Abstract
Conjugal DNA transfer occurs by an atypical mechanism in Mycobacterium smegmatis. The transfer system is chromosomally encoded and requires recipient recombination functions for both chromosome and plasmid transfer. Cis-acting sequences have been identified that confer mobility on nontransferable plasmids, but these are larger and have different properties to canonical oriT sites found in bacterial plasmids. To identify trans-acting factors required for mediating DNA transfer, a library of transposon insertion mutants was generated in the donor strain, and individual mutants were screened for their effect on transfer. From this screen, a collection of insertion mutants was isolated that increased conjugation frequencies relative to wild type. Remarkably, the mutations map to a 25-kb region of the M. smegmatis chromosome that is syntenous with the RD1 region of Mycobacterium tuberculosis, which is considered to be the primary attenuating deletion in the related vaccine strain Mycobacterium bovis bacillus Calmette–Guérin. The genes of the RD1 region encode a secretory apparatus responsible for exporting Cfp10- and Esat-6, both potent antigens and virulence factors. In crosses using two M. smegmatis donors, we show that wild-type cells can suppress the elevated transfer phenotype of mutant donors, which is consistent with the secretion of a factor that suppresses conjugation. Most importantly, the RD1 region of M. tuberculosis complements the conjugation phenotype of the RD1 mutants in M. smegmatis. Our results indicate that the M. tuberculosis and M. smegmatis RD1 regions are functionally equivalent and provide a unique perspective on the role of this critical secretion apparatus.
The leading cause of death by an infectious agent is Mycobacterium tuberculosis. It is estimated that 2 million people die of tuberculosis every year and that one-third of the world's population is infected with M. tuberculosis (1). The global health problems associated with the disease have been exacerbated by its deadly synergistic association with HIV and the appearance of multidrug-resistant strains. A comprehensive understanding of the biology of this organism is critical for the identification of novel drug targets, the development of vaccines, and determining how it evades the host immune system; this requires the development of basic molecular techniques to determine the genetic and biochemical basis of pathogenesis and drug resistance (2, 3). Our recent efforts have been focused on the characterization of a conjugation system and its application for use in mycobacterial genetics (4–7).
Bacterial conjugation plays a major role in the horizontal transfer of genetic material, especially because many of the plasmids mediating transfer have a broad host range, allowing them to transfer into, and establish themselves in, a wide range of organisms. Conjugation is also an important molecular tool that, along with transformation and transduction, is essential for the genetic analysis of bacteria. Conjugation systems are generally plasmid-borne and require the synthesis of conjugative pili, which are responsible for mating-pair formation (8). These proteins also encode the pore through which DNA is exported into the recipient. More recent studies have shown that this conjugal DNA transport apparatus belongs to a larger family of transport apparatus called type IV secretion systems (9–11). Many of these type IV systems are encoded by pathogens and are intimately involved in pathogenesis as they secrete proteins that modify their eukaryotic host. Other plasmid-encoded proteins recognize a unique cis-acting site on the plasmid from which transfer is initiated, and this is called the origin of transfer (oriT) (8).
Recently, we have described a chromosomally encoded transfer system in Mycobacterium smegmatis, which operates by a previously undescribed mechanism (6, 7). M. smegmatis chromosome transfer requires extended contact between two viable bacteria (a donor and a recipient), is unidirectional, and is resistant to DNase I; thus, it meets the working definition of conjugal transfer (12). However, unlike other conjugation systems described in both Gram-positive and -negative bacteria, no mobile or integrative genetic elements are implicated in the conjugation system of M. smegmatis. In addition, we have thus far failed to identify typical transfer-associated genes either genetically or computationally. Cis-acting regions have been identified that confer mobility on nonmobilizable plasmids (called bom sites), but they are larger and more complex than the single oriT site found in plasmid systems (6). Moreover, we have shown that there are multiple bom sequences distributed around the M. smegmatis chromosome. Our current model posits that transfer is initiated via a break in the chromosome and, after transfer into the recipient, the transferred DNA is integrated into the recipient chromosome (5).
We have initiated a genetic screen to identify the trans-acting factors necessary for DNA transfer. In this initial screen, we failed to identify proteins essential for transfer from the donor; however, we did isolate insertion mutants that had acquired a hypertransfer phenotype. Here, we describe the isolation and characterization of these mutations and provide a model as to how they negatively regulate DNA transfer.
