Abstract
GRM3, a metabotropic glutamate receptor-modulating synaptic glutamate, is a promising schizophrenia candidate gene. In a family-based association study, a common GRM3 haplotype was strongly associated with schizophrenia (P = 0.0001). Within this haplotype, the A allele of single-nucleotide polymorphism (SNP) 4 (hCV11245618) in intron 2 was slightly overtransmitted to probands (P = 0.02). We studied the effects of this SNP on neurobiological traits related to risk for schizophrenia and glutamate neurotransmission. The SNP4 A allele was associated with poorer performance on several cognitive tests of prefrontal and hippocampal function. The physiological basis of this effect was assessed with functional MRI, which showed relatively deleterious activation patterns in both cortical regions in control subjects homozygous for the SNP4 A allele. We next looked at SNP4's effects on two indirect measures of prefrontal glutamate neurotransmission. Prefrontal N-acetylaspartate, an in vivo MRI measure related to synaptic activity and closely correlated with tissue glutamate, was lower in SNP4 AA homozygotes. In postmortem human prefrontal cortex, AA homozygotes had lower mRNA levels of the glial glutamate transporter EAAT2, a protein regulated by GRM3 that critically modulates synaptic glutamate. Effects of SNP4 on prefrontal GRM3 mRNA and protein levels were marginal. Resequencing revealed no missense or splice-site SNPs, suggesting that the intronic SNP4 or related haplotypes may exert subtle regulatory effects on GRM3 transcription. These convergent data point to a specific molecular pathway by which GRM3 genotype alters glutamate neurotransmission, prefrontal and hippocampal physiology and cognition, and thereby increased risk for schizophrenia.
Schizophrenia is a complex genetic disorder involving dysfunction in several brain regions, particularly the prefrontal and mesial temporal cortices and neurotransmitter systems, such as glutamate. Evidence for cortical dysfunction has been observed by a variety of techniques (1). In prefrontal cortex, for example, patients exhibit cognitive (2), physiological (3), and molecular (4) alterations. Glutamatergic abnormalities have been inferred from several observations. In postmortem prefrontal cortex, alterations in several glutamate-related measures have been reported (e.g., refs. 5–7), including reduced mRNA levels of excitatory amino acid transporter 2 (EAAT2) (8), also known as the glial glutamate transporter, which plays an essential role in synaptic glutamate removal. These convergent findings suggest that some aspects of glutamate neurotransmission may be altered in prefrontal and mesial temporal regions in patients with schizophrenia and that these abnormalities could be partly genetic.
Genes regulating glutamate neurotransmission have thus become attractive candidates for genetic association studies. These include the metabotropic glutamate receptor GRM3 (9). In animal models, GRM2/3 agonists block the behavioral and prefrontal cognitive abnormalities induced by the psychosis-producing drug ketamine (10), suggesting a possible deficit in GMR3 function in psychotic disorders like schizophrenia. GRM3 is expressed in astroglial cells, where it regulates expression of the glial glutamate transporter EAAT2 (11). EAAT2-mediated glutamate uptake is critical in regulating glutamate neurotransmission (12). GRM3 has been mapped to 7q21.1, spans 220 kb, and contains six exons. One linkage study of schizophrenia reported a significant logarithm of odds score of 3.18 at 7q22 (13), whereas two others have been suggestive (14, 15). Remarkably, two prior studies found a significant association between GRM3 and schizophrenia (16, 17) with single-nucleotide polymorphisms (SNPs) 17 kb apart, in exon 3 and intron 3, respectively, although one negative association sample was also reported (16).
Significant associations for complex genetic disorders such as schizophrenia often are difficult to replicate. This difficulty is not surprising given their presumed heterogeneity and the small effects of individual genes. Support for such associations can be enhanced by providing convergent biological and molecular evidence for functional effects of genes on pathophysiological mechanisms (18). In this study, we examined the effects of GRM3 genotype on risk for schizophrenia and on intermediate cognitive, physiological, and molecular phenotypes related to schizophrenia and glutamate function. Our goal was to examine a specific pathway by which this gene might affect risk for schizophrenia, from DNA sequence to gene expression, to cellular effects on glutamate neurotransmission, to cortical effects on physiological response, and, finally, to characteristic cognitive impairments. We report the second replication of a significant association with GRM3 and now also show association with many elements in this putative pathophysiological pathway. In particular, a SNP marker associated with increased risk for schizophrenia adversely affects (i) measures of prefrontal and hippocampal cognition, (ii) physiological activation of both regions assayed with functional MRI (fMRI), and (iii) prefrontal N-acetylaspartate (NAA), an in vivo measure related to neuronal synaptic activity and highly correlated with tissue glutamate. In human brain tissue, the high-risk GRM3 allele was associated with marked reduction in mRNA levels of the glial glutamate transporter EAAT2, a critical determinant of extracellular glutamate modulated by GRM3 agonists. In contrast, evidence for effects of GRM3 genotype on GRM3 mRNA levels and GRM2/3 immunoreactivity was weak. Thus, the impact of intronic SNP4 or related haplotypes on regulation of GRM3 transcription or translation remains unclear. Nevertheless, these convergent data implicate GRM3 as a susceptibility gene for schizophrenia and suggest that it does so by altering prefrontal glutamate neurotransmission, prefrontal and mesial temporal physiology, and related prefrontal/mesial temporal cognitive capacities.
Methods
Human Subjects. Subjects were recruited for the Clinical Brain Disorders Branch “Sibling Study,” a study of neurobiological abnormalities related to genetic risk for schizophrenia (19). (Details are available on request.) In brief, subjects included patients with schizophrenia-spectrum disorders, their primarily unaffected siblings, and controls. Participants were from 18 to 60 years of age and gave informed consent. Ethnicity was 6.3% African American and 89.8% European American; to avoid stratification, only data from the European Americans were analyzed. DNA was available for 217 patients, 311 siblings, 362 parents, and 136 controls. Other measures were acquired for subgroups, as described below. A replication sample for association studies with the diagnosis of schizophrenia was obtained from the National Institute of Mental Health Genetics Initiative (NIMHGI) and included African American (n = 51) and Caucasian (n = 67) families. This sample included all nuclear families (average size, 2.92 and 3.04, respectively) with at least one parent and one to two siblings (average, 2.1 and 1.9, respectively) with schizophrenia or schizoaffective disorder (depressed). Details of the NIMHGI have been described (14).
