Abstract
Ryanodine receptor 1 (RyR1, the sarcoplasmic reticulum Ca2+ release channel) and α1Sdihydropyridine receptor (DHPR, the surface membrane voltage sensor) of skeletal muscle belong to separate membrane systems but are functionally and structurally linked. Four α1SDHPRs associated with the four identical subunits of a RyR form a tetrad. We treated skeletal muscle cell lines with ryanodine, at concentrations that block RyRs, and determined whether this treatment affects the distance between DHPRs in the tetrad. We find a substantial (≈2-nm) shift in the α1SDHPR positions, indicating that ryanodine induces large conformational changes in the RyR1 cytoplasmic domain and that the α1SDHPR-RyR complex acts as a unit.
Keywords: dihydropyridine receptors, ryanodine receptors, skeletal muscle, ryanodine, E4032A mutant ryanodine receptor
The events that link surface membrane depolarization to the release of a messenger Ca2+ from the sarcoplasmic reticulum (SR) in muscle cells are collectively known as excitation-contraction coupling. This process is mediated by two calcium channels that are physically and functionally linked within junctional complexes called calcium-release units (CRUs) (1). The L-type calcium channel of the surface membrane and transverse (T) tubules, also called the dihydropyridine receptor (DHPR), acts as a voltage sensor. In response to depolarization of the surface membrane, DHPRs change conformation and activate the other calcium channel, the type 1 ryanodine receptor (RyR1) or Ca2+ release channel of the SR, through a molecule-to-molecule interaction. The functional interaction is bidirectional, because DHPRs control gating of the RyR channels and the RyRs in turn affect DHPR channel kinetics (2, 3).
DHPRs are heteropentamers with a total molecular mass of ≈430 kDa. The α1 subunit forms the channel, is essential for targeting to CRUs (4, 5), and interacts with RyRs (3), with some contribution from other subunits, particularly the small β (6). RyRs are homotetrameric channels of exceptionally large dimensions (2.3 MDa). Each of the four equal subunits has a small intramembrane domain in the channel region and a larger cytoplasmic domain that bridges the gap between the SR and exterior membranes and thus is available for interaction with the DHPR.
The positioning of DHPRs is detected by the technique of freeze-fracture, which reveals a rough outline of the channel complex by splitting the phospholipid bilayer and forming a bump or particle wherever one of the channels is present. By this technique, DHPRs are seen to be located at the corners of squares that are superimposed on the outlines of the RyR1 (7-9), and such grouping of four RyR-linked DHPRs has been called a tetrad. Tetrads indicate a symmetrical location of DHPRs relative to the center of the RyR1 channel and a well defined position relative to the cytoplasmic domains of the four identical RyR1 subunits. This arrangement suggests the hypothesis that the four DHPRs interact in a stereospecific manner with the RyR subunits. The tetradic disposition of skeletal muscle DHPRs requires an interaction with RyR1 and is necessary for the bidirectional coupling between the two molecules (10, 11).
Ryanodine is a plant alkaloid that modifies the SR calcium channel activity. Binding to high-affinity sites at nanomolar concentration (Kd = 1-100 nM), ryanodine activates the channel, locking it into a persistent subconductance open state. At higher concentration (100 μM or more), it inhibits the channel irreversibly in a concentration- and time-dependent manner, binding to low-affinity sites (Kd = 0.5-3.0 μM) (12, 13). When exposed to an appropriate ryanodine concentration and for a sufficient length of time, all channels are in the same nonconducting conformation.
We have measured the spacing between DHPRs within tetrads in cells that were either untreated or treated with high micro-molar concentration of ryanodine for a prolonged time, to test whether conformational changes in the RyR affect DHPR positioning. In addition, we have tested the RyR-DHPR interaction in RyR1 knockout cells (1B5s) expressing a RyR1 mutant (E4032A) that normally does not respond to any physiological stimulation (14, 15) but that can be restored functionally by treatment with high concentrations of ryanodine.