Materials and Methods
Transposon Mutagenesis. A library of insertion mutations was generated in a hygromycin-resistant (Hygr) derivative of the donor strain, mc2155 (MKD158), in which a Hygr gene is integrated at the L5 attB site (13). A mariner transposon encoding kanamycin resistance (Kmr), mobilized by the C9Himar1 hyperactive transposase, was delivered by means of the temperature-sensitive mycobacteriophage φMycoMarT7 (14, 15). Previous studies have shown that this transposon exhibits no strong regional preferences, and thus a random distribution of mutations was expected, with the exception that insertions into essential genes would not be detected (14, 16). The randomness of our library was confirmed by Southern blot and DNA sequencing analyses (data not shown). The location of each insertion was identified by DNA sequence analysis after the isolation of plasmid DNA containing the transposon and flanking chromosomal DNA. These plasmid clones were isolated by taking advantage of the R6K plasmid origin of replication, which is contained within the mariner transposon. The sequence was then compared to the unannotated M. smegmatis genome sequence. Preliminary sequence data were obtained from The Institute for Genomic Research (www.tigr.org). ORFs were assigned putative functions based on blast searches and analyses using the pfam and tuberculist databases (17–19).
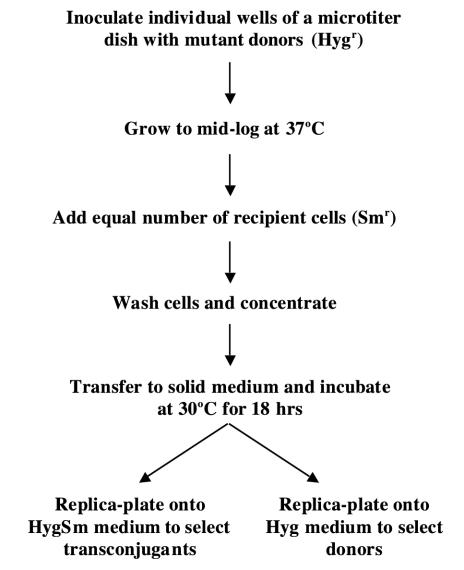
Microtiter Mating Screen. Individual colonies containing transposon insertions were picked and used to inoculate 200 μl of trypticase soy broth, containing 0.05% Tween 80 in 96-well microtiter dishes, and grown with gentle shaking at 37°C for 2 days; 200 μl of a mid-log culture of recipient was added to each well, and the cells were pelleted by brief centrifugation (Fig. 1). The supernatant was removed by aspiration, and the cells were washed three times in trypticase soy broth to remove Tween 80 and antibiotics before resuspending in a minimal volume (3–5 μl) of trypticase soy broth. The resuspended cells were spotted in arrays onto trypticase soy agar and incubated at 30°C for 18 h to allow transfer to take place. The cells were then replicated onto media selective for transconjugants [containing Hyg and streptomycin (HygSm)] or donors (containing Hyg) and incubated for 3–4 days at 37°C before scoring for relative transfer efficiencies by comparison with appropriate controls (Fig. 2).
Fig. 1.
A schematic outline of the microtiter mating procedure used to screen for insertion mutants affecting DNA transfer in M. smegmatis.
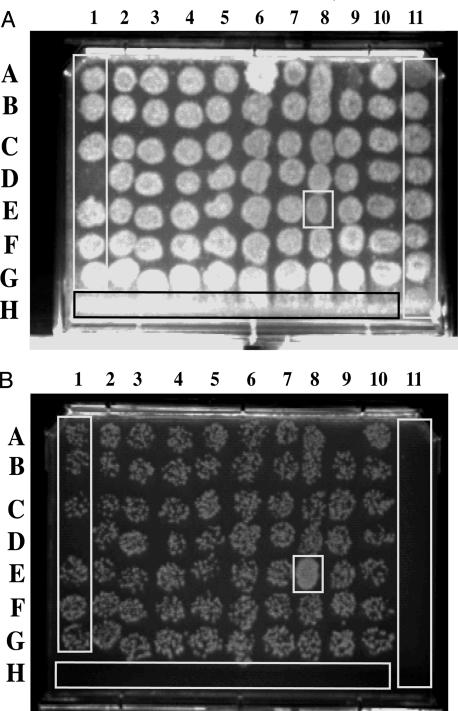
Fig. 2.
