Abstract
By employing the nuclear DNA of the African rice Oryza glaberrima as a reference genome, the timing, natures, mechanisms, and specificities of recent sequence evolution in the indica and japonica subspecies of Oryza sativa were identified. The data indicate that the genome sizes of both indica and japonica have increased substantially, >2% and >6%, respectively, since their divergence from a common ancestor, mainly because of the amplification of LTR-retrotransposons. However, losses of all classes of DNA sequence through unequal homologous recombination and illegitimate recombination have attenuated the growth of the rice genome. Small deletions have been particularly frequent throughout the genome. In >1 Mb of orthologous regions that we analyzed, no cases of complete gene acquisition or loss from either indica or japonica were found, nor was any example of precise transposon excision detected. The sequences between genes were observed to have a very high rate of divergence, indicating a molecular clock for transposable elements that is at least 2-fold more rapid than synonymous base substitutions within genes. We found that regions prone to frequent insertions and deletions also exhibit higher levels of point mutation. These results indicate a highly dynamic rice genome with competing processes for the generation and removal of genetic variation.
Comparisons of orthologous genomic sequences from multiple grass species have revealed numerous genic rearrangements (e.g., gene insertions, deletions, duplications, and/or translocations) under the umbrella of overall genomic colinearity (1). Most of these studies have featured barley, maize, rice, sorghum, and wheat, all of which are grasses that diverged from common ancestors ≈10–60 million years ago (mya) (2–4). Although some conserved noncoding sequences (CNSs) have been detected (5, 6), these cereal genomes show very little sequence homology in intergenic regions. Rates of genome change outside genes are so rapid that most individual events cannot be precisely defined in species that have evolved independently for these long times. Only a subset of intragenic changes are observed because of the filtration applied by natural selection. Hence, very little is known about the nature or mechanisms of recent genomic sequence variation.
With the near completion of Arabidopsis and rice genome sequences (7–11), LTR-retrotransposons were used to explore the rates and mechanisms of recent DNA loss in these two species (12–14). These studies indicated that homologous unequal recombination and illegitimate recombination are primarily responsible for removal of DNA from LTR-retrotransposons (15). In fact, an accumulation of small deletions was estimated to be responsible for the removal of at least 194 Mb of LTR-retrotransposon sequences from the genome of Oryza sativa subspecies japonica over the last 5 million years (14). These processes were also detected as major forces behind DNA removal in other plant species (16–18). However, whether they affect more than just transposable elements has not been determined.
Analysis of orthologous genomic sequences from closely related subspecies and/or species could provide clues to recent genome variation. A sequence comparisons of large colinear regions from two cultivated subspecies of Asian rice (O. sativa L.), indica and japonica, has been very informative (9, 19). Feng et al. (9) found numerous single nucleotide polymorphisms (SNPs) and small indels (insertions or deletions) within repetitive DNA and genes in ≈2.3 Mb of orthologous regions from indica and japonica. Because the compared regions were larger overall in japonica than indica, Han and Xue (19) proposed that the japonica genome was expanding relative to indica. However, these studies could rarely identify the precise natures of specific DNA sequence changes, because it was usually not clear whether the changes occurred in the indica or japonica lineage.
Oryza glaberrima is a cultivated rice species that was domesticated in Africa ≈2,000–3,000 years ago (20). Phylogenetic studies have consistently demonstrated that O. glaberrima is a close sister species to O. sativa (21–25) and exhibits the least intraspecies genetic variation compared with other Oryza species that have been investigated (23, 25). Hence, we believe that O. glaberrima is an appropriate reference to characterize the nature of DNA sequence variation between the indica and japonica subspecies of O. sativa.
We used comparisons to O. glaberrima to analyze sequence variation across >1 Mb of genomic DNA from indica and japonica. We report here natures, rates, lineages, and apparent mechanisms of DNA rearrangements and point mutations in these two subspecies.
Materials and Methods
DNA Isolation, PCR, and Sequencing. O. glaberrima seed (accession no. PI232853) was provided by Scott Jackson (Purdue University, West Lafayette, IN). DNA isolation, PCR primer design, PCR amplification, and sequencing of PCR fragments were conducted as described in Supporting Text, which is published as supporting information on the PNAS web site.
Sequence Alignments and Comparisons. The bacterial artificial chromosome (BAC) sequences from rice subspecies indica (cv. GLA4), which were deposited in GenBank by December 13, 2002, were used in blastn (National Center for Biotechnology Information, BLAST 2.0) searches against the rice nonredundant genomic sequence database to determine their orthologous regions from the japonica subspecies (cv. Nipponbare). The orthologous regions identified between indica and japonica were extracted from the complete BAC sequences. The conserved segments, indels, and substitution mutations in the orthologous regions of indica and japonica were identified by using the blast2 (26), cross_match (www.phrap.org/phrap.docs/general.html), gap (Wisconsin Package Version 10.1, Genetics Computer Group, Madison, WI), and clustalx (27) programs.