Methods
Cell Culture and Transfection. BC3H1 cells were obtained from the American Type Culture Collection. The cells were grown in a growth medium containing low-glucose DMEM (GIBCO/BRL), 20% FBS, 0.5% chicken embryo extract, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Cells were plated on Matrigel- (Collaborative Biomedical Products, Bedford, MA) covered Thermanox (Nunc). At ≈70% confluence, the growth medium was replaced by a differentiation medium containing 0.5% FBS and no chicken embryo extract, and the cells were fixed 3-8 days later. 1B5 cells were grown as above but at higher CO2 levels (11). At ≈70% confluence, the cells were differentiated in a no-serum medium containing 5% (vol/vol) heat-inactivated horse serum (Gemini Biological Products, Calabasas, CA) to induce differentiation and fixed 4-6 days later. Media were changed at 2-day intervals. 1B5 cells were transduced after differentiation with either wild-type RyR1 or RyR1 E4032A cDNA packaged in helper-free herpes simplex 1 virions (16).
Ryanodine Treatment. BC3H1 and 1B5 myotubes (the latter expressing either wild-type RyR1 or E4032A) were treated with 500 μM ryanodine for 24 h in one experiment and for 2 h for two other experiments. The length of the ryanodine treatment did not affect the intratetrad spacing, but the cells looked healthier after the short exposure. After exposure to the alkaloid the cells were fixed for electron microscopy experiments. Ryanodine-treated and control cultures were prepared in parallel for each experiment. Electron Microscopy and Immunocytochemistry. The cells were washed twice in PBS at 37°C, fixed in 3.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, stored in fixative for 1-4 wk, and briefly infiltrated in 30% glycerol. A small piece of the coverslip was mounted with the cells facing a droplet of 30% glycerol/20% polyvinyl alcohol on a gold holder, and then frozen in liquid nitrogen-cooled propane (17, 18). The coverslip was flipped off to produce a fracture that followed the culture surface originally facing it. The fractured surfaces were shadowed with platinum at 25° while rotating, and then replicated with carbon in a freeze-fracture machine (model BFA 400, Balzers Spa, Milan). Replicas were photographed in a 410 electron microscope (Philips Electron Optics, Mahwah, NJ). The specimen holder was positioned in the eucentric position to minimize variations in electron optical magnification.
Parallel culture dishes were immunolabeled with 34C, an anti-RyR antibody (19), to check for expression and differentiation. 34C was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City.
Measurements. The distances between the pale center of each black ring of platinum (representing a DHPR) and its nearest neighbors in a tetrad were measured by using nih image software.
Data from BC3H1 cells were standardized against the data from 1B5 cells by individually multiplying each BC3H1 measurement by the ratio of the mean intratetrad spacing of RyR1-expressing 1B5 cells and of BC3H1 cells. For control cells we used the ratios of the control values and for ryanodine-treated cells we used the ratios of treated values.
Results
The experiments were performed on cultured cells from two cell lines, which offer the significant advantage of forming DHPR-RyR interactions within CRUs located at the surface membrane. The first cell line, BC3H1, derived from a mouse brain tumor (20), expresses skeletal type RyR1 and α1SDHPR (21). The cells are identified as having a skeletal muscle origin, and the arrays of DHPR tetrads in their plasmalemma have been studied in detail (22). The second cell line, 1B5, is derived from mouse embryonic stem cells with an engineered null mutation for RyR1 and is differentiated under the influence of MyoD. Differentiated 1B5 cells express several CRU proteins (e.g., triadin and calsequestrin) and α1SDHPR, but do not express any RyR isoforms (RyR1, RyR2, or RyR3) (23). Transfection of 1B5 cells with herpes simplex virus 1 virions containing the cDNAs encoding for either wild-type RyR1 or a mutant of RyR1, E4032A, restores CRUs, and these show arrays of DHPR tetrads (11, 14).