Microtiter replica plates used to identify transfer-up mutants. The example shown is the result of a single 96-well mating assay replica-plated onto trypticase soy agar plates. (A) Hyg-containing trypticase soy agar medium selecting for donors. (B) HygSm-containing trypticase soy agar medium selecting for transconjugants. The spot boxed at 8E is an example of an up-mutant. Note the confluent growth of 8E transconjugants compared with the speckled growth of other spots. Controls are indicated in rectangles and were as follows. Column 11 contains wild-type donor cells only, and consequently growth is only observed on plate A. In row H, wells 1–10 contain recipient cells only, which cannot grow on either plate A or B. In column 1, rows A–G contain wild-type donor and recipient and represent normal levels of Hygr transfer on plate B.
Quantitative Filter Matings. Conjugation frequencies were determined in filter matings as previously described (12). Two-donor matings were carried out with a mixed population of donors. The donor fraction consisted of a 10-fold excess of MKD158 (a wild-type donor, Hygr) over a Kmr up-mutant donor. In all cases, the mating mix consisted of an equal mixture of donor and recipient cells. MKD10 (ref. 12; wild-type donor, Kmr) was used as a control for the effect of a 10-fold excess of MKD158 on DNA transfer. For each of the up-mutants, the mutation (Kmr) was transduced into the wild-type mc2155 background to separate it from the Hygr gene, which was used in the original mutant screen (see above). The transduction was carried out by using a generalized transducing mycobacteriophage Bxz1 (S. Lee, J. Kriakov, and W. Jacobs, personal communication). Successful transduction of each mutation was confirmed by Southern analysis comparing original and transduced chromosomal DNAs (data not shown) and by a mating-out assay that showed the inheritance of the elevated-transfer phenotype.
Results and Discussion
Isolation of Insertion Mutants with an Elevated-Transfer Frequency. To identify genes responsible for conjugation, a library of transposon insertions was generated in the donor strain mc2155. A Kmr mariner transposon, mobilized by the C9Himar1 transposase gene, was delivered into donor cells via the temperature-sensitive mycobacteriophage, φMycoMarT7 (14). To facilitate the screen, a high-throughput microtiter-based mating scheme was developed (Fig. 1). In this screen, transfer of an Hygr gene, located at the attB locus, into the Sm-resistant (Smr) recipient MKD8 was measured for each mutant. Thus, any variation in DNA transfer frequency can be attributed to the mutation rather than to the location of the selectable marker. From a library of >20,000 mariner insertion mutants, we identified 20 insertion mutants that increased DNA transfer (Fig. 2). Transfer of each of the mutants, identified from the screen, was assayed in a more quantitative fashion by a filter-mating assay. The transfer frequency for each mutant was much higher than that for the wild-type donor, by a factor of between 23-fold and several thousandfold (Fig. 3).
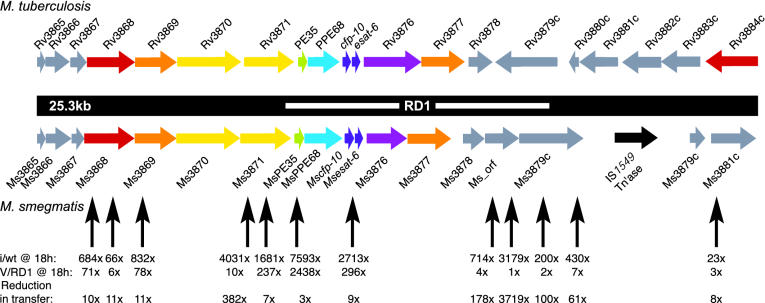
Fig. 3.