Amplified fragments from O. glaberrima were used in blastn searches against the rice nonredundant genomic sequence database to determine and confirm their orthologous relationships to the targeted regions (spanned by primer pairs) in indica and japonica. Only the O. glaberrima fragments with the highest degree of homology to the targeted regions of indica or japonica were considered to be orthologs and further analyzed. The O. glaberrima sequences and their orthologous segments from indica and japonica were extracted and aligned by using clustalx to determine the natures of small DNA rearrangements (e.g., insertion or deletion). The substitution mutations in these regions were identified by using the mega2 program (28) and confirmed by manual inspection. The O. glaberrima sequence alignments were individually inspected and, if needed, edited manually. When O. glaberrima was used as a reference to identify substitution mutations in the orthologous regions of indica and japonica, only regions with at least two sequences generated for each PCR fragment were chosen and only identical bases in these sequence data were used to minimize possible errors from sequencing PCR products.
Sequence Annotation. Sequence annotations were performed as described (29), with some modifications as described in Supporting Text.
Dating Rice Divergence and LTR-Retrotransposon Insertions. The strategies for dating rice divergence and LTR-retrotransposon insertions are described in Supporting Text.
Statistical Analysis. We randomly dissected 76 nonredundant orthologous subregions of indica and japonica (≈10 kb of common sites per subregion) to investigate the statistical significance regarding differences of levels of point mutations and frequency of unequal homologous recombinations between indica and japonica genomic regions. A detailed description of this analysis is provided in Supporting Text.
Results
Recent DNA Variation in Orthologous Regions of indica and japonica. All finished BAC sequences from indica cultivar GLA4 (deposited in GenBank by December 13, 2002; no more BAC sequences from GLA4 have been deposited in GenBank) were used to anchor their orthologous regions from japonica cultivar Nipponbare. Twelve orthologous regions composed of 14 finished BACs from indica and 15 finished BACs from japonica were identified, which constitute 1.15 Mb of indica and 1.20 Mb of japonica DNA, respectively.
We investigated the base substitution (transition and transversion) mutations in these conserved regions between indica and japonica. In 972 kb of common sites analyzed (the rest differed by insertions or deletions), we found 9,383 substitutions (6,293 transitions and 3,090 transversions). These data indicate that the overall sequence identity between the investigated regions from indica and japonica is 99.04% if the indels are excluded.
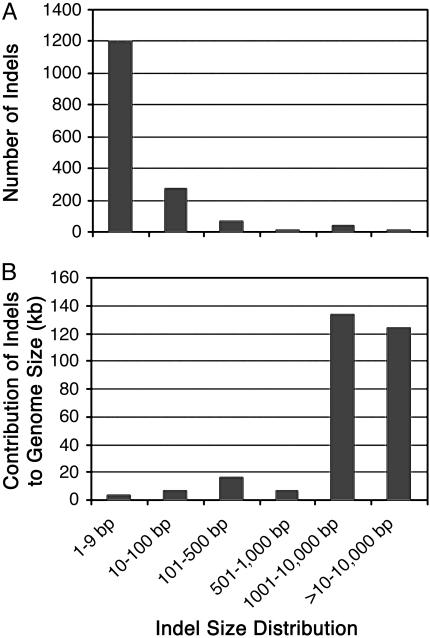
In these orthologous regions, we identified a total of 1,587 indels based on sequence alignments. Of these indels, 792 appear to be insertions in indica relative to japonica, whereas 795 appear to be insertions in japonica relative to indica, accounting for 124,787 and 174,255 bp of DNA sequence, respectively. Hence, ≈13% of the DNA sequence in the compared regions are different between indica and japonica. Large indels with sizes ≥1 kb are rare but are primarily responsible for the different sizes of orthologous regions between indica and japonica (Fig. 1). Small indels, most with sizes of 1 or 2 bp, far outnumber the larger ones, but contribute little to overall genome size variation in these regions (Fig. 1).
Fig. 1.
Indels and their contribution to genome size in orthologous regions of indica and japonica. Indels identified in indica and japonica with specified sizes were pooled into six groups, as shown on the x axis. Bars indicate number of indels (A) and their contribution to genome size (B).
Nucleotide Substitution in Genic and Intergenic Regions. To date the divergence of indica and japonica subspecies, we calculated the level of nucleotide substitution in 24 protein-encoding genes in the orthologous regions of indica and japonica investigated in this study. These 24 randomly chosen genes exhibited an average degree of nucleotide substitution in genic regions (0.29%) that was significantly lower than observed in overall orthologous regions (0.96%) (P < 0.001, Fisher's exact test) (see Table 3, which is published as supporting information on the PNAS web site). In contrast, the intergenic regions analyzed in this study exhibited relatively rapid sequence divergence. The average level of site substitution in intergenic regions was 1.18%, ≈4 times higher than in genic regions (see Table 3).