Cognitive Testing. Several measures of cognition are impaired in schizophrenia and appear to be related to genetic risk (e.g., see ref. 20). Therefore, we examined the effects of GRM3 genotype on these phenotypes, including (i) three tests of episodic memory (from the Wechsler Memory Scale-Revised, and California Verbal Learning Test), (ii) three measures of working memory (from the Wisconsin Card Sorting and N back tests), (iii) one measure of attention (d' from the Gordon continuance performance test), (iv) verbal fluency for letters, and (v) Trails B t scores. IQ (from the Wechsler Adult Intelligence Scale-Revised) was also included to control for levels of general intelligence.
Functional Magnetic Resonance Neuroimaging. To test the hypothesis that GRM3 has an impact on physiological processes in prefrontal and mesial temporal cortices, brain regions prominently implicated in the pathophysiology of schizophrenia (1), we used fMRI to study healthy controls performing cognitive tasks shown to activate these regions. For the prefrontal task, we administered the N back working memory task (see ref. 3) to 65 controls divided into two groups based on GRM3 SNP4 genotype: A/A homozygotes (n = 39) and G carriers (n = 26). Subjects with an A/GorG/G genotype were lumped together because of small numbers. To remove effects based on performance per se, groups were matched on performance and reaction time (Table 6, which is published as supporting information on the PNAS web site).
The second task was an episodic memory task previously shown to produce activation of the human hippocampal formation (21). Sixty-four healthy controls were divided equally between two groups of A/A homozygotes and G carriers matched for age, gender, and performance (Table 7, which is published as supporting information on the PNAS web site). In brief, the task required incidental encoding and subsequent recognition (“retrieval”) of novel visual scenes (for details, see Tables 6 and 7 and ref. 22).
1H Magnetic Resonance Spectroscopic Imaging (MRSI). To assess the affect of GRM3 genotype on glutamate neurotransmission, we first examined its relationship to an indirect in vivo measure, NAA by using 1H MRSI as described (23, 24). In brief, four MRI slices were acquired as 32 × 32 arrays of spectra oriented parallel to the sylvian fissure. Each volume element (“voxel”) had nominal dimensions of 7.5 × 7.5 × 15 mm (0.84 ml). To produce metabolite maps, location and integration of the signal strength of NAA compounds, creatine plus phosphocreatine, and choline-containing compounds in all brain voxels was automatically computed. Metabolite signals were reported as ratios of the area under the peaks NAA/creatine plus phosphocreatine. Prefrontal and hippocampal regions of interest were drawn blindly with reference to standard anatomical atlases by two raters on coplanar structural MRI scans (24).
Postmortem Studies. Brain tissue was obtained after securing consent from family members. The cohort consisted of normal (n = 10–19) and schizophrenic (n = 14–18) subjects for four experiments. These experiments included Western blot analysis of GRM2/3 protein and in situ hybridization analyses of mRNA for GRM3, the glial glutamate transporter (EAAT2), and the neuron-specific transporter EAAT3 with tissue from dorsolateral prefrontal cortex. EAAT3, which has a negligible role in removing extracellular glutamate, was used as a potential negative control. Methodological details and effect of diagnosis (i.e., schizophrenia) are described elsewhere (T.M.H., C.S.-W., P. Shashidharan, S. E. Bachus, M. H. Herman, and J.E.K., and S. Ghose, J.M.C., C. L. Bartus, T.M.H., J.E.K., and M.A., unpublished work, and ref. 25; see also legends for Figs. 3 and 4, which are published as supporting information on the PNAS web site). In brief, for in situ hybridization, fresh-frozen sections were taken from Brodmann's area 46. Sense and antisense ribonucleotide probes were radiolabeled with 35S-UTP. Slide preparation and mRNA quantification were performed with standard methods. Measures were obtained for two adjacent sections and averaged. Levels of glutamate transporter mRNAs were measured for combined superficial (I, II, and III) and deep lamina (IV, V, and VI), to be consistent with T.M.H., C.S.-W., P. Shashidharan, S. E. Bachus, M. H. Herman, and J.E.K. (unpublished work) and based on different afferent and efferent connectivity between these lamina. GRM3 mRNA levels were measured within each cortical layer independently to be consistent with S. Ghose, J.M.C., C. L. Bartus, T.M.H., J.E.K., and M.A. (unpublished work), who showed some heterogeneity across layers. For immunoblot studies, tissue samples were homogenized at 4°C and centrifuged; pellets were washed three times and analyzed for protein content. Two samples per subject underwent SDS/PAGE and were transferred by electroblotting to nitrocellulose membrane. Blots were washed, incubated with an mGLUR2/3 antibody, and treated with secondary antibody. The immunoreactive proteins were detected by using enhanced chemiluminescense (25).
For analyses of molecular markers based on genotype, we focused on GRM3 SNP4 (see below). Because sample sizes were small, we combined A/G with G/G subjects (“G carriers”). Because schizophrenic subjects had taken neuroleptics, which may affect some measures [e.g., EAAT2 (26) and GRM3 (27)] we focused first on controls. We also report analyses including both controls and patients, where group and genotype are main effects. As described previously, age, race, gender, postmortem interval, or tissue pH were included as covariates when significant.
