Abstract
The RhaS and RhaR proteins are transcription activators that respond to the availability of l-rhamnose and activate transcription of the operons in the Escherichia coli l-rhamnose catabolic regulon. RhaR activates transcription of rhaSR, and RhaS activates transcription of the operon that encodes the l-rhamnose catabolic enzymes, rhaBAD, as well as the operon that encodes the l-rhamnose transport protein, rhaT. RhaS is 30% identical to RhaR at the amino acid level, and both are members of the AraC/XylS family of transcription activators. The RhaS and RhaR binding sites overlap the −35 hexamers of the promoters they regulate, suggesting they may contact the σ70 subunit of RNA polymerase as part of their mechanisms of transcription activation. In support of this hypothesis, our lab previously identified an interaction between RhaS residue D241 and σ70 residue R599. In the present study, we first identified two positively charged amino acids in σ70, K593 and R599, and three negatively charged amino acids in RhaR, D276, E284, and D285, that were important for RhaR-mediated transcription activation of the rhaSR operon. Using a genetic loss-of-contact approach we have obtained evidence for a specific contact between RhaR D276 and σ70 R599. Finally, previous results from our lab separately showed that RhaS D250A and σ70 K593A were defective at the rhaBAD promoter. Our genetic loss-of-contact analysis of these residues indicates that they identify a second site of contact between RhaS and σ70.
Transcription activation in Escherichia coli often involves the interaction of a DNA-binding activator protein with one of the subunits of RNA polymerase (RNAP), most often the sigma (σ) or alpha (α) subunit. Transcription activators that bind immediately upstream and adjacent to RNAP, in some cases overlapping the −35 promoter hexamer, may interact with the C-terminal domain (domain 4) of the σ subunit of RNAP (8, 27). The cI protein of bacteriophage λ is required for the establishment and maintenance of lysogeny and is perhaps the best-characterized example of a transcription activator that contacts σ70. The λ cI protein activates transcription of the PRM promoter when bound at the OR2 operator site, which overlaps the PRM −35 hexamer by 2 bp (30). Current evidence suggests that σ70 residues R588, K593, and R596 are required for activation by λ cI (23, 26, 35). Genetic and molecular modeling studies, as well as the recent structure of a ternary λ cI-σ domain 4-DNA complex, indicate that λ cI D38 contacts both σ70 K593 (σA K418) and R596 (σA R421) and λ cI E34 contacts σ70 R588 (σA R413) (8, 19, 26, 35). Prior to the identification of the ternary complex structure, a molecular model of the interaction indicated that σ70 K593 (σA K418) contacts DNA but was not positioned to contact λ cI (6, 8, 35). However, the ternary structure showed that the σA residue that aligns with σ70 K593 has moved away from the DNA (relative to the model of the interaction) and instead makes a protein-protein contact with λ cI D38 (19).
There is also evidence that activation by several transcription activators in the AraC/XylS family involves σ70 domain 4. Our lab previously identified two σ70 residues, K593 and R599, which are required for full activation by RhaS and further obtained genetic evidence that σ70 R599 is directly involved in a contact with RhaS D241 (4). These genetic results are also strongly supported by molecular modeling of the RhaS-σ70 complex on DNA (4). Evidence for AraC interactions with σ70 come from early σ70 mutations (eventually identified at R596) that increased araBAD expression in the absence of activation by cyclic AMP receptor protein (CRP) (18, 39, 44), as well as the finding that araBAD expression in a Δcya strain was increased by σ70 E591A and R596A and decreased by σ70 K593A (27). In addition, with a DNA that mimicked an open complex, a small amount of DNA-binding cooperativity could be detected between AraC and σ70 (7). At the melAB promoter, genetic evidence indicated that σ70 R596 interacts with MelR D261 and T265 while σ70 R599 also interacts with MelR D261, which aligns with RhaS D241 (16). The Ada protein has two activation domains, one of which is an AraC/XylS family domain which is required to activate transcription of the alkA operon (33). Alanine substitutions of σ70 residues K593, K597, and R603 each led to significant defects in Ada-dependent alkA transcription in vivo and in vitro (24).
The transcription activator RhaS, and the closely related RhaR protein, activate transcription of the E. coli l-rhamnose catabolic operons in the presence of the sugar l-rhamnose (10, 11, 42). RhaS activates transcription of the rhaBAD and rhaT operons by binding as a dimer to sites that overlap the −35 hexamers of the promoters by 4 bp and extend upstream to −81 and −82, respectively (see Fig. 1 for the rhaBAD promoter) (10, 45). Similarly, RhaR activates transcription of the rhaSR operon by binding as a dimer to a site that overlaps the RNAP binding site by 4 bp and extends upstream to −82 (Fig. 1) (43). The long binding sites for RhaS and RhaR each consist of two 17-bp imperfect inverted repeat half sites that are separated by 16 or 17 bp of uncontacted DNA (10, 43, 45). Each RhaS and RhaR monomer is predicted to contain two helix-turn-helix DNA-binding motifs and thereby contact two consecutive major grooves of DNA (38). CRP also activates transcription at all three of the rha promoters. At the rhaBAD and rhaT promoters, the CRP binding site is located immediately upstream of the RhaS binding site and is centered at −92.5 and −93.5, respectively (see Fig. 1 for the rhaBAD promoter) (11, 45). The CRP site required for full activation at rhaSR is located upstream but not adjacent to the RhaR binding site and is centered at −111.5 (Fig. 1) (17).
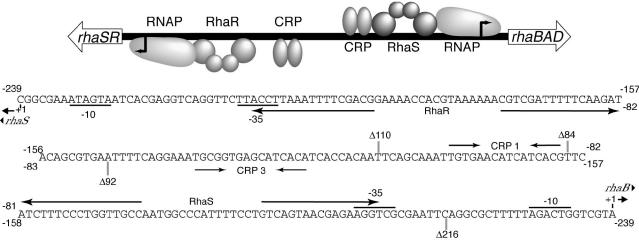
FIG. 1.
(Top) Representation of the divergent rhaSR and rhaBAD promoter regions, showing the approximate positions of the transcription activators RhaS, RhaR, and CRP, as well as RNAP at each promoter. (Bottom) Three consecutive lines of DNA sequence extending from the rhaSR transcription start point to the rhaBAD transcription start point. Binding sites for RhaS, RhaR, and CRP are shown by arrows, and the −35 and −10 hexamers of each promoter are indicated. The upstream end points of promoter fusions used in this study are marked by Δs. Deletion end points, protein binding sites, and numbering relative to the rhaSR promoter are shown below the DNA sequence, while deletion end points, protein binding sites, and numbering relative to the rhaBAD promoter are shown above the DNA sequence.