We further calculated synonymous and nonsynonymous substitution levels in coding regions of the 24 genes by the method of Nei-Gojobori (30) using the Jukes–Cantor correction. These genes exhibited variable levels of synonymous substitution and nonsynonymous substitution, ranging from 0 to 2.5% and from 0 to 0.8%, with average rates of 0.58% and 0.15%, respectively (see Table 4 and Fig. 3, which are published as supporting information on the PNAS web site). This degree of variation is primarily caused by the small number of scorable sites per gene. Hence, the combined average for all 24 analyzed genes was used to determine overall divergence. A few genes exhibited exceptionally high levels of nonsynonymous substitutions (genes 5, 17, and 19), suggesting possible diversifying selection. Other genes, for instance 23 and 24, exhibited an apparent higher-than-average rate of synonymous substitution. This summing of degrees of synonymous divergence for 24 genes should minimize the inaccuracy in a molecular clock based on any single gene that might have an unusual rate of synonymous site change (31). By employing an average substitution rate of 6.5 × 10–9 mutations per synonymous site per year determined for the coding regions of adh1 and adh2 genes in grasses (32), we estimate that the indica and japonica genomes in this study last shared a common ancestor ≈0.44 mya.
Sequencing Targeted Genomic Regions from O. glaberrima. Based only on sequence comparisons of these two O. sativa subspecies, one cannot determine whether the nucleotide substitution mutations or indels identified by sequence alignments occurred in indica or japonica. That is, if japonica has a 300-bp insertion relative to indica, it is not clear whether this was an insertion in japonica or a deletion from indica. Hence, determination of the exact nature of point mutations and DNA rearrangement events requires additional information. For this purpose, we used the genome of O. glaberrima, a close relative of O. sativa (21–25). PCR primers were designed according to the consensus sequences flanking indels that were 10 bp or larger detected in our indica::japonica comparisons. Some pairs of primers were designed to span two or more adjacent indels. Indels <10 bp were not included in this study because >60% of them are associated with simple sequence repeats or short tandem duplication (data not shown). Moreover, these tiny indels accounted for a small portion of the differences in DNA quantity in any region, despite their high frequency (Fig. 1).
PCR amplifications were conducted in O. glaberrima by using 263 pairs of primers. We successfully amplified 196 (75%) “single-band” products as shown on agarose gels. These fragments would be expected to be orthologous to the corresponding indel regions investigated in indica and japonica and thus were purified and directly sequenced with the primers used in the corresponding PCRs. In addition, 26 “multiple-band” products were obtained, suggesting that paralogous sequences, repetitive DNA or some other regions sharing the same primer sites in the O. glaberrima genome may have been amplified. These 26 products (10%) were excluded from further sequence analysis. The other 41 attempted amplifications (15%) failed to generate any distinct fragments, perhaps because of sequence divergence at primer sites and/or large DNA rearrangements in the targeted regions of O. glaberrima.
The 196 “single-band” products amplified from O. glaberrima were sequenced. These sequences were blast-searched against the rice genomic sequences produced by the International Rice Genome Sequencing Program to check their best matches. We found that 168 of these 196 O. glabberima sequences were appropriately aligned to the expected indel regions in indica and japonica, demonstrating that these aligned sequences were orthologous. The other 28 O. glaberrima sequences exhibited better matches to other regions of the indica and japonica genomes. These apparent paralogous amplification products were not used for further analysis.
Phylogenetic Analysis of indica, japonica, and O. glaberrima. To validate the use of O. glaberrima as an outgroup to identify the natures of recent DNA changes detected in indica and japonica, we investigated sequence divergence of the orthologous regions of indica, japonica, and O. glaberrima. To minimize possible errors from sequencing O. glaberrima PCR fragments, regions were analyzed only when at least two sequences were generated for each PCR fragment. Any differences between these overlapping reads of a single PCR fragment were attributed to sequencing errors and were excluded from sequence divergence analysis.
In a total of 36,952 bp of common sites investigated across O. glaberrima and the two O. sativa subspecies, we identified 519 substitutions between indica and japonica, 764 substitutions between indica and O. glaberrima, and 701 substitutions between japonica and O. glaberrima. The corresponding percentages of sequence divergence are 1.4%, 2.1%, and 1.9%, respectively. It is apparent that indica and japonica share the highest sequence homology among these three subspecies/species in the orthologous regions investigated. As expected, O. glaberrima was found to be about equally distant from both indica and japonica. These data support the use of the nuclear DNA of O. glaberrima as a reference genome to investigate DNA changes in indica and japonica.
As calculated above, the indica and japonica haplotypes characterized in this study have experienced independent variation for ≈0.44 million years. By this same molecular clock approach, we calculate that O. glaberrima diverged from a common ancestor with indica and japonica ≈0.64 mya (see Fig. 4, which is published as supporting information on the PNAS web site).
Different Rates of Point Mutation in indica and japonica. By using the O. glaberrima sequence as a reference, the rates of point mutation in the shared 36,952 bp of the indica and japonica genomes were analyzed. Of 519 substitution mutations identified between indica and japonica, 291 (188 transitions and 103 transversions) were found in indica, whereas the other 228 mutations (158 transitions and 70 transversions) were found in japonica (see Table 5, which is published as supporting information on the PNAS web site). The average rate of site substitution in the indica regions is significantly higher than in the corresponding japonica regions (P < 0.05, Fisher's exact test).
In both subspecies, transitions are more frequent than transversions. The average ratio of transition to transversions is ≈2:1. As demonstrated in the adh1 region of maize, the extensive deoxycytidine-methylation of repetitive DNA and spontaneous deamination of 5′ methylcytosine could be largely responsible for the higher rate of transitions relative to transversions (33).