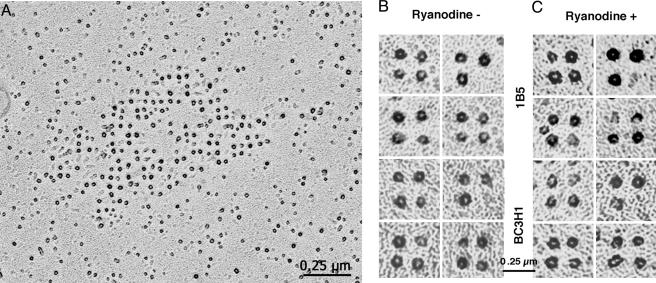
In our experiments, the position of DHPRs was determined in rotary-shadowed freeze-fracture replicas. In these, the highest points of the fractured DHPRs (or particles) appear in the electron micrographs as pale circles surrounded by dark rings of metal (Fig. 1A). The center of the circle presumably represents the position of the same molecular details for each channel, and thus it provides determination of the molecule's position. Tetrads, groups of four DHPRs, are composed of four such particles delimiting a square. Often tetrads miss one component, but the three or two remaining particles maintain the same disposition (Fig. 1B).
Fig. 1.
Freeze-fracture and rotary shadowing of BC3H1 cells. Freeze-fracture replicas of surface membranes from differentiated BC3H1 and RyR1-expressing 1B5 cells. (A) Patch of membrane containing an array of tetrads in a BC3H1 cell. Each DHPR particle is coated by platinum on all sides, and thus it appears as a dark ring with a lighter region at its peak. (B and C) Galleries of selected tetrads from control and ryanodine treated cells at higher magnification. Tetrads may be complete (i.e., composed of four particles), or incomplete (i.e., they lack one or more particles), due either to the breaking of particles during fracturing or to a missing DHPR.
In the process of fracturing, the proteins are subjected to a variable amount of distortions, resulting in tetrads that have lost their exact symmetry. To minimize the effect of this distortion on our measurements, we used strict criteria in selecting the tetrads to be measured. The selected tetrads belonged to ordered arrays; they had at least three of the four component particles, and they showed angles of 90° (or very close to it) between three adjacent particles. We estimate that approximately one-third of the three- and four-particle tetrads were discarded for not fitting these criteria. We assumed that large structural deviations due to distortions during fracturing are the same in control and in experimental samples. Fig. 1B and C shows galleries of tetrads that fitted the selection criteria for both control and 500 μM ryanodine-treated samples. Care was taken to minimize the effect of tilting of the image by measuring only the areas of the cells' surfaces that were flat, as indicated by details of the shadowing. Selection and measurements were done on “blind” data, i.e., without the operator knowing whether the images were from control or treated cells.
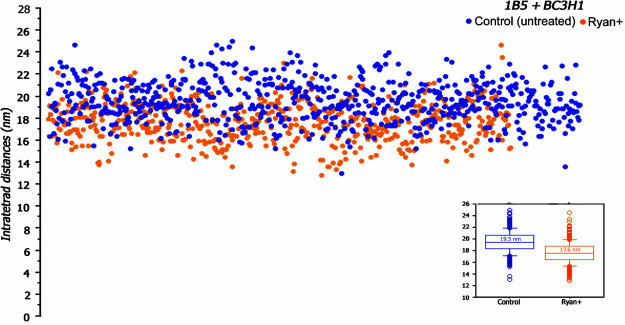
Despite this selection, the measured intratetrad distance, or center-to-center distance, between two adjacent particles in the tetrads is somewhat variable, because of the distortions that are visible in the detailed images of Fig. 1B and C. Fig. 2 presents a scattergram of intratetrad spacing (i.e., the center-to-center distance between adjacent particles in the tetrad), in untreated BC3H1 and RyR1-expressing 1B5 cells. Each dot represents one measurement, and the distance is plotted on the ordinate. The data from the cells of the two cell lines were standardized as defined in Methods. Blue dots are the data from untreated cells, and orange dots are the data from ryanodine-treated cells. The average intratetrad distance for untreated cells is 19.5 ± 1.9 nm (mean ± SD, from n = 169 measurements in one culture of BC3H1 cells and n = 497 measurements in two separate cultures of 1B5 cells); see Fig. 2 Inset.
Fig. 2.
