the same way as the DNase I digestions. For indirect labeling of the ends of MNase sites, very brief digestions were performed by incubating human T-cell nuclei with 100 U of MNase per ml for 2.5 min, and transgenic mouse T-cell nuclei were incubated with 40 U of MNase per ml at 22°C. To generate mononucleosomes and oligonucleosomes appropriate for directly probing with internal probes, T-cell nuclei were digested with MNase at concentrations ranging from 150 to 500 U per ml for 3 to 15 min at 22°C. To digest nuclei predominantly to mononucleosome-length fragments for LM-PCR mapping of nucleosome boundaries, human T-cell nuclei were digested with 500 U of MNase per ml for 20 min at 22°C, and DNA fragments of approximately 146 and 167 bp were then purified after electrophoresis on a 3% agarose gel.
Restriction enzyme accessibility assays were performed by digesting nuclei with 1 U of restriction enzyme per ml for 30 min at 37°C, with the exceptions that ApaI digestion was performed at 25°C and HaeIII digestion was conducted for 20 min. Note that because the salt concentration strongly influences the degree of chromatin condensation, each digestion was performed in nucleus digestion buffer rather than restriction enzyme buffer to maintain uniform conditions. Samples were processed as described above for DNase I digestion (11).
LM-PCR assays of nuclease cleavage sites were performed essentially as described above. For the DNase I analyses, the LM-PCR procedure commenced at the point where a biotinylated primer is used to synthesize DNA to the cleavage sites before ligation of the linker and binding to magnetic beads. Control digestion products were also assayed; control digestion products consisted of purified DNA digested with DNase I to an average size comparable to that of the nucleus digestion products. For LM-PCR analysis of MNase digestion products, DNA was first phosphorylated with polynucleotide kinase and then ligated to the LP25-LP21 linker duplex, the biotinylated primer was used to synthesize DNA to the end of the ligated linker, and the products were bound to magnetic beads. An MNase control digestion product of purified DNA was also included. Two sets of primers were designed specifically to assess nucleosome boundary positions. The primers in these two primer sets follow (sequences, boundary positions, and annealing temperatures shown in parentheses): for the 2R2 primers, EB2R2 (GCCCAAGTCAGCACAAAC, positions 507 to 490, 60°C), EP2R (GTCAGCACAAACAGGACAGAAATC, positions 501 to 478, 62°C), EL2R2 (AGGACAGAAATCCATGGGTTTGGTGATG, positions 489 to 461, 60°C), with 1.4 M betaine included in the labeling step only; for the 4F primers, EB4F (GCCCACAAACCTCATCACCAAACCC, positions 450 to 474, 58°C), EP4F (GATTTCTGTCCTGTTTGTGCTGAC, positions 478 to 501, 60°C), EL4F (CCTGTTTGTGCTGACTTGGGC, positions 487 to 507, 60°C), with 1.4 M betaine and 5% dimethyl sulfoxide included in both the PCR and labeling steps.
RESULTS
Induction of NFAT/AP-1 promotes cooperative transcription factor binding in vivo.
As the first step in defining basic mechanisms of GM-CSF enhancer activation, we employed DMS in vivo footprinting to identify regulatory elements that become occupied in cells expressing GM-CSF (Fig. 2). For a model system, we adopted proliferating human peripheral blood T cells recruited from the quiescent state by a single round of activation with phytohemagglutinin and then expanded by culture in the presence of IL-2 for several generations in the absence of any additional stimulus. This procedure yields highly enriched cultures of T cells (∼97% as assessed by staining for surface CD3) that are more readily activated than newly isolated resting T cells that normally exist in a dormant state. Cells were either left unstimulated or stimulated for 4 h with the phorbol ester PMA and the calcium ionophore A23187 to directly activate the TCR signaling pathways known to induce GM-CSF gene expression.
FIG. 2.
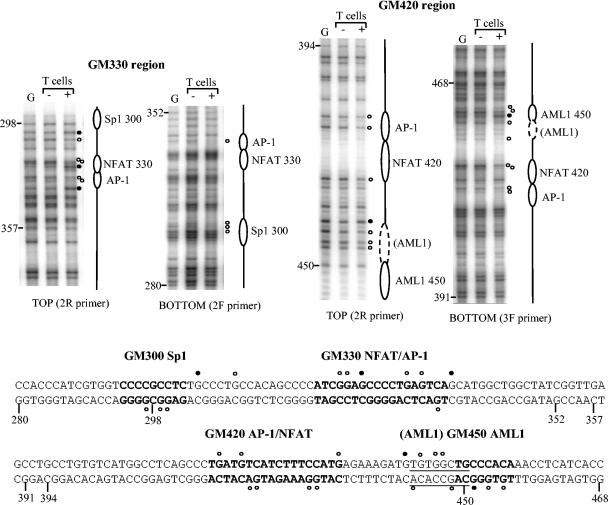
In vivo footprinting of the GM-CSF enhancer in human T cells. Cultured human peripheral blood T cells were either left unstimulated (−) or stimulated for 4 h with 20 ng of PMA per ml and 2 μM A23187 (+). Cells were then incubated with DMS, and sites of DMS-mediated DNA methylation for both DNA strands were identified by LM-PCR. DMS-methylated purified genomic DNA (G) was also included to indicate DMS reactivity towards protein-free DNA. The GM330 region of the enhancer was assayed using the 2R primer set for the upper strand and the 2F primer set for the lower strand. The GM420 region was assayed using the 2R primer set for the upper strand and the 3F primer set for the lower strand. Note that no data are displayed for the GC-rich region of the upper strand just upstream of position 298, because the LP25 linker primer was not able to amplify DNA modified within this section. Bases protected from methylation by DMS (open circles) and bases with enhanced DMS reactivity (closed circles) are indicated. Previously identified transcription factor binding sites are shown in bold type. A novel potential AML1 site is underlined. Bases are numbered relative to the upstream BglII site.
Intact cells were then treated with DMS, which methylates G bases at N7, which lies within the major groove of DNA. Sites bound by factors appear as regions of DNA where the extent of G methylation is either reduced by proteins binding within the major groove or enhanced by local perturbation of the DNA structure by proteins binding either adjacent to G bases or contacting the minor groove instead of the major groove. Methylated G bases were identified by the LM-PCR procedure, using DMS-treated genomic DNA as a control for protein-free DNA. LM-PCR probes were designed so that we could study interactions with both the upper and lower strands of the GM330 and GM420 regions of the enhancer that are located within the inducible DH site.
The patterns obtained for genomic DNA and nonstimulated T cells were identical, indicating that even the constitutively expressed factors Sp1 and AML1 are unable to gain significant access to the enhancer in the inactive state (Fig. 2). However, upon activation of the kinase and Ca2+ signaling pathways that induce AP-1 and NFAT, each of the known transcription factor binding sites located within the DH site exhibited either protection or enhancement of DMS reactivity, indicating that factors had bound to these regions. The sites occupied included the AP-1 and NFAT motifs of the GM330 and GM420 elements, the GM450 AML1 site, and the GM300 Sp1 site. We also observed modest changes in a second AML1-like element (Fig. 2) that overlaps the previously defined GM450 AML1 site. Each of these changes in DMS sensitivity was verified in independent experiments (data not shown).
The GM170 and GM550 regions outside the DH site were also examined using additional sets of LM-PCR probes, but no binding of any factors either upstream of the Sp1 site at position 300 or downstream of the AML1 site at position 450 was detected (data not shown).
Fine structure and DNase I footprints within the GM-CSF enhancer DH site.