RhaS and RhaR are members of a subset of the AraC/XylS family that share amino acid sequence similarity with AraC over its entire length (9, 13, 28). Based on this similarity, RhaS and RhaR are predicted to consist of two domains connected by a flexible linker (5, 12, 29, 41). The N-terminal domains are predicted to be responsible for l-rhamnose binding and dimerization, while the C-terminal domains contain the 99-amino-acid region that classifies them as members of the AraC/XylS family. In all studied cases of AraC/XylS family members, including RhaS and RhaR, the characteristic 99-amino-acid region constitutes a DNA-binding domain (3, 10, 43). This DNA-binding domain has also been shown to be involved in transcription activation in a number of AraC/XylS family proteins including Ada, RhaS, AraC, MelR, MarA, SoxS, XylS, and UreR (1, 4, 5, 15, 16, 20-22, 37).
In this study, we further explored the mechanisms of transcription activation by RhaR and RhaS. We identified amino acid residues in the C-terminal domain of σ70 and in RhaR that are important for RhaR-mediated transcription activation at the rhaSR promoter. We then used a genetic loss-of-contact approach to identify an interaction between RhaR D276 and σ70 R599 that is required for RhaR-mediated activation. We also extended the previous studies by Bhende and Egan (4) of RhaS-mediated transcription activation at rhaBAD. Here we identified a second interaction between RhaS and σ70, in this case, RhaS D250 and σ70 K593.
MATERIALS AND METHODS
Culture media and conditions.
E. coli cultures for β-galactosidase assays were grown in morpholinepropanesulfonic acid-buffered medium (4, 34). Tryptone-yeast extract liquid medium (0.8% tryptone, 0.05% yeast extract, 0.05% NaCl) was used to grow cells for most other experiments. SacB selection medium (1% tryptone, 0.5% yeast extract, 1.5% agar, 5% sucrose [pH 7.8]) was used to select against sacB+ strains (14). Antibiotics were used as indicated at the following concentrations: ampicillin (200 μg/ml), chloramphenicol (25 μg/ml), kanamycin (25 μg/ml), and tetracycline (20 μg/ml).
General methods.
Standard methods were used for restriction endonuclease digestion and ligation using restriction endonucleases and T4 DNA ligase purchased from New England Biolabs (Beverly, Mass.). Transformation was carried out using chemically induced competent cells of E. coli, and plasmid DNA was purified by alkaline lysis. DNA sequencing reactions were carried out using custom-synthesized IRD41 dye-labeled primers (Table 1) from LI-COR Inc. (Lincoln, Nebr.) and the Thermo Sequenase primer cycle sequencing kit from Amersham Life Sciences (Arlington Heights, Ill.). DNA sequences were analyzed by automated dideoxy sequencing on a LI-COR 4000L sequencer (University of Kansas Biochemical Research Service Laboratory). The Expand High Fidelity PCR system (Roche, Indianapolis, Ind.) was used to amplify DNA fragments for cloning as well as to generate templates for DNA sequencing from rhaS and rhaR alleles that were recombined into the chromosome. The DNA sequences of both strands were determined for the entire cloned region of all cloned, mutagenized, and recombined DNA fragments. The QIAquick PCR Purification kit (QIAGEN, Chatsworth, Calif.) was used to clean up PCR products.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence, 5′-3′a | Use |
|---|---|---|
| 1170 | CCGGAATTCTTGTGGTGATGTGATGCTCAC | Amplify rhaSRT′ (within rhaBAD promoter) |
| 2068 | ATGACCGTATTACATAGTGTGGAT | IRD41b labeled, rhaS sequencing |
| 2069 | TTATTGCAGAAAGCCATCCCGTCC | IRD41 labeled, rhaS sequencing |
| 2074 | TGGTTGCACAGATGGAACAGC | IRD41 labeled, rhaS sequencing |
| 2075 | GTTGAGACGTGATGCGCTGTT | IRD41 labeled, rhaS sequencing |
| 2097 | CGCGAATTCAAGGGTATGGTTTTGCAG | Amplify rhaSRT from chromosome (within rhaT promoter) |
| 2204 | GGTCACCGCGTGATATTCG | IRD41 labeled, rhaR sequencing |
| 2205 | ATTCCGGGATTTAACGCCAG | IRD41 labeled, rhaR sequencing |
| 2206 | TTAATCTTTCTGCGAATTGAG | IRD41 labeled, rhaR sequencing |
| 2207 | CAAACGGCACATGCTGACTA | IRD41 labeled, rhaR sequencing |
| 2208 | CGGTCGAAATTGCACTGATT | rhaR D276A mutagenesis |
| 2210 | AATAGTTACTTGCTTCAAAGCCAC | rhaR D285A mutagenesis |
| 2292 | CCTGGATCCCCGCAAAAGTGAA | Amplify rhaSRT′ from plasmids (within rhaT) |
| 2297 | GGTAAGATCTCGGTCATACTGGCCTCCTGATG | Long-way-around PCR to delete rhaSR |
| 2298 | GGTAAGATCTTTAATTCGCCATGCCGATGCCGA | Long-way-around PCR to delete rhaSR |
| 2299 | GGTAAGATCTCGGACCGGGTCGAATTTGC | Amplify cat-sac cassette |
| 2300 | GGTAAGATCTATATCGGCATTTTCTTTTGCG | Amplify cat-sac cassette |
| 2381 | ATTCTCTTCGTTGCGGATAGTAACTATTTTTCGGTGGTG | rhaR E284A mutagenesis |
Regions of oligonucleotides not complementary to the wild-type DNA sequence (encode restriction endonuclease sites and flanking DNA or mutations) are underlined.
IRD41, infrared dye from LI-COR.
Strains, plasmids, and phages.