Scattergram showing the distribution of intratetrad distances in control and ryanodine-treated 1B5 and BC3H1 cells. Each spot represents a single measurement of the distance between the light centers of the tetrad particles (or intratetrad distance, in nm). Data from control (blue dots) and ryanodine-treated (orange dots) cells are from three matched experiments: one involving BC3H1 cells and two involving RyR1-infected 1B5 cells. The data were combined after standardizing against the mean for 1B5 cells, separately for control and ryanodine-treated cells. The intratetrad distances within each group vary over a fairly wide range. The range of variability is approximately the same for control and ryanodine-treated cells and is mostly due to distortion of the protein during the fracturing process. However, the intratetrad distances are clearly shifted to lower values in ryanodine-treated versus control cells. (Inset) The median and the 10th (box) and 25th (crossbar) percentiles for the two sets of data. The mean for control distances is ≈2 nm larger than for the treated cells, and the difference is statistically significant (P < 0.0001).
At first sight, tetrads in control and ryanodine-treated cells do not appear different (Fig. 1B and C). The treatment induces no apparent distortion of the tetrad arrays. Measurements of intratetrad spacings, however, show a definite shift after the ryanodine treatment. The orange dots in Fig. 2 represent the data for ryanodine-treated cells, internally standardized as for the controls. The values of intratetrad spacing vary over approximately the same range in control and ryanodine-treated cells. However, ryanodine treatment results in a clear downward shift (by an average of ≈2 nm) in the intratetrad spacing. The average distances between the adjacent DHPRs within tetrads of ryanodine-treated cells is 17.6 ± 1.8 nm (from n = 264 measurement in one BC3H1 culture and n = 316 measurements in two 1B5 cultures done in parallel to the control ones). This is significantly different from the value for control cells (see Fig. 2 Inset; Student's t test, P < 0.0001).
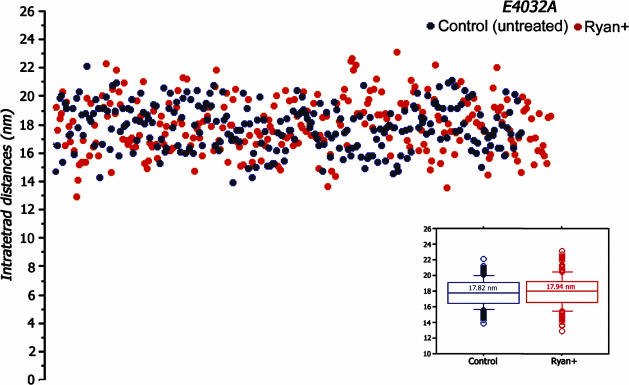
The RyR1 E4032A point mutant channel does not participate in excitation-contraction coupling nor does it respond to caffeine, and it is not blocked by high concentrations of ryanodine. Instead, treatment with high concentrations of ryanodine (100-500 μM) restores its ability to participate in excitation-contraction coupling and respond to RyR agonists such as caffeine and 4-chloro-m-cresol (14). The intratetrad spacing for the E4032A is 17.8 ± 1.6 nm for cells untreated by ryanodine and 17.9 ± 1.9 nm for cells exposed to ryanodine (Fig. 3, black and red dots). Both measurements are close to those of the ryanodine-treated cells expressing the wild-type RyR (17.6 ± 1.8 nm).
Fig. 3.
Tetrads in cell expressing the RyR point mutant (E4032A) have are smaller and not affected by ryanodine. Tetrads from 1B5 cells expressing the RyR point mutant (E4032A) are smaller than those in RyR1-expressing cells but show the same intratetrad distance variability. The intratetrad spacing for control (black dots) and ryanodine-treated (red dots) cells are not significantly different and both are similar to those in ryanodine-treated RyR1-expressing BC3H1 and 1B5 cells. As in Fig. 2, the Inset shows the medians and the 10th and 25th percentiles for the two sets of measurements.