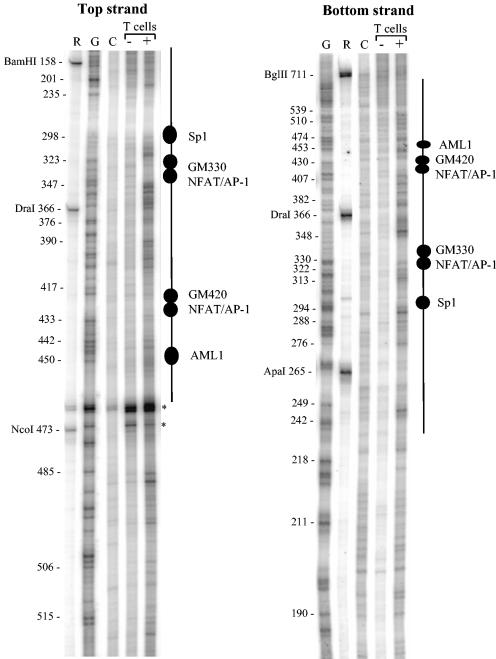
The LM-PCR assay can also be used to map DNase I cleavage sites within the enhancer at single-base resolution to provide information about induction of both DNase I accessibility and transcription factor binding to the enhancer. To this end, we assayed DNase I cleavage sites within nuclei prepared from T cells either before or after activation as in the above in vivo footprinting analyses (Fig. 3). DNA samples were carefully selected for this assay from a DNase I titration series on the basis that they support optimal detection of DH sites in other assays. This approach revealed that the inducible DH site extends from about positions 220 to 520 within the enhancer, with several distinct clusters of hypersensitive regions. Low-level DNase I cutting was seen throughout the enhancer region in nonstimulated cells, but in stimulated cells, enhanced cutting was observed at sites flanking each of the transcription factor binding sites known to exist within the enhancer core. Furthermore, each site existed as a protected footprinted region, suggesting that each of these sites is indeed occupied in stimulated cells. From the results of this analysis, it was evident that stimulation increased accessibility to nucleases, and by inference to transcription factors, throughout the enhancer core region containing the GM330 and GM420 elements.
FIG. 3.
Fine structure of the DH site within the GM-CSF enhancer. Human T cells were either left unstimulated (−) or stimulated for 5 h with 20 ng of PMA per ml and 2 μM A23187 (+). Nuclei were isolated and briefly digested with 15 (−) or 12 (+) μg of DNase I per ml. These samples were selected for LM-PCR analysis from a DNase I titration series, because they had equivalent levels of overall DNase I digestion and were optimal for the detection of DH sites in other assays. Cleavage sites were identified by LM-PCR using the 1R primer set for the top strand and the 1F primer set for the lower strand. The positions of transcription factor binding were identified by aligning the DNase I pattern with the positions of G bases determined using DMS-treated DNA (G). To align the G reaction, we also included a DNA sample partially digested with BglII, BamHI, ApaI, DraI, and NcoI (R). We included a DNase I digestion product of purified DNA for a control (C). Note that the region immediately upstream of the Sp1 site on the upper strand represents a region not amplified by the LP25 primer, and therefore appears as a blank region. The positions of fragments digested by different restriction endonucleases are indicated to the left of the gels. Asterisks indicate background artifacts. Bases are numbered relative to the upstream BglII site.
The GM330 and GM420 NFAT/AP-1 sites are essential for GM-CSF enhancer function.
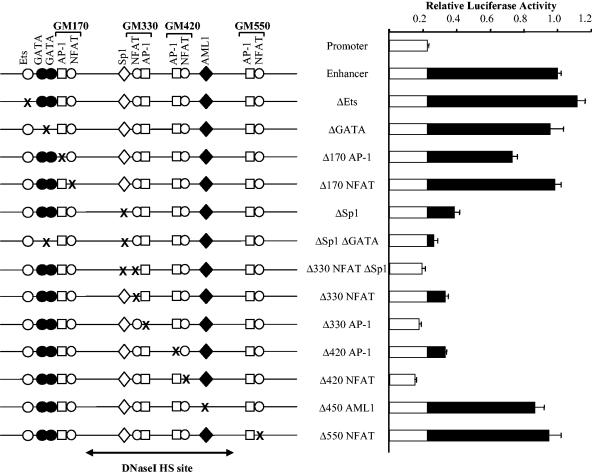
The next stage in defining mechanisms of NFAT-dependent enhancer activation was to determine which elements were actually required for enhancer function and the extent to which different elements acted synergistically. Site-directed mutagenesis was employed to determine the role of each of the known and predicted transcription factor binding sites within the GM-CSF enhancer. In this analysis, we also included two known GATA binding sites (B. V. Johnson and P. N. Cockerill, unpublished data) and a potential Ets site which exist upstream of the array of NFAT sites. This upstream region was investigated, because it exists as a distinct DH site present in cells, such as K562 cells, which express high levels of GATA-1 but not in most T cells (13).
Transient-transfection assays were performed on luciferase reporter gene plasmids containing the 717-bp BglII fragment of the GM-CSF enhancer upstream of the region of the human GM-CSF promoter from positions −627 to +28. This plasmid (pXPG-GM627-B717) is induced approximately 100-fold upon stimulation (data not shown) (13) and is constructed from the novel luciferase vector pXPG, which has much lower background activities than some commonly used reporter gene plasmids, such as pGL3, which appear to be affected by read-through transcription (5).
In stimulated Jurkat T cells, the intact enhancer increased the activity of the promoter fivefold (Fig. 4). No significant decrease in this level of activity was seen for any of the mutations of either the upstream or downstream regions flanking the enhancer core. In stark contrast, however, mutation of either the AP-1 or NFAT motif within either the GM330 or GM420 NFAT/AP-1 elements in each case essentially abolished all enhancer activity. These results suggested a high degree of cooperation not only between NFAT and AP-1 within composite elements but also between distinct composite NFAT/AP-1 elements. Mutation of the Sp1 site also substantially reduced but did not eliminate enhancer activity. However, there was some indication that the upstream GATA sites may in part compensate for the loss of the Sp1 site, because a double mutation of both the Sp1 and GATA sites did abolish enhancer activity. Mutation of the GM450 AML1 site had no effect, but it is possible that either a second potential overlapping AML1 site is used in its absence or that it does not function in the context of transient-transfection assays. We also confirmed that the entire flanking region upstream of the Sp1 site was dispensable for activity in T cells by demonstrating that a 317-bp ApaI-MscI fragment of the enhancer, extending from positions 260 to 586 had essentially the same activity as the 717-bp BglII fragment (data not shown). Hence, the regions of the enhancer required for function are directly equivalent to the regions where in vivo footprints were detected.
FIG. 4.
Mutational analysis of the GM-CSF enhancer in transient-transfection assays. Luciferase reporter gene plasmids containing the luciferase vector pXPG and a segment of the human GM-CSF promoter covering positions −627 bp to +28 bp relative to the transcription start site and an upstream 717-bp BglII segment of the human GM-CSF enhancer were transiently transfected into Jurkat T cells and assayed for luciferase activity after stimulation for 9 h with 20 ng of PMA per ml and 1 μM calcium ionophore A23187. The assays included plasmids containing the promoter lacking the enhancer (pXPG-GM627) or containing the intact enhancer (pXPG-GM627-B717) or the enhancer carrying the specified mutations to destroy the indicated binding sites. At least two independent preparations of each plasmid were assayed, and each plasmid was assayed an average of 34 times to maximize reliability of the data. The luciferase activity of each plasmid is expressed relative to the plasmid containing the intact enhancer. Note that as reported previously (13), in the absence of stimulation the luciferase activity of each plasmid was approximately 1% of the activity obtained after stimulation, and these data are not presented here. Error bars represent the standard errors of the means. HS, hypersensitive. White bars indicate the activity of the promster alone, black bars indicate the additional activity supported by the enhancer.
Kinetics of GM-CSF gene and enhancer activation.
To gain further insights into which factors might be required for either chromatin remodelling or gene activation, we performed a temporal analysis of activation of the GM-CSF locus and induction of the transcription factors that control its expression in cultured T cells. This analysis was designed to take advantage of the fact that induction of NFAT nuclear translocation is complete within a few minutes, whereas de novo AP-1 induction takes considerably longer.