Natures of DNA Rearrangement in Orthologous Regions of indica and japonica. Amplified sequences from O. glaberrima were aligned to their orthologous regions of indica and japonica to determine the nature of the ≥10-bp indels that were identified. By this approach, a total of 62 insertions, 85 deletions, 17 tandem duplications, and 28 changes of simple sequence repeats were identified (Table 1).
Table 1. Amount of DNA involved in small DNA arrangements.
| Subspecies | Small DNA rearrangements* | Number | Total size, bp |
|---|---|---|---|
| indica | Insertions† | 41 | 81,202 |
| Insertions of LTR-retrotransposons | 13 | 56,775 | |
| Insertions of MITEs | 16 | 4,353 | |
| Insertions of other DNA transposons | 2 | 15,627 | |
| Insertions (unknown) | 10 | 4,447 | |
| Deletions | 51 | 19814‡ | |
| Unclear indels§ | 92 | 38,735 | |
| Tandem duplications | 8 | 298 | |
| Amplification of SSRs | 9 | 563 | |
| japonica | Insertions† | 42 | 111,127 |
| Insertions of LTR-retrotransposons | 15 | 95,065 | |
| Insertions of MITEs | 16 | 4,085 | |
| Insertions of other DNA transposons | 1 | 11,480 | |
| Insertions (unknown) | 10 | 4,431 | |
| Deletions | 38 | 2487 | |
| Unclear indels¶ | 84 | 40,885 | |
| Tandem duplications | 9 | 309 | |
| Amplification of SSRs | 19 | 611 |
A total of 604 and 590 small indels <10 bp are excluded from this table, accounting for 1,502 and 1,509 bp of indica and japonica genomic sequences, respectively.
Thirteen of 28 insertions of LTR-retrotransposons and 8 of 32 insertions of MITEs were judged by structural analysis.
Loss of 5,135 bp of DNA from a LTR-retrotransposon by homologous recombination and a deletion of a 12,749-bp fragment from a LTR-retrotransposon were identified based on sequence alignments relative to known LTR-retrotransposons.
Insertions in indica relative to japonica.
Insertions in japonica relative to indica.
Of the 62 inserted sequences, 15 were LTR-retrotransposons, 24 were miniature inverted repeat transposable elements (MITEs), and 3 were other DNA transposons. All of these elements show typical structural features of DNA or RNA transposons (34, 35). The origins of 20 other insertions remain unclear because they neither show structural features of transposable elements nor share significant sequence homology with known transposable elements.
We also analyzed the 85 deletions and their flanking sequences. Of these deletions, 64 were found in single-copy segments of the rice genome, including three deletions within introns of three separate genes and one deletion within an exon of another gene. We did not find any deletions of complete genes in all regions of indica and japonica that we investigated. The other 21 deletions were harbored within transposable elements and uncharacterized repetitive DNA, but no case of complete transposon excision was detected.
Recent Growth of the indica and japonica Genomes. To shed additional light on the direction of possible recent genome size change in rice, we attempted to identify insertions or deletions that were not characterized because they did not amplify from orthologous segments of the O. glaberrima genome. One approach used, as described (12, 14), was to define deletion or insertion events by aligning LTR-retrotransposons harboring indels to multiple elements belonging to the same family. Through this approach, two deletions of large fragments within two LTR-retrotransposons were identified. Another technique used was to analyze the structures of indels. We regarded an indel as an insertion if it had the structure of an LTR-retrotransposon or DNA transposon and was bounded by a target-site duplication. Based on this analysis, an additional 13 insertions of LTR-retrotransposons, eight insertions of MITEs, and four deletions of partial sequences from LTR-retrotransposons were identified in the orthologous regions of indica and japonica (Table 1) among the regions that did not amplify by PCR in O. glaberrima.
In total, we found 13 insertions of LTR-retrotransposons in indica and 15 insertions of LTR-retrotransposons in japonica, which make up 57 and 95 kb of genomic DNA, corresponding to 5% and 8% of the respective regions investigated. These data indicate that LTR-retrotransposons have played a central role in expanding the indica and japonica genomes. In addition, 28 and 27 insertions of MITEs, other DNA transposons, and unknown small insertions were found in indica and japonica, respectively (Table 1).
In contrast to insertions, 51 and 38 deletions ≥10 bp were detected in the orthologous regions of indica and japonica, respectively. Although the deletions outnumber insertions in both indica and japonica, the majority of these deletions are relatively small (10–450 bp), accounting for a small fraction of all rearranged sequences. However, a 9- and a 5-kb deletion were identified in the indica regions through structural analysis of LTR-retrotransposons, indicating that individual deletions of relatively large fragments can occur.
Because of a lack of PCR amplification in O. glaberrima, 92 and 84 indels remain unclear, constituting 39 kb of sequence in indica and 41 kb of sequence in japonica. However, given that the large sizes of insertions of LTR-retrotransposons provided more DNA than the total sizes of deletions and unclear indels combined, it is apparent that both indica and japonica have substantially expanded their genomes in the orthologous regions investigated since their divergence from a common ancestral species. Even if all of the unclear indels were proposed to be deletions, we still calculate ≈2% and 6% increases in the genomic sizes of indica and japonica, respectively, since their divergence from a common ancestor with O. glaberrima, in the investigated regions (Table 1).