Genetic Analysis. DNA was extracted from white blood cells or brain tissue. We initially selected seven SNPs from databases based on position and allele frequencies (Table 1). These SNPs included those reported to be associated with schizophrenia [SNP5 (16) and SNP6 (17)]. Genotyping was performed by using the Taqman 5′-exonuclease assay (details are available upon request). Genotype reproducibility was assessed by regenotyping 1,092 subjects; eight were discordant (0.74%). Genotyping errors were also detected as Mendelization errors with transmit and for haplotype inconsistency with merlin (28). We resequenced all exons and splice sites in 45 probands and found four SNPs with frequencies >1%: two in the 5′-UTR, a Gly/Asp (frequency, 2.4%) in exon 4, and one common (frequency, >5%) SNP (T/G) in intron 1 (chromosomal location, 85,872,423, UCSC Genome Bioinformatics, www.genome.ucsc.edu/index.html, freeze date April 2003) for which all subjects were also genotyped.
Table 1. GRM3 SNP markers.
| SNP | Coding, strand* | Frequency allele 2 | dbSNP or celera ID | Distance from M1† |
|---|---|---|---|---|
| 1 | T/G | 0.32 | rs187993 | 0 |
| 2 | C/T | 0.17 | hCV2627921 | 50,740 |
| 3 | C/T | 0.30 | rs917071 | 90,101 |
| 4 | A/G | 0.27 | hCV11245618 | 139,795 |
| 5 | C/T | 0.07 | N/A | 152,307 |
| 6 | A/T | 0.27 | rs1468412 | 169,771 |
| 7 | G/A | 0.25 | hCV2536213 | 211,987 |
Allele 2 frequencies are from Caucasian controls. Within each group, genotypes (not shown) for each marker were in Hardy-Weinberg equilibrium. Moderate linkage disequilibrium was observed across this region.
Coding strand format, e.g., SNP1 T coding-common-1 allele, which means, for SNP1, the T allele is the common allele on the coding strand and is referred to as allele 1.
UCSC Genome Bioinformatics, www.genome.ucsc.edu/index.html, freeze date April 2003.
Statistical Analyses. The effects of GRM3 genotype on cognitive and NAA measures were assessed by using mixed-effects ANOVA, where genotype was a fixed effect and family membership was a random effect, permitting the inclusion of related subjects (probands and sibs). Potential confounding factors (demographic and clinical variables) were also examined but were not significant. For patients with schizophrenia, genotype had no effects on neuroleptic dose, clinical symptoms, or global function, nor did dose correlate with clinical dependent measures reported below. When variances were unequal, nonparametric comparisons were performed. The effect of GRM3 on risk for schizophrenia was analyzed by using transmit. For three marker haplotype analyses, we included markers with >10% frequency for the uncommon allele and haplotypes with >3% frequency. Linkage disequilibrium between markers was determined by gold (29).
A critical issue in assessing the significance of association with phenotypic measures is the likelihood of type I errors. Many genotypes and phenotypes will be examined in this data set, but Bonferroni correction for all combinations seems overly stringent. We selectively analyzed one polymorphism, the most positive based on the transmission disequilibrium test results, against a small number of cognitive phenotypes related to genetic risk for schizophrenia and to abnormal glutamate function (20, 30). We next looked for convergent evidence of abnormal glutamate synaptic activity by using related neuroimaging and postmortem phenotypes. P values are therefore uncorrected, unless indicated.
Results
GRM3 Genotype and Risk for Schizophrenia. To test for association with schizophrenia, we performed family-based analyses, which look at transmissions of alleles from parents to affected offspring. In the Sibling Study (Caucasian only), the A allele of the marker SNP4 was slightly overtransmitted to probands (P = 0.02) (Table 2). We observed trends for overtransmission of two previously reported markers (SNP5 and 6) (16, 17). Combining SNPs to create three- and five-marker haplotypes (Table 3), we found overtransmission of a common haplotype that included the SNP4 A allele. This haplotype also included the two SNPs previously reported to be associated with schizophrenia. The haplotype contained only intronic SNPs. To look further for functional SNPs, we sequenced all exons and splice sites for 45 probands and found only one common (>4% frequency) synonymous SNP, which did not show distorted transmission to probands. Despite the marginal significance of individual SNP results, these data suggest that a common GRM3 haplotype increases risk for schizophrenia, not by changing mRNA splice sites or protein structure but by altering more subtle aspects of regulation of gene expression.
Table 2. Transmission disequilibrium test results for GRM3 SNPs.
| Sib Study (Caucasian)
|
NIMHGI (Caucasian)
|
|||
|---|---|---|---|---|
| SNP | Allele 1 observed/expected | P* | Allele 1 observed/expected | P* |
| 1 | 286/297 | 0.06 | 152/150 | 0.43 |
| 2 | 350/342 | 0.19 | 189/191 | 0.51 |
| 3 | 292/280 | 0.06 | 100/95 | 0.07 |
| 4 | 317/304 | 0.02 | 175/179 | 0.27 |
| 5† | 408/401 | 0.08 | 208/211 | 0.16 |
| 6‡ | 307/298 | 0.12 | 161/161 | 0.98 |
| 7 | 318/312 | 0.30 | 179/182 | 0.52 |
Table 3. Transmission disequilibrium test results for three- and five-marker haplotypes.
| SNP | Sib study (Caucasian) haplotypes | ||||
|---|---|---|---|---|---|
| 1 | 2 | ||||
| 2 | 1 | 1 | 1 | ||
| 3 | 1 | 1 | 1 | 1 | |
| 4 | 1 | 1 | 1 | 1 | |
| 6 | 1 | 1 | 1 | ||
| 7 | 1 | 1 | |||
| Global P value | 0.23 | 0.05 | 0.01 | 0.01 | 0.006 |
| Best P value | 0.01 | 0.01 | 0.007 | 0.0001 | 0.002 |
| Frequency | 0.29 | 0.57 | 0.62 | 0.61 | 0.53 |
| Transmitted | 14 over | 15 over | 15 over | 18 over | 21 over |
SNP5 is omitted because its frequency is <0.10. “Global P value” refers to 1 χ2 analysis testing for an altered transmission for all common haplotypes. “Best P value” refers to the most significant individual result for a specific haplotype, shown in the table. Because rare haplotypes (<3%) were excluded, they cannot account for this association.