Upon stimulation with PMA and A23187, GM-CSF mRNA expression was rapidly induced. Significant expression could be detected after 40 min, and mRNA levels continued to rise over a 4-h period (Fig. 5A). To compare the kinetics of chromatin reorganization and transcription activation, we also performed an assay to determine whether DH sites were present in the same cell cultures. As in our previous studies, we detected strong induction of a DH site within the GM-CSF enhancer, but here we demonstrated for the first time that this DH site was almost fully formed within just 20 min (Fig. 5B). In this analysis, we can also detect a constitutive DH site in the GM-CSF promoter; these results can be used to confirm that the overall levels of DNase I cutting are indeed similar for the different samples analyzed and that no DH site exists within the enhancer prior to activation.
FIG. 5.
Kinetics of GM-CSF gene activation. (A) GM-CSF mRNA expression in T cells stimulated for the indicated times with 20 ng of PMA per ml and 2 μM A23187 (PMA/I). RNA expression was assayed by real-time PCR and expressed relative to GAPDH expression where the value for unstimulated cells was set at 1. (B) Assay of DH sites upstream of the GM-CSF gene in T cells from the same cultures assayed in panel A. For each preparation of T-cell nuclei, aliquots of nuclei were digested with DNase I in the range of 4.5 to 8 μg of DNase I per ml and only the optimally digested samples are displayed. DH sites were mapped by indirect end labeling of EcoRI-digested DNA using an upstream 1.4-kb BamHI fragment of DNA as a probe as indicated in Fig. 1. The positions of the promoter (P) and GM-CSF enhancer (E) are indicated to the right of the gel. (C) EMSA with oligonucleotides carrying binding sites for the indicated transcription factors (see Materials and Methods) using nuclear extracts prepared from the same cultures shown in panels A and B.
To determine which factors might be responsible for the timing of the above patterns of gene activation and DH site formation, we also performed EMSAs of nuclear extracts prepared from the same cultures (Fig. 5C). Here we assayed for induction of the four classes of transcription factor known to associate with the GM-CSF enhancer core region within the DH site (Fig. 1 and 2). NFAT was rapidly induced to maximal levels in the nucleus within 20 min. This complex contained approximately equal proportions of NFATc1 and NFATc2, as assessed by incorporating specific antibodies in EMSAs (data not shown). AP-1 was detected at very low levels after 20 min and was significantly induced from 40 min onwards. No induction was observed for the preexisting constitutively expressed factors Sp1 and AML1, which can therefore be used to control for the levels of NFAT and AP-1. It was apparent from this analysis that the kinetics of GM-CSF mRNA induction most closely mirrored AP-1 levels, whereas the kinetics of DH site induction most closely reflected NFAT levels.
Strong NFAT sites bind NFAT independently of AP-1 but rely on cooperativity for function.
Although it is normally assumed that NFAT and AP-1 act synergistically, there have in fact been no detailed studies investigating mechanisms of cooperation between these factors other than studies demonstrating cooperative binding. To define the extent to which these factors cooperate and to study mechanisms of cooperation that might be distinct from cooperative binding, we chose to focus on the function of the GM420 NFAT/AP-1 element. Of the two critical NFAT sites in the enhancer, the GM420 NFAT/AP-1 element is the only motif that can bind NFAT with a high affinity independently of AP-1 (12).
The binding properties of the GM420 element were investigated in EMSAs of recombinant proteins using the Rel homology domain of NFATc2 as a source of NFAT and a heterodimer of the DNA-binding and leucine zipper domains of c-Fos and c-Jun as a source of AP-1 (Fig. 6A) as in our previous studies (12). Using the intact GM420 element as a probe, we observed strong binding of NFAT alone and a moderate increase in the overall amount of bound NFAT as a NFAT/AP-1 complex when both species were included. AP-1 alone gave moderate binding, and the AP-1 complex was mostly shifted to an AP-1/NFAT complex upon the addition of NFAT. It was evident that the binding of NFAT and AP-1 in this assay was not as rigidly cooperative as we have observed for other composite sites in previous studies (12). (Note that the GM420 element has an atypical 8-bp AP-1-like element in which the first half-site [TGA] is one base closer to the NFAT motif than in typical composite NFAT/AP-1 elements [Fig. 1] [12], and this might not be optimal for cooperative binding.)
FIG. 6.
Cooperative transcription factor binding and function of the GM420 NFAT/AP-1 site. (A) EMSA with recombinant NFAT, AP-1, and GM420 duplex oligonucleotides with or without the indicated mutations. (B) Transient-transfection assays of pXPG-GM55 luciferase reporter gene plasmids containing the minimal GM-CSF promoter and tandem arrays of three copies of the GM420 elements with and without the indicated mutations. Transfected Jurkat T cells were stimulated for 9 h with 20 ng of PMA per ml and 1 μM calcium ionophore A23187. Luciferase activities are shown as the means ± standard errors of the means (error bars) for 12 transfection assays, employing two separate preparations of each plasmid construct, and are expressed relative to the luciferase activity of pXPG-GM55, which was set at 1.
To develop additional tools for studying NFAT and AP-1 functions, we engineered mutations in the GM420 element that either abolished NFAT binding but did not affect AP-1 binding (Δ-NFAT) or abolished AP-1 binding without affecting NFAT binding (Δ-AP-1) (Fig. 6A). Because no higher-order NFAT/AP-1 complex was detected with either of these mutated probes, this analysis also demonstrated that the NFAT/AP-1 complex is not preformed in solution and that it requires intact NFAT and AP-1 sites in order to be assembled directly on DNA. A double mutation of both the AP-1 and NFAT sites abolished formation of all complexes containing NFAT and AP-1 (Fig. 6A).
Each of the GM420 elements examined in the above EMSA were also studied to determine their function as enhancer elements linked to a minimal GM-CSF promoter from positions −55 to +28 in luciferase reporter gene plasmids in transient-transfection assays of Jurkat T cells. To mimic the organization seen in most NFAT-regulated enhancers, we tested the GM420 element as an array of three linked head-to-tail copies. In stimulated Jurkat cells, the minimal promoter alone had a modest activity, but this was dwarfed by the huge activity of the plasmid containing the GM420 element, which was 500 times more active than the promoter alone (Fig. 6B). Hence, the GM420 site functions as an extraordinarily powerful enhancer element, and in parallel studies, it was found to be 2.5 times more active than the GM550 NFAT/AP-1 site and 7 times more active than the high-affinity AP-1 site from the stromelysin gene (J. Borrows, A. W. Bert, and P. N. Cockerill, data not shown). The function of the GM420 element was completely abolished, however, by the mutation of the NFAT site, as the activity of the GM420 Δ-NFAT plasmid was identical to the activity of the plasmid containing the promoter alone. The GM420 Δ-AP-1 plasmid retains three high-affinity NFAT sites, but nevertheless, mutation of the AP-1 sites also nearly abolished its activity. Furthermore, even this low level of activity seen with the GM420 Δ-AP-1 plasmid may involve cooperation with a low-affinity AP-1 site that exists approximately 60 bp downstream of the array of NFAT sites within the minimal GM-CSF promoter (32). Hence, not only is the NFAT component of the GM420 element essential for function, but it is also unable to contribute any significant transcription activation function in the absence of an AP-1 site.
NFAT elements can create an inducible DH site independently of AP-1 elements.
To attempt to understand why NFAT and AP-1 should be so interdependent, we opted to investigate the potential functions of NFAT by studying the high-affinity GM420 NFAT motif in a chromatin context in the absence of any linked AP-1 motif. To this end, we created clones of cell lines stably transfected with plasmids containing either the intact GM-CSF enhancer or three copies of just the NFAT-binding segment of the GM420 NFAT site, inserted upstream of the GM-CSF promoter (Fig. 7). We assayed each of these cell lines for the presence of inducible DH sites and confirmed that the transfected GM-CSF enhancer did indeed form a DH site upon stimulation. Significantly, we found that the GM420 NFAT site alone was also sufficient to form an inducible DH site (Fig. 7).
FIG. 7.
Identification of DH sites encompassing NFAT elements in stably transfected cells. DH sites were assayed in single clones of Jurkat T cells transfected with pHGM0.6 GM-CSF promoter plus chloramphenicol acetyltransferase (CAT) reporter gene plasmids (14) containing either the intact GM-CSF enhancer or three copies of just the NFAT-binding region of the GM420 NFAT site. Cells were either left unstimulated (−) or stimulated for 6 h with 20 ng of PMA per ml and 2 μM A23187 (PMA/I) (+). DH sites were identified by indirect end labeling using a 0.7-kb SacI-ScaI fragment of pHGM0.6 as a probe.