Discussion
These results are quite significant both in terms of the α1SDHPR-RyR1 interaction and in terms of the effect of ryanodine on the RyR. The fact that the effect of ryanodine is transmitted from the RyR (on which it acts) to the tetrads (composed of DHPRs) indicates a conformational coupling between the two Ca2+ channels, requiring a very specific intermolecular interaction. The coupling can be achieved only if the two channels are in molecular contact, even though this may be through an intermediary protein. The specificity of this interaction is also indicated by the reciprocal functional signaling between RyR1 and α1SDHPR (3) and by the fact that defined RyR1 and α1SDHPR domains present only in the skeletal isoforms are required for the formation of tetrads (ref. 24 and H. Takekura, C.P., C.F.-A., M. Grabner, and B. E. Flucher, unpublished work).
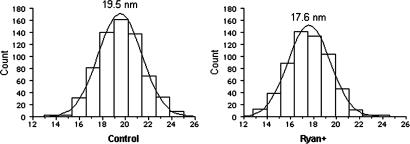
The data raised the question whether the changes in intratetrad spacing reflect the equal movement of all four DHPRs or the unequal shift of one or more tetrad components. To answer this question we compared the distribution of intratetrad spacing in control and ryanodine-treated cells (Fig. 4). In both cases the spacings fall within a classic Gaussian distribution with a single maximum, indicating that the four sides of the tetrad are equally changed. Ryanodine binds to RyRs in the open state, and there is some indication that channel closing is accompanied by, and perhaps due to, conformational changes in the cytoplasmic domain (13). The shift in DHPR tetrad size induced by ryanodine (2 nm) is quite large even in terms of the RyR cytoplasmic domain dimensions (≈26 nm on the side), and it seems to affect all four DHPRs. This finding indicates major rearrangements of all four subunits in the molecule, consistent with the hypothesis that the ryanodine effect is mediated by an allosteric influence of high concentrations of ryanodine on RyR1 conformation (12, 25). A single high-affinity binding site for ryanodine has been localized to the transmembrane assembly between Arg-4475 and the carboxyl terminus by using tryptic digestion of [3H]ryanodine-labeled RyR1 and photoaffinity techniques (26, 27). More recently Gln-4863 within putative transmembrane 10 of RyR2 was shown to be critical for high-affinity binding of [3H]ryanodine (28). These data indicate that a consequence of ryanodine binding to high-affinity sites is the partial or entire occlusion of the conduction pore. However the present results indicate that ryanodine at high concentrations induces major rearrangements of all four subunits within the RyR1 oligomer. These results are consistent with the hypothesis that ryanodine can induce changes in RyR1 mediated by more complex allosteric interactions of the alkaloid with binding sites outside the permeation pore (14, 29, 30).
Fig. 4.
Size distribution of intratetrad distances. The distance distributions are almost exactly Gaussian for both control and treated cells, indicating that all intratetrad distances are equally affected by the treatment.
The observed shift in tetrad size is comparable in magnitude although not exactly the same as that seen in the clamp region of channels observed in the open and closed configurations under more physiological conditions (31). Interestingly, the clamp region of RyR is in the cytoplasmic domain, at some distance from the intramembrane channel-forming domain (32, 33), but in proximity to the DHPR (34, 35). This location indicates that ryanodine may block the channel through some indirect effect over large molecular distances (see also ref. 14) and that the block may affect the RyR domain that is likely to interact with α1SDHPR. The long-range molecular interactions required for this rearrangement are not unlike those directly observed for other macromolecules: the SR Ca2+ ATPase (36), type II myosin (37, 38), and the acetylcholine receptor (39).
Results obtained from 1B5 cells expressing the RyR1 E4032A point mutant give additional insights into these data. By the indirect measurement of tetrad spacing, the mutant may seem to assume the ryanodine-closed conformation in the absence of the alkaloid (when it is unresponsive) as well as in its presence, thus maintaining a very stable conformation within the cytoplasmic domains of the RyR involved in interaction with DHPRs (40, 41). Ryanodine, which interacts at sites near the RyR1 transmembrane assembly (26), may uncouple the functional communication between the junctional foot and transmembrane assembly. Ryanodine treatment of the mutant may not induce enough of a conformational change to be detected as a change in tetrad spacing even though the alkaloid can restore function of the mutant.