We attempted to replicate our association in small samples from the NIMHGI (Tables 2 and 8, which is published as supporting information on the PNAS web site). In the single-marker analysis, both Caucasian and African American cohorts showed weak statistical evidence for distorted transmission for markers in introns 1 and 3. The African American cohort showed marginal overtransmission of the SNP6 T allele (P = 0.03), reported to be less frequent in a Japanese case/control study (17). Despite the small sample sizes and, in African Americans, different alleles, these data provide further albeit weak support for the hypothesis that variation in GRM3 increases risk for schizophrenia. To control for possible artifactual transmission distortions based on statistical reconstruction of missing genotypes by transmit, we performed the same analyses with the unaffected sibs designated as the index cases. No positive associations were found.
Effect of GRM3 Genotype on Cognition. Intermediate phenotypes may improve power in genetic studies because they are closer to the effects of genes. Focusing on the most positive SNP (SNP4) in this cohort, we studied its effects on putative cognitive intermediate phenotypes (20). We found a significant effect of genotype for measures of verbal list learning F = 3.98, df = 2,498, P = 0.02) and verbal fluency (for letters) (F = 4.71, df = 2,519, P = 0.009) (Table 4). Verbal list learning involves remembering and repeating back a list of 16 words for five successive times. The verbal fluency task asks subjects to say as many words beginning with a specified letter in a 1-min period. Both tasks involve prefrontal and temporal lobe function and are impaired in subjects given glutamate antagonists (31, 32). Post hoc comparisons showed that G/G homozygotes had higher list learning scores than both other groups (P < 0.007). For verbal fluency, G/G homozygotes had higher scores than A/A homozygotes (P < 0.004) and heterozygotes (P = 0.07). Group effects were significant for both verbal fluency (df = 2,519, F = 19.9, P < 0.0001) and list learning (df = 2,498, F = 43.42, P < 0.0001). Neither showed a group by genotype interaction, although in the sibling group little difference appears between genotype groups.
Table 4. Effect in three groups of SNP4 genotype on verbal list learning (California Verbal Learning Test, 1–5 summary scores, CVLT 1–5 SS) and verbal fluency (for letters) scores.
| SNP4 | Verbal fluency | CVLT1-5SS | Number |
|---|---|---|---|
| Index A/A | 33.2 (11.8) | 28.8 (18.2) | 104 |
| Index A/G | 34.0 (12.0) | 30.4 (18.7) | 77 |
| Index G/G | 40.7 (14.0) | 43.1 (14.3) | 12 |
| sib A/A | 41.6 (11.3) | 51.6 (13.3) | 123 |
| sib A/G | 42.1 (10.9) | 49.2 (12.1) | 79 |
| sib G/G | 42.8 (11.8) | 49.8 (16.5) | 22 |
| Controls A/A | 43.5 (8.6) | 50.6 (11.4) | 73 |
| Controls A/G | 47.5 (10.2) | 50.5 (11.7) | 45 |
| Controls G/G | 48.5 (9.1) | 56.9 (9.5) | 13 |
| All groups | 39.9 (12.0) | 43.6 (17.8) | 548 |
SDs are given in parentheses. Genotype groups were well matched for age, gender, IQ, and education.
Exploratory analyses revealed that alleles in two other SNPs on the overtransmitted haplotype also were associated with poorer cognition. Allele 1 for SNP5 predicted poorer performance for verbal fluency (F = 5.79, df = 2,510, P = 0.003), whereas allele 1 for SNP6 predicted poorer performance on both verbal fluency (F = 11.38, df = 2,522, P = 0.00001) and list learning (F = 3.55, df = 2,522, P = 0.03).
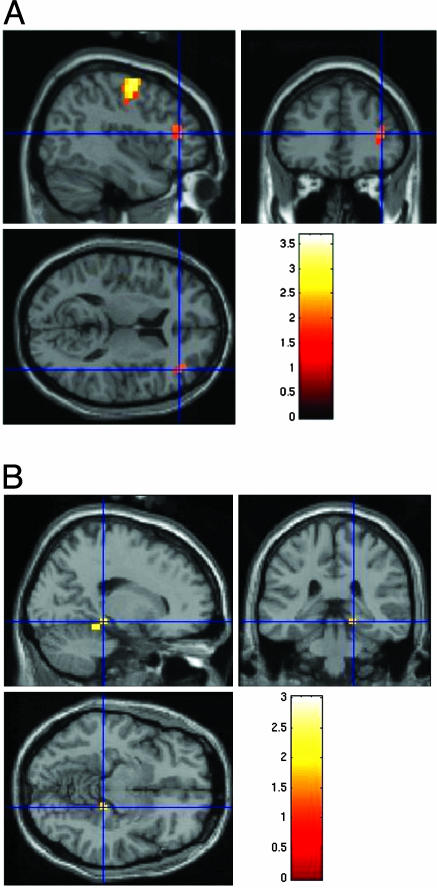
Effect of GRM3 Genotype on Prefrontal and Hippocampal Physiology. Because the overtransmitted SNP4 A allele was associated with impaired performance on cognitive tasks related to frontal and temporal lobe function, we predicted that SNP4 genotype would affect physiological measures, assayed with fMRI, that are related to prefrontal and temporal lobe information processing even in healthy subjects. During the working memory “N back” task, which actives prefrontal regions, healthy controls with two A alleles showed greater activation in a dorsolateral prefrontal cortex locale (Brodmann Area 46) (Talairach coordinates are 41, 37, 9; cluster size = 10 voxels; t score = 2.14; P < 0.02) compared with control subjects with a G allele (Fig. 1A). Because genotype groups had been matched on actual performance, this overactivation indicates that the SNP4 G allele is associated with relatively more “efficient” processing in the prefrontal cortex, and A/A subjects were relatively inefficient. This pattern of dorsolateral prefrontal inefficiency has been observed in prior studies of patients with schizophrenia and their healthy siblings, suggesting it is a physiological phenotype related to genetic risk (33). The SNP4 A allele contributes to this signature pattern of prefrontal inefficiency even in healthy controls.