The GM-CSF enhancer DH site exists within an ordered array of nucleosomes.
Because NFAT function is so closely linked to the creation of DH sites and because the nature of DH sites is in general still poorly understood, we undertook a detailed chromatin structure analysis of the GM-CSF enhancer before and after activation. In the first instance, we needed to define the locations of nucleosomes within the enhancer, and then we needed to establish what becomes of nucleosomes after the DH site has formed. MNase, which cuts preferentially in linker regions, was employed to define the positions of nucleosomes. These analyses were performed using two distinct model systems: (i) cultured human T cells, as used in many of the above studies, and (ii) cultured splenic T cells obtained from a line of transgenic mice containing approximately 100 copies of the human GM-CSF gene. It became necessary to employ transgenic mouse T cells to overcome certain technical limitations of studies of single-copy genes and to obtain much higher levels of resolution and sensitivity. For example, we were able to overcome severe lane-to-lane differences in overall DNA mobility that occurred when 10 μg of human DNA was loaded (Fig. 8B and C) by loading just 1 μg of transgenic mouse DNA in each lane (Fig. 8D and E) while still increasing sensitivity. Also note that we have previously shown that the GM-CSF gene is correctly regulated in a copy number-dependent manner in this transgenic mouse model (13).
FIG. 8.
Chromatin architecture of the GM-CSF enhancer. (A to E) Indirect end-labeling assays of MNase and DNase I sites within the GM-CSF enhancer in nuclei prepared from cells that were either left unstimulated (nil) or stimulated for 4 h with 20 ng of PMA per ml and 2 μM A23187 (PMA/I). Analyses were performed by Southern blot hybridization using either 10 μg of human T-cell DNA per lane (A to C) or 1 μg of Gm-CSF transgenic mouse T-cell DNA per lane (D and E). The positions of nucleosomes N1 and N2 and the DH site (DHS) are indicated by brackets. Band positions are calculated relative to the upstream BglII site at position 1 in the 717-bp BglII enhancer sequence. Asterisks indicate the positions of new inducible MNase sites at positions 260 and 470. Panel A depicts the ethidium bromide staining pattern for the analyses performed in panels B and C and the positions of bands containing two to six nucleosomes (indicated to the left of the gel). The restriction enzyme marker in panels B and C consists of DNA partially digested with BglII, PstI, and ApaI, with the positions of these sites depicted on the right. (F) Map of the major nuclease cleavage sites within the 1,660-bp BglI fragment encompassing the GM-CSF enhancer, showing the predicted positions of nucleosomes, the DH site, and the indirect end-labeling probes used to map cleavage sites. HS, hypersensitive. (G) LM-PCR mapping of nucleosome boundaries in nucleosome length DNA fragments purified from human T cells that were either left nonstimulated (nil) or stimulated for 4 h with 20 ng of PMA per ml and 2 μM A23187 (PMA/I). The leftmost panel depicts agarose gel electrophoresis of 146-bp nucleosome-length DNA (N) and 167-bp chromatosome length DNA (C) purified from MNase digestion products of T-cell nuclei. Twenty-five nanograms of purified nucleosomal DNA was assayed by LM-PCR using the 2R2 and 4F primer sets. DMS-treated DNA samples to align the sequence (G) and a control MNase digest of purified DNA (M) were also included. The map shows the relative positions of the primer sets used to map nucleosome boundaries.
Figure 8A shows the MNase cleavage pattern generated by digestion of nuclei prepared from nonstimulated and stimulated human T cells. Similar digestions, yielding equivalent ladders of MNase cleavage products were also performed on the transgenic mouse T cells (data not shown). The locations of MNase cleavage sites were mapped by indirect labeling of the ends of BglI digestion products in Southern blots using probes on either side of the enhancer (Fig. 8B to F). DNA from DNase I-digested activated T-cell nuclei was assayed in parallel to align the DH site with the nucleosome ladder. To provide an internal DNA size marker, a DNA sample partially digested with a mixture of BglII, ApaI, and PstI was also included in each assay (Fig. 8B and C; not shown in Fig. 8D and E).
MNase digestion of nonactivated T-cell nuclei produced a ladder of bands, which suggested that nucleosomes adopt preferred sites within an ordered array of nucleosomes across the GM-CSF enhancer (Fig. 8B to E). Similar results were also obtained with Jurkat T cells, Ball-1 B cells, and HeLa cells (data not shown). The positions of MNase cleavage sites and the likely locations of nucleosomes are illustrated in Fig. 8F where the positions of MNase sites are numbered relative to their distance from the upstream BglII site. MNase cleavage sites clustered at intervals of approximately 150 to 200 bp apart, which is consistent with the standard size of a nucleosome, which is typically 146 bp but can vary from ∼100 to 170 bp (54). From this analysis, it would appear that the critical GM330 and GM420 NFAT/AP-1 elements lie within distinct adjacent nucleosomes, defined here as nucleosomes N1 and N2, located approximately between positions 220 and 540, with a linker centered at position 370. Note that although we obtained essentially identical data from both T-cell model systems, our analysis of the data was performed principally on the transgenic T cells (Fig. 8D and E) for which we could perform more accurate DNA size estimations.
After the cells were stimulated, we observed the same basic ladder of bands with MNase digestion products of nuclei prepared from activated T cells as was seen with nonstimulated cells (Fig. 8B to E). Significantly, however, we observed that cleavage within regions centered at positions 190, 370, and 540 became more intense upon stimulation. This suggested that there was a partial unravelling of the chromatin fiber in a region spanning two nucleosomes whereby the linker regions flanking nucleosomes N1 and N2 became more accessible to MNase. It was also apparent that some degree of nucleosome mobilization or disruption was also occurring within the N1/N2 region upon stimulation, because new inducible cleavage sites appeared at positions 260 and 470 (Fig. 8D and E), which was not consistent with the maintenance of two fixed 146-bp nucleosome particles at the N1 and N2 positions.
Parallel analysis of DNase I cleavage sites within the DH site in transgenic mouse T cells revealed that the DH site had a discrete peak at position 370, coincident with the strongest MNase peak and the predicted N1/N2 nucleosome linker region, and extended from about positions 140 to 520 (Fig. 8D and E). In the case of our analyses of the human DNA samples, the highest MNase and DNase I peaks also appeared to coincide, but the lane-to-lane differences in DNA mobility seen for MNase and DNase I digestion products (e.g., compare the mobility of the BglI fragments) did not allow direct alignment of these peaks (Fig. 8B and C).
Nucleosome boundaries within the enhancer do not change upon activation.
Because it was possible that nucleosome sliding could account for some of the inducible changes in the MNase pattern, we mapped the boundaries of the nucleosomes within the enhancer before and after stimulation at high resolution using LM-PCR (Fig. 8G). To this end, we purified two discrete fractions of DNA from MNase-digested nuclei from stimulated and nonstimulated human T cells: (i) a 146-bp species that represents the predominant minimal nucleosome core particle and (ii) a 167-bp species that represents chromatosome length DNA which is generated either when nucleosomes are associated with additional proteins, such as histone H1 or HMG proteins, or if more than 146 bp of DNA wraps around the histone octamer (54) (Fig. 8G, leftmost gel). Results are presented for two primer sets mapping the 5′ and 3′ boundaries of nucleosome N2. Due to technical difficulties, our analyses of nucleosome N1 were inconclusive, and these data are not presented. The analyses of nucleosome-length DNA revealed nucleosome boundaries consistent with the nucleosome positions predicted by the above indirect end-labeling assays. Most importantly, these boundaries did not change significantly after stimulation (Fig. 8G). The products detected with the 4F and 2R2 primers defined a region encompassing nucleosome N2 between approximately positions 373 to 392 at the 5′ end and positions 527 to 535 at the 3′ end. Similar results were obtained with chromatosome length DNA, but in this case we also detected slightly larger products with the 2R2 primer sets, consistent with an approximately 20-bp increase in the length of DNA associated with the histone core particle.