Alignment of the M. tuberculosis and M. smegmatis RD1 regions. ORFs are colored according to their putative functions, which were derived from the pfam and tuberculist databases (18, 19), as follows: red ORFs encode AAA domains (pfam accession no. PF0004); orange ORFs encode putative transmembrane proteins; yellow ORFs encode FtsK/SpoIIIE domains (PF01580, note that the FtsK/SpoIIIE and AAA domains are specific examples of the AAA+ domain); green ORFs encode PE (PF00934) proteins; cyan ORFs encode PPE (PF00823) proteins; purple ORFs encode members of COG0455 (35), ATPases involved in chromosome partitioning are shown. The functions of the gray ORFs are unclear. An ORF corresponding to the transposase of IS1549 is black. M. smegmatis ORFs (Ms) were given names reflective of the closest M. tuberculosis (H37Rv) homologue. Note that the genes downstream of Ms3878 and Rv3878, although conserved, are not in the same orientation. This finding contradicts previous computational predictions of the region (27) but is in agreement with sequencing analyses from both our laboratory and the tigr database. Ms-orf is a predicted gene with no known M. tuberculosis homologue. The white bar indicates the RD1 deletion originally defined in BCG, which also attenuates M. bovis and M. tuberculosis. Black vertical arrows below the maps represent mariner/Kmr insertions, which increase DNA transfer. For clarity, a third insertion in Ms3871 and an insertion 2.6 kb downstream of Ms3881c, which also elevates DNA transfer, are not shown. The fold increase (i/wt) in DNA transfer for each insertion mutant (i) compared with wild type (wt), after an 18-h mating, is shown immediately below each arrow. In these experiments, the wild-type transfer frequency was 1 × 10–5 events per donor cell. Mutant transfer frequencies were determined from five independent crosses. V/RD1 indicates the fold increase in DNA transfer for the mutants when complemented with an integrated M. tuberculosis cosmid encompassing the entire region. These frequencies are compared with just the cosmid vector (v) integrated into the attB site of M. smegmatis. The bottom row indicates the overall reduction in transfer frequency (i.e., the extent of complementation).
Insertion Mutations Map to the RD1 Region of M. smegmatis. The location of each mariner insertion was determined by DNA sequence analysis and compared to the M. smegmatis genome sequence. Significantly, 14 of the 20 insertion mutations were mapped to an ≈30-kb locus, indicating that this region plays an important role in regulating DNA transfer in M. smegmatis (Fig. 3). Even more surprisingly, this locus is syntenous with the RD1 region of M. tuberculosis, a region shown to be critical for full virulence of this pathogen (Table 1 and refs. 20–22).
Table 1. M. smegmatis RD1 gene products and their M. tuberculosis homologues.
| M. smegmatis gene | No. of insertions | M. tuberculosis gene | Percentage identity | e value | Putative function |
|---|---|---|---|---|---|
| Ms3868 | 3 | Rv3868 | 73 | 0 | AAA ATPase |
| Ms3869 | 1 | Rv3869 | 60 | e-154 | Membrane protein |
| Ms3871 | 2 | Rv3871 | 75 | 0 | AAA ATPase |
| MsPE35 | 1 | PE35/Rv3870 | 51 | 0.17 | Unknown |
| Mscfp10 | 1 | Cfp10/Rv3874 | 52 | 8e-12 | Secreted antigen |
| Ms-orf1 | 2 | No homologue | - | - | Unknown |
| Ms3879c | 2 | Rv3879c | 43 | 2e-42 | Unknown |
| Ms3881c | 1 | Rv3881c | 18 | 4e-16 | Unknown |
| Ms3882c | 1 | Rv3882c | 66 | 0 | Unknown |
M. smegmatis genes discussed in the text and Fig. 3 that contain mariner insertions are listed along with their M. tuberculosis homologue. Ms3882c is not shown in Fig. 3 but is located 2.6 kb downstream of Ms3881c. Values for percentage identity and e values were taken from blast alignments between the two sequences (17).
RD1 is one of several regions of difference identified by genetic comparisons of virulent strains of Mycobacterium bovis and the attenuated vaccine strain, M. bovis bacillus Calmette–Guérin (BCG) (23). The RD1 locus is deleted in all BCG strains when sequences are compared with those of virulent M. bovis and M. tuberculosis strains and is thought to be the primary attenuating mutation (23, 24). The RD1 deletion in BCG is 9.5 kb and includes M. tuberculosis genes Rv3871-3979. A related deletion has been described in Mycobacterium microti, which causes disease in voles but not in humans (25). At least one of the roles of this region is the secretion of two highly antigenic proteins, Esat-6 and Cfp-10, which are encoded from within RD1 (Fig. 3). Computational analyses of the genes in the vicinity of RD1 led to the hypothesis that RD1 encodes an ATP-dependent secretory apparatus (26, 27), and this prediction has been borne out in recent studies. M. tuberculosis mutants with mutations in genes Rv3870-3871 and Rv3876-3877 fail to export Esat-6 and are attenuated for virulence (20, 28). Also, complementation of the RD1 deletions in BCG and M. microti with the RD1 region from M. tuberculosis restores secretion of both Cfp-10 and Esat-6 (22). Preliminary data suggest that Esat-6 plays a role in lysis of alveolar epithelial cells and in escape from macrophages (20, 28).