Fig. 1.
Effects of GRM3 genotype on prefrontal and hippocampal physiological responses. (A) Brain map showing locales where SNP4 genotype groups differed in blood oxygenation measured with fMRI during a working memory task (the N back). Locales marked in red are groups of voxels where subjects with the A/A genotype showed higher activation and were significantly different when compared with a combined group of A/G and G/G subjects. One large cluster (k = 10) is in the dorsolateral prefrontal cortex (Brodmann Area 46) for A/A>A/G and G/G. (B) Brain map showing locales in hippocampus where SNP4 genotype differed in blood oxygenation during an episodic memory task. The region marked in red is an activation cluster (k = 9) where healthy subjects with the A/A genotype showed reduced activation relative to A/G and G/G subjects.
In healthy subjects performing an episodic memory task, which typically activates mesial temporal lobe structures, A/A subjects had relatively decreased activation during encoding in one locale of the right hippocampus (Talairach coordinates are 22, –38, –15; cluster size = 9 voxels; Z = 2.91; P < 0.05, corrected) relative to G allele carriers (see Fig. 1B). No genotype effect was seen during retrieval, suggesting that the effects of GRM3 on cognitive measures, such as list learning, may be related in part to relatively poor neural encoding of the material. In contrast to physiological responses in prefrontal cortex during working memory, greater hippocampal activation during declarative memory encoding has been associated with improved performance during retrieval, presumably because it reflects more effective encoding of the information (21). Here again, the SNP4 A allele predicts a relatively deleterious physiological response, although the character of the abnormal response varies with the information processing paradigm. GRM3 genotype is thus associated with variation in physiological responses involving two specific cortical regions implicated in the pathophysiology of schizophrenia, even in healthy subjects matched for levels of performance.
Effect of GRM3 Genotype on NAA. We examined the relationship between SNP4 genotype and NAA, an in vivo measure highly correlated with synaptic abundance and tissue glutamate (34) and which is reduced in these regions in schizophrenia (24). Based on evidence that GRM3 modulates synaptic glutamate and that the SNP4 A allele affects prefrontal function, we predicted it would be associated with reduced NAA. The results showed a genotype effect for left (F = 2.80, df = 2,239, P = 0.03, one-tailed) and right (F = 2.71, df = 2,238, P = 0.02, one-tailed) dorsolateral prefrontal cortex. Consistent with the cognitive and fMRI results, the G/G group showed higher NAA than the other genotype groups (Table 5). In left dorsolateral prefrontal cortex, A/A subjects had lower levels than G/G subjects (P = 0.02). G/A subjects had (P = 0.03) higher levels than A/A homozygotes (Table 5). Although group by genotype interaction was not significant, siblings seemed to show little effect of genotype.
Table 5. Effect of SNP4 genotype on NAA levels in left (L) and right (R) dorsolateral prefrontal cortex (DLPFC).
| NAA
|
|||
|---|---|---|---|
| SNP4 genotype | L DLPFC | R DLPFC | Valid number |
| Index A/A | 2.34 (0.34) | 2.39 (0.33) | 49 |
| Index A/G | 2.44 (0.38) | 2.35 (0.39) | 47 |
| Index G/G | 2.58 (0.50) | 2.68 (0.4.4) | 8 |
| sib A/A | 2.40 (0.31) | 2.40 (0.35) | 50 |
| sib A/G | 2.40 (0.45) | 2.45 (0.38) | 40 |
| sib G/G | 2.42 (0.47) | 2.37 (0.40) | 12 |
| Controls A/A | 2.30 (0.26) | 2.22 (0.26) | 23 |
| Controls A/G | 2.47 (0.43) | 2.42 (0.37) | 18 |
| Controls G/G | 2.99 (0.60) | 2.73 (0.11) | 3 |
| All groups | 2.40 (0.38) | 2.39 (0.36) | 250 |
Subjects with an A allele have lower levels than G/G homozygotes. Although we did not see a significant group by genotype interaction, genotype seemed to have little effect in the sibling group. SDs are given in parentheses. Genotype groups were well matched for age, gender, IQ, and education.
Because of the small sample sizes in G/G groups and unequal variances, we also used nonparametric statistics and collapsed A/G and G/G groups. We again found a significant difference between A/A genotype and those with a G allele for left (Mann–Whitney U test, Z = –2.01, P = 0.02, one-tailed) and right dorsolateral prefrontal cortex (Z = –2.18, P = 0.015, one-tailed). In the hippocampal formation, parametric analyses indicated a significant effect of genotype; however, nonparametric analyses did not. Overall, these results suggest that the SNP4 A allele, associated with increased risk for schizophrenia, poorer cognition, and impaired physiological responses, is also associated with reduced prefrontal NAA.
Effect of GRM3 Genotype on Postmortem Glutamate Measures. Although the clinical phenotypic associations with SNP4 offer convergent support for GRM3's role in schizophrenia, the mechanism by which SNP4 and related haplotypes do so is unclear. Because we found no SNPs that alter splice sites or coding sequences, we hypothesized that SNP4-related haplotypes might alter subtle aspects of gene expression. We therefore measured GRM3 mRNA and protein levels in human prefrontal cortical tissue. Furthermore, if SNP4-related haplotypes are functional, they should alter GRM3's critical functions, such as its regulation of downstream genes like the glutamate transporter EAAT2. GRM3 protein in glia regulates EAAT2 expression (11), which in turn regulates (and is highly correlated with) glutamate neurotransmission (12). We thus also measured mRNA levels of EAAT2 and, as a negative control, the neuronal glutamate transporter (EAAT3) mRNA, which is not regulated by GRM3 and does not correlate with extracellular glutamate (12).