Recent Unequal Homologous Recombination in Orthologous Regions of indica and japonica. More precise analysis was made of the structures of LTR-retrotransposons inserted into these orthologous regions. In indica, we found that 9 of the 13 LTR-retrotransposons are solo LTRs, whereas 8 of 15 LTR-retrotransposons are solo LTRs in japonica (Table 2). A solo LTR is the product of unequal homologous recombination between the two LTRs of a single element. These data demonstrate that >50% of LTR-retrotransposons inserted into the orthologous regions of indica and japonica rice after their divergence from a common ancestor have undergone unequal homologous recombination, leading to >70 kb of DNA loss from these elements in the indica and japonica regions, respectively (data not shown). Hence, the competitive processes of insertion and subsequent unequal homologous recombination of LTR-retrotransposons are major determinants of genome size variation in rice.
Table 2. Structures and ages of LTR-retrotransposons identified in orthologous regions of indica and japonica.
| Structures of LTR-retrotransposons
|
Number of LTR-retrotransposons
|
Age,* mya
|
|
|---|---|---|---|
| Range | Average | ||
| Present in indica only | |||
| Solo LTR | 9 | ||
| Intact element | 4 | 0.12-0.31 | 0.22 |
| Present in japonica only | |||
| Solo LTR | 8 | ||
| Intact element | 7 | 0.00-0.31 | 0.16 |
| Present in indica and japonica† | |||
| Solo LTR | 13 and 12 | ||
| Intact element | 1 and 2 | 0.69-2.04 | 1.37 |
Ages of LTR-retrotransposons were estimated by employing a substitution rate of 1.3 × 10-8 mutations per site per year.
Thirteen and 12 of 14 conserved LTR-retrotransposons between indica and japonica have undergone unequal homologous recombination, respectively.
Measurements of sequence divergences of two LTRs from single retrotransposons have been used to estimate the time of LTR-retrotransposon insertion in grasses (14, 33, 36). The dates calculated in these studies used a gene-based molecular clock, although it was expected that LTR-retrotransposons diverged more rapidly than genes (14, 33).
The average level of nucleotide substitution in intergenic regions (1.18%) was ≈2-fold higher than that of synonymous substitution in coding regions of genes (0.58%), as demonstrated above. Hence, we propose that a substitution rate of 1.3 × 10–8 mutations per site per year, 2-fold higher than determined for the coding regions of adh1 and adh2 genes in grasses (32), is appropriate to date the insertions of LTR-retrotransposons. When this rate was used, all 11 intact LTR-retrotransposons that were uniquely present in either indica or japonica were found to be younger than 0.31 million years, with average ages of <0.22 million years (Table 2). This observation strongly supports the conclusion that these 11 elements inserted into indica or japonica after their divergence from a common ancestor.
We also identified 14 conserved LTR-retrotransposons in both studied regions of indica and japonica. Of these 14 elements, 1 is an intact LTR-retrotransposons and 13 are solo LTRs in indica, whereas 2 are intact elements and 12 are solo LTRs in japonica. In contrast to the LTR-retrotransposons uniquely present in the orthologous regions of indica or japonica, which were composed of 61% solo LTRs, the conserved LTR-retrotransposons contain a significantly higher percentage of solo LTRs (89%) (P < 0.05, Fisher's exact test). This finding agrees with the previous observation that older insertions are more commonly found as solo LTRs (14). The insertions of two intact elements shared by indica and japonica were dated to ≈0.7 and 2 mya (Table 2).
Recent Illegitimate Recombination in Orthologous Regions of indica and japonica. We examined the breakpoints of all ≥10-bp deletions not associated with solo LTR generation in this study. Of 88 deletions, 78 (89%) were bounded by short flanking repeats (FRs) of 2–21 bp. This phenomenon has been previously observed within LTR-retrotransposons in the Arabidopsis, wheat, and rice genomes (12, 14, 18) and is a hallmark of illegitimate recombination. Thus, our study suggests that illegitimate recombination is a common process in rice that actively deletes all classes of nuclear DNA sequence (see Table 6, which is published as supporting information on the PNAS web site).
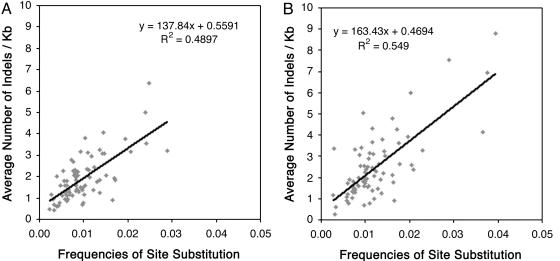
Correlation Between Levels of Point Mutation and Small DNA Rearrangement. Because different levels of point mutation and DNA rearrangement were detected between indica and japonica, we wondered whether the number of point mutations in different regions correlated with the abundance of small DNA rearrangements in the corresponding regions. For this purpose, we randomly dissected the orthologous regions of indica and japonica into 76 nonredundant subregions (≈10 kb of common sites per subregion) and subsequently counted the numbers of point mutations and all indels in each subregion based on sequence alignments. We found that levels of point mutation and small DNA rearrangement in different subregions were extremely variable, regardless of whether the predicted genes were included (Fig. 2A) or excluded (Fig. 2B) from the analysis. This indicates that different classes of DNA sequences or regions within a genome can have distinct evolutionary clocks. Of special interest is a significant linear correlation observed between rates of point mutation and small DNA rearrangement [two-tailed Pearson's correlation coefficient, R = 0.6998, P < 0.001 (Fig. 2 A); R = 0.7409, P < 0.001 (Fig. 2B)].