The structural effect of ryanodine treatment described here represents a direct confirmation of the conformational coupling between the SR calcium channel (RyR1) and the transversetubule voltage sensor (α1SDHPR). This is an example of a unique interaction between molecules belonging to two membrane systems that may serve as a model for elucidating the operation of intermembrane signaling.
Acknowledgments
We thank N. Glaser for technical support and F. Protasi for critical reading of the manuscript. We thank P. D. Allen (Brigham and Women's Hospital) for the RyR1-carrying herpes simplex virus virions. This work was supported by National Institutes of Health Grant PO1AR17605 (to C.F.-A. and I.N.P.).
Abbreviations: CRU, calcium-release unit; DHPR, dihydropyridine receptor; RyR, ryanodine receptor; SR, sarcoplasmic reticulum.
References
- 1.Flucher, B. E. & Franzini-Armstrong, C. (1996) Proc. Natl. Acad. Sci. USA 93, 265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakai, J., Dirksen, R. T., Nguyen, H. T., Pessah, I. N., Beam, K. G. & Allen, P. D. (1996) Nature 380, 72-75. [DOI] [PubMed] [Google Scholar]
- 3.Grabner, M., Dirksen, R. T., Suda, N. & Beam, K. G. (1999) J. Biol. Chem. 274, 21913-21919. [DOI] [PubMed] [Google Scholar]
- 4.Flucher, B. E., Weiss, R. G. & Grabner, M. (2002) Proc. Natl. Acad. Sci. USA 99, 10167-10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flucher, B. E., Phillips, J. L. & Powell, J. A. (1991) J. Cell Biol. 115, 1345-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beurg, M., Ahern, C. A., Vallejo, P., Conklin, M. W., Powers, P. A., Gregg, R. G. & Coronado, R. (1999) Biophys. J. 77, 2953-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzini-Armstrong, C. & Nunzi, G. (1983) J. Muscle Res. Cell Motil. 4, 233-252. [DOI] [PubMed] [Google Scholar]
- 8.Block, B. A., Imagawa, T., Campbell, K. P. & Franzini-Armstrong, C. (1988) J. Cell Biol. 107, 2587-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takekura, H., Bennett, L., Tanabe, T., Beam, K. G. & Franzini-Armstrong, C. (1994) Biophys. J. 67, 793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Protasi, F., Takekura, H., Wang, Y., Chen. S. R. W., Meissner, G., Allen, P. D. & Franzini-Armstrong, C. (2000) Biophys. J. 79, 2494-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Protasi, F., Paolini, C., Nakai, J., Beam, K. G., Franzini-Armstrong, C. & Allen, P. D. (2002) Biophys. J. 83, 3230-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck, E., Zimanyi, I., Abramson, J. J. & Pessah, I. N. (1992) J. Biol. Chem. 267, 23560-23567. [PubMed] [Google Scholar]
- 13.Sutko, J. L., Airey, J. A., Welch, W. & Ruest, L. (1997) Pharmacol. Rev. 49, 53-98. [PubMed] [Google Scholar]
- 14.Fessenden, J. D., Chen. L., Wang, Y., Paolini, C., Franzini-Armstrong, C., Allen, P. D. & Pessah, I. N. (2001) Proc. Natl. Acad. Sci. USA 98, 2865-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien, J. J., Feng, W., Allen, P. D., Chen, S. R. W., Pessah, I. N. & Beam, K. G. (2002) Biophys. J. 82, 2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Y., Fraefel, C., Protasi, F., Moore, R. A., Fessenden, J. D., Pessah, I. N., DiFrancesco, A., Breakefield, X. & Allen, P. D. (2000) Am. J. Physiol. 47, C619-C623. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, S. A. & Pumplin, D. W. (1979) J. Cell Biol. 82, 494-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osame, M., Engel, A. G., Rebouche, C. J. & Scott, R. E. (1981) Neurol. 31, 972-979. [DOI] [PubMed] [Google Scholar]