Table 2 lists the strains, phages, and plasmids used in this study. All strains used in β-galactosidase assays were derived from ECL116 (2) and carried lacZ fusions in a single copy on λ phage integrated at attλ (40). P1 phage-mediated generalized transduction was used to move Δ(recC ptr recB recD)::Plac-bet exo kan (from KM22) into SME1216 (selecting for kanamycin resistance) to make SME2417. SME2495 was made using P1 transduction to move Δ(rhaSR)::kan zih-35::Tn10 (from SME2800) into SME1217, selecting for tetracycline resistance and then screening for a Rha− phenotype. SME2496 was made from SME2416 by transformation with a PCR product containing Δ(rhaSR)::cat-sac (amplified from pSE254, described below), which was recombined onto the chromosome by using the recombination genes of bacteriophage λ (encoded by Δ(recC ptr recB recD)::Plac-bet exo kan) (31). The plasmid pSE250 was made by restriction endonuclease digestion of pSE101 with BamHI at the rhaT′ end of the clone and EcoRI (a natural site within the rhaBAD promoter), creating the fragment rhaSRT′, which was purified from an agarose gel by using QIAGEN′s QIAEX Gel Extraction kit. The rhaSRT′ fragment was then ligated to pUC18 (46), which had been digested with BamHI and EcoRI. To make pSE254, long-way-around PCR using pSE250 as the template and primers 2297 and 2298 amplified all of the pSE250 sequence except rhaSR and added a BglII site at each end. Then, PCR with primers 2299 and 2300, using a PCR product containing the cat-sac cassette (provided by Kenan Murphy) (32) as the template, generated a product that was ligated to the BglII sites of the long-way-around PCR product to create pSE254. Plasmids pML148 to -169 (containing mutations in the rpoD gene) were obtained from the laboratory of Carol Gross and were sequenced to ensure that they still carried the expected mutations. Several of the rpoD alleles were initially found to be wild type. Assays involving these alleles were repeated upon obtaining true mutants.
TABLE 2.
Strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| KM22 | Δ(recC ptr recB recD)::Plac-bet exo kan | 31 |
| ECL116 | F− ΔlacU169 endA hsdR thi | 2 |
| SME1074 | ECL116 λ SME106 | Laboratory collection |
| SME1216 | ECL116 λ SME103 | Laboratory collection |
| SME1851 | ECL116 λ SME104 | Laboratory collection |
| SME2416 | SME2417 zih-35::Tn10 | Laboratory collection |
| SME2417 | SME1216 Δ(recC ptr recB recD)::Plac-bet exo kan | This study |
| SME2495 | SME2417 ΔrhaSR::kan zih-35::Tn10 | This study |
| SME2496 | SME2416 ΔrhaSR::cat-sacB | This study |
| SME2508 | ECL116 λ SME114 recA::cat | Laboratory collection |
| SME2515 | ECL116 λ SME114 | Laboratory collection |
| SME2608 | SME1851 rhaS(wild type) recA::kan | Laboratory collection |
| SME2689 | SME1851 rhaS(D250A) zih-35::Tn10 recA::kan | This study |
| SME2691 | SME2515 rhaR(D276A) zih-35::Tn10 recA::kan | This study |
| SME2692 | SME2515 rhaR(D285A) zih-35::Tn10 recA::kan | This study |
| SME2693 | SME2515 rhaR(wild type) recA::kan | This study |
| SME2800 | SME1074 ΔrhaSR::kan zih-35::Tn10 | This study |
| SME2933 | SME2515 rhaR(E284A) zih-35::Tn10 recA::kan | This study |
| Phages | ||
| λ RS45 | bla′-lacZscatt+imm21ind+ | 40 |
| λ SME103 | λ RS45 φ(rhaB-lacZ)Δ110 | 11 |
| λ SME104 | λ RS45 φ(rhaB-lacZ)Δ84 | 11 |
| λ SME106 | λ RS45 φ(rhaS-lacZ)Δ216 | 11 |
| λ SME114 | λ RS45 φ(rhaS-lacZ)Δ92 | 17 |
| Plasmids | ||
| pGEX-2T σ70 | AprrpoD, wild type | 27 |
| pML148 to -169 | pGEX-2T, rpoD substitutions | 27 |
| pSE101 | pTZ18R AprrhaSR rhaBA′ | Laboratory collection |
| pSE172 | Tetr pALTER-1 rhaS D250A | Laboratory collection |
| pSE249 | pSE101 rhaS D250A | This study |
| pSE250 | pUC18 rhaSRT′, wild type | This study |
| pSE251 | pUC18 rhaSRT′ rhaR D276A | This study |
| pSE252 | pUC18 rhaSRT′ rhaR E284A | This study |
| pSE253 | pUC18 rhaSRT′ rhaR D285A | This study |
| pSE254 | pUC18 ΔrhaSR::cat-sac | This study |
| pUC18 | AprlacZα | 46 |
Mutagenesis of rhaS and rhaR.
The mutant rhaS D250A allele was moved from pSE172 into the context of pSE101 by digesting pSE172 with BstEII and BglII (both sites are native to the wild-type rhaS gene) to create a fragment encoding the RhaS D250A substitution. This fragment was then ligated to similarly digested pSE101 to make pSE249. Genes encoding alanine substitution derivatives of RhaR D276A and RhaR D285A were constructed by oligonucleotide-directed mutagenesis of rhaR in pGEM-11Zf(+) (Promega, Madison, Wis.), using the GeneEditor kit (Promega) and oligonucleotides 2208 and 2210. The mutant rhaR alleles were then subcloned into pSE250, using NheI and SmaI restriction endonuclease sites (both sites occur naturally within rhaR), to make pSE251 and pSE253, respectively. The rhaR E284A mutagenesis was performed using PCR to make oligonucleotide-directed mutations at the desired position with primer 2381, which also contained the recognition sequence for the EarI restriction endonuclease. Second, a nonmutagenized PCR fragment was made, also using a primer with EarI restriction sites. Finally, ligation of the mutant and wild-type DNA fragments allowed seamless reconstruction (25) of the full-length rhaR E284A gene in the context of rhaSRT′ to make pSE252. Oligos Etc (Wilsonville, Oreg.), Integrated DNA Technologies (Coralville, Iowa), and MWG-Biotech (High Point, N.C.) synthesized oligonucleotide primers used in mutagenesis (Table 1). Mutations were initially identified by diagnostic PCR using the following method. The very 3′ nucleotides of the diagnostic oligonucleotide contained the desired substitution(s), such that amplification was possible only (in combination with a suitable downstream primer) when the template DNA carried the desired mutation. Putative mutants identified by this method were confirmed by DNA sequencing of both strands of the entire cloned region (see Table 1 for sequencing oligonucleotides).
Recombination of rhaS and rhaR alleles onto the chromosome.