The phenotypes of the M. smegmatis donor mutants are consistent with either, or both, of the following two hypotheses. Secreted proteins may decorate the surfaces of the bacteria and interfere with the formation of mating pairs. Alternatively, the secreted factor may act as an extracellular pheromone, repressing transfer until conditions are suitable for it to occur. We assume that, in each case, repression is lifted through some environmental or physiological trigger. This assumption is in agreement with our observation that crosses must be carried out for at least 18 h before transconjugants can be detected. Ms3870 and Ms3871 encode putative homologues of FtsK/SpoIIIE, which are proteins involved in chromosome partitioning and translocation, and this suggests a direct role in DNA transfer (29). Indeed, a SpoIIIE homologue is required for DNA transfer of the plasmid pSAM2 from Streptomyces (30). However, the insertion mutants have an effect opposite to that expected for a knockout, and the FtsK/SpoIIIE homologies include ATPase motifs common to macromolecular motors. It has been hypothesized, therefore, that Rv3870 and Rv3871 are more likely to provide energy for secretion rather than the translocation of DNA (26).
Evidence for Transfer Regulation by a Secreted Factor. To determine whether the inhibition of transfer is due to a factor secreted by the wild-type donor, we carried out the following two-donor experiments. Donor and recipient strains were mixed at a ratio of 1:1, but with the donor fraction of the mating mix consisting of a 10:1 mix of wild-type: up-mutant. We reasoned that if the wild-type donor supplied a secreted factor, transfer from the up-mutant should be reduced. A Kmr RD1 insertion was mixed with the wild-type donor MKD158 (Hygr), and transfer was monitored into a Smr recipient. As a control, the effect of a 10-fold excess of MKD158 on transfer of a second wild-type donor, MKD10 (Kmr), was measured. Transfer from MKD10 was reduced <2-fold by a 10-fold excess of MKD158 (Table 2). By contrast, Kmr transfer from four different up-mutants was reduced by as much as 102-fold for an insertion in Ms3868-10 (Table 2). The reduction of Kmr transfer is entirely consistent with the inhibition of the elevated transfer frequency phenotype of the RD1 mutants by a secreted trans-acting factor from the wild-type donor.
Table 2. An excess of wild-type donor cells reduces the transfer frequency from up-mutant donors.
| Gene disrupted by mariner/Kmr | Tenfold excess of MKD158 | Average transfer frequency, no. of Kmr Smr transconjugants per donor | Fold decrease in transfer with addition of MKD158 |
|---|---|---|---|
| Wild-type MKD10 | 1.43 ± 0.6 × 10-5 | ||
| MKD10 | + | 1.26 ± 0.16 × 10-5 | 1.14 |
| Ms3868 | 3.17 ± 4.2 × 10-3 | ||
| Ms3868 | + | 3.1 ± 1.8 × 10-5 | 102 |
| Ms3869 | 6 ± 5.5 × 10-3 | ||
| Ms3869 | + | 6.2 ± 2.3 × 10-5 | 97 |
| Ms3871 | 3.01 ± 1.6 × 10-3 | ||
| Ms3871 | + | 3.44 ± 0.97 × 10-5 | 88 |
| Mscfp-10 | 2.00 ± 0.71 × 10-4 | ||
| Mscfp-10 | + | 4.35 ± 1.3 × 10-6 | 46 |
Transfer of the Kmr marker was measured into the Smr recipient MKD8. Transfer is expressed as Kmr Smr transconjugants per donor and is expressed as the average of five experiments. In control experiments, we showed that transfer of Hygr from MKD158 was unaffected. Matings were carried out for 18 h at 30°C. Note that transfer frequencies and ratios differ slightly from those in Fig. 3, because we are monitoring transfer of Kmr from within the RD1 locus and comparing it with transfer of a wild-type Kmr donor rather than comparison with transfer of Hygr from the attB site. Thus, although the absolute frequencies vary between Fig. 3 and this table, the reduction in transfer caused by excess wild-type donor is clear and reproducible.