In controls, a trend for an effect of SNP4 genotype was seen on GRM3 mRNA levels in layers 4 (F = 3.90, df = 1,6, P = 0.096) and 5(F = 5.46, df = 1,6, P = 0.058), with A/A subjects showing slightly lower levels than G carriers (Fig. 3). When patients with schizophrenia were included in this analysis, genotype effect was not significant. GRM3 protein levels did not differ according to genotype in the control group alone (F = 1.62, df = 1,14, P = 0.22), although A/A individuals had a mean decrease of 36.5% compared with G carriers. This reduction was nominally significant in the entire sample (F = 4.40, df = 1,32, P = 0.04) (Fig. 4). These marginally significant results, although suggestive, do not confirm that SNP4 or related haplotypes alter levels of the full-length GRM3 mRNA or protein.
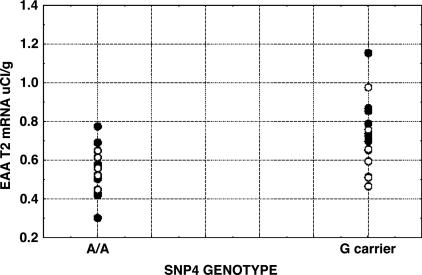
SNP4 genotype did have a significant effect on EAAT2 mRNA levels in the healthy group in both superficial (F = 12.2, df = 1,10, P = 0.006) and deep (F = 6.13, df = 1,10, P = 0.03) layers (Fig. 2 and Fig. 5, which is published as supporting information on the PNAS web site). A/A subjects had lower levels than G carriers. Similarly, when schizophrenic samples were added, a genotype effect was observed in superficial layers (F = 10.6, df = 1,24, P = 0.003) with weaker effects in deep layers (F = 3.03, df = 1,24, P = 0.095). Although we did not see a main effect of group or a group by genotype interaction, our sample sizes were too small [e.g., compared with Hyde et al.(unpublished data)] to draw definitive conclusions about possible differences of genotype effects between groups. Genotype effects were also seen without covariates (e.g., superficial, F = 13.8, df = 1,24, P = 0.001). Groups were well matched on other variables, such as age, postmortem interval, pH, ethnicity, and gender. In contrast, mRNA levels for the neuronal transporter, EAAT3, showed no effects of GRM3 genotype (P > 0.7), suggesting the EAAT2 results were not due to reduced neuropil or nonspecific tissue effects.
Fig. 2.
Distribution of EAAT2 mRNA levels (μCi/g) from the dorsolateral prefrontal cortex (superficial layers) by GRM3 SNP4 genotype (see text for statistics). •, controls; ○, probands.
Discussion
This study describes the effects of variation in GRM3 on risk for schizophrenia and several related biological phenotypes. We observed strong overtransmission of a common haplotype to patients in a family data set, suggesting that variation in this gene increases risk for schizophrenia. Using the most significant GRM3 marker, SNP4, we found that the overtransmitted A allele predicted poorer performance on cognitive phenotypes dependent on prefrontal and hippocampal function. These phenotypes are related to genetic risk for schizophrenia and can be induced in healthy individuals by glutamate antagonists. Using fMRI to assay prefrontal and hippocampal physiological responses in normal individuals, we found that the A allele was associated with relatively abnormal activation patterns. The SNP4 A allele also predicted lower levels in vivo of NAA in prefrontal cortex, suggesting reduced tissue glutamate levels and synaptic abundance. Finally, in postmortem prefrontal cortex, the SNP4 A allele predicted lower mRNA levels of the glial glutamate transporter EAAT2, an astrocytic protein regulated by GMR3 (11). Evidence for an effect of GRM3 genotype on GRM3 mRNA and protein levels was weak, leaving unclear how GRM3 sequence variants have an impact on transcription or translation. Nevertheless, these convergent data suggest that GRM3 affects prefrontal and hippocampal physiology, cognition, and risk for schizophrenia by altering glutamate neurotransmission.
The effect of GRM3 on risk for schizophrenia appears weak, as might be expected for a polygenic disorder. Similar weak effects have been reported for other putative schizophrenia genes (35). It has been suggested that at least three positive association samples are required to have preliminary confidence that a gene is related to risk for a complex disorder (36). After two initial reports of associations between GRM3 SNPs in exon 3 and intron 3 and schizophrenia (16, 17), we now report weak evidence of association for these and nearby SNPs in several new cohorts and statistically more robust associations with a haplotype that includes these SNPs. Furthermore, we have added physiological and molecular data, implicating a potential mechanism. The implicated SNPs in each cohort are physically close and in linkage disequilibrium but are not identical. Furthermore, the allele associated with increased risk differs between some cohorts [e.g., SNP 6, the A allele is enriched in Japanese probands (17), but this allele is marginally undertransmitted in the NIMHGI African American cohort]. Such differences are not unusual and may be due to allelic heterogeneity in different ethnic groups (35). Of note, the more common SNP4 allele and related haplotypes are associated with increased risk, rather than the less common alleles, which may be considered as relatively protective. This finding has been reported with several other genetic associations to complex disorders, such as CAPN10 in diabetes (37). The determinants of allele frequencies, which may include competition in several biological domains, are undoubtedly complex. The deleterious A allele presumably is common in humans because it provides a counterbalancing advantage, perhaps related to reduced glutamate. Finally, although the less common SNP4 G allele may be protective, it is unclear how less common G- or A-containing haplotypes affect risk.
For analyses with intermediate phenotypes, we focused on SNP4 because it gave the strongest association with diagnosis; using haplotypes is impractical for molecular and physiological studies. SNP4 is intronic and not obviously functional, but it may be in linkage disequilibrium with a causative SNP. Despite resequencing all exons and splice sites, we were unable to identify a more compelling variant. It is possible that SNP4 is monitoring a functional DNA element that has an impact in trans on another gene, but, given the convergent associations with multiple aspects of glutamate-related biology, this explanation seems improbable.
GRM3 appears to have an impact on aspects of cognition dependent on hippocampal and prefrontal cortical function even in healthy controls. The allele associated with increased risk was associated with worse cognitive performance and the physiological responses characteristic of worse performance. Not all cognitive measures dependent on prefrontal and hippocampal function showed a significant relationship to GRM3 genotype. The reasons for this are unclear; possibly some aspect of information processing shared by verbal fluency and list learning (e.g., verbal retrieval) are more sensitive to the GRM3 genotype effects than other tasks of executive function or episodic memory.