Nucleosome reorganization across an extensive region of the GM-CSF locus.
The indirect evidence for nucleosome disruption made it necessary to employ a more quantitative approach to directly assay for the continued presence of nucleosomes within the enhancer after stimulation. To achieve this, we probed Southern blots of digestion products of more extensive MNase digestions of T-cell nuclei with internal probes encompassing regions both within the enhancer and across the GM-CSF locus (Fig. 9). Because of sensitivity problems encountered with our analyses of human DNA with short probes, we employed our transgenic mouse T cells for this purpose. Nuclei isolated from stimulated and nonstimulated T cells were digested with increasing amounts of MNase, yielding equivalent series of nucleosomal ladders in each case (Fig. 9A). The sizes of the MNase products were calculated using a HaeIII-digested φX174 DNA size marker (Fig. 9A), and the specificity of the hybridization was confirmed by including a DNA sample digested with BamHI, HincII, and PstI in the leftmost lanes (Fig. 9A to H). The relative amounts of DNA remaining as nucleosomal DNA and approximate nucleosome repeat lengths across the nucleosome array were calculated by densitometry of lanes having the greatest degree of MNase cleavage, as depicted below each panel.
FIG. 9.
Nucleosome destabilization and mobilization in the GM-CSF locus. Southern blot hybridization analysis of DNA purified from MNase-digested nuclei isolated from GM-CSF transgenic mouse T cells that were either left unstimulated (nil) or stimulated for 4 h with 20 ng of PMA per ml and 2 μM A23187 (PMA/I). The three sets of MNase digestion conditions employed were, from left to right, 250 U of MNase/ml for 3 min, 150 U of MNase/ml for 15 min, and 500 U of MNase/ml for 15 min. The latter was chosen for performing densitometry of the hybridization patterns, as depicted below each panel, with nonstimulated T-cell samples in grey and stimulated T-cell samples in black, and the average nucleosome repeat length was calculated for nonstimulated (−) and stimulated (+) cells. DNA was analyzed on a total of four replica 2% agarose gels, using 3 μg of DNA per lane. HaeIII-digested φX174 DNA was employed as a size marker in the rightmost lanes, and as can still be seen in panel H, the positions of the marker and the major 146-bp nucleosome bands were marked on the gel and filter with a needle and Indian ink to enable accurate size estimations. The leftmost lanes contain 3 μg of DNA digested with BamHI, HincII, and PstI. Panels B, C, and D were reprobed, and the results are shown in panels E, G, and H, respectively. The map depicts the 10.5-kb XhoI-HindIII fragment used as a transgene and the positions of the GM-CSF probes used in panels B to G. Probe B extended from positions 219 to 390, and probe C extended from positions 386 to 559 within the GM-CSF enhancer. Pr., promoter. Panel H was probed with a 17-kb EcoRI DNA fragment spanning the Cɛ region of the mouse heavy-chain immunoglobulin (IgH) gene, which is inactive in T cells.
Using short nucleosome-length probes spanning nucleosomes N1 and N2, we detected regular arrays of nucleosomes in nonstimulated cells and the expected 198-bp BamHI-HincII and 158-bp HincII-PstI restriction enzyme fragments, which confirmed specificity (Fig. 9B and C). The ladders of oligonucleosomes in this region were indistinguishable from those detected for bulk genomic DNA (Fig. 9A). After the cells were stimulated, there was significant evidence for both nucleosome disruption within the enhancer and for mobilization of nucleosomes across the array. The regular array had become much less distinct, with considerable blurring of the bands, and much less material remained as mononucleosome-length DNA.
There was also evidence that the nucleosomal repeat length across the enhancer region had become compressed by about 35 bp per nucleosome from approximately 187 to 190 bp before stimulation to 151 to 153 bp after stimulation. Significantly, the same degree of nucleosome mobilization and compression was detected up to 2 kb downstream of the enhancer, when a probe located from −2 to −0.6 kb was employed (Fig. 9D). Similar results were obtained with a 450-bp BglI-BamHI DNA probe located immediately upstream of nucleosome N1 that extends up to −3.6 kb (data not shown). This indicated that a region of approximately 3 kb had been reorganized upon activation of the enhancer, generating a chromatin structure with a repeat length not much larger than the 146 bp of DNA occupied by a single nucleosome and significantly less that the 167 bp predicted for chromatosome-length DNA.
In contrast, little change in nucleosomal organization was detected within a region 1.4 kb downstream of the gene that maintained a repeat length of approximately 187 bp after stimulation (Fig. 9G), and no change in chromatin structure was detected within an inactive immunoglobulin gene (Fig. 9H). Within the GM-CSF coding region, there was also some mobilization of the nucleosome array, and the nucleosome repeat length was compressed by approximately 20 bp to 160 bp upon activation of transcription (Fig. 9F). The GM-CSF promoter has previously been reported to exist as a positioned nucleosome that becomes disrupted upon activation of transcription (30), and using a probe extending from −110 to +28 we also found that this region was disrupted to an extent at least as great as that seen for the enhancer (Fig. 9E). However, it was evident that the region encompassing the enhancer experienced the highest level of long-range nucleosome reorganization within the GM-CSF locus.
The entire enhancer core becomes more accessible to restriction enzyme digestion after stimulation.
The above studies firmly suggested that there was significant destabilization and increase in accessibility of the compact nucleosome array upon activation of the enhancer. To test this model directly, we employed a restriction enzyme accessibility assay of activated human T cells (Fig. 10). Restriction enzymes typically bind DNA as dimers from both sides of the double helix and generate double-strand cuts. Such assays therefore represent a rigorous means of determining whether the DNA helix has adopted an open confirmation likely to bind transcription factors without hindrance from extensive close contacts with nucleosomal histones.
FIG. 10.
Restriction enzyme accessibility assays of the GM-CSF enhancer. Assays were performed on nuclei isolated from cultured human T cells that were either left unstimulated (−) or stimulated for 4 h (A and B) or 6 h (E) with 20 ng of PMA per ml and 2 μM A23187 (PMA/I) (+). (A and B) Nuclei were digested with the restriction enzymes indicated above each lane. Purified DNA was then redigested with either BglII (the ApaI, HincII, NcoI, BamHI, and PstI lanes and the first G lane) or BglI (the BglII lanes and the rightmost G lane) prior to electrophoresis on a 2% agarose gel and Southern blot hybridization analysis using either the 717-bp BglII fragment of the enhancer (A) or a BglII-BamHI fragment downstream of the enhancer (B) as probes. ApaI, HincII, PstI, and BamHI each cut the enhancer just once. Because NcoI and BglII cut the enhancer region two or three times, the migration positions of complete and partial digestion products are shown on the righthand side of panel A and on each side of panel B, and a map of these digestion products is provided in panel D. The size markers to the left of the gel in panel A indicate the migration positions of selected bands of HaeIII-digested φX174 DNA. (C) Locations of nuclease cleavage sites in the GM-CSF enhancer, showing the total percentage of DNA cut at each site before (nil) and after stimulation with 20 ng of PMA per ml and 2 μM A23187 (PMA/I). (D) Summary of the products detected by probes A and B after digestion with either BglII followed by BglI or NcoI followed by BglII. The positions of sites cleaved by the enzymes used in the accessibility assays are indicated by asterisks. (E) Nuclei from nonstimulated (0 lanes) and stimulated cells (P/I lanes) were digested with HaeIII, and cleavage at the indicated sites or positions was assayed by LM-PCR. The fold increase in cleavage products detected is shown below each panel. The map below shows the locations of HaeIII sites in the enhancer and the positions of LM-PCR primers used to detect cleavage at these sites.