- 19.Airey, J. A., Baring, M. D. & Sutko, J. L. (1991) Dev. Biol. 148, 365-374. [DOI] [PubMed] [Google Scholar]
- 20.Schubert, D., Harris, C. E., Devine, C. E. & Heinemann, S. (1974) J. Cell Biol. 61, 398-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks, A. R., Taubman, M. B., Saito, A., Dai, Y. & Fleisher, S. (1991) J. Cell Biol. 114, 303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Protasi, F., Franzini-Armstrong, C. & Flucher, B. E. (1997) J. Cell Biol. 137, 859-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, R. A., Nguyen, H., Galceran, J., Pessah, I. N. & Allen, P. D. (1998) J. Cell Biol. 140, 843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Protasi, F. (2002) Front. Biosci. 7, d650-d658. [DOI] [PubMed] [Google Scholar]
- 25.Pessah, I. N. & Zimanyi, I. (1991) Mol. Pharmacol. 39, 679-689. [PubMed] [Google Scholar]
- 26.Witcher, D. R., McPherson, P. S., Kahl, S. D., Lewis, T., Bentley, P., Mullinnix, M. J., Windass, J. D. & Campbell, K. P. (1994) J. Biol. Chem. 269, 13076-13079. [PubMed] [Google Scholar]
- 27.Callaway, C., Seryshev, A., Wang, J. P., Slavik, K. J., Needleman, D. H., Cantu, C., III, Wu, Y., Jayaraman, T., Marks, A. R. & Hamilton, S. L. (1994) J. Biol. Chem. 269, 15876-15884. [PubMed] [Google Scholar]
- 28.Wang, R., Zhang, L., Bolstad, J., Diao, N., Brown, C., Ruest, L., Welch, W., Williams, A. J. & Chen, S. R. W. (2003) J. Biol. Chem. 278, 51557-51565. [DOI] [PubMed] [Google Scholar]
- 29.Du, G. G., Guo, X., Khanna, V. K. & MacLennan, D. H. (2001) Proc. Natl. Acad. Sci. USA 98, 13625-13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bidasee, K. R., Xu, L., Meissner, G. & Besch, H. R., Jr. (2003) J. Biol. Chem. 278, 14237-14248. [DOI] [PubMed] [Google Scholar]
- 31.Orlova, E. V., Serysheva, I. I., van Heel, M., Hamilton, S. L. & Chiu, W. (1996) Nat. Struct. Biol. 6, 547-552. [DOI] [PubMed] [Google Scholar]
- 32.Serysheva, I. I., Orlova, E. V., Chiu, W., Sherman, M. B., Hamilton, S. L. & van Heel, M. (1995) Nat. Struct. Biol. 2, 18-24. [DOI] [PubMed] [Google Scholar]
- 33.Radermacher, M., Rao, V., Grassucci, R., Frank, J., Timerman, A. P., Fleischer, S. & Wagenknecht, T. (1994) J. Cell Biol. 127, 411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serysheva, I. I., Ludtke, S. J., Baker, M. R., Chiu, W. & Hamilton, S. L. (2002) Proc. Natl. Acad. Sci. USA 99, 10370-10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf, M., Eberhart, A., Glossmann, H., Striessnig, J. & Grigorieff, N. (2003) J. Mol. Biol. 332, 171-182. [DOI] [PubMed] [Google Scholar]
- 36.Toyoshima, C. & Nomura, H. (2002) Nature 418, 605-611. [DOI] [PubMed] [Google Scholar]
- 37.Rayment, I., Holden, H. M., Whittaker, M., Yohn, C. B., Lorenz, M., Holmes, K. C. & Milligan, R. A. (1993) Science 261, 50-58. [DOI] [PubMed] [Google Scholar]
- 38.Dominguez, R., Freyzon, Y., Trybus, K. M. & Cohen, C. (1998) Cell 94, 559-571. [DOI] [PubMed] [Google Scholar]
- 39.Unwin, N. (2000) Philos. Trans. R. Soc. London B 1404, 1813-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez, C. F., Mukherjee, S. & Allen, P. D. (2003) J. Biol. Chem. 278, 39644-39652. [DOI] [PubMed] [Google Scholar]
- 41.Perez, C. F., Voss, A., Pessah, I. N. & Allen, P. D. (2003) Biophys. J. 84, 2655-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]