Fig. 2.
Correlation between frequencies of point mutations and frequencies of indels. A total of 76 redundant subregions, each with ≈10 kb of common sites from the orthologous regions of indica and japonica, were dissected and aligned. The frequency of substitution in each subregion was determined by the mega2 program, and indels in each subregion were identified manually. Pearson's correlation coefficient was used in linear correlation analysis. The significance of the slope of the regression line is determined from the t statistic. (A) Predicted genes were included in each subregion. (B) Predicted genes were excluded in each subregion.
Discussion
Comparative genomic studies have begun to generate data regarding sequence divergence and local genome rearrangement in plants, but we still know very little about the nature and molecular mechanisms of such sequence changes. Most of the organisms previously investigated diverged from ancient common ancestors so long ago that the detected differences were usually the products of multiple overlapping mutational events. Very recently, several comparative sequence analyses have focused on closely related species (18, 37), subspecies (9), and haplotypes (38) to reveal recent genomic sequence variation. In the present study, comparative analysis of finished BAC sequences from two subspecies of O. sativa, indica and japonica, revealed numerous base substitution mutations and small DNA rearrangements in >1.1 Mb of orthologous regions. By using African rice (O. glabberima) as a reference, the timing, nature, mechanisms, and specificities of recent DNA changes that have led to rapid and dramatic divergence of the indica and japonica genomes could be and were identified.
O. glaberrima was chosen for this study based on the phylogenetic relationships of Oryza species inferred by molecular marker analyses (22–25) and gene sequence comparisons (21). These phylogenetic studies consistently place O. glaberrima as a very close sister group to O. sativa. Our present data, at the large-scale genomic sequence level, strongly support these previous observations. Although African rice is currently grown in regions of Africa where O. sativa has become the major rice crop (39), our data do not support extensive gene flow from O. sativa into the accession of O. glaberrima that we used. All transposon insertions that differentiate indica and japonica were found to be absent from O. glaberrima. Moreover, the point mutations that differentiate O. glaberrima from either indica or japonica were not unevenly distributed as would be expected if segments from one of the O. sativa subspecies had been introgressed into the African rice. Hence, we believe that O. glaberrima serves as an appropriate outgroup for these studies. As a secondary outcome of our studies, we found that the divergence of the indica and japonica subspecies dates to ≈0.44 mya, long before the domestication of these two crops. Therefore, our data support models suggesting independent domestication of indica and japonica rice (40, 41).
One intriguing finding is that indica exhibits higher sequence variation than japonica in the orthologous regions investigated in our study. This can be inferred based on three facts. First, the rate of site substitution mutations detected in indica was higher than detected in japonica (P < 0.001, Fisher's exact test) (see Table 5), providing the most direct evidence regarding differentiation of clock-like rates of sequence divergence in indica and japonica. Second, a higher ratio of solo LTRs to intact LTR-retrotransposons was identified in indica than in japonica (Table 2), indicating a higher frequency of unequal homologous recombination in indica. Third, more deletions were found in the indica genome than in the japonica genome (Table 1). Together, these observations suggest that indica has changed more rapidly than japonica in the investigated regions. Interestingly, the ratio of solo LTRs to intact LTR-retrotransposons observed in the japonica regions investigated in this study is very close to that previously identified in the whole genome of japonica (14), suggesting that these regions are a representative sample of the rice genome. Currently, we do not know what factors are responsible for accelerated or decelerated rates of sequence divergence. Different effective population sizes, environmental histories or inherited differences in genes involved in DNA replication, DNA repair, or transposon silencing are likely factors (15, 38, 42, 43).
Beyond the different molecular clocks in indica and japonica, we also found evidence that different components or regions of the same genome exhibited dramatic variation in rates of sequence divergence. Given that intergenic regions diverged ≈4-fold faster than genes in rice as observed in this study, it is easy to understand why only gene islands in the vast genome oceans, in general, show recognizable sequence similarities among all grass species investigated to date (1). Interestingly, the rates of sequence divergence in different intergenic regions also vary greatly, and such variations were found to be associated with the frequencies of small DNA rearrangements in corresponding regions. We do not know whether this indicates different de novo rates of mutation in different adjacent regions or whether it could be caused by variable levels of selection on these regions.
We found that insertions of LTR-retrotransposons are primarily responsible for genome expansion in both indica and japonica. However, the degree of expansion has also been affected by the frequency of subsequent unequal homologous recombination within LTR-retrotransposons. Although unequal homologous recombination has been found to be an active mechanism to remove DNA from LTR-retrotransposons inserted into the rice genome, the residual solo LTRs still yield a net increase in genome size.