We chose a panel of restriction enzymes to probe sites throughout the enhancer within nuclei prepared from stimulated and nonstimulated human T cells (Fig. 10A). Accessibility to ApaI, HincII, NcoI, PstI, and BamHI was mapped within the 717-bp BglII fragment, whereas BglII accessibility was mapped within a larger 1,825-bp BglI fragment spanning the enhancer. In each case, the 717-bp BglII fragment was used as a Southern blot hybridization probe. Note that NcoI and BglII each cut at two or three sites within the enhancer, and Fig. 10D provides a map of the potential mixture of fragments detected by the 717-bp BglII probe (probe A) for these digestion products.
Figure 10C provides a summary of the restriction enzyme accessibility data generated in Fig. 10A, with the percentage cutting before and after activation depicted above each site. This analysis revealed that both the ApaI site within nucleosome N1 and the HincII site within the predicted linker between nucleosomes N1 and N2 were inaccessible in T-cell nuclei before stimulation but highly accessible after stimulation. A NcoI site within nucleosome N2 also became much more accessible upon stimulation, and the NcoI site at position 94 increased twofold in accessibility. A BamHI site in the linker at the 5′ boundary of the enhancer core and a PstI site at position 514 just inside the predicted 3′ boundary of nucleosome N2 were already partly accessible before stimulation but became more accessible after stimulation. Three BglII sites that exist in the regions flanking the enhancer core were in each case partly accessible before stimulation, and their accessibility did not change after stimulation.
To control for variations in the way in which different enzymes might act, we reprobed the filter used in Fig. 10A with a second probe downstream of the enhancer (probe B [Fig. 10D]). In contrast to the enhancer core region, ApaI, HincII, and NcoI generated a maximum of about 2% cleavage in this downstream region either before or after stimulation (Fig. 10B). The BglII cleavage pattern in Fig. 10B served to confirm the conclusions drawn from Fig. 10A. BamHI and PstI again gave a significant degree of cleavage within the downstream region, but in this instance there was no change in accessibility upon stimulation. The results of this assay, and the patterns observed for bulk DNA (data not shown), may also suggest that BamHI, BglII, and PstI have an intrinsically greater ability to cleave chromatin than ApaI, NcoI, and HincII have.
These experiments were verified by examining HaeIII accessibility in human T cells (Fig. 10E). Because of the large number of HaeIII sites, this semiquantitative analysis was performed by LM-PCR using probes as close as possible to each site. This analysis again showed that sites throughout the enhancer core, sites at positions 118, 263, 406, and 516, all become more accessible upon stimulation, whereas sites upstream of base 118 or downstream of base 516 did not exhibit any change in accessibility.
A striking feature of these analyses it that they indicate that sites predicted to exist within either nucleosome N1 or N2 or the intervening linker each become more accessible after stimulation, and the extent to which accessibility increases is even greater than that seen for sites within or close to flanking nucleosome linker regions.
DISCUSSION
The DH site defines an essential enhancer core that functions as an integral unit.
This study represents the first in vivo analysis of the binding of transcription factors to the GM-CSF enhancer in activated cells. The results of our in vivo footprinting studies revealed that none of the preexisting nuclear factors, such as Sp1 and AML1, can gain access to the enhancer without induction and that each of the predicted binding sites were bound by factors only after activation. Furthermore, most of the occupied sites were essential for enhancer function. This activation-dependent binding of a whole assembly of transcription factors is reminiscent of the enhanceosome-like complexes that have been shown to assemble on a number of genes (43, 51). For example, the TCRα enhancer was similarly shown to assemble transcription factors in vivo in a highly cooperative manner, and no stable binding of CREB or AML1 could be detected if the adjacent TCF or Ets site were deleted (27). Other NFAT-dependent regulatory elements also share this property. The IL-2 promoter does not exhibit any detectable in vivo binding of transcription factors in T cells prior to activation but binds an extensive array of factors upon stimulation (22). These in vivo studies demonstrate that tissue-specific and inducible factors play an important role in regulating gene expression in part by controlling the binding of other ubiquitous factors to promoters and enhancers. The basis for this cooperativity may stem from the fact that transcription factors display an intrinsic cooperativity in their binding in a chromatin context (1, 3, 26, 38). For example, it is known that the binding of some transcription factors to inefficiently accessed sites just inside nucleosomes is enhanced by transcription factors that bind to DNA and disrupt histone contacts at adjacent sites closer to the point where DNA exits the nucleosome (1, 3, 38).
NFAT and AP-1 cooperate at a functional level.
The results of our kinetic studies (Fig. 5) suggested that activation of GM-CSF enhancer function was dependent on the induction of both AP-1 and NFAT. Site-directed mutagenesis revealed a high degree of cooperativity between the functional elements of the enhancer core and confirmed the importance of NFAT and AP-1 sites for enhancer activity. Furthermore, we obtained evidence that NFAT and AP-1 cooperativity cannot always be accounted for simply by their ability to bind DNA cooperatively. The GM420 NFAT/AP-1 element has a high-affinity NFAT site that efficiently binds NFAT independently of AP-1, but nevertheless, it is a true composite element and was essentially inactive as an enhancer element if the NFAT or AP-1 site was deleted. We gained new insights into the basis of NFAT cooperation when we demonstrated that high-affinity NFAT elements were nevertheless sufficient to form DH sites, thereby implicating NFAT as a factor that can initiate chromatin remodelling. In preliminary efforts to identify the mechanisms underlying this phenomenon, we have performed cotransfection assays overexpressing Brg-1, a component of mammalian SWI/SNF-like complexes. In these studies, we found that the GM-CSF enhancer was transactivated by expression of Brg-1 (data not shown), but further studies will be required to determine whether Brg-1 plays a direct role in NFAT-dependent enhancer activation and chromatin remodelling.
Assembly of transcription factors on enhancer elements.
It is also worth considering the order in which transcription factors assemble, which can be very dependent on the context, and is unlikely to be the same for all NFAT/AP-1 elements. For example, the GM420 element carrying the high-affinity NFAT site is likely to bind NFAT before AP-1, whereas published EMSAs (12) indicate that the GM330 element, which has a low-affinity NFAT site linked to a high-affinity AP-1 site, must bind AP-1 before it can recruit NFAT. Furthermore, the GM330 and GM420 elements defined two clusters of sites located within adjacent nucleosomes, where binding of NFAT and AP-1 appeared necessary before closely linked Sp1 and AML1 sites could form stable complexes at sites located more centrally within these nucleosomes. It is known that Sp1 and AML1 can bind to nucleosomes but at greatly reduced efficiencies compared to that of free DNA; therefore, they are likely to rely on additional factors for stable binding (3, 24, 37, 38). However, AML1 appears to be able to bind independently to the osteocalcin promoter if its binding site is located close to the end of nucleosomal DNA (24). Furthermore, AML1 may be functioning rather like an initiator factor (3) at the TCRδ enhancer where AML1 promotes binding of cMyb in a chromatin context (26). In contrast, Sp1 is not thought to be able to bind to inducible promoters in the absence of induction of other factors (3).
Two specific nucleosomes within the GM-CSF enhancer become more accessible upon induction of NFAT and AP-1.
In this study, we investigated the chromatin architecture of the GM-CSF enhancer. Prior to this study, essentially nothing was known as to what becomes of the underlying nucleosomes when an NFAT-dependent DH site is created. Our chromatin structure studies revealed that two essential composite NFAT/AP-1 elements existed on distinct nucleosomes. Nucleosome N1 encompassed the GM330 region, and nucleosome N2 encompassed the GM420 region. We found that the resident nucleosomes at the GM-CSF enhancer underwent a significant degree of remodelling after activation. However, we did not detect any changes in actual nucleosome boundaries.
Nucleosomes within bulk chromatin are normally closely packed in a helical zigzag confirmation, and it may be assumed that many sites on DNA would be quite inaccessible within this structure (3, 38, 52, 54). We propose that the DH site formed upon binding of NFAT/AP-1 complexes in the GM-CSF enhancer represents a localized unfolding and remodelling of nucleosomes N1 and N2. From the nuclease accessibility assay results presented here, it is likely that nucleosomes N1 and N2 are closely stacked before stimulation, but after activation, this close stacking becomes disrupted, freeing up both the linker regions and the DNA on the surface of the nucleosome. We clearly demonstrate that the linker region separating nucleosomes N1 and N2 is only moderately accessible to either DNase I (Fig. 3), MNase (Fig. 8A), or HincII (Fig. 9A) before stimulation but becomes highly accessible after stimulation. These observations are not entirely unexpected, however, as some disruption of the chromatin fiber is inevitable when a 220-kDa NFAT/AP-1 complex interacts with a chain of nucleosomes assembled from 108-kDa histone octamers.