Consistent with the clinical results, findings from postmortem brain suggest that GRM3 genotype may have an impact on prefrontal function through its effects on glutamate neurotransmission. We focused on three measures: (i) GRM3 mRNA levels, (ii) GRM2/3 protein levels, and (iii) mRNA levels of the glial glutamate transporter EAAT2. Measures of GRM3 expression provided only marginal evidence that GRM3 genotype exerts a functional effect. However, our mRNA analysis used a “pan” probe that would not have identified changes in expression of novel GRM3 transcripts. Because we used an antibody that binds monomers of both GRM2 and GRM3, we may have missed more subtle changes, such as counterbalancing changes in GRM2 and GRM3 or in monomer and dimers, or in a novel protein species. More robust evidence was seen with a downstream measure of glutamate transmission, the glial glutamate transporter EAAT2, whose expression is regulated by GRM3. The association between SNP4 A/A genotype and reduced EAAT2 mRNA strongly suggests that some variant in GRM3 is indeed functional, even though the mechanism by which it affects EAAT2 is uncertain. One possibility is that SNP4-related haplotypes alter GRM3 expression in glia, which then down-regulates EAAT2 (11). Alternatively, GRM3 genotype could exert its effects indirectly through other mechanisms. Because EAAT2 is important in regulating extracellular glutamate, altered expression is likely to have significant effects on glutamate neurotransmission, although these effects may be complex and not readily reduced to a simple model of too much or too little glutamate.
The effects of GRM3 on intermediate phenotypes appear weak, in particular, when one considers the number of statistical tests performed. This problem of multiple testing in studies of complex genetic disorders is unavoidable. Large databases, which include many phenotypes, will of necessity be queried repeatedly by using many SNPs whose effects are likely to be weak and difficult to replicate. Furthermore, although we did not see significant interactions between genotype and group, Tables 4 and 5 suggest that the GRM3 genotype may have little effect in siblings, for unclear reasons. Despite these caveats, our replication of prior significant associations, combined with these convergent biological data implicating altered prefrontal glutamate function, substantially enhance the plausibility that these associations are true positives.
In conclusion, we provide evidence that the GRM3 genotype affects risk for schizophrenia perhaps by means of altered EAAT2 expression and glutamate neurotransmission. These changes in turn may affect prefrontal and mesial temporal physiological responses and several cognitive operations dependent on them. Finally, because the GRM3 genotype affects EAAT2 expression and thereby likely has an impact on extracellular glutamate, it may exert effects on other neuropsychiatric disorders in which glutamate has been implicated.
Supplementary Material
Abbreviations: SNP, single-nucleotide polymorphism; EAAT, excitatory amino acid transporter; fMRI, functional MRI; NAA, N-acetylaspartate; NIMHGI, National Institute of Mental Health Genetics Initiative.
References
- 1.Weinberger, D. R., Egan, M. F., Bertolino, A., Callicott, J. H., Mattay, V. S., Lipska, B. K., Berman, K. F. & Goldberg, T. E. (2001) Biol. Psychiatry 50, 825–844. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg, T. E. & Weinberger, D. R. (1988) Schizophr. Bull. 14, 179–183. [DOI] [PubMed] [Google Scholar]
- 3.Callicott, J. H., Bertolino, A., Mattay, V. S., Langheim, F. J. P., Duyn, J., Coppola, R., Goldberg, T. E. & Weinberger, D. R. (2000) Cereb. Cortex 10, 1078–1092. [DOI] [PubMed] [Google Scholar]
- 4.Selemon, L. D. & Goldman-Rakic, P. S. (1999) Biol. Psychiatry 45, 17–25. [DOI] [PubMed] [Google Scholar]
- 5.Deakin, J. F., Slater, P., Simpson, M. D., Gilchrist, A. C., Skan, W. J., Royston, M. C., Reynolds, G. P. & Cross, A. J. (1989) J. Neurochem. 52, 1781–1786. [DOI] [PubMed] [Google Scholar]
- 6.Nishikawa, T., Takashima, M. & Toru, M. (1983) Neurosci. Lett. 40, 245–250. [DOI] [PubMed] [Google Scholar]
- 7.Tsai, G., Passani, L. A., Slusher, B. S., Carter, R., Baer, L., Kleinman, J. E. & Coyle, J. T. (1995) Arch. Gen. Psychiatry 52, 829–836. [DOI] [PubMed] [Google Scholar]
- 8.Ohnuma, T., Tessler, S., Arai, H., Faull, R. L., McKenna, P. J. & Emson, P. C. (2000) Brain Res. Mol. Brain Res. 85, 24–31. [DOI] [PubMed] [Google Scholar]
- 9.Cartmell, J. & Schoepp, D. D. (2000) J. Neurochem. 75, 889–907. [DOI] [PubMed] [Google Scholar]
- 10.Moghaddam, B. & Adams, B. W. (1998) Science 281, 1349–1352. [DOI] [PubMed] [Google Scholar]