One dramatic feature in these comparisons was the great numerical excess of small deletions compared to insertions. These deletions did not make a very large quantitative contribution to local genome size because of their usual small size. However, they were identified in all classes of DNA in both indica and japonica regions. Perhaps most interesting from a mechanistic perspective, these deletions were usually flanked by short sequence repeats. Hence, the association of these deletions with illegitimate recombination, previously identified by structural analyses of LTR-retrotransposons in Arabidopsis (12), is not limited to mobile DNAs. Illegitimate recombination has caused deletions in introns and exons of genes, transposable elements, and other single-, low-, medium-, and high-copy-number DNA sequences, suggesting a common mechanism(s) that can delete all classes of DNA in the rice genome.
Although small indels were detected in both introns and exons, we did not find any case of differential presence/absence of a complete protein-encoding gene in either indica or japonica. This result differs from the previous conclusion obtained by comparison of 2.3 Mb of orthologous regions between indica and japonica (9, 19). In their study, Feng et al. (9) predicted 388 genes in indica regions and 415 genes in japonica regions. They thus proposed numerous exceptions to microcolinearity at the gene level between these two subspecies (19). However, incomplete genomic sequences and automated annotation of the rice genome (44) were used in their investigation. Almost all LTR-retrotransposons, including solo LTRs, identified in our studies were predicted as genes by the gene-finding program fgenesh (data not shown). We found many fewer true genes in the common regions that we compared than did Feng et al. (9), and all confirmed genes were present in both indica and japonica in the orthologous regions. Hence, fine annotation and precise characterization of sequence data are extremely important to unveil the natures of genome organization, structure, and evolution in flowering plants.
There are several currently unsolved methodological issues regarding our indel and point mutation analyses. Although O. glaberrima has been confirmed to be a close sister species to O. sativa, we cannot guarantee that all DNA changes identified by sequence alignments occurred after the divergence of indica and japonica lineages from a common ancestor. Some observed point mutations or DNA rearrangements may have existed as polymorphisms in a shared ancestor of indica and japonica. However, as we described above, all conserved LTR-retrotransposons between the orthologous regions of indica and japonica were far older than 0.44 million years, whereas all LTR-retrotransposons that differed in presence/absence between the regions of indica or japonica were much younger than 0.44 million years (the estimated divergence time of indica and japonica). These data indicate that the unconserved LTR-retrotransposons inserted into the current sites in the orthologous regions of indica or japonica some time after their divergence.
In addition, we cannot tell whether the rapid DNA changes that we observed are general in the indica and japonica lineages or whether they are specific to these two varieties or their direct ancestors. In a previous study, Fu and coworkers sequenced and compared bz1 regions from two maize inbreds (38, 45). They detected exceptional haplotype variation in the investigated regions of maize, including insertions of distinct retrotransposons and deletions of genes from one of the two inbreds investigated, demonstrating that both components and sizes of plant genomes could change rapidly and dramatically. Similarly, further sequence comparisons among multiple rice species, subspecies, and accessions are needed to enrich our understanding of dynamic variation in the rice nuclear genome.
Supplementary Material
Acknowledgments
We thank Scott Jackson for providing seeds of O. glaberrima (accession no. PI232853), Phillip SanMiguel for assistance in DNA sequencing, and Katrien Devos and Susan Wessler for helpful discussions. This research was supported by the Plant Genome Program at the National Science Foundation (Grant 9975793).
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: BAC, bacterial artificial chromosome; mya, million years ago; MITE, miniature inverted repeat transposable element.
Data deposition: The sequence data reported in this study were deposited in the GenBank database (accession nos. CL440284–CL440620, AJ254900, AJ243961, AL117264, AL117265, AL442007, AL442110–AL442115, AL512542, AL512544, AL512546, AL606444, AL606445, AL606455, AL606594, AL606603, AL606606, AL606629, AL606635, AL606648, AL606686, AL606692, AL606998, AL662938, AL662959, and AL662970).
See accompanying Biography on page 12402.