Long-range nucleosome reorganization upstream of the GM-CSF gene.
A surprising finding of this study was the observation that nucleosome reorganization is not restricted to the immediate vicinity of the GM-CSF enhancer core. Using a range of probes, we observed that nucleosomes were mobilized across most of the locus after stimulation, leading to alterations in the normally regular spacing of nucleosomes. We observed that the average nucleosome repeat length was greatly reduced from the typical range of 180 to 200 bp prior to activation to approximately 150 to 153 bp after activation across a 3-kb region spanning the enhancer upstream of the GM-CSF gene. This is reminiscent of observations by Cavalli and Thoma (8) who found that the nucleosomal repeat length was compressed from 180 to 160 bp when a galactose-inducible gene was activated in Saccharomyces cerevisiae. Chromatin compaction in the active mouse kappa immunoglobulin gene has also been observed; this gene had a nucleosome repeat length of approximately 155 bp, compared to 175 bp for an inactive globin gene (53). In contrast, similar studies of the β-globin LCR HS2 enhancer element (33) demonstrated that although nucleosomes encompassing this enhancer were disrupted, the surrounding nucleosome array was not mobilized.
Conventional models of chromatin organization dictate that a nucleosome occupies 146 to 168 bp of DNA and that a linker of 10 to 40 bp is required to enable nucleosomes to zigzag across a 25- to 30-nm diameter chromatin fiber (52, 54). The repeat length of 150 to 153 bp that we observe for mobilized nucleosomes does not conform to these strict rules. However, Zlatanova et al. have proposed that chromatin fibers may assemble from nucleosomes containing as little as 120 to 130 bp of DNA (54). In practice, a shorter repeat length could arise by a variety of mechanisms, such as (i) a collapse of the folded 30-nm-diameter chromatin fiber to an extended 10-nm-diameter conformation, (ii) a reduction in the average of 1.7 turns of DNA present on each nucleosome, (iii) an alteration in the composition of the nucleosomes, (iv) a decrease in the level of association of histone H1 and HMG proteins with nucleosomes, or (v) the replacement of nucleosomes by alternate complexes. Cavalli and Thoma suggested that nucleosome compression might occur when nucleosomes are displaced by a passing polymerase and that they reassociate behind the polymerase (8). It will obviously be of great interest to determine whether intergenic transcription occurs within the GM-CSF enhancer, as this is one of the mechanisms that could account for the observed mobilization of nucleosomes upstream of the GM-CSF gene.
Mechanisms of DH site induction and chromatin reorganization.
At this time, we do not know whether NFAT/AP-1 complexes alter nucleosome organization by an intrinsic mechanism or by recruiting remodelling complexes, and there are several potential mechanisms. On the one hand, factors that interfere with the histone-histone contacts that mediate nucleosome stacking are likely to increase DNA accessibility within chromatin. For example, HNF3 is thought to function as a pioneer factor by this type of mechanism, as it makes contacts with histones H3 and H4 and disrupts associations between nucleosomes within histone H1-compacted chromatin fibers (10). The cooperative binding of transcription factors to arrays of sites is also known to be sufficient to disrupt nucleosomal organization without always necessarily dislodging histone octamers (3, 7, 38). Alternatively, recent studies now show that enhanced nuclease sensitivity within nucleosomes can also arise via a SWI/SNF-dependent remodelling activity that remodels nucleosomes (19), but not via an ISWI-type activity which would instead be expected to reposition nucleosomes (19, 20). SWI/SNF has the capacity to create loops on the surface of the nucleosome and expose DNA sequences accessible to nucleases at multiple sites (19). There is already some indication that SWI/SNF-dependent remodelling occurs at the GM-CSF promoter, which contains a positioned nucleosome that is destabilized upon activation (32). In these studies, it was demonstrated that the association of NF-κB and Sp1 to the promoter was accompanied by recruitment of Brg-1.
The actual mechanisms of remodelling and the order of recruitment of the different factors leading to the formation of a DH site will be important avenues of investigation in future studies but are beyond the scope of this investigation. NFAT and AP-1 are both known to recruit histone acetyltransferases (2, 21), which is likely to contribute to the overall level of chromatin accessibility at the GM-CSF enhancer. However, complexes containing Brg-1 remain strong candidates for factors that could destabilize nucleosomes within the GM-CSF enhancer in an NFAT-dependent manner.
Other NFAT-dependent activities in a chromatin context.
NFAT is known to associate with inducible DH sites at many NFAT-dependent enhancers where it may promote chromatin remodelling. NFAT also appears to be required for formation of these DH sites, because they do not form when NFAT induction is suppressed by CsA (15, 18, 25, 35, 49, 50). Although NFAT does not appear to activate any genes by itself (17), it is known to cooperate with many classes of transcription factors in addition to AP-1; these include Oct-1, c-Maf, GATA, Sp1, and EGR (17, 18, 29). It has also been established that cooperation between NFAT and AP-1 is required only for a subset of NFAT-dependent genes (39). We propose that NFAT functions within many genes, at least in part by creating access to adjacent transcription factor binding sites. It remains to be determined at what level NFAT functions and whether it is a true pioneer factor that can disrupt condensed chromatin arrays containing histone H1.
Acknowledgments
We thank C. Bonifer and H. Tagoh for assistance in establishing in vivo footprinting assays, F. Ponchel for assistance in establishing real-time PCR assays, S. Harvey and J. Burrows for assistance with plasmid constructions, J. Gasson for providing a GM-CSF promoter plasmid, A. Rao for providing NFAT antibodies and a NFAT expression vector, D. Cohen and T. Curran for providing AP-1 expression vectors, G. Crabtree for providing a Brg-1 expression vector, T. Honjo for providing a clone of the mouse immunoglobulin H locus, and C. Bonifer for assistance revising the manuscript. We also thank M. Vadas and A. Markham for their support, and M. F. Shannon for her input in our earlier collaborations that led up to this study.
This work was supported in part by the NH&MRC, the South Australian Anti-Cancer Foundation, and Yorkshire Cancer Research.