- 11.Aronica, E., Gorter, J. A., Ijlst-Keizers, H., Rozemuller, A. J., Yankaya, B., Leenstra, S. & Troost, D. (2003) Eur. J. Neurosci. 17, 2106–2118. [DOI] [PubMed] [Google Scholar]
- 12.Rothstein, J. D., Dykes-Hoberg, M., Pardo, C. A., Bristol, L. A., Jin, L., Kuncl, R. W., Kanai, Y., Hediger, M. A., Wang, Y., Schielke, J. P. & Welty, D. F. (1996) Neuron 16, 675–686. [DOI] [PubMed] [Google Scholar]
- 13.Ekelund, J., Lichtermann, D., Hovatta, I., Ellonen, P., Suvisaari, J., Terwilliger, J. D., Juvonen, H., Varilo, T., Arajarvi, R., Kokko-Sahin, M. L., et al. (2000) Hum. Mol. Genet. 9, 1049–1057. [DOI] [PubMed] [Google Scholar]
- 14.Faraone, S. V., Matise, T., Svrakic, D., Pepple, J., Malaspina, D., Suarez, B., Hampe, C., Zambuto, C. T., Schmitt, K., Meyer, J., et al. (1998) Am. J. Med. Genet. 81, 290–295. [PubMed] [Google Scholar]
- 15.Blouin, J. L., Dombroski, B. A., Nath, S. K., Lasseter, V. K., Wolyniec, P. S., Nestadt, G., Thornquist, M., Ullrich, G., McGrath, J., Kasch, L., et al. (1998) Nat. Genet. 20, 70–73. [DOI] [PubMed] [Google Scholar]
- 16.Marti, S. B., Cichon, S., Propping, P. & Nothen, M. (2002) Am. J. Med. Genet. 114, 46–50. [DOI] [PubMed] [Google Scholar]
- 17.Fujii, Y., Shibata, H., Kikuta, R., Makino, C., Tani, A., Hirata, N., Shibata, A., Ninomiya, H., Tashiro, N. & Fukumaki, Y. (2003) Psychiatr. Genet. 13, 71–76. [DOI] [PubMed] [Google Scholar]
- 18.Weiss, K. M. & Terwilliger, J. D. (2000) Nat. Genet. 26, 151–157. [DOI] [PubMed] [Google Scholar]
- 19.Egan, M., Goldberg, T., Gscheidle, T., Weirick, M., Bigelow, L. & Weinberger, D. (2000) Am. J. Psychiatry 157, 1309–1316. [DOI] [PubMed] [Google Scholar]
- 20.Egan, M., Goldberg, T., Gscheidle, T., Bigelow, L., Hyde, T. & Weinberger, D. (2001) Biol. Psychiatry 50, 98–107. [DOI] [PubMed] [Google Scholar]
- 21.Gabrieli, J. D., Brewer, J. B., Desmond, J. E. & Glover, G. H. (1997) Science 276, 264–266. [DOI] [PubMed] [Google Scholar]
- 22.Hariri, A. R., Goldberg, T. E., Mattay, V. S., Kolachana, B. S., Callicott, J. H., Egan, M. F. & Weinberger, D. R. (2003) J. Neurosci. 23, 6690–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duyn, J. H., Gillen, J., Sobering, G., van Zijl, P. C. & Moonen, C. T. (1993) Radiology 188, 277–282. [DOI] [PubMed] [Google Scholar]
- 24.Bertolino, A., Nawroz, S., Mattay, V. S., Barnett, A. S., Duyn, J. H., Moonen, C. T., Frank, J. A., Tedeschi, G. & Weinberger, D. R. (1996) Am. J. Psychiatry 153, 1554–1563. [DOI] [PubMed] [Google Scholar]
- 25.Crook, J. M., Akil, M., Law, B. C., Hyde, T. M. & Kleinman, J. E. (2002) Mol. Psychiatry 7, 157–164. [DOI] [PubMed] [Google Scholar]
- 26.Melone, M., Vitellaro-Zuccarello, L., Vallejo-Illarramendi, A., Perez-Samartin, A., Matute, C., Cozzi, A., Pellegrini-Giampietro, D. E., Rothstein, J. D. & Conti, F. (2001) Mol. Psychiatry 6, 380–386. [DOI] [PubMed] [Google Scholar]
- 27.Tascedda, F., Blom, J. M., Brunello, N., Zolin, K., Gennarelli, M., Colzi, A., Bravi, D., Carra, S., Racagni, G. & Riva, M. A. (2001) Biol. Psychiatry 50, 117–122. [DOI] [PubMed] [Google Scholar]
- 28.Abecasis, G. R., Cherny, S. S., Cookson, W. O. & Cardon, L. R. (2002) Nat. Genet. 30, 97–101. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis, G. R. & Cookson, W. O. (2000) Bioinformatics 16, 182–183. [DOI] [PubMed] [Google Scholar]
- 30.Callicott, J. H., Egan, M. F., Bertolino, A., Mattay, V. S., Langheim, F. J., Frank, J. A. & Weinberger, D. R. (1998) Biol. Psychiatry 44, 941–950. [DOI] [PubMed] [Google Scholar]
- 31.Malhotra, A. K., Pinals, D. A., Weingartner, H., Sirocco, K., Missar, C. D., Pickar, D. & Breier, A. (1996) Neuropsychopharmacology 14, 301–307. [DOI] [PubMed] [Google Scholar]
- 32.Krystal, J. H., Karper, L. P., Seibyl, J. P., Freeman, G. K., Delaney, R., Bremner, J. D., Heninger, G. R., Bowers, M. B., Jr., & Charney, D. S. (1994) Arch. Gen. Psychiatry 51, 199–214. [DOI] [PubMed] [Google Scholar]
- 33.Callicott, J. H., Egan, M. F., Mattay, V. S., Bertolino, A., Bone, A. D., Verchinksi, B. & Weinberger, D. R. (2003) Am. J. Psychiatry 160, 709–719. [DOI] [PubMed] [Google Scholar]
- 34.Petroff, O. A., Errante, L. D., Rothman, D. L., Kim, J. H. & Spencer, D. D. (2002) Ann. Neurol. 52, 635–642. [DOI] [PubMed] [Google Scholar]
- 35.Harrison, P. J. & Weinberger, D. R. (July 20, 2004) Mol. Psychiatry, 10.1038/sj.mp.4001558.
- 36.Lohmueller, K. E., Pearce, C. L., Pike, M., Lander, E. S. & Hirschhorn, J. N. (2003) Nat. Genet. 33, 177–182. [DOI] [PubMed] [Google Scholar]
- 37.Horikawa, Y., Oda, N., Cox, N. J., Li, X., Orho-Melander, M., Hara, M., Hinokio, Y., Lindner, T. H., Mashima, H., Schwarz, P. E., et al. (2000) Nat. Genet. 26, 163–175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.