The mutant rhaS and rhaR alleles constructed as described above were recombined onto the E. coli chromosome such that they replaced the wild-type rhaS or rhaR allele by the following methods. Each rhaS or rhaR mutant was present on a plasmid in the context of rhaSRT′ (pSE249, pSE251, pSE252, or pSE253). Oligonucleotides 1170 and 2292 were used to amplify each mutant rhaSRT′ region by high-fidelity PCR. Approximately 500 ng of the rhaSRT′ PCR product carrying a mutant allele was used to transform either SME2495 [λ Φ(rhaB-lacZ)Δ110 Δ(recC ptr recB recD)::Plac-bet exo kan Δ(rhaSR)::kan, zih-35::Tn10] or SME2496 [λ Φ(rhaB-lacZ)Δ110 Δ(recC ptr recB recD)::Plac-bet exo kan Δ(rhaSR)::cat-sac, zih-35::Tn10]. Since SME2495 and SME2496 both contained Plac-bet exo, which encodes the λ phage recombination proteins, the frequency of homologous recombination was much higher in these strains than in wild-type E. coli strains (31). The transformants were screened by spread plating on media containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml) and l-rhamnose (0.2%) to identify functional, or partially functional, rhaSR genes that replaced the ΔrhaSR allele. There was no selection for successful recombinants in SME2495; rather we screened for blue colonies among a lawn of white colonies. When transforming SME2496, which contains a cat-sacB cassette (32) in place of rhaSR, we selected for the sucrose resistance of cells that had lost the sacB gene (which confers sucrose sensitivity) by homologous recombination. However, due to a significant background of spontaneous sucrose-resistant mutants, the transformants were also screened for at least a partially functional rhaS or rhaR gene by adding X-Gal (40 μg/ml) and l-rhamnose (0.2%) to the SacB selection plates. We found that sucrose inhibition of the sacB+ cells worked most reproducibly at room temperature, although it took about 3 days for the cells to grow. Phage P1-mediated generalized transduction was then used to transfer the rhaS or rhaR allele of interest (linked to zih-35::Tn10) to either SME1851 [λ Φ(rhaB-lacZ)Δ84] or SME2515 [λ Φ(rhaS-lacZ)Δ92] by selecting for the tetracycline resistance conferred by zih-35::Tn10. Diagnostic PCR, as described above, was used to initially identify transductants that contained the rhaS or rhaR mutation of interest. High-fidelity PCR was then used to amplify rhaSRT′ from the chromosome, using oligonucleotides 2097 and 1170, and the entire 3-kb PCR product was sequenced, as described above, to verify the presence of the desired mutation with no additional mutations. Phage P1-mediated transduction was then used to introduce recA::kan into each strain to make SME2689, -2691, -2692, and -2933. Finally, competent cells of each strain were made and transformed with plasmids containing either the wild type-, K593A-, L595A-, R599A-, or R608A-encoding σ70 gene for β-galactosidase assays.
β-Galactosidase assay.
β-Galactosidase assays were performed as previously described (3). In all cases, chromosomal rpoD was expressed from its own promoter, not the trp promoter described by Lonetto et al. (27), and the plasmid-encoded σ70 derivatives were expressed in the absence of isopropyl-β-d-thiogalactopyranoside. Under these conditions, the σ70 derivatives are expected to account for approximately 50% of the total σ70 in the cells (27). Specific activities were averaged from at least three independent assays with two replicates in each assay. The assays were performed on at least two different days, with independent cell growth steps (starter tryptone-yeast extract culture, overnight culture, and final growth culture) for each assay.
RESULTS
σ70 derivatives at the rhaSR promoter.
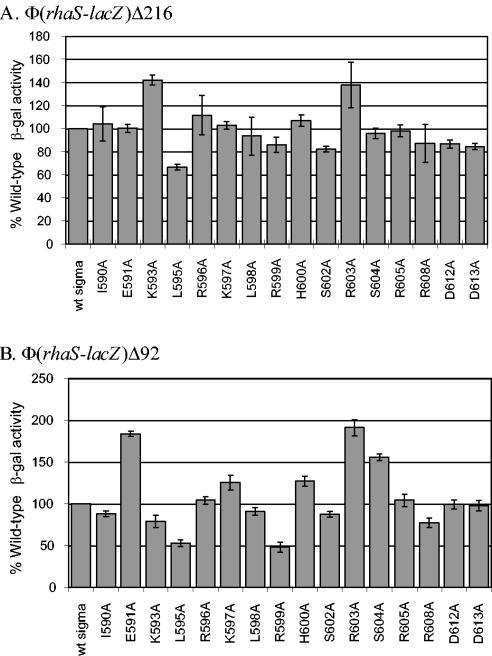
We wished to determine whether any residues near the C-terminal end of σ70 were important for transcription activation by RhaR. Lonetto et al. (27) constructed a library of alanine substitutions at 17 different positions near the C terminus of σ70 and found that some substitutions resulted in defects at class II activator-dependent promoters. Previous work from our lab found that two of the alanine substitutions in this library were defective at a truncated rhaBAD promoter where RhaS was the only transcription activator (4). We assayed this library of alanine substitutions in σ70 at two RhaR-activated single-copy translational fusions, Φ(rhaS-lacZ)Δ216 and Φ(rhaS-lacZ)Δ92. The Φ(rhaS-lacZ)Δ216 promoter contained the RhaR binding site as well as upstream CRP sites, while the Φ(rhaS-lacZ)Δ92 promoter contained only the RhaR binding site (Fig. 1). Since the assays were carried out with a strain that also expressed wild-type σ70 from the chromosome (27), we considered values below 80% of wild-type activity to be significant defects. At Φ(rhaS-lacZ)Δ216, σ70 derivative L595A had 66% of the activity of wild-type σ70 (Fig. 2A) while the remaining σ70 derivatives were not significantly defective. When the same sigma derivatives were assayed at Φ(rhaS-lacZ)Δ92, L595A was still significantly defective, with 53% activity compared to wild-type σ70 (Fig. 2B). In addition, three alanine substitutions of positively charged amino acid residues, K593A (79%), R599A (48%), and R608A (77%) were defective at Φ(rhaS-lacZ)Δ92. These results suggested that σ70 residues K593, L595, R599, and R608 might make protein-protein contacts with RhaR that are required for transcription activation. The lack of defect from σ70 K593A, R599A, or R608A at the Φ(rhaS-lacZ)Δ216 promoter is similar to previous findings with RhaS and AraC that substitutions at some σ70 residues were defective only in the absence of CRP activation (4, 27).
FIG. 2.
Alanine substitutions within the C-terminal domain of the σ70 subunit of RNAP assayed at two rhaS-lacZ fusions, Φ(rhaS-lacZ)Δ216 in SME1074 (A) and Φ(rhaS-lacZ)Δ92 in SME2508 (B). The σ70 alanine substitutions were encoded on plasmids, and the rhaS-lacZ fusions were in the chromosome as single-copy λ lysogens. In each panel, the values obtained with wild-type σ70 were set to 100% and the activity of each σ70 derivative is represented as a percentage of the wild-type σ70 value. In panel A, the activity of wild-type σ70 was 86 Miller units for the I590A, R596A, L598A, R603A, and R608A derivatives, while the wild-type σ70 activity for the other derivatives was 87 Miller units. In panel B, the wild-type σ70 activity was 3.6 Miller units for the I590A, R596A, L598A, R603A, and R608A derivatives and 2.2 Miller units for the other derivatives. β-gal, β-galactosidase.