References
- 1.Bennetzen, J. L. & Ma, J. (2003) Curr. Opin. Plant Biol. 6, 128–133. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe, K. H., Gouy, M., Yang, Y.-M., Sharp, P. M. & Li, W.-H. (1989) Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gale, M. D. & Devos, K. M. (1998) Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller, B. & Feuillet, C. (2000) Trends Plant Sci. 5, 246–251. [DOI] [PubMed] [Google Scholar]
- 5.Kaplinsky, N. J., Braun, D. M., Penterman, J., Goff, S. A. & Freeling, M. (2002) Proc. Natl. Acad. Sci. USA 99, 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo, H. & Moose, S. P. (2003) Plant Cell 15, 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki, T., Matsumoto, T., Yamamoto, K., Sakata, K., Baba, T., Katayose, Y., Wu, J., Niimura, Y., Cheng, Z., Nagamura, Y., et al. (2002) Nature 420, 312–316. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Q., Zhang, Y., Hao, P., Wang, S., Fu, G., Huang, Y., Li, Y., Zhu, J., Liu, Y., Hu, X., et al. (2002) Nature 420, 316–320. [DOI] [PubMed] [Google Scholar]
- 10.Rice Chromosome 10 Sequencing Consortium (2003) Science 300, 1566–1569. [DOI] [PubMed] [Google Scholar]
- 11.Nagaki, K., Cheng, Z., Ouyang, S, Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R. & Jiang, J. (2004) Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- 12.Devos, K. M., Brown, J. K. & Bennetzen, J. L. (2002) Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitte, C. & Panaud, O. (2003) Mol. Biol. Evol. 20, 528–540. [DOI] [PubMed] [Google Scholar]
- 14.Ma, J., Devos, K. M. & Bennetzen, J. L. (2004) Genome Res. 14, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennetzen, J. L., Ma, J. & Devos, K. (2004) Annals Bot., in press. [DOI] [PMC free article] [PubMed]
- 16.SanMiguel, P., Tikhonov, A., Jin, Y.-K., Motchoulskaia, N., Zakharov, D., Melake Berhan, A., Springer, P. S., Edwards, K. J., Avramova, Z. & Bennetzen, J. L. (1996) Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- 17.Shirasu, K., Schulman, A. H., Lahaye, T. & Schulze-Lefert, P. (2000) Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wicker, T., Yahiaoui, N., Guyot, R., Schlagenhauf, E., Liu, Z. D., Dubcovsky, J. & Keller, B. (2003) Plant Cell 15, 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, B. & Xue, Y. (2003) Plant Biol. 6, 134–138. [DOI] [PubMed] [Google Scholar]
- 20.Portères, R. (1976) in The Origin of African Plant Domestication, eds. Harlan, J. R., de Wet, J. M. J. & Stemler, A. B. L. (Mouton, The Hague), pp. 409–452.
- 21.Ge, S., Sang, T., Lu, B. R. & Hong, D. Y. (1999) Proc. Natl. Acad. Sci. USA 96, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii, T., Xu, Y. & McCouch, S. R. (2001) Genome 44, 658–666. [PubMed] [Google Scholar]
- 23.Cheng, C., Tsuchimoto, S., Ohtsubo, H. & Ohtsubo, E. (2002) Genes Genet. Syst. 77, 323–334. [DOI] [PubMed] [Google Scholar]
- 24.Ren, F., Lu, B. R., Li, S., Huang, J. & Zhu, Y. (2003) Theor. Appl. Genet. 108, 113–120. [DOI] [PubMed] [Google Scholar]
- 25.Park, K. C., Kim, N. H., Cho, Y. S., Kang, K. H., Lee, J. K. & Kim, N. S. (2003) Theor. Appl. Genet. 107, 203–209. [DOI] [PubMed] [Google Scholar]
- 26.Tatusova, T. A. & Madden, T. L. (1999) FEMS Microbiol. Lett. 174, 247–250. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 29.Dubcovsky, J., Ramakrishna, W., SanMiguel, P. J., Busso, C. S., Yan, L., Shiloff, B. A. & Bennetzen, J. L. (2001) Plant Physiol. 125, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei, M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L., Vision, T. J. & Gaut, B. S. (2002) Mol. Biol. Evol. 19, 1464–1473. [DOI] [PubMed] [Google Scholar]
- 32.Gaut, B. S., Morton, B. R., McCaig, B. C. & Clegg, M. T. (1996) Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SanMiguel, P., Gaut, B. S., Tikhonov, A., Nakajima, Y. & Bennetzen, J. L. (1998) Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, A. & Bennetzen, J. L. (1999) Annu. Rev. Genet. 33, 479–532. [DOI] [PubMed] [Google Scholar]
- 35.Wessler, S. R., Bureau, T. E. & White, S. E. (1995) Curr. Opin. Genet. Dev. 5, 814–821. [DOI] [PubMed] [Google Scholar]
- 36.SanMiguel, P. J., Ramakrishna, W., Bennetzen, J. L., Busso, C. S. & Dubcovsky, J. (2002) Funct. Integr. Genomics 2, 51–59. [DOI] [PubMed] [Google Scholar]
- 37.Gu, Y. Q., Coleman-Derr, D., Kong, X. & Anderson, O. D. (2004) Plant Physiol. 135, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu, H. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linares, O. F. (2002) Proc. Natl. Acad. Sci. USA 99, 16360–16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai, H. & Morishima, H. (2002) Theor. Appl. Genet. 104, 1217–1228. [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka, S., Nakamura I., Watanabe, K. N. & Sato, Y. I. (2004) Theor. Appl. Genet. 180, 1200–1204. [DOI] [PubMed] [Google Scholar]
- 42.Vicient, C. M., Suoniemi, A., Anamthawat-Jónsson, K., Tanskanen, J., Beharav, A., Nevo, E. & Schulman, A. H. (1999) Plant Cell 11, 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalendar, R., Tanskanen, J., Immonen, S., Nevo, E. & Schulman, A. H. (2000) Proc. Natl. Acad. Sci. USA 97, 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoof, H. & Karlowski, W. M. (2003) Curr. Opin. Plant Biol. 6, 106–112. [DOI] [PubMed] [Google Scholar]
- 45.Bennetzen, J. L. & Ramakrishna, W. (2002) Proc. Natl. Acad. Sci. USA 99, 9093–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.