The RD1 Regions of M. tuberculosis and M. smegmatis Are Functionally Equivalent. The similar genetic organizations (Fig. 3) and the high degree of homology between the RD1 regions of M. tuberculosis and M. smegmatis (Table 1) suggest that the locus performs similar functions in the two species, despite their very different lifestyles and habitats. This inference is supported by the results above, which show that M. smegmatis RD1 mutants can be suppressed by a secreted factor. To determine the significance of the homologies between these two related regions, we used a plasmid (pRD1–2F9; ref. 31) encompassing the entire region from M. tuberculosis (Rv3860–Rv3885) to complement the M. smegmatis insertion mutants. Transfer of each insertion mutant was reduced by the presence of the plasmid, indicating that the regions are functionally equivalent (Fig. 3). Although the degree of complementation varied and was only reduced to wild-type levels in a few cases, the overall trend is clear and reproducible. We suspect that this incomplete complementation is a consequence of the fact that these experiments were designed to suppress, rather than rescue, the transfer phenotype, and it may also reflect the natural variations between the two secretion systems. For example, mutations of Ms3879c were complemented by the cosmid much more efficiently than were mutations mapping in Ms3868, suggesting that, of the two, the function of Ms3879 is more highly conserved between the two species. Note that insertions in Ms3881c, and also in a downstream gene not shown in Fig. 1, map outside the putative secretion apparatus genes and have an elevated transfer phenotype, which implies that the extent of functionality extends beyond that defined by current computational and deletion studies.
RD1 Regulation and DNA Transfer. How does RD1 regulate transfer in M. smegmatis and facilitate virulence in M. tuberculosis? Our observation that this region regulates transfer and involves a secretion apparatus, along with the finding by others that at least Esat-6 and Cfp-10 are secreted by genes in this region (20, 22), suggests two possibilities compatible with each species. First, the secreted proteins might coat the cell surface and thereby interfere with formation of specific cell–cell contacts necessary for DNA transfer in M. smegmatis, whereas they may protect M. tuberculosis cells from the hostile macrophage environment. This hypothesis is supported by the observation that, when RD1 is expressed in M. bovis BCG, Esat-6 and Cfp-10 are localized in cell wall fractions, and transformants exhibit an altered colony–morphology phenotype (31). The second hypothesis is that the secreted proteins act as sex pheromones, which negatively regulate transfer until appropriate conditions are met. Similarly, in the macrophage, the secreted proteins may act as a quorum-sensing mechanism, sensing the minimum number of cells before triggering lysis and reinfection of macrophages. This idea is consistent with the observation that attenuated M. tuberculosis RD1 mutants replicate and accumulate within macrophages but are unable to spread to uninfected cells (28). Important future goals will be to establish which proteins are secreted by the RD1 apparatus, whether that protein–secretion profile varies among species, and, if so, how the different spectrum of proteins accommodate the biology of the mycobacteria. A role for Esat-6 in tissue necrosis has recently been posited, but it is still unknown whether the effect is direct or indirect (20, 28). Our results are more consistent with an indirect effect, because the influence of the secreted protein(s) on transfer is observed in the absence of macrophages, although it is possible that M. smegmatis-secreted proteins have specifically evolved to facilitate DNA transfer. Many of these possibilities can be addressed by using DNA transfer in M. smegmatis as a genetic reporter for RD1-mediated secretion.
An obvious question arises from our studies: Is M. tuberculosis active in conjugal transfer, and, if so, is transfer elevated in RD1 mutants? The possibility of DNA transfer among clinical M. tuberculosis certainly has sinister overtones. However, we have not detected transfer between slow-growing mycobacteria, and given the predominantly clonal nature of M. tuberculosis infections (36), the possibility for it occurring naturally seems unlikely. Demonstrating such transfer in the laboratory is also not straightforward, because it first requires isolation of donor and recipient species, assuming they exist. This latter point also makes BCG, naturally defective for RD1, a poor choice because it is derived from the same isolate, and therefore all derivatives will most likely be either donor or recipient. Indeed, these caveats underlie the premise of using M. smegmatis as a model organism to understand DNA exchange before applying that knowledge to the more recalcitrant mycobacterial pathogens.