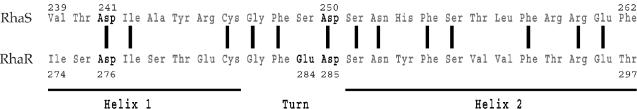
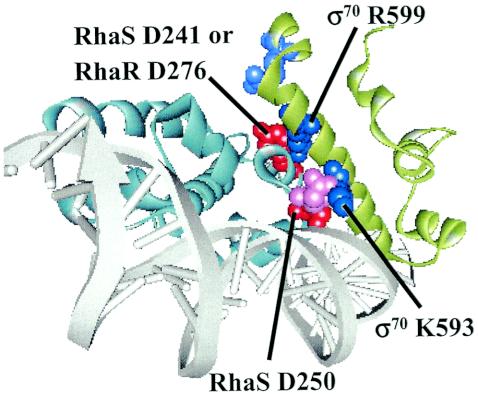
Based on the previous finding in our lab that a contact between RhaS residue D241 and σ70 residue R599 is required for full transcription activation by RhaS (4), we predicted that RhaR D276, which aligns with RhaS D241 (Fig. 3), might contact σ70 R599 at the rhaSR promoter. Molecular modeling of the RhaR-σ70 interaction (Fig. 4) indicates that the negatively charged RhaR residue D276 is very close to the positively charged σ70 residue R599. RhaR D276 is also near σ70 R608, although in the model they do not appear close enough to interact. The molecular model further shows that two adjacent negatively charged RhaR residues, E284 and D285, are located near σ70 K593. Based on these pieces of evidence, we hypothesized that contacts between some or all of the RhaR residues D276, E284, and D285 and σ70 might be required for maximal transcription activation by RhaR. We therefore tested alanine substitutions at these positions in RhaR for defects in transcription activation.
FIG. 3.
Alignment of the amino acid sequences of the second helix-turn-helix DNA-binding motifs of RhaS and RhaR. Amino acids shown in bold are those tested in this work for possible interactions with the σ70 subunit of RNAP. Identical amino acids are indicated by vertical lines between the two sequences. The boundaries of the first helix (Helix 1), the turn, and the recognition helix (Helix 2) are based on the structure of MarA (38) and alignments between MarA and RhaS and RhaR. The numbers of the first and last residues shown, as well as those of the residues tested for interactions with σ70, are indicated.
FIG. 4.
Model of RhaS or RhaR interactions with σ70 domain 4. The model of the RhaS or RhaR C-terminal domain (aqua) is based on the crystal structure of the MarA-DNA complex (38), while the model of σ70 domain 4 (green) is based on the crystal structure of the same domain of σA from Thermus aquaticus on DNA (6). Only the DNA from the MarA structure is shown (white). Amino acid residues in RhaS or RhaR and σ70 that are implicated in interactions are shown in a space-filling model and labeled, with the RhaR or RhaS residues colored red and the σ70 residues colored dark blue. The unlabeled space filling residues are RhaR E284 (pink), RhaR D285 (which is at the same position as RhaS D250), and σ70 R608 (light blue). Since σ70 sits in front of RhaS or RhaR when the DNA is shown parallel to the page, the model has been rotated somewhat around the vertical axis to allow a view between the interacting proteins. The modeling was performed using the program Insight II (Accelrys, Inc.) by first manually superimposing the DNAs in the PDF files of MarA on DNA (Protein Data Bank file 1BL0) and σA domain 4 on DNA (Protein Data Bank file 1KU7) such that the base pairs that corresponded to the −35 region of each were aligned as closely as possible. The σA model was then rotated to minimize clashes with MarA while maintaining the DNA superimposition. Finally, the residues implicated in interactions were highlighted. A second σA domain 4 molecule in the 1KU7 structure which does not make specific contacts with the DNA is not shown.
RhaR residues D276, E284, and D285 are important for rhaSR transcription activation.
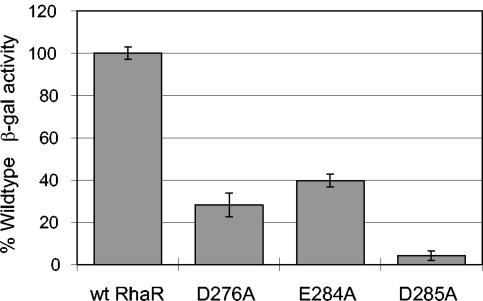
In order to test whether the side chains of RhaR residues D276, E284, and D285 might play a role in transcription activation by RhaR, we constructed alanine substitutions at each of these positions. If the amino acid residues at these positions are required for transcription activation, the alanine substitution should result in a significant decrease in activation of rhaSR transcription. To assay the RhaR derivatives, the mutant rhaR alleles on plasmids were first recombined onto the chromosome such that they replaced the wild-type rhaR gene (see Materials and Methods). The wild-type and mutant rhaR alleles were then assayed for activation of Φ(rhaS-lacZ)Δ92 (Fig. 5). The results showed that all three of the alanine substitutions in RhaR were significantly defective, highlighting the importance of the wild-type residues at those positions. The especially large defect of RhaR D285A may be partly due to a role in DNA binding based on its alignment with D250 in RhaS (Fig. 3), which makes base-specific contacts with DNA (3). However, a role in DNA binding for RhaR D285 does not rule out interactions with σ70; therefore, all three of these RhaR residues are candidates for specific contacts with σ70.
FIG. 5.
Transcription activation by RhaR derivatives. β-Galactosidase activity was assayed from a single-copy fusion of the rhaSR promoter with lacZ that included the RhaR binding site but not the CRP binding sites [Φ(rhaS-lacZ)Δ92]. In each case, wild-type RhaR or the alanine substitutions in RhaR were encoded in the chromosome at the natural rhaR locus (strains SME2691, -2692, -2693, and -2933). The value obtained with wild-type RhaR (3.3 Miller units) was set to 100%, and the activity of each RhaR derivative is represented as a percentage of that value.
Evidence for an interaction between RhaR D276 and σ70 R599.