REFERENCES
- 1.Adams, C. C., and J. L. Workman. 1995. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol. Cell. Biol. 15:1405-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. [DOI] [PubMed] [Google Scholar]
- 3.Beato, M., and K. Eisfeld. 1997. Transcription factor access to chromatin. Nucleic Acids Res. 25:3559-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bert, A. G., J. Burrows, A. Hawwari, M. A. Vadas, and P. N. Cockerill. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646-5655. [DOI] [PubMed] [Google Scholar]
- 5.Bert, A. G., J. Burrows, C. S. Osborne, and P. N. Cockerill. 2000. Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high copy replication. Plasmid 44:173-182. [DOI] [PubMed] [Google Scholar]
- 6.Boise, L. H., B. Petryniak, X. Mao, C. H. June, C. Y. Wang, T. Lindsten, R. Bravo, K. Kovary, J. M. Leiden, and C. B. Thompson. 1993. The NFAT-1 DNA binding complex in activated T cells contains Fra-1 and JunB. Mol. Cell. Biol. 13:1911-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyes, J., J. Omichinski, D. Clark, M. Pikaart, and G. Felsenfeld. 1998. Perturbation of nucleosome structure by the erythroid transcription factor GATA-1. J. Mol. Biol. 279:529-544. [DOI] [PubMed] [Google Scholar]
- 8.Cavalli, G., and F. Thoma. 1993. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 12:4603-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., J. N. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392:42-48. [DOI] [PubMed] [Google Scholar]
- 10.Cirillo, L. A., F. R. Lin, I. Cuesta, D. Friedman, M. Jarnik, and K. S. Zaret. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9:279-289. [DOI] [PubMed] [Google Scholar]
- 11.Cockerill, P. N. 2000. Identification of DNase I hypersensitive sites within nuclei. Methods Mol. Biol. 130:29-48. [DOI] [PubMed] [Google Scholar]
- 12.Cockerill, P. N., A. G. Bert, F. Jenkins, G. R. Ryan, M. F. Shannon, and M. A. Vadas. 1995. Human granulocyte-macrophage colony-stimulating factor enhancer function is associated with cooperative interactions between AP-1 and NFATp/c. Mol. Cell. Biol. 15:2071-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockerill, P. N., A. G. Bert, D. Roberts, and M. A. Vadas. 1999. The human granulocyte-macrophage colony-stimulating factor gene is autonomously regulated in vivo by an inducible tissue-specific enhancer. Proc. Natl. Acad. Sci. USA 96:15097-15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cockerill, P. N., C. S. Osborne, A. G. Bert, and R. J. M. Grotto. 1996. Regulation of GM-CSF gene transcription by core-binding factor. Cell Growth Differ. 7:917-922. [PubMed] [Google Scholar]
- 15.Cockerill, P. N., M. F. Shannon, A. G. Bert, G. R. Ryan, and M. A. Vadas. 1993. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A sensitive enhancer. Proc. Natl. Acad. Sci. USA 90:2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabtree, G. R. 1999. Generic signals and specific outcomes: signalling through Ca2+, calcineurin, and NF-AT. Cell 96:611-614. [DOI] [PubMed] [Google Scholar]
- 17.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109 (Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 18.Duncliffe, K. D., A. G. Bert, M. A. Vadas, and P. N. Cockerill. 1997. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity 6:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Fan, H.-Y., X. He, R. E. Kingston, and G. J. Narlikar. 2003. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11:1311-1322. [DOI] [PubMed] [Google Scholar]
- 20.Fazzio, T. G., and T. Tsukiyama. 2003. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell 12:1333-1340. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrity, P. A., D. Chen, E. V. Rothenberg, and B. J. Wold. 1994. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell. Biol. 14:2159-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasson, J. C. 1991. Molecular physiology of granulocyte-macrophage colony-stimulating activity. Blood 77:1131-1145. [PubMed] [Google Scholar]
- 24.Gutierrez, J., J. Sierra, R. Medina, M. Puchi, M. Imschenetzky, A. van Wijnen, J. Lian, G. Stein, J. Stein, and M. Montecino. 2000. Interaction of CBF alpha/AML/PEBP2 alpha transcription factors with nucleosomes containing promoter sequences requires flexibility in the translational positioning of the histone octamer and exposure of the CBF alpha site. Biochemistry 39:13565-13574. [DOI] [PubMed] [Google Scholar]
- 25.Hawwari, A., J. Burrows, M. A. Vadas, and P. N. Cockerill. 2002. The human IL-3 locus is regulated cooperatively by two NFAT-dependent enhancers that have distinct tissue-specific activities. J. Immunol. 169:1876-1886. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez-Munain, C., and M. S. Krangel. 2002. Distinct roles for c-Myb and core binding factor/polyoma enhancer-binding protein 2 in the assembly and function of a multiprotein complex on the TCR delta enhancer in vivo. J. Immunol. 169:4362-4369. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez-Munain, C., J. L. Roberts, and M. S. Krangel. 1998. Cooperation among multiple transcription factors is required for access to minimal T-cell receptor α-enhancer chromatin in vivo. Mol. Cell. Biol. 18:3223-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hershkovitz, M., and A. D. Riggs. 1997. Ligation-mediated PCR for chromatin-structure analysis of interphase and metaphase chromatin. Methods 11:253-263. [DOI] [PubMed] [Google Scholar]
- 29.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205-2232. [DOI] [PubMed] [Google Scholar]
- 30.Holloway, A. F., S. Rao, X. Chen, and M. F. Shannon. 2003. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor κB proteins. J. Exp. Med. 197:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain, J., P. G. McCaffrey, Z. Miner, T. K. Kerpola, J. N. Lambert, G. L. Verdine, T. Curran, and A. Rao. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365:352-355. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, F., P. N. Cockerill, D. Bohman, and M. F. Shannon. 1995. Multiple signals are required for function of the human granulocyte-macrophage colony-stimulating factor gene promoter in T cells. J. Immunol. 155:1240-1251. [PubMed] [Google Scholar]
- 33.Kim, A., and A. Dean. 2003. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 23:8099-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontaraki, J., H. H. Chen, A. Riggs, and C. Bonifer. 2000. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 14:2106-2122. [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, D. U., O. Avni, L. Chen, and A. Rao. 2004. A distal enhancer in the IFN-γ locus revealed by genome sequence comparison. J. Biol. Chem. 279:4802-4810. (First published 8 November 2003; 10.1074/jbc.M307904200.) [DOI] [PubMed]
- 36.Lee, H. J., E. S. Masuda, N. Arai, K. Arai, and T. Yokota. 1995. Definition of cis-regulatory elements of the mouse interleukin-5 gene promoter. Involvement of nuclear factor of activated T cell-related factors in interleukin-5 expression. J. Biol. Chem. 270:17541-17550. [DOI] [PubMed] [Google Scholar]
- 37.Li, B., C. C. Adams, and J. L. Workman. 1994. Nucleosome binding by the constitutive transcription factor Sp1. J. Biol. Chem. 269:7756-7763. [PubMed] [Google Scholar]
- 38.Li, Q., O. Wrange, and P. Eriksson. 1997. The role of chromatin in transcriptional regulation. Int. J. Cell Biol. 29:731-742. [DOI] [PubMed] [Google Scholar]
- 39.Macian, F., C. Garcia-Rodriguez, and A. Rao. 2000. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19:4783-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda, E. S., Y. Naito, K. Arai, and N. Arai. 1993. Expression of lymphokine genes in T cells. Immunologist 1:198-203. [Google Scholar]
- 41.Mattila, P. S., K. S. Ullman, S. Fiering, E. A. Emmel, M. McCutcheon, G. R. Crabtree, and L. A. Herzenberg. 1990. The actions of cyclosporin A and FK506 suggest a novel step in the activation of T lymphocytes. EMBO J. 9:4425-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 44.Nicola, N. A. 1989. Hemopoietic cell growth factors and their receptors. Annu. Rev. Biochem. 58:45-77. [DOI] [PubMed] [Google Scholar]
- 45.Rao, A. 1994. NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol. Today 15:274-281. [DOI] [PubMed] [Google Scholar]
- 46.Rao, A., C. Luo, and P. G. Hogan. 1997. Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15:707-747. [DOI] [PubMed] [Google Scholar]
- 47.Rooney, J. W., Y. L. Sun, L.H. Glimcher, and T. Hoey. 1995. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol. Cell. Biol. 15:6299-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon, M. F., L. S. Coles, M. A. Vadas, and P. N. Cockerill. 1997. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit. Rev. Immunol. 17:301-323. [DOI] [PubMed] [Google Scholar]
- 49.Siebenlist, U., D. B. Durand, P. Bressler, N. J. Holbrook, C. A. Norris, M. Kamoun, J. A. Kant, and G. R. Crabtree. 1986. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol. Cell. Biol. 6:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solymar, D. C., S. Agarwal, C. H. Bassing, F. W. Alt, and A. Rao. 2002. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity 17:41-50. [DOI] [PubMed] [Google Scholar]
- 51.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 52.Woodcock, C. F. L., L.-L. Y. Frado, and J. B. Rattner. 1984. The higher order structure of chromatin. J. Cell Biol. 99:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, M., M. B. Barnard, S. M. Rose, P. N. Cockerill, S. Y. Huang, and W. T. Garrard. 1986. Transcription termination and chromatin structure of the active immunoglobulin kappa gene locus. J. Biol. Chem. 261:3838-3845. [PubMed] [Google Scholar]
- 54.Zlatanova, J., S. H. Leuba, and K. van Holde. 1999. Chromatin structure revisited. Crit. Rev. Eukaryot. Gene Expr. 9:245-255. [DOI] [PubMed] [Google Scholar]