Secretion, DNA Transfer, and Virulence. This is not the first example of a link between DNA transfer and bacterial virulence. Conjugative pili belong to a larger class of type IV secretion systems. In the past decade, other type IV secretion systems have been demonstrated to export both DNA (between donor and recipient cells) and virulence proteins (including those encoded by Agrobacterium and Legionella; refs. 9, 10, 32, and 33). Our results provide an example of the adaptation of an unrelated secretion system for similar functions (DNA transfer and virulence) and thus emphasize how evolutionary pressures encourage the utilization of existing cellular machinery for multiple functions. Several of the type IV secretion systems are encoded by pathogenicity islands (11, 34). Intriguingly, sequences related to RD1 are found in multiple copies in M. tuberculosis and are found in both other mycobacteria (including M. smegmatis) and organisms with a high G+C content such as Streptomyces coelicolor and Corynebacterium diphtheriae (26, 27). The presence of RD1-like regions in different organisms and in multiple copies suggests that the locus may also have been mobile in the past. Our demonstration that the RD1 locus regulates DNA transfer strengthens this connection and raises the possibility that RD1 may have been responsible for its own dissemination.
The most important insight provided by this work is that a secretory apparatus is functionally conserved between M. smegmatis and its pathogenic cousins. Our studies demonstrate that at least one of the roles of this locus is to regulate DNA transfer in M. smegmatis, raising the intriguing possibility that DNA transfer can occur between pathogenic mycobacteria, including M. tuberculosis. Moreover, the ability to characterize the mechanism of RD1-mediated secretion in the genetically facile M. smegmatis should provide critical insights into this locus's contributions to both virulence and regulation of DNA transfer.
Acknowledgments
We thank Victoria Derbyshire and Kathleen McDonough for critically reading the manuscript, the Wadsworth Molecular Genetics Core for DNA sequence analysis, and Bill Jacobs (Albert Einstein College of Medicine, Bronx, NY) for supplying pRD1-2F9. This work was supported by National Institutes of Health Grant AI42308 (to K.M.D.).
Abbreviations: BCG, bacillus Calmette–Guérin; Kmr, kanamycin resistance; Hyg, hygromycin; Hygr, Hyg resistance; Sm, streptomycin; Smr, Sm resistance.
References
- 1.Corbett, E. L., Watt, C. J., Walker, N., Maher, D., Williams, B. G., Raviglione, M. C. & Dye, C. (2003) Arch. Intern. Med. 163, 1009–1021. [DOI] [PubMed] [Google Scholar]
- 2.Hatfull, G. F. (1996) Curr. Top. Microbiol. Immunol. 215, 29–47. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs, W. R. & Bloom, B. R. (1994) in Tuberculosis, ed. Bloom, B. R. (Am. Soc. Microbiol., Washington, DC), pp. 253–270.
- 4.Derbyshire, K. M. & Bardarov, S. (2000) in Molecular Genetics of Mycobacteria, eds. Hatfull, G. F. & Jacobs, W. R. (Am. Soc. Microbiol., Washington, DC), pp. 93–107.
- 5.Wang, J. & Derbyshire, K. M. (2004) Mol. Microbiol. 53, 1233–1242. [DOI] [PubMed] [Google Scholar]
- 6.Wang, J., Parsons, L. M. & Derbyshire, K. M. (2003) Nat. Genet. 34, 80–84. [DOI] [PubMed] [Google Scholar]
- 7.Bhatt, A. & Jacobs, W. R., Jr. (2003) Nat. Genet. 34, 3–4. [DOI] [PubMed] [Google Scholar]
- 8.Zechner, E. L., de la Cruz, F., Eisenbrandt, R., Grahn, A. M., Koraimann, G., Lanka, E., Muth, G., Pansegrau, W., Thomas, C. M., Wilkins, B. M. & Zatyka, M. (2000) in The Horizontal Gene Pool, Bacterial Plasmids and Gene Spread, ed. Thomas, C. M. (Harwood, London), pp. 87–175.