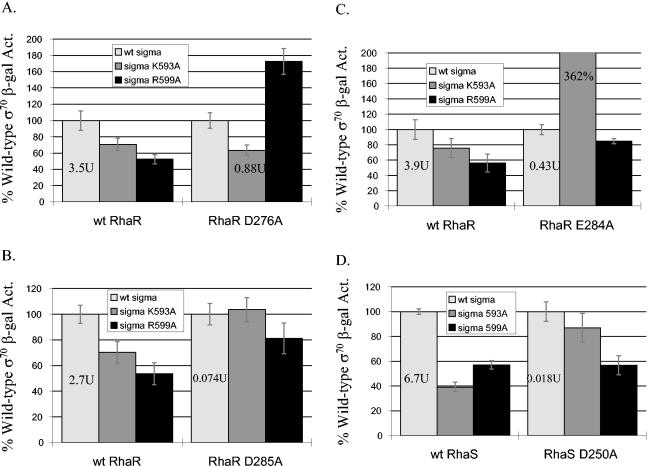
We used a genetic loss-of-contact approach to test potential interactions between RhaR D276 and σ70 K593 or R599. Using this approach, we separately combined wild-type RhaR or the RhaR D276A derivative with each of three plasmids encoding either σ70 wild type, K593A, or R599A in a strain carrying a single copy of Φ(rhaS-lacZ)Δ92. The results shown in Fig. 6A were plotted so that the activity with wild-type σ70 was set to 100% for each RhaR derivative, thereby illustrating the relative effects of each σ70 derivative. On this graph, therefore, a σ70 derivative that does not define a site of interaction with a given RhaR derivative is expected to have the same relative defect when combined with the indicated RhaR derivative as it does with wild-type RhaR, since the defects will be independent of each other. On the other hand, if a σ70 derivative does define a site of interaction with a given RhaR derivative, the σ70 derivative will confer no further defect when combined with the indicated RhaR derivative, since the interaction would already have been lost with the RhaR derivative. Using this method to analyze the results in Fig. 6A, our first conclusion is that there is no interaction between RhaR D276 and σ70 K593 since the σ70 K593A derivative had approximately the same relative defect in combination with either wild-type RhaR or RhaR D276A. Therefore, the defects of σ70 K593A and RhaR D276A are independent. These results and those in Fig. 2B also show that the σ70 R599A derivative by itself (in a wild-type rhaR strain) had approximately 50% activity compared to wild-type σ70. However, when σ70 R599A was combined with RhaR D276A, the σ70 R599A derivative conferred no further defect upon RhaR D276A. In fact, the strain with the combination of σ70 R599A and RhaR D276A had approximately 1.7-fold-higher activity than the strain with wild-type σ70 and RhaR D276A. These results fit the criteria for an allele-specific contact between σ70 R599 and RhaR D276. As mentioned above, molecular modeling is consistent with this interaction since RhaR D276 is in close proximity to σ70 R599 in the model (Fig. 4). We also tested the σ70 R608A derivative in combination with RhaR D276A and found that it had the same relative defective as it did with wild-type RhaR (79% of wild-type σ70 activity in both cases [data not shown]); therefore, there was no indication of an interaction between these two residues.
FIG. 6.
Combinations of RhaR or RhaS derivatives with σ70 derivatives. Plasmid-encoded σ70 alanine substitutions were combined with chromosomally encoded alanine substitutions in RhaR or RhaS (strains SME2689, -2691, -2692, -2693, and -2933), and β-galactosidase activity was measured from Φ(rhaS-lacZ)Δ92 (A, B, and C), or Φ(rhaB-lacZ)Δ84 (D). The activity of wild-type σ70 in combination with each RhaR or RhaS derivative in Miller units is shown on the corresponding bar in each graph and was set to 100% in each case. This representation allows the relative defects of the σ70 derivatives to be directly compared. The value for σ70 K593A in combination with RhaR E284A was 362% of the wild-type σ70 value and is drawn off scale to prevent compression of the remaining bars.
RhaR E284 and D285 and σ70.
Using the same genetic loss-of-contact approach, we also tested for potential interactions between σ70 and RhaR E284 and D285. The results in Fig. 6B show that the K593A σ70 derivative was not defective in combination with RhaR D285A (104% of wild-type σ70 activity) but the R599A σ70 derivative also became less defective (81% of wild-type σ70 activity). In the absence of the results obtained for σ70 599A, one might conclude that RhaR D285 contacts σ70 K593, since σ70 K593A had no significant defect when combined RhaR D285A; however, the lack of strict allele specificity sheds doubt on this conclusion. To further investigate the non-allele-specific defects of σ70 substitutions in combination with RhaR D285A, we tested σ70 L595A and R608A derivatives, which were both defective in a wild-type rhaR strain, as shown in Fig. 2B. When combined with RhaR D285A, the σ70 L595A and R608A derivatives were not significantly defective, with 86 and 87% of wild-type σ70 activity, respectively (data not shown). These results suggest that RhaR D285A may reduce the ability of RhaR to interact with σ70 in a non-allele-specific manner; therefore, we can't conclude whether RhaR D285 contacts any of these σ70 residues.
Figure 6C shows the results of assays to identify potential interactions involving RhaR E284. Our results showed that neither σ70 K593A nor R599A conferred a significant defect on RhaR E284A. In fact, the σ70 K593A-RhaR 284A combination gave much higher activity (362%) than the RhaR 284A derivative with wild-type σ70. Therefore, as described above, we tested the σ70 L595A and R608A derivatives in the rhaR E285A strain and found 139 and 93% activity, respectively, compared to wild-type σ70 (data not shown). Thus, we again found that all four of the σ70 derivatives that were defective in the wild-type rhaR strain were no longer significantly defective in the rhaR E284A strain. These results suggest that, similar to the RhaR D285A derivative, the RhaR E284A derivative may reduce the ability of RhaR to interact with σ70 in a non-allele-specific manner. One explanation for the very high relative activity of RhaR E284A in combination with σ70 K593A is that a new interaction may have been created in this case.
Evidence for a specific interaction between RhaS D250 and σ70.
Our lab previously identified an interaction between RhaS D241 and σ70 R599, and we also found that σ70 K593A was defective at Φ(rhaB-lacZ)Δ84 but did not identify an amino acid in RhaS that might contact σ70 K593 (4). The molecular model in Fig. 4 shows that the only negatively charged RhaS residue that is in close proximity to the positively charged σ70 K593 is RhaS D250, suggesting that these two residues might make a contact. Previous results from our lab showed that RhaS D250A was 12-fold defective for Φ(rhaB-lacZ)Δ84 activation; however, they also indicated that this residue participates in a base-specific DNA contact (3). In contrast to other approaches to identify positive-control mutants, the genetic loss-of-contact approach does not require that the protein have wild-type DNA-binding capability; hence it has the potential to identify residues that have dual DNA-binding and transcription activation functions. We therefore used the genetic loss-of-contact approach to test whether RhaS D250 and σ70 K593 might be involved in an interaction. The results (Fig. 6D) support the hypothesis of an interaction between RhaS D250 and σ70 K593 since the K593A derivative was not significantly defective when combined with RhaS D250A (87% activity compared to wild-type σ70). However, when σ70 R599A was combined with RhaS D250A, it maintained approximately the same relative defect as it had with wild-type RhaS. These results suggest that there is an allele-specific interaction between RhaS D250 and σ70 K593. Molecular modeling is consistent with this interaction since, as mentioned above, RhaS D250 is in close proximity to σ70 K593 in the model (Fig. 4).