- 9.Lessl, M. & Lanka, E. (1994) Cell 77, 321–324. [DOI] [PubMed] [Google Scholar]
- 10.Winans, S. C., Burns, D. L. & Christie, P. J. (1996) Trends Microbiol. 65, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsons, L. M., Jankowski, C. S. & Derbyshire, K. M. (1998) Mol. Microbiol. 28, 571–582. [DOI] [PubMed] [Google Scholar]
- 13.Lee, M. H., Pascopella, L., Jacobs, W. R., Jr., & Hatfull, G. F. (1991) Proc. Natl. Acad. Sci. USA 88, 3111–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2001) Proc. Natl. Acad. Sci. USA 98, 12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardarov, S., Kriakov, J., Carriere, C., Yu, S., Vaamonde, C., McAdam, R. A., Bloom, B. R., Hatfull, G. F. & Jacobs, W. R. J. (1997) Proc. Natl. Acad. Sci. USA 94, 10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2003) Mol. Microbiol. 48, 77–84. [DOI] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E. L., et al. (2004) Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole, S. T. (1999) FEBS Lett. 452, 7–10. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, T., Hingley-Wilson, S. M., Chen, B., Chen, M., Dai, A. Z., Morin, P. M., Marks, C. B., Padiyar, J., Goulding, C., Gingery, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12420–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, K. N., Liao, R., Guinn, K. M., Hickey, M. J., Smith, S., Behr, M. A. & Sherman, D. R. (2003) J. Infect. Dis. 187, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pym, A. S., Brodin, P., Majlessi, L., Brosch, R., Demangel, C., Williams, A., Griffiths, K. E., Marchal, G., Leclerc, C. & Cole, S. T. (2003) Nat. Med. 9, 533–539. [DOI] [PubMed] [Google Scholar]
- 23.Mahairas, G. G., Sabo, P. J., Hickey, M. J., Singh, D. C. & Stover, C. K. (1996) J. Bacteriol. 178, 1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behr, M. A., Wilson, M. A., Gill, W. P., Salamon, H., Schoolnik, G. K., Rane, S. & Small, P. M. (1999) Science 284, 1520–1523. [DOI] [PubMed] [Google Scholar]
- 25.Brodin, P., Eiglmeier, K., Marmiesse, M., Billault, A., Garnier, T., Niemann, S., Cole, S. T. & Brosch, R. (2002) Infect. Immun. 70, 5568–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallen, M. J. (2002) Trends Microbiol. 10, 209–212. [DOI] [PubMed] [Google Scholar]
- 27.Gey Van Pittius, N. C., Gamieldien, J., Hide, W., Brown, G. D., Siezen, R. J. & Beyers, A. D. (2001) Genome Biol. 2, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinn, K. M., Hickey, M. J., Mathur, S. K., Zakel, K. L., Grotzke, J. E., Lewinsohn, D. M., Smith, S. & Sherman, D. R. (2004) Mol. Microbiol. 51, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, L. J., Lewis, P. J., Allmansberger, R., Hauser, P. M. & Errington, J. (1995) Genes Dev. 9, 1316–1326. [DOI] [PubMed] [Google Scholar]
- 30.Possoz, C., Ribard, C., Gagnat, J., Pernodet, J. L. & Guerineau, M. (2001) Mol. Microbiol. 42, 159–166. [DOI] [PubMed] [Google Scholar]
- 31.Pym, A. S., Brodin, P., Brosch, R., Huerre, M. & Cole, S. T. (2002) Mol. Microbiol. 46, 709–717. [DOI] [PubMed] [Google Scholar]
- 32.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873–876. [DOI] [PubMed] [Google Scholar]
- 33.Ding, Z., Atmakuri, K. & Christie, P. J. (2003) Trends Microbiol. 11, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Censini, S., Lange, C., Xiang, Z., Crabtree, J. E., Ghiara, P., Borodovsky, M., Rappuoli, R. & Covacci, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., Krylov, D. M., Mazumder, R., Mekhedov, S. L., Nikolskaya, A. N., et al. (2003) BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren, R. M., Victor, T. C., Streicher, E. M., Richardson, M., Beyers, N., Gey van Pittus, N. C. & Helden, P. D. (2004) Am. J. Respir. Care Med. 169, 610–614. [DOI] [PubMed] [Google Scholar]