DISCUSSION
The C terminus of σ70 is important for RhaS- and RhaR-mediated transcription activation.
The binding sites for both RhaS and RhaR overlap the −35 region of their respective core promoters by 4 bp, placing them in ideal positions to interact with the σ70 subunit of RNAP. Previous results reported by Bhende and Egan (4) identified two amino acid residues in σ70, K593 and R599, that were important for RhaS-mediated transcription activation at rhaBAD and rhaT. In the present study, we identified four amino acid residues in σ70, K593, L595, R599, and R608, which were important for RhaR-mediated transcription activation of rhaSR (Fig. 2B). Two of the alanine substitutions in σ70, K593A and R599A, were defective at all three of the rha promoters, suggesting similar mechanisms of activation by RhaS and RhaR.
The results reported in this paper (Fig. 2), as well as those from a previous study (4), showed that σ70 K593A and R599A were defective only at truncated rha promoters that did not include the upstream CRP binding sites. This is similar to the findings obtained for several other promoters that require multiple activators, such as araBAD, uhpT, and narG (18, 27, 36, 39). Two possible explanations for this trend are that the second activator increases the total number of interactions such that the relative importance of each individual interaction decreases or that the second activator creates redundancies in activation that mask the importance of other interactions. A third possibility is that the second activator alters the orientation of the first activator relative to σ70 such that the primary activator is no longer in an ideal position to interact with σ70. In the first two models, the activator interaction with σ70 occurs in both the presence and absence of the second activator but can be detected only in its absence, whereas in the third model, the interaction between the first activator and σ70 occurs only in the absence of the second activator. Further experiments will be needed to distinguish these models. At the rhaSR promoter, the σ70 L595A derivative was unique in that it was defective in both the presence and absence of the second activator, CRP, but it's role in RhaR-mediated transcription activation is not yet known.
Specific amino acid contacts between σ70 and RhaR.
Previous results showing an interaction between RhaS D241 and σ70 R599 at rhaBAD (4) led us to investigate whether an interaction between RhaR D276 and σ70 R599 might be required for RhaR activation at rhaSR. We also used a molecular model of the RhaR-σ70 domain 4 interaction in which the structure of MarA (38) represented RhaR (Fig. 4) to identify the only two negatively charged RhaR residues, E284 and D285, that were near σ70 K593. RhaR E284 and D285 were therefore considered candidates for residues that might interact with σ70 K593. After determining that alanine substitutions at RhaR residues D276, E284, and D285 were all defective for rhaSR activation (Fig. 5), we used a genetic loss-of-contact approach to test for specific amino acid interactions between σ70 and RhaR.
To carry out a loss-of-contact analysis, one must first identify defective derivatives of each of the potentially interacting proteins. In the simplest case, the full defect of both of the two interacting residues is due to loss of the interaction—in other words, the only role of the two residues is the interaction. The rationale behind this approach in this simple case is that mutation of one or the other of the interacting residues will eliminate the interaction; therefore, the phenotype of a strain carrying both mutations will be the same as the phenotype of the strains carrying the individual mutations. If one of the residues has a second role in addition to the interaction, then the strain carrying both mutations will have a phenotype that is no worse than the more defective of the strains carrying the two individual mutations. This analysis does not provide conclusive results if both residues have roles in addition to the interaction. It is also expected that the predicted interactions will be allele specific. The majority of combinations of defective derivatives are not expected to identify interacting residues, and in these cases the defects resulting from each of the two mutations will at least be additive.
Using this rationale to interpret our genetic loss-of-contact assays, the results in Fig. 6A provide evidence for an interaction between σ70 R599 and RhaR D276. This result is similar to previous results from our lab (4) that indicate an interaction between σ70 R599 and RhaS D241 and evidence from Grainger et al. that σ70 R599 interacts with MelR D261 (16), which aligns with RhaS D241 and RhaR D276. Molecular modeling of the RhaR-σ70 complex (Fig. 4) shows that σ70 R599 and RhaR D276 are in close proximity, consistent with our interpretation that these two residues interact. Our genetic loss-of-contact results do not provide evidence for an interaction between σ70 K593 and RhaR E284 or D285 (Fig. 6C). Instead, our results indicate that alanine substitutions at RhaR E284 and D285 result in non-allele-specific decreases in the defects of all of the σ70 alleles tested. One hypothesis is that RhaR E284A and D285A alter the details of the RhaR DNA interaction such that RhaR is no longer in an ideal position to interact with σ70 domain 4.
The role of σ70 K593 in transcription activation by RhaS.
Residue K593 of σ70 has been found to be important for several transcription activators, including AraC, UhpA, λ cI, FNR, Ada, RhaR (this study), and RhaS (4, 24, 27, 35, 36). With the exception of λ cI and RhaS (this study), evidence that σ70 K593 directly contacts an activator has not been obtained. Our results indicate that σ70 K593 contacts RhaS D250 as a part of the mechanism of activation by RhaS (Fig. 6D). Our molecular model of the RhaS-σ70 interaction shows that σ70 K593 and RhaS D250 are in close proximity, consistent with this result (Fig. 4). While the binary complex of Taq σA domain 4 and DNA shows that the residue that corresponds to σ70 K593 contacts DNA, in the λ cI-σ domain 4-DNA ternary complex, this residue participates in a protein-protein contact with λ cI instead (6, 19). These findings indicate that σ70 K593 is capable of interacting with an appropriately positioned transcription activator and are consistent with our proposal that σ70 K593 may contact RhaS D250.
Comparison of transcription activation by RhaS and RhaR.
In this study we identified an interaction between RhaR D276 and σ70 R599 that is equivalent to our previously identified interaction of RhaS D241 and σ70 R599. Further, although a RhaR equivalent of the RhaS D250 interaction with σ70 K593 was not identified, our results do not rule out that such an interaction occurs with RhaR. Therefore, our current evidence suggests that the RhaS-σ70 interface is similar to the RhaR-σ70 interface. We certainly expect, however, that not all aspects of RhaS activation and RhaR activation will be identical. For example, we know that the CRP site at rhaBAD is centered at position −92.5, whereas the most important CRP site at rhaSR is centered at position −111.5. It is not possible to draw conclusions about how or whether differences in the RhaS-σ70 and RhaR-σ70 interfaces might relate to this difference in CRP binding site position since all but one of the σ70 derivatives tested were defective only in the absence of CRP. However, it is likely that there is a difference in the mechanisms of RhaS and RhaR activation that relates to this difference in the positions of the CRP binding sites.
Acknowledgments
We thank Carol Gross for the library of alanine substitutions in σ70, Richard Wolf for alerting us that some of the σ70 mutants had reverted to the wild type, Kenan Murphy for providing strain KM22 and the cat-sacB cassette, Jeff Urbauer for assistance with the modeling of σ70 domain 4 in the MarA-DNA structure, and Vydehi Rao for performing the assays of the σ70 library in the strain containing Φ(rhaS-lacZ)Δ92.
This work was supported by Public Health Service grant GM55099 from the National Institute of General Medical Sciences and NIH Grant RR-P20 RR17708 from the Institutional Development Award Program of the National Center for Research Resources, both to S.M.E.
REFERENCES
- 1.Akimaru, H., K. Sakumi, T. Yoshikai, M. Anai, and M. Sekiguchi. 1990. Positive and negative regulation of transcription by a cleavage product of Ada protein. J. Mol. Biol. 216:261-273. [DOI] [PubMed] [Google Scholar]
- 2.Backman, K., Y.-M. Chen, and B. Magasanik. 1981. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 78:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 181:5185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhende, P. M., and S. M. Egan. 2000. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J. Bacteriol. 182:4959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 90:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 7.Dhiman, A., and R. Schleif. 2000. Recognition of overlapping nucleotides by AraC and the sigma subunit of RNA polymerase. J. Bacteriol. 182:5076-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dove, S. L., S. A. Darst, and A. Hochschild. 2003. Region 4 of sigma as a target for transcription regulation. Mol. Microbiol. 48:863-874. [DOI] [PubMed] [Google Scholar]
- 9.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan, S. M., and R. F. Schleif. 1994. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J. Mol. Biol. 243:821-829. [DOI] [PubMed] [Google Scholar]
- 11.Egan, S. M., and R. F. Schleif. 1993. A regulatory cascade in the induction of rhaBAD. J. Mol. Biol. 234:87-98. [DOI] [PubMed] [Google Scholar]
- 12.Eustance, R. J., S. A. Bustos, and R. E. Schleif. 1994. Reaching out: locating and lengthening the interdomain linker in AraC protein. J. Mol. Biol. 242:330-338. [DOI] [PubMed] [Google Scholar]
- 13.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grainger, D. C., T. A. Belyaeva, D. J. Lee, E. I. Hyde, and S. J. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase alpha subunit. Mol Microbiol. 51:1311-1320. [DOI] [PubMed] [Google Scholar]
- 16.Grainger, D. C., C. L. Webster, T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol Microbiol. 51:1297-1309. [DOI] [PubMed] [Google Scholar]
- 17.Holcroft, C. C., and S. M. Egan. 2000. Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain and RhaR. J. Bacteriol. 182:6774-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, J. C., and C. A. Gross. 1985. Mutations in the sigma subunit of E. coli RNA polymerase which affect positive control of transcription. Mol. Gen. Genet. 199:7-13. [DOI] [PubMed] [Google Scholar]
- 19.Jain, D., B. E. Nickels, L. Sun, A. Hochschild, and S. A. Darst. 2004. Structure of a ternary transcription activation complex. Mol. Cell 13:45-53. [DOI] [PubMed] [Google Scholar]
- 20.Jair, K., R. G. Martin, J. L. Rosner, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 177:7100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jair, K.-W., W. P. Fawcett, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 19:307-317. [DOI] [PubMed] [Google Scholar]
- 22.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 182:1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuldell, N., and A. Hochschild. 1994. Amino acid substitutions in the −35 recognition motif of σ70 that result in defects in phage λ repressor-stimulated transcription. J. Bacteriol. 176:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landini, P., and S. J. Busby. 1999. The Escherichia coli Ada protein can interact with two distinct determinants in the σ70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J. Bacteriol. 181:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPointe, C. F., and R. K. Taylor. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502-1510. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., H. Moyle, and M. M. Susskind. 1994. Target of the trancriptional activation function of phage λ cI protein. Science 263:75-77. [DOI] [PubMed] [Google Scholar]
- 27.Lonetto, M. A., V. Rhodius, K. Lamberg, P. Kiley, S. Busby, and C. Gross. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284:1353-1365. [DOI] [PubMed] [Google Scholar]
- 28.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 29.Menon, K. P., and N. L. Lee. 1990. Activation of ara operons by a truncated AraC protein does not require inducer. Proc. Natl. Acad. Sci. USA 87:3708-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer, B. J., R. Maurer, and M. Ptashne. 1980. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J. Mol. Biol. 139:163-194. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 33.Nakabeppu, Y., and M. Sekiguchi. 1986. Regulatory mechanisms for induction of synthesis of repair enzymes in response to alkylating agents: ada protein acts as a transcriptional regulator. Proc. Natl. Acad. Sci. USA 83:6297-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickels, B. E., S. L. Dove, K. S. Murakami, S. A. Darst, and A. Hochschild. 2002. Protein-protein and protein-DNA interactions of sigma70 region 4 involved in transcription activation by lambdacI. J. Mol. Biol. 324:17-34. [DOI] [PubMed] [Google Scholar]
- 36.Olekhnovich, I. N., and R. J. Kadner. 1999. RNA polymerase α and σ70 subunits participate in transcription of the Escherichia coli uhpT promoter. J. Bacteriol. 181:7266-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poore, C. A., C. Coker, J. D. Dattelbaum, and H. L. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverstone, A. E., M. Goman, and J. G. Scaife. 1972. ALT: a new factor involved in the synthesis of RNA by Escherichia coli. Mol. Gen. Genet. 118:223-234. [DOI] [PubMed] [Google Scholar]
- 40.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 41.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 42.Tobin, J. F., and R. F. Schleif. 1987. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J. Mol. Biol. 196:789-799. [DOI] [PubMed] [Google Scholar]
- 43.Tobin, J. F., and R. F. Schleif. 1990. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli. J. Mol. Biol. 211:75-89. [DOI] [PubMed] [Google Scholar]
- 44.Travers, A. 1974. RNA polymerase-promoter interactions: some general principles. Cell 3:97-104. [DOI] [PubMed] [Google Scholar]
- 45.Via, P., J. Badia, L. Baldoma, N. Obradors, and J. Aguilar. 1996. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology 142:1833-